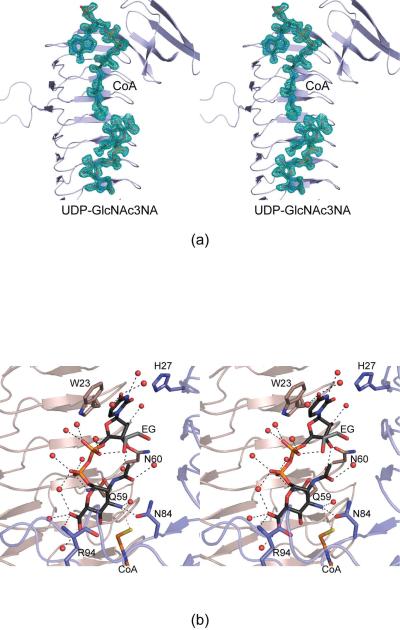
Figure 4.
Active site architecture for the WlbB/CoA/UDP-GlcNAc3NA complex model. Shown in (a) is the initial electron density corresponding to the bound ligands (before they were included in the model). The map was contoured at ~1.5σ and calculated with coefficients of the form (2Fo - Fc), where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude. Those amino acid residues lying within ~3.2 Å of UDP-GlcNAc3NA are shown in (b). Water molecules are depicted as red spheres, and potential hydrogen bonds are indicated by the dashed lines. Note that the water molecule located within hydrogen bonding distance of the sugar C-3' amino group is displaced when acetyl-CoA is bound in the active site.