Abstract
During feline leukemia virus (FeLV) infection in the domestic cat, viruses with a novel envelope gene arise by recombination between endogenous FeLV-related elements and the exogenous infecting species. These recombinant viruses (FeLV-B) are of uncertain disease association, but have been linked to the induction of thymic lymphoma. To assess the role of FeLV-B in the induction of multicentric lymphoma and other non-T-cell disease, the frequency of occurrence and nature of FeLV-B was examined in diseased tissues from a large collection of FeLV-infected animals. Diseased tissues were examined by Southern blot and PCR amplification to detect the presence of FeLV-B. Further analysis was performed to establish the recombination junctions and infectivity of FeLV-B in diseased tissues. The results confirmed the frequent association of FeLV-B with thymic lymphoma but showed infrequent generation, low levels and lack of infectivity of FeLV-B in non-T-cell diseases including multicentric lymphoma.
Keywords: feline leukemia virus, FeLV-B, FeLV-945, multicentric lymphoma, recombinant envelope gene
Introduction
Feline leukemia virus (FeLV) is a naturally occurring, horizontally transmitted gammaretrovirus associated with malignant, proliferative and degenerative diseases in the domestic cat. FeLV occurs in nature not as a single genomic species but as a genetically complex family of closely related viruses. Genetic variation in FeLV is generated during virus replication through error-prone reverse transcription and by recombination with endogenous FeLV-related sequences in the domestic cat genome. Such variation has led to four naturally occurring FeLV subgroups, designated A, B, C, and T, that are distinguished genetically by sequence differences in the surface glycoprotein gene (SU) and functionally by interaction with distinct host cell receptors for entry. The weakly pathogenic FeLV subtype A (FeLV-A) is thought to represent the predominant agent spread horizontally cat-to-cat in nature, from which FeLV-B, -C and -T arise de novo in the infected animal by envelope (env) gene recombination, mutation or insertion events (Levy, 2008; Overbaugh and Bangham, 2001). While FeLV-A is associated with prolonged asymptomatic infection in the cat that may lead to malignant disease, typically a T-cell lymphoma of the thymus (Neil et al., 1991; Rezanka, Rojko, and Neil, 1992), the FeLV-B, -C and -T subgroups facilitate and/or redirect disease outcome to lymphoma, anemia or immunodeficiency disease, respectively (Levy, 2008; Overbaugh and Bangham, 2001).
Of particular interest in the present study is the generation of FeLV-B, a polytropic virus that arises by recombination in vivo between the infecting exogenous species and endogenous FeLV-related sequences in the cat genome. As a consequence of recombination, FeLV-B viruses contain a novel env gene with SU sequences containing variable representation of endogenous FeLV-derived DNA (Neil et al., 1991; Roy-Burman, 1995; Stewart et al., 1986). The endogenous, FeLV-related elements in the domestic cat occur at 9 – 16 copies per haploid genome (Pontius et al., 2007; Roca et al., 2005), differ in load among domestic cats of different breeds (Tandon et al., 2007) and represent a genetic set of fluid, polymorphic composition (Roca, Pecon-Slattery, and O’Brien, 2004). The endogenous FeLV-related elements are defective and do not encode infectious particles, although some elements remain transcriptionally active and may be expressed in leukemic cells and in lymphoid tissues from healthy animals (McDougall et al., 1994; Roy-Burman, 1995). Recombination between endogenous and exogenous FeLV env sequences is thought to occur in a manner analogous to the generation of MCF recombinant viruses during MuLV infection (Fan, 1997). Unlike MCF viruses, which are known to represent the proximal leukemogen in MuLV infection (Fan, 1997), the disease association of FeLV-B infection remains unclear. While FeLV-B can be identified in 30% – 50% of natural infections, always in conjunction with FeLV-A (Coelho et al., 2008; Neil et al., 1991; Roy-Burman, 1995), FeLV-B is overrepresented in the diseased tissues of animals with lymphoma as compared to asymptomatic, FeLV-infected cats, and has been identified in the majority of thymic lymphomas in particular (Jarrett, 1980; Jarrett, 1992; Roy-Burman, 1995; Sheets et al., 1993; Tsatsanis et al., 1994). In a recent study, however, FeLV-B recombinants were identified only infrequently by PCR or Southern blot analysis in a set of unusual multicentric lymphomas of B-cell origin induced experimentally by viruses containing the unique sequence elements of a natural variant designated FeLV-945 (Chandhasin, Coan, and Levy, 2005). FeLV-945 had been identified originally in a geographic and temporal cohort of naturally infected animals in which non-T-cell disease, including multicentric lymphoma, was the condition most commonly identified. As the predominant isolate in non-T-cell diseases in the cohort, FeLV-945 was shown to contain unique sequence elements in the LTR and SU gene. The substitution of these elements into prototype FeLV-A significantly reduced the latency to disease and shifted the disease spectrum entirely to recapitulate the multicentric lymphoma from which FeLV-945 was originally identified (Chandhasin et al., 2005). Considering the relatively rapid disease induction in animals infected with chimeric viruses containing elements of FeLV-945, the infrequent detection of FeLV-B in preliminary analysis (Chandhasin, Coan, and Levy, 2005; Chandhasin et al., 2005) suggested that the generation of FeLV-B recombinants may not be involved in the induction of multicentric lymphoma, or alternatively, may occur infrequently or inefficiently when FeLV-945 is the exogenously infecting species. To address this question further, the frequency of occurrence and nature of FeLV-B recombinants was examined in a large collection of diseased tissues from naturally and experimentally infected cats. Twenty-two naturally infected animals were examined, representing the geographic and temporal cohort from which FeLV-945 was originally identified (Chandhasin, Lobelle-Rich, and Levy, 2004; Levesque, Bonham, and Levy, 1990). Twenty experimentally infected animals were examined from several previous studies (Chandhasin, Coan, and Levy, 2005; Chandhasin et al., 2005; Levy, Fish, and Baskin, 1988) in which neonatal cats were inoculated with prototype FeLV-A/61E, with chimeric viruses in which the unique LTR and/or SU gene of FeLV-945 were substituted into FeLV-A/61E, or with the myc-oncogene containing LC-FeLV isolate. Diseased tissues were examined by Southern blot and PCR amplification to detect the presence of FeLV-B. Further analysis was performed to establish the recombination junctions and infectivity of FeLV-B in diseased tissues.
Material and Methods
Tissues from natural and experimental infections in the domestic cat
The tissues of twenty-two naturally infected animals were examined, representing the geographic and temporal cohort from which FeLV-945 was originally identified (Chandhasin, Lobelle-Rich, and Levy, 2004; Levesque, Bonham, and Levy, 1990) and a gift of Dr. Murray Gardner. The cohort included twelve cases of multicentric lymphoma, four cases of thymic lymphoma, one case of myeloproliferative disease, one case of mast cell leukemia, one case of feline infectious peritonitis, one case of chronic glomerulonephritis, and two asymptomatic cases of FeLV-infected, healthy animals. An additional animal from the cohort was shown to be FeLV-negative and healthy, and was examined as a negative control. Twenty experimentally infected animals were examined from several previous studies in which neonatal cats were inoculated with one of the following viruses: (1) FeLV-A/61E, a natural virus isolated from a complex of viruses linked to immunodeficiency disease when inoculated together (Donahue et al., 1991) but causative of thymic lymphoma when inoculated alone (Chandhasin, Coan, and Levy, 2005; Chandhasin et al., 2005); (2) LC-FeLV, a defective natural isolate from a thymic lymphoma, demonstrated to contain the myc oncogene (Levy, Gardner, and Casey, 1984). The defective nature of LC-FeLV was complemented by growth in the presence of infectious helper viruses FeLV-A/Glasgow-1 and FeLV-B/Gardner-Arnstein {an inoculum designated LC-FeLV (FeLV-A+B)} (Levy, Fish, and Baskin, 1988); (3) Infectious FeLV-A+B helper viruses in the absence of LC-FeLV (Levy, Fish, and Baskin, 1988); or (4) chimeric viruses in which the LTR or the LTR and SU gene of the natural isolate, FeLV-945, were substituted into FeLV-A/61E {designated 61E/945L or 61E/945SL, respectively} (Chandhasin, Coan, and Levy, 2005; Chandhasin et al., 2005). Tissues from two mock-infected, healthy animals were also examined as negative controls.
Southern blot analysis for integrated FeLV proviruses
High molecular weight genomic DNA was prepared from cat tissues and used for Southern blot analysis as previously described (Chandhasin et al., 2005). Briefly, DNA (8μg) was digested with KpnI, electrophoresed in a 0.8% agarose gel and transferred to nitrocellulose. Filters were hybridized for 18 hours at 67°C to radiolabeled probe B/S, a Sau3A fragment from the env gene of FeLV-B/Gardner-Arnstein specific for the major classes of endogenous FeLV that serve as substrates for recombination. By this approach, FeLV-B is visualized in genomic DNA as a distinctive hybridizing fragment of ~3.6-kb (Chandhasin et al., 2005; Tsatsanis et al., 1994). Filters were similarly hybridized to a radiolabeled probe representing the U3 region of the LTR of exogenous FeLV. By this approach, clonally integrated exogenous proviruses are visualized as host-virus junction fragments in tumor DNA. Endogenous FeLV-related elements in feline DNA are weakly recognized as well (Levy, Gardner, and Casey, 1984).
PCR amplification of recombinant proviral DNA
Genomic DNA samples were amplified using forward (5′-AACCCTGACAGACACCTTCCCTAC) and reverse (5′ GAGTGACCTACCATCAACCCGA) primers specific for the endogenous FeLV-related CFE-6 element (GenBank M25425.1) and exogenous FeLV-945 (GenBank AY662447), respectively. PCR was performed using 300 ng of genomic DNA in a 50 μl reaction with 200 ng of each primer. An initial denaturation step at 95°C for 5 min was followed by 30 cycles of denaturation at 94°C for 45s, 62°C for 45s, and 72°C for 45s with a final extension for 10 min. PCR products were analyzed by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining. To obtain sufficient material for nucleotide sequence analysis, gel-purified PCR products were further amplified using internal primers (5′-ATGTAGTGGAGGTGGTTGGTG and 5- GGGGGGTTTGTTTGGTTGCTG) under the amplification conditions described above except that the annealing temperature was 57°C. Amplification products were verified by nucleotide sequencing directly, without molecular cloning. Nucleotide sequences thus obtained have been deposited in GenBank (Accession numbers GU434305 – GU434315). Sequence data were compared for analysis to the published sequences of FeLV-945 (AY662447), FeLV-A/61E (M18248), FeLV-B/Gardner-Arnstein (K01209), and endogenous element CFE-6 (M25425). Genomic DNA from FEA embryonic feline fibroblasts was used as a negative control for amplification.
DNA-mediated transfection and assay for productive infection
Canine D-17 cells (American Type Culture Collection, CCL-183) were maintained in modified Eagle’s medium (MEM) with non-essential amino acids and 10% fetal bovine serum. Cells were grown to 40–50% confluency in 6-well culture plates containing 2 ml of culture medium. Genomic DNA (5 μg) of five experimentally-induced thymic lymphomas, three experimentally-induced multicentric lymphomas, one natural multicentric lymphoma, one natural thymic lymphoma and one case of feline infectious peritonitis was introduced by Lipofectamine Plus-mediated transfection (Invitrogen Corp; Carlsbad, California, USA) for four hours into D-17 cells. Genomic DNA from D-17 cells infected with FeLV-B/Gardner-Arnstein was included as a positive control. Cells were harvested two days later and transferred to a T-75 culture flask. Culture supernatant was collected at regular intervals for 15 days thereafter and examined by p27Gag ELISA (Synbiotics Corporation, San Diego, California) for the presence of FeLV particles. A positive result was defined in this analysis as an optical density (A450) at least twice that of the negative control.
Results and Discussion
Demonstration of integrated FeLV-B proviral DNA in diseased tissues by Southern blot
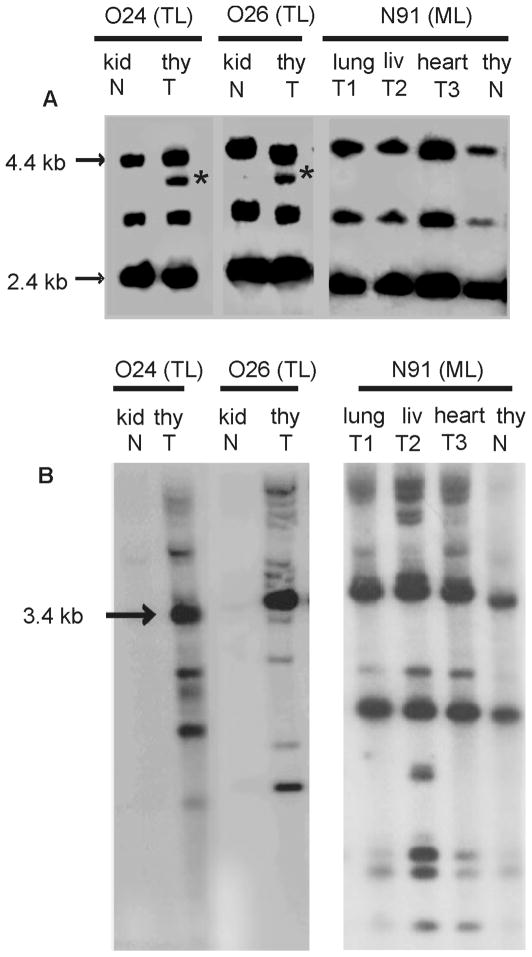
To evaluate the frequency with which integrated FeLV-B proviral DNA could be detected in diseased tissues, large molecular weight genomic DNA (8μg) was digested with KpnI and examined by Southern blot analysis. The blot was hybridized to probe B/S, a Sau3A fragment from the env gene of FeLV-B/Gardner-Arnstein specific for the major classes of endogenous FeLV that serve as substrates for recombination (Chandhasin et al., 2005; Tsatsanis et al., 1994). Analysis revealed the 3.6-kb fragment distinctive of FeLV-B proviral DNA in thymic lymphoma samples, but not in samples of multicentric lymphoma or in uninvolved tissues of the same animals (Figure 1A). By comparison, hybridization of KpnI-digested DNA with a probe for the LTR of exogenous FeLV clearly demonstrated the pattern of clonally integrated proviruses distinctive of the monoclonal expansion in all tumor samples examined, but not in uninvolved tissues of the same animals (Figure 1B). By applying this approach to the entire collection, FeLV-B provirus was detected in the DNA of diseased tissues from 5 of 22 naturally infected animals, including 2 of 4 cases of thymic lymphoma and 3 of 12 cases of multicentric lymphoma. FeLV-B was not detected in myeloproliferative disease, mast cell leukemia, feline infectious peritonitis, chronic glomerulonephritis, or in healthy, FeLV-infected animals (Table 1). In experimentally infected animals, FeLV-B was detected in 9 of 13 cases of thymic lymphoma and in two additional animals for which FeLV-B was included in the original inoculum. FeLV-B was not detected by this measure in multicentric lymphoma, in an animal inoculated with myc-containing FeLV in the presence of FeLV-B in the inoculum, or in mock-infected animals (Table 2). Considering all cases together, the incidence of FeLV-B in thymic lymphoma (11 of 17) was significantly higher than in multicentric lymphoma (3 of 15) as measured by Fisher Exact Test (p = 0.015).
Figure 1.
Southern blot analysis of genomic DNA from tumor (T) or paired normal (N) tissue from FeLV-infected cats O24, O26 and N91 bearing thymic lymphoma (TL) or multicentric lymphoma (ML) (Chandhasin et al., 2005). A. DNA samples (8 μg) were digested with KpnI and hybridized to probe B/S, a Sau3A fragment from the env gene of FeLV-B/Gardner-Arnstein specific for the major classes of endogenous FeLV that serve as substrates for recombination. The distinctive hybridizing fragment of ~3.6-kb (*) indicates recombinant FeLV-B provirus in genomic DNA (Chandhasin et al., 2005; Tsatsanis et al., 1994). B. KpnI-digested DNA samples were also hybridized to a probe representing the U3 region of the LTR of exogenous FeLV. By this analysis, clonally integrated proviruses are visualized as host-virus junction fragments in tumor DNA (Levy, Gardner, and Casey, 1984).
Table 1.
Detection of FeLV-B proviral DNA in diseased tissues from a cohort of naturally infected cats.a
| Animal | Diseaseb | S. Blot | PCR |
|---|---|---|---|
| 922 | Multicentric lymphoma | − | − |
| 925 | Multicentric lymphoma | − | − |
| 934 | Multicentric lymphoma | − | − |
| 945 | Multicentric lymphoma | − | − |
| 1011 | Multicentric lymphoma | − | − |
| 1043 | Multicentric lymphoma | + | + |
| 1046 | Multicentric lymphoma | − | − |
| 1049 | Multicentric lymphoma | + | + |
| 1112 | Multicentric lymphoma | − | − |
| 1316 | Multicentric lymphoma | − | − |
| 1325 | Multicentric lymphoma | − | − |
| 1345 | Multicentric lymphoma | + | + |
| 931 | Thymic lymphoma | − | − |
| 1037 | Thymic lymphoma | + | + |
| 1100 | Thymic lymphoma | − | − |
| 1110 | Thymic lymphoma | + | + |
| 903 | Myeloproliferative disease | − | − |
| 905 | Mast cell leukemia | − | − |
| 924 | Feline infectious peritonitis | − | − |
| 1002 | Chronic glomerulonephritis | − | − |
| 936 | Healthy, FeLV-negative | − | − |
| 1223 | Healthy, FeLV-infected | − | − |
| 942 | Healthy, FeLV-infected | − | − |
Presence (+) or absence (−) of FeLV-B proviral DNA as detected by Southern blot analysis or polymerase chain reaction (PCR) amplification.
Tumor tissues were analyzed from animals with lymphoma. Mesenteric lymph node, bone marrow, thymus or kidney was examined from animals 903, 905, 1223 or 942, respectively. Spleen was examined from animals 924, 1002 and 936.
Table 2.
Detection of FeLV-B proviral DNA in diseased tissues from experimentally infected cats.a
| Animal | Diseaseb | Inoculumc | Reference | S. Blot | PCR |
|---|---|---|---|---|---|
| O3 | Thymic lymphoma | FeLV-A/61E | Chandhasin et al., 2005 | + | + |
| O4 | Thymic lymphoma | FeLV-A/61E | Chandhasin et al., 2005 | + | + |
| O5 | Thymic lymphoma | FeLV-A/61E | Chandhasin et al., 2005 | + | + |
| O6 | Thymic lymphoma | FeLV-A/61E | Chandhasin et al., 2005 | + | + |
| O23 | Thymic lymphoma | 61E945L | Chandhasin et al., 2005 | + | + |
| O24 | Thymic lymphoma | 61E945L | Chandhasin et al., 2005 | + | + |
| O25 | Thymic lymphoma | 61E945L | Chandhasin et al., 2005 | + | + |
| O26 | Thymic lymphoma | 61E945L | Chandhasin et al., 2005 | + | + |
| 981-2 | Thymic lymphoma | LC-FeLV (FeLV-A+B) | Levy, Fish & Baskin, 1988 | − | − |
| 981-3 | Thymic lymphoma | LC-FeLV (FeLV-A+B) | Levy, Fish & Baskin, 1988 | + | + |
| J5-1 | Thymic lymphoma | LC-FeLV (FeLV-A+B) | Levy, Fish & Baskin, 1988 | − | − |
| N65 | Thymic lymphoma | 61E/945SL | Chandhasin et al., 2005 | − | + |
| N67 | Thymic lymphoma | 61E/945SL | Chandhasin et al., 2005 | − | + |
| N89 | Unknown | 61E/945SL | Chandhasin et al., 2005 | − | ± |
| N90 | Multicentric lymphoma | 61E/945SL | Chandhasin et al., 2005 | − | ± |
| N91 | Multicentric lymphoma | 61E/945SL | Chandhasin et al., 2005 | − | ± |
| N92 | Multicentric lymphoma | 61E/945SL | Chandhasin et al., 2005 | − | ± |
| 586-4 | Feline infectious peritonitis | FeLV-A+B | Levy, Fish & Baskin, 1988 | + | + |
| 585-2 | Transient lymphadenopathy | LC-FeLV (FeLV-A+B) | Levy, Fish & Baskin, 1988 | − | + |
| 586-3 | Healthy | LC-FeLV (FeLV-A+B) | Levy, Fish & Baskin, 1988 | + | + |
| 586-5 | Healthy | Mock | Levy, Fish & Baskin, 1988 | − | − |
| 585-1 | Healthy | Mock | Levy, Fish & Baskin, 1988 | − | − |
Presence (+) or absence (−) of FeLV-B proviral DNA as detected by Southern blot analysis or polymerase chain reaction (PCR) amplification. ± = weakly detectable.
Tumor tissues were analyzed from animals with lymphoma. Lymph node was examined from animals 586-3 and 586-4. PBMC were examined from animals 585-1, 585-2 and 586-5. Lung, lymph node and liver was examined from N89.
Animals were inoculated experimentally with FeLV-A/61E, a synthetic recombinant of FeLV-A/61E containing either the LTR of FeLV-945 (61E945L) or the SU gene and LTR of FeLV-945 (61E/945SL), a natural myc-containing isolate of FeLV (LC-FeLV) in the presence of helper viruses FeLV-A and FeLV-B (FeLV A+B), or the helper viruses alone (FeLV-A+B). All animals were inoculated intraperitoneally except N65 and N67, which were inoculated intradermally.
PCR amplification of FeLV-B proviral DNA from diseased tissues
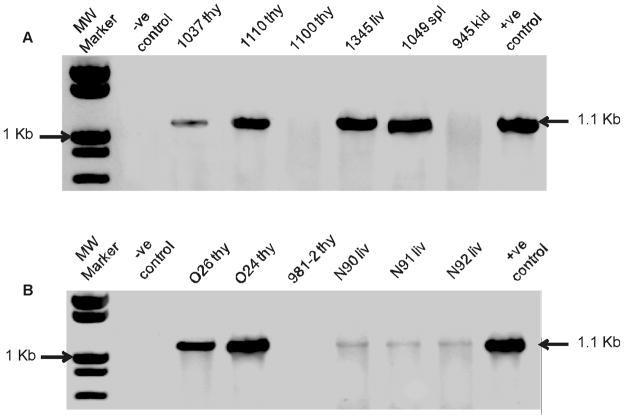
PCR amplification was then used to validate the results and to extend the findings to cases in which FeLV-B integrations may have occurred at a frequency in the tumor mass below the level of detection by Southern blot analysis. Genomic DNA samples were amplified using forward and reverse primers specific for the endogenous FeLV-related CFE-6 element and exogenous FeLV-945, respectively. CFE-6 was selected for this purpose because it represents the major class of nearly full-length proviruses in the cat genome and contains a complete env gene with open reading frame (Kumar, Berry, and Roy-Burman, 1989). Specificity of the amplification reaction was demonstrated by the absence of a product from DNA of uninfected FEA cells and the ready detection of a product of the predicted size (1.1-kb) from canine D-17 cells infected with FeLV-B/Gardner-Arnstein (indicated in Figure 2 as negative and positive control, respectively). When this strategy was applied to the collection of diseased tissues from naturally infected animals, the predicted amplification product was detected from 2 of 4 cases of thymic lymphoma (50%) and 3 of 12 cases of multicentric lymphoma (25%) (Figure 2A), thus recapitulating the findings from Southern blot analysis (Table 1). Likewise consistent with the results of Southern blot analysis, FeLV-B sequences were not detected in myeloproliferative disease, mast cell leukemia, feline infectious peritonitis, chronic glomerulonephritis, or in healthy, FeLV-infected animals (Table 1). Thus, in the natural cohort in which FeLV-945 predominates, FeLV-B is identified in thymic lymphomas at a frequency consistent with previous reports, but is relatively infrequent in non-T-cell diseases including multicentric lymphoma. Application of the PCR strategy to diseased tissues of experimentally infected animals demonstrated the predicted amplification product in 11 of 13 cases of thymic lymphoma (85%), in two of which FeLV-B was not evident by Southern blot analysis (N65 and N67; Figure 2B, Table 2). Thus, the frequency of detection of FeLV-B was higher in experimental as compared to natural thymic lymphomas (85% vs. 50%). While the basis for this disparity is not understood, the difference may reflect characteristics of the challenge virus. It is noteworthy that the experimental thymic lymphomas resulted from infection with FeLV-A/61E, or with a chimeric virus (61E/945L) in which only the LTR of FeLV-945 was substituted into FeLV-A/61E. By comparison, the naturally infected animals represented a cohort in which FeLV-945 was the predominant isolate (Chandhasin, Lobelle-Rich, and Levy, 2004). Thus, the difference in frequency of FeLV-B in natural vs. experimental thymic lymphomas may reflect a reduced frequency or efficiency of recombination in animals infected with FeLV-945, among other variables including age or route of inoculation.
Figure 2.
PCR amplification of the recombinant env gene of FeLV-B from the genomic DNA of naturally and experimentally infected cats. Genomic DNA samples were amplified by PCR using forward and reverse primers specific for the endogenous FeLV-related CFE-6 element and exogenous FeLV-945, respectively. A. Genomic DNA was examined from naturally occurring thymic lymphomas (1037 thy, 1110 thy and 1100 thy) and multicentric lymphomas (1345 liver, 1049 spleen, and 945 kidney). B. Genomic DNA was examined from experimentally induced thymic lymphomas (O26 thy, O24 thy, and 981-2 thy) and multicentric lymphomas (N90 liver, N91 liver, and N92 liver). Indicated by the arrow is the predicted amplification product of ~1.1-kb. Genomic DNA from uninfected FEA cells, and from canine D-17 cells infected with FeLV-B/Gardner-Arnstein were examined as negative and positive controls, respectively.
In 3 of 3 cases of experimental multicentric lymphoma (100%), the predicted amplification product was weakly demonstrated (Figure 2B), although no such demonstration was evident by Southern blot. PCR amplification further demonstrated the predicted product in feline infectious peritonitis and in two healthy animals inoculated with myc-containing FeLV in the presence of FeLV-B (Table 2). The weak detection of FeLV-B in multicentric lymphomas suggests that FeLV-B occurs at subclonal levels in those tumors, and may indeed represent a passenger rather than a causal agent of the disease. Considering natural and experimental multicentric lymphomas together, FeLV-B was detected by Southern blot in only 3 of 15 cases and was weakly demonstrated by PCR in 3 additional cases. In contrast, when considering natural and experimental thymic lymphomas together, FeLV-B was detected by Southern blot in 11 of 17 cases and was demonstrated by PCR in 2 additional cases. Thus, the frequency of detection of FeLV-B was significantly lower in multicentric lymphoma and other non-T-cell disease than in thymic lymphoma in both naturally- and experimentally-infected animals (Tables 1 and 2). This difference may reflect characteristics of the challenge virus, since multicentric lymphomas were induced by FeLV-945 (in natural infections) or by virus containing the envelope gene of FeLV-945 (in experimental infections). By contrast, thymic lymphomas were induced by experimental infection with FeLV-A/61E or virus containing the envelope gene of FeLV-A/61E (Tables 1 and 2). The FeLV genome present in thymic lymphomas in the natural cohort has not yet been characterized. An alternative explanation is that the difference may reflect the generally permissive environment for recombination in T-cells, and/or the increased expression of endogenous FeLV as recombination substrate in those cells (McDougall et al., 1994). Taken together, these findings are consistent with other reports in the literature regarding the frequent association of FeLV-B with thymic lymphoma, and by implication, with a role for FeLV-B in the induction of that disease (Bechtel et al., 1998; Jarrett, 1980; Jarrett, 1992; Roy-Burman, 1995; Sheets et al., 1993; Tsatsanis et al., 1994). In contract, the infrequent detection and low levels of FeLV-B in multicentric lymphoma do not support a role in the induction of that disease.
Nucleotide sequence analysis of FeLV-B recombinants and identification of recombination junctions
Gel-purified PCR products were further amplified using internal primers, and the nucleotide sequence of products thus amplified was determined from 5 experimentally induced thymic lymphomas (O3, O4, O6, O23, O24), 3 experimentally induced multicentric lymphomas (N90, N91, N92), a naturally occurring multicentric lymphoma (1049), a naturally occurring thymic lymphoma (1110), and an experimentally infected animal with feline infectious peritonitis (586-4) (Figure 3). The sequence of the amplification products was determined directly, without molecular cloning; thus, the reported sequences represent the predominant species in each sample. Other variants may indeed occur that were not detected by this analysis. Of note, identical or nearly identical sequences were detected as predominant among multiple, independent tumors of the same type. For example, the predominant sequences from thymic lymphomas O3, O4, O6, O23 are identical or nearly identical, as are those from natural and experimental multicentric lymphomas. The sequence from thymic lymphoma O24 more closely resembled those from multicentric lymphomas. For this reason, multiple independent amplifications of O24 DNA were submitted for sequencing with confirmatory results. To determine the recombination junctions utilized for the generation of FeLV-B, sequences of the amplification products were compared to the endogenous FeLV element CFE-6 and to the sequences of FeLV-B/Gardner-Arnstein, FeLV-A/61E and FeLV-945. The identification of 34 nucleotide positions at which the amplification products differed in sequence from CFE-6 was used for identification of the recombination junctions (Figure 3). This analysis was facilitated by a previous study in which a limited number of sites were shown to be preferentially utilized for recombination to generate FeLV-B during infection in vitro. The preferential recombination sites, designated A – G (as shown in Figure 3), are located within a 275-bp region of the SU gene (Roy-Burman, 1995; Sheets et al., 1992).
Figure 3.
Nucleotide sequence of the PCR amplification products from tumor DNA of the indicated animals aligned with the sequence of endogenous FeLV-related provirus CFE-6. The previously reported sequences of FeLV-945, FeLV-A/61E and FeLV-B/Gardner-Arnstein are aligned for comparison. Nucleotide sequence differences as compared to CFE-6 are indicated; (…) indicates sequence identity with CFE-6; (____) indicates deletion relative to CFE-6; (//) indicates a break in the sequence shown. Indicated above the sequence are the positions of 34 nucleotide sequence differences in the amplification products as compared to CFE-6. Those indicated with an asterisk (*) result in amino acid change relative to CFE-6. Also indicated about the sequence are recombination sites A – G, previously reported to be preferred sites for crossover in the generation of FeLV-B (Sheets et al., 1992).
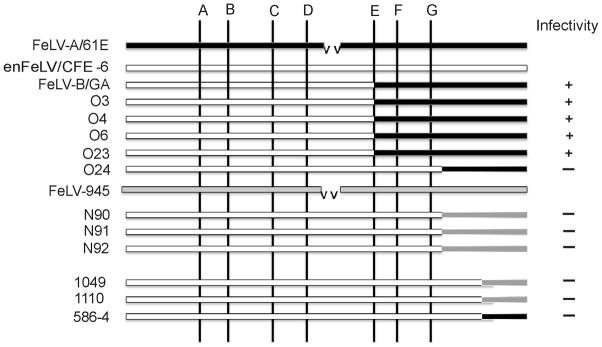
The present analysis demonstrated that FeLV-B/Gardner-Arnstein resulted from recombination at the previously designated site E, as did 4 of 5 amplification products from experimentally-induced thymic lymphoma samples (Figure 3, position 8). The recombination junction site used in thymic lymphoma O24 and multicentric lymphomas N90, N91 and N92 was detected further downstream, at 47 nucleotides beyond previously identified site G (Fig 3, position 20). The recombination site used in naturally occurring thymic lymphoma 1110 and multicentric lymphoma 1049, and in experimentally infected cat 586-4 was identified at the 3′ end of the SU gene, 96 nucleotides downstream from site G (Figure 3, position 34). Thus, FeLV-B proviruses from 4 of 5 thymic lymphomas utilized an upstream recombination junction relative to those from multicentric lymphomas and therefore contained significantly more DNA from the exogenous FeLV partner (Figure 4). In the endogenous FeLV-derived region upstream of the recombination sites, the analysis also demonstrated seven nucleotide sequence changes in FeLV-B as compared to CFE-6 (Figure 3, sites 1 – 7), five of which resulted in amino acid substitutions (Site 1, Asn to Ser; Site 2, Thr to Ile; Site 4, Pro to Leu; Site 5, Ile to Thr; Site 6, Pro to Ser). These sequence differences may represent mutational changes that were selected in the generation of FeLV-B. It is noteworthy that the amino acid changes are identical to those identified in an earlier analysis of FeLV-B generation. In that study, the selection of these particular amino acid changes was reported in natural and experimentally derived FeLV-B isolates from diverse populations and geographic locations (Bechtel et al., 1998). Taken together with the earlier study, our findings support the hypothesis that recombinant viruses harboring the specific amino acid changes are strongly selected during the disease course.
Figure 4.
Schematic representation of the recombination junctions identified in the SU genes of FeLV-B viruses amplified from tumor DNA of the indicated animals. Depicted for comparison are the SU genes of exogenous viruses FeLV-A/61E (solid bar) and FeLV-945 (gray bar) and endogenous FeLV-related provirus CFE-6 (open bar). A deletion within FeLV-A/61E relative to CFE-6 is indicated (vv). Shown diagrammatically are the positions of previously identified preferred recombination sites A – G (Sheets et al., 1992). Also indicated is the infectivity of genomic DNA for canine-D17 cells, presumably representing polytropic FeLV-B replication.
Infectivity of integrated FeLV-B proviruses in tumor DNA
The results of recombination site analysis demonstrated that FeLV-B recombinants in thymic lymphomas contain less endogenous FeLV sequence than do recombinants in multicentric lymphomas (Figure 4). As endogenous FeLV-related elements are defective and do not encode infectious particles (McDougall et al., 1994), these observations suggest that the inclusion of increasing amounts of endogenous FeLV-derived sequence in the recombinants in multicentric lymphomas may have reduced their infectivity. To test this possibility, genomic DNA (5 μg) of five experimentally-induced thymic lymphomas, three experimentally-induced multicentric lymphomas, one natural multicentric lymphoma, one natural thymic lymphoma and one case of feline infectious peritonitis was introduced by Lipofectamine Plus-mediated transfection for four hours into canine D-17 cells. Genomic DNA from D-17 cells infected with FeLV-B/Gardner-Arnstein was included as a positive control. At regular intervals thereafter for fifteen days, the culture supernatant was examined by p27Gag ELISA for the presence of FeLV particles. The logic of this approach is that exogenous FeLV-A proviruses present in genomic DNA samples would not be expected to replicate in canine cells because of their ecotropic host range. Rather, considering the expanded host range of FeLV-B that includes canine cells (Neil et al., 1991), FeLV replication in this assay is assumed to result from infectious FeLV-B proviruses in genomic DNA. The results demonstrated infectious FeLV-B in FeLV-B/Gardner-Arnstein-infected D-17 cells and from four cases of thymic lymphoma, all of which utilized recombination site E. Of the remaining samples, all of which utilized recombination sites downstream of site G, none demonstrated infectious virus (Figure 4). These findings are consistent with an earlier report that associated increasing endogenous SU content with reduced infectivity of FeLV-B. Indeed, analysis of chimeric virus constructs demonstrated lack of infectivity when most of the recombinant SU gene was derived from CFE-6 (Pandey et al., 1991). Among the natural recombinants examined in the present study was that from thymic lymphoma 1110, which was the original source of the myc oncogene-containing FeLV isolate designated LC-FeLV (Levy, Gardner, and Casey, 1984). In this case, the rapid induction of lymphoma 1110 by an oncogene-containing virus may have obviated the need for FeLV-B in pathogenesis. In the remaining cases, including all multicentric lymphomas examined, the lack of infectivity indicates that the FeLV-B proviruses are defective and unlikely to have played a causal role in disease induction.
In summary, the data presented here confirm previous demonstrations of the frequent association of FeLV-B recombinants with thymic lymphoma in the domestic cat (Jarrett, 1980; Jarrett, 1992; Roy-Burman, 1995; Sheets et al., 1993; Tsatsanis et al., 1994), but show further that FeLV-B recombinants occur infrequently and at low levels in non-T-cell diseases including multicentric lymphoma (Figures 1 and 2; Tables 1 and 2). It is noteworthy that the multicentric lymphomas examined in the present study were induced by FeLV-945, or by chimeric viruses containing the LTR and SU gene of FeLV-945; thus, the absence of FeLV-B may reflect infrequent or inefficient recombination during FeLV-945 infection. Further, while FeLV-B proviruses in the genomic DNA of thymic lymphomas were generally infectious, those in the DNA of multicentric lymphomas were not. The lack of infectivity may reflect the relatively distal recombination junction and consequent abundance of endogenously-derived sequence in the FeLV-B genomes (Figures 3 and 4). The findings indicate that the generation of recombinant FeLV-B is unlikely to play a significant role in the induction of non-T-cell disease including multicentric lymphoma as evidenced by the infrequent detection and low levels of FeLV-B and their lack of infectivity. Of note in this regard is a fatal epidemic of FeLV infection recently described in the Florida panther, in which clinical findings were consistent with FeLV-related diseases of non-T-cell origin. The virus isolate, designated FeLV-Pco, was shown to be closely related and perhaps identical to FeLV-945. Indeed, it was suggested that the outbreak may have been caused by cross-species transmission from a domestic cat, perhaps one infected with FeLV-945 (Brown et al., 2008). If recombination with endogenous FeLV-related elements is not required for disease induction by FeLV-945, then wild felids lacking such sequences in the genome may indeed be susceptible to infection and disease by this virus.
Acknowledgments
Statistical support from Dr. Sudesh Srivastav and technical contributions of Patricia Lobelle-Rich are acknowledged with gratitude. This work was supported by NIH grant CA083823 from the National Cancer Institute and by support from the Louisiana Cancer Research Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shamim Ahmad, Email: sahmad1@tulane.edu.
Laura S. Levy, Email: llevy@tulane.edu.
References
- Bechtel MK, Mathes LE, Hayes KA, Phipps AJ, Roy-Burman P. In vivo evolution and selection of recombinant feline leukemia virus species. Virus Res. 1998;54(1):71–86. doi: 10.1016/s0168-1702(98)00015-x. [DOI] [PubMed] [Google Scholar]
- Brown MA, Cunningham MW, Roca AL, Troyer JL, Johnson WE, O’Brien SJ. Genetic characterization of feline leukemia virus from Florida panthers. Emerg Infect Dis. 2008;14(2):252–9. doi: 10.3201/eid1402.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandhasin C, Coan PN, Levy LS. Subtle mutational changes in the SU protein of a natural feline leukemia virus subgroup A isolate alter disease spectrum. J Virol. 2005;79(3):1351–60. doi: 10.1128/JVI.79.3.1351-1360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandhasin C, Coan PN, Pandrea I, Grant CK, Lobelle-Rich PA, Puetter A, Levy LS. Unique long terminal repeat and surface glycoprotein gene sequences of feline leukemia virus as determinants of disease outcome. J Virol. 2005;79(9):5278–87. doi: 10.1128/JVI.79.9.5278-5287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandhasin C, Lobelle-Rich P, Levy LS. Feline leukaemia virus LTR variation and disease association in a geographical and temporal cluster. J Gen Virol. 2004;85(Pt 10):2937–42. doi: 10.1099/vir.0.80149-0. [DOI] [PubMed] [Google Scholar]
- Coelho FM, Bomfim MR, de Andrade Caxito F, Ribeiro NA, Luppi MM, Costa EA, Oliveira ME, Da Fonseca FG, Resende M. Naturally occurring feline leukemia virus subgroup A and B infections in urban domestic cats. J Gen Virol. 2008;89(Pt 11):2799–805. doi: 10.1099/vir.0.2008/003855-0. [DOI] [PubMed] [Google Scholar]
- Donahue PR, Quackenbush SL, Gallo MV, deNoronha CM, Overbaugh J, Hoover EA, Mullins JI. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65(8):4461–9. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 1997;5(2):74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- Jarrett O. Natural Occurrence of Subgroups of Feline Leukemia Virus. In: Essex M, Todaro G, zur Hausen H, editors. Viruses in Naturally Occurring Cancers. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1980. pp. 603–611. [Google Scholar]
- Jarrett O. Pathogenicity of feline leukemia virus is commonly associated with variant viruses. Leukemia. 1992;6(Suppl 3):153S–154S. [PubMed] [Google Scholar]
- Kumar DV, Berry BT, Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989;63(5):2379–84. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque KS, Bonham L, Levy LS. flvi-1, a common integration domain of feline leukemia virus in naturally occurring lymphomas of a particular type. J Virol. 1990;64(7):3455–62. doi: 10.1128/jvi.64.7.3455-3462.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy LS. Advances in understanding molecular determinants in FeLV pathology. Vet Immunol Immunopathol. 2008;123(1–2):14–22. doi: 10.1016/j.vetimm.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy LS, Fish RE, Baskin GB. Tumorigenic potential of a myc-containing strain of feline leukemia virus in vivo in domestic cats. J Virol. 1988;62(12):4770–3. doi: 10.1128/jvi.62.12.4770-4773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy LS, Gardner MB, Casey JW. Isolation of a feline leukaemia provirus containing the oncogene myc from a feline lymphosarcoma. Nature. 1984;308(5962):853–6. doi: 10.1038/308853a0. [DOI] [PubMed] [Google Scholar]
- McDougall AS, Terry A, Tzavaras T, Cheney C, Rojko J, Neil JC. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol. 1994;68(4):2151–60. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil JC, Fulton R, Rigby M, Stewart M. Feline leukaemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol. 1991;171:67–93. doi: 10.1007/978-3-642-76524-7_4. [DOI] [PubMed] [Google Scholar]
- Overbaugh J, Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292(5519):1106–9. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- Pandey R, Ghosh AK, Kumar DV, Bachman BA, Shibata D, Roy-Burman P. Recombination between feline leukemia virus subgroup B or C and endogenous env elements alters the in vitro biological activities of the viruses. J Virol. 1991;65(12):6495–508. doi: 10.1128/jvi.65.12.6495-6508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schaffer AA, Agarwala R, Narfstrom K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, O’Brien SJ. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17(11):1675–89. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezanka LJ, Rojko JL, Neil JC. Feline leukemia virus: pathogenesis of neoplastic disease. Cancer Invest. 1992;10(5):371–89. doi: 10.3109/07357909209024796. [DOI] [PubMed] [Google Scholar]
- Roca AL, Nash WG, Menninger JC, Murphy WJ, O’Brien SJ. Insertional polymorphisms of endogenous feline leukemia viruses. J Virol. 2005;79(7):3979–86. doi: 10.1128/JVI.79.7.3979-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AL, Pecon-Slattery J, O’Brien SJ. Genomically intact endogenous feline leukemia viruses of recent origin. J Virol. 2004;78(8):4370–5. doi: 10.1128/JVI.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Burman P. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes. 1995;11(2–3):147–61. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- Sheets RL, Pandey R, Jen WC, Roy-Burman P. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol. 1993;67(6):3118–25. doi: 10.1128/jvi.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets RL, Pandey R, Klement V, Grant CK, Roy-Burman P. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology. 1992;190(2):849–55. doi: 10.1016/0042-6822(92)90924-e. [DOI] [PubMed] [Google Scholar]
- Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58(3):825–34. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R, Cattori V, Willi B, Meli ML, Gomes-Keller MA, Lutz H, Hofmann-Lehmann R. Copy number polymorphism of endogenous feline leukemia virus-like sequences. Mol Cell Probes. 2007;21(4):257–66. doi: 10.1016/j.mcp.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tsatsanis C, Fulton R, Nishigaki K, Tsujimoto H, Levy L, Terry A, Spandidos D, Onions D, Neil JC. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J Virol. 1994;68(12):8296–303. doi: 10.1128/jvi.68.12.8296-8303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]