Abstract
Specific activation of amino acids by aminoacyl-tRNA synthetases is essential for maintaining translational fidelity. Here, we present crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase (sTrpRS) in apo form and in complexes with various ligands. In each complex, there is a sulfate ion bound at the active site which mimics the α- or β-phosphate group of ATP during tryptophan activation. In particular, in one monomer of the sTrpRS–TrpNH2O complex, the sulfate ion appears to capture a snapshot of the α-phosphate of ATP during its movement towards tryptophan. Simulation study of a human TrpRS–Trp–ATP model shows that during the catalytic process the α-phosphate of ATP is driven to an intermediate position equivalent to that of the sulfate ion, then moves further and eventually fluctuates at around 2 Å from the nucleophile. A conserved Arg may interact with the oxygen in the scissile bond at the transition state, indicating its critical role in the nucleophilic substitution. Taken together, eukaryotic TrpRSs may adopt an associative mechanism for tryptophan activation in contrast to a dissociative mechanism proposed for bacterial TrpRSs. In addition, structural analysis of the apo sTrpRS reveals a unique feature of fungal TrpRSs, which could be exploited in rational antifungal drug design.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) are an ancient family of enzymes that play a central role in protein synthesis. They specifically catalyze the transfer of amino acids to their cognate tRNAs, which is critical for maintaining the fidelity of the translation process. Basically, the aminoacylation reaction proceeds in two steps including the phosphoryl transfer from ATP to the amino acid to generate an aminoacyl-AMP intermediate (the amino acid activation step) and the subsequent attachment of the aminoacyl moiety of the activated amino acid to the acceptor stem of the cognate tRNA (the acyl-transfer step) (1). According to the sequence homology and structural similarity, aaRSs can be divided into two classes (2–5). Class I aaRSs have two highly conserved motifs (HIGH and KMSKS) at the catalytic domain which contains a Rossmann fold (RF) composed of parallel β-strands and α-helices; Class II aaRSs are characterized by three conserved sequence motifs and the catalytic domain exhibits a unique fold of anti-parallel β-strands flanked by α-helices. Each class can be further grouped into three subclasses with the synthetases within the same subclass sharing common sequence and structural features (6,7). Tryptophanyl-tRNA synthetase (TrpRS) belongs to subclass Ic, containing an additional GXDQ motif besides the HIGH and KMSKS motifs.
Due to the importance of the phosphoryl transfer reaction in a vast variety of biological processes, the mechanisms underlying nucleophilic substitution at phosphoryl groups have received much attention (8,9). For these reactions, two mechanisms (dissociative and associative) have been proposed: in a reaction proceeding with a dissociative mechanism, before the nucleophillic attack the bond to the leaving group is already weakened or broken, leading to formation of a hydrated PO3O− anion; while in that with an associative mechanism, the nucleophile approaches Pα before the bond breaks and a pentavalent phosphorus forms at the transition state (10). However, the differences between the associative and dissociative mechanisms are subtle with the energetics of the two mechanisms in solution being comparable, suggesting that phosphoryl transfer reactions could proceed with either mechanism (10).
Recently, structural and computational studies of TrpRSs from Baccillus stearothermophilus (bTrpRS) (11–19) and Homo sapiens (hTrpRS) (20–26) have been performed to investigate how the enzymes catalyze the phosphoryl transfer during the Trp activation step. Comparison of the eukaryotic TrpRS with the bacterial TrpRS demonstrates marked differences in various aspects. Specifically, structural comparison of the bTrpRS–TrpNH2O–ATP complex (representing the pre-transition state) and the bTrpRS–TrpAMP complex (representing the product state) revealed an average movement of the Cα atoms of the KMSKS loop by 1.3 Å, indicating that the phosphoryl transfer in Trp activation occurs with a rearrangement of the KMSKS loop (13,14), which is supported by further simulation study of the bTrpRS–Trp–ATP–Mg2+ complex (15). In contrast, analysis of the corresponding hTrpRS structures shows a similar configuration of the equivalent KMSAS loop without significant positional displacement (26). Recently, Retailleau et al. proposed that bTrpRS might utilize a dissociative mechanism in the Trp activation step (16–18) mainly based on the structure of bTrpRS in complex with an ATP analog, adenosine-5′ tetraphosphate (AQP) and a transition-state model derived from this structure (17). In particular, at the active site, two lysine residues (Lys111 in an insertion and Lys195 of the KMSKS loop) are suggested to play important roles in the Trp activation reaction, which is confirmed by the recent mutational studies by Weinreb et al. (19). However, in eukaryotic TrpRSs, Lys111 is missing and Lys195 is replaced by an Ala which obviously is incapable of exerting similar functions as Lys195 of bTrpRS. Taken together, the differences in the configurations of the key KMSAS motif observed in the binary complexes and the absence of the critical Lys residues suggest that the tryptophan activation reaction of eukaryotic TrpRS might proceed with an alternative mechanism from that of bacterial TrpRS.
Here, we report the crystal structures of Saccharomyces cerevisiae TrpRS (sTrpRS) in the apo form and in complexes with Trp, TrpNH2O (a Trp analog), and TrpAMP, respectively. Intriguingly, in each complex a sulfate ion is bound at the active site, and in particular, in one monomer of the sTrpRS–TrpNH2O complex, the sulfate ion occupies a unique position and appears to mimic an intermediate state of the α-phosphate of ATP during the nucleophilic substitution reaction. We also present the results of simulation studies of the hTrpRS–TrpNH2O–ATP structure and the modeled hTrpRS–Trp–ATP and hTrpRS–ATP complexes. Our structural and computational studies not only reveal the key structural elements and residues of sTrpRS involved in the recognition and binding of the ligands, but also provide new insights into the catalytic mechanism of Trp activation by eukaryotic TrpRSs. Additionally, by detailed comparison of the structure of the apo sTrpRS with that of hTrpRS, we identify a potential drug-targeting site of fungal TrpRSs.
MATERIALS ANS METHODS
Cloning, expression and purification of sTrpRS
The yeast trpS gene fragment was amplified by PCR using genomic DNA of S. cerevisiae as the template and subsequently inserted into the NcoI and XhoI restriction sites of the pET3E expression vector (Novagen) which adds a hexahistidine tag to the C-terminus of the protein product. The plasmid was transformed to the Escherichia coli strain BL21 (DE3) and when the transformed cells reached an OD600 of 0.6–0.8, 1 mM IPTG was added to induce the expression of sTrpRS. After 4 more hours’ culture at 37°C, the E. coli cells were harvested and lysed by sonication in the lysis buffer (50 mM NaH2PO4/Na2HPO4, pH 8.0, 300 mM NaCl, 5 mM 2-mercaptoethanol and 1 mM PMSF).
Purification of the sTrpRS protein was carried out with Nickel affinity chromatography using an Ni-NTA agarose column (Qiagen) following the standard protocol. The supernatant fraction of the lysate was applied onto an Ni-affinity column pre-equilibrated with the lysis buffer, and the column was loaded with the washing and elution buffers (the lysis buffer supplemented with 20 mM and 200 mM imidazole, respectively) in turn. The elution fractions containing the purified sTrpRS protein were pooled, followed by dialyzation against the storage buffer (20 mM Tris–HCl, pH 8.0, 2 mM MgCl2, 5 mM 2-mercaptoethanol and 1 mM PMSF). The protein solution was then concentrated to 15 mg/ml for crystallization experiments.
Crystallization and diffraction data collection
Screening of crystallization conditions was carried out at 20°C using the hanging drop vapor diffusion method. The drops were prepared by mixing equal volumes (2 µl each) of the protein solution and the crystallization solution. Crystals of the apo sTrpRS were obtained from drops with the crystallization solution containing 0.1 M MES buffer, pH 6.5, 0.05 M CsCl and 30% v/v Jeffamine M-600 Reagent. The crystals grew to maximum dimensions of 0.2 × 0.2 × 0.2 mm3 within 2 days. Crystals of sTrpRS in complexes with Trp or Trp analog TrpNH2O were obtained from drops with the crystallization solution [0.1 M HEPES, pH 5.9, and 2.2 M (NH4)2SO4] supplemented with 0.5 µl of 20 mM Trp or TrpNH2O. Both types of crystals grew to maximum dimensions of 0.3 × 0.3 × 0.2 mm3 within a week. Crystals of sTrpRS in complex with TrpAMP were obtained by adding 0.5 µl of 100 mM ATP to a drop containing the pregrown crystals of the sTrpRS–Trp complex, as it has been shown by the previous study that in the presence of Trp and ATP, the Trp activation reaction took place in the crystallization solution and crystals, leading to the formation of TrpAMP (26). Data collection was performed at about 48 h later to allow the completeness of the reaction. The X-ray diffraction data of the apo sTrpRS and the sTrpRS complexes were collected from flash-cooled crystals at beamlines BL6A and NW12 of Photon Factory, Japan, respectively. The diffraction data were processed using the HKL2000 program package (27), and the crystal parameters and the data-processing statistics are summarized in Table 1.
Table 1.
X-ray diffraction data and structure refinement statistics
| E | E-Trp | E-TrpNH2O | E-TrpAMP | ||
|---|---|---|---|---|---|
| Statistics of diffraction data | |||||
| Space group | P43212 | P61 | P61 | P61 | |
| Cell parameters | |||||
| a = b (Å) | 55.9 | 252.6 | 252.3 | 251.8 | |
| c (Å) | 313.8 | 111.8 | 111.4 | 111.1 | |
| Resolution range (Å) | 50.0–2.1 | 50.0–2.8 | 50.0–3.0 | 50.0–2.6 | |
| (2.18–2.10)a | (2.90–2.80) | (3.11–3.00) | (2.69–2.60) | ||
| No. of observed reflections | 268 709 | 665 661 | 450 584 | 532 795 | |
| No. of unique reflections (I/σ(I)> 0) | 29 469 | 99 766 | 79 287 | 118 348 | |
| Average redundancy | 9.1 (9.5) | 6.7 (5.1) | 5.7 (5.4) | 4.5 (2.3) | |
| I/σ(I) | 24.1 (4.0) | 21.4 (3.3) | 16.0 (2.3) | 18.8 (3.2) | |
| Completeness (%) | 96.6 (98.5) | 99.6 (98.7) | 97.9 (95.1) | 96.3 (83.4) | |
| Rmerge (%)b | 10.2 (58.6) | 8.6 (36.3) | 11.7 (50.7) | 5.6 (34.4) | |
| Statistics of refinement and model | |||||
| No. of reflections (Fo ≥ 0σ(Fo)) | 29 427 | 99 520 | 79 131 | 118 158 | |
| Working set | 27 956 | 94 506 | 75 143 | 112 240 | |
| Free Rset | 1471 | 5014 | 3988 | 5918 | |
| Rfactor (%)c | 20.7 | 21.6 | 24.7 | 21.4 | |
| Free Rfactor (%) | 23.8 | 24.7 | 27.3 | 23.7 | |
| No. of protein residues | 373 | 1630 | 1628 | 1630 | |
| Average B factor of all atoms (Å2) | 37.4 | 67.2 | 80.6 | 53.4 | |
| Protein main-chain atoms | 36.2 | 66.6 | 80.7 | 53.0 | |
| Protein side-chain atoms | 38.5 | 67.7 | 80.6 | 53.8 | |
| Ligand atoms | – | 59.9 | 74.0 | 33.8 | |
| RMS bond lengths (Å) | 0.008 | 0.007 | 0.006 | 0.006 | |
| RMS bond angles (°) | 1.1 | 1.0 | 0.9 | 1.0 | |
| Ramachandran plot (%) | |||||
| Most favored regions | 95.7 | 93.5 | 93.0 | 93.9 | |
| Allowed regions | 4.3 | 6.5 | 6.9 | 5.9 | |
| Generously allowed | 0.0 | 0.0 | 0.1 | 0.1 |
aNumbers in parentheses refer to the highest resolution shell.
bRmerge = ||I0| − <I>|/<I>.
cRfactor = ||Fo| − |Fc||/|Fo.
In an attempt to obtain the structure of sTrpRS in complex with TrpNH2O and ATP, crystallization trials were set out by co-crystallization of the enzyme with TrpNH2O and ATP. Crystals grew in drops containing 2 µl each of the protein solution and the crystallization solution [0.1 M HEPES, pH 5.9 and 2.2 M (NH4)2SO4] supplemented with 0.5 µl of 20 mM TrpNH2O and 100 mM ATP. However, structure determination indicated that no ATP is bound at the active site of the structure, while TrpNH2O and a sulfate ion are bound at the active site in the same way as in the TrpNH2O binary complex. The binding of the sulfate ion instead of ATP is mostly likely due to the presence of ammonium sulfate as the precipitant.
Structure determination and refinement
The structure of the apo sTrpRS was solved by the molecular replacement method implemented in the program CNS (28) using the structure of the apo hTrpRS (PDB code 1O5T) (22) as the search model. The structures of the sTrpRS–Trp, sTrpRS–TrpNH2O and sTrpRS–TrpAMP complexes were determined by the molecular replacement method implemented in the program PHASER (29) of the CCP4 suite (30) using the structure of the apo sTrpRS as the starting model. Initial structure refinement was carried out with the program CNS following the standard protocol and final structure refinement was performed using the maximum likelihood algorithm implemented in the program REFMAC5 (31). Model building was performed manually with the programs COOT (32) and O (33). For the sTrpRS complexes, there are two homodimers in an asymmetric unit and the two dimers were refined independently. Throughout the refinement, 5% of randomly chosen reflections were set aside for free R factor monitor, a bulk solvent correction was applied and PROCHECK (34) was used to check the stereochemical quality of these structural models. A summary of the statistics of the structure refinement is listed in Table 1.
Molecular modeling and dynamics simulation
Due to the impossibility of obtaining a crystal structure of the transition-state complex, computational studies of the catalytic process were carried out with construction of an hTrpRS–Trp–ATP model and subsequent molecular dynamics (MD) simulations of the complex. This model was built based on the structure of the hTrpRS–TrpNH2O–ATP complex by replacement of TrpNH2O with Trp. Due to the unavailability of an Mg2+-containing structure of eukaryotic TrpRS, an Mg2+ ion was docked to the complex based on the bTrpRS–ATP–Mg2+ structure (13) and simulations of the hTrpRS–Trp–ATP complex were performed in the presence or absence of the ion. It should be noted that, in the eukaryotic TrpRS the metal ion may take a configuration different from that of the docked Mg2+ ion, as the electrostatic microenvironment at the active site and the conformational changes associated with the catalysis are different between the eukaryotic and bacterial TrpRSs. For comparison, a model of the hTrpRS–ATP complex was constructed by removal of the TrpNH2O molecule from the hTrpRS–TrpNH2O–ATP structure, and simulations of the hTrpRS–ATP model and the hTrpRS–TrpNH2O–ATP complex were also performed. MD simulations were carried out using the software package Groningen Machine for Chemical Simulations (GROMACS) 3.3 (35) with the GROMOS 53a6 force field (36). The topologies and parameters of Trp, TrpNH2O and ATP were defined by the PRODRG server using the GROMOS 96.1 force field (37). The starting structures were first subjected to energy minimization, and subsequently placed in the center of a box which extended a minimum of 10 Å from the protein surface and solvated with Simple Point Charge (SPC) water molecules (38). In addition to the proteins and ligands, the system contains about 14 000 water molecules and 7 or 8 Na+ ions to neutralize the charges in the system. A short equilibration run of 20 ps was then performed with the protein atoms being restrained and the system gradually heated to 300 K, followed by 5 ns of the production run with the temperature maintained at 300 K and the system pressure coupled using the Berendsen method (39). The electrostatic interactions were evaluated using the Particle-Mesh Ewald method (40,41) with a real space cut-off of 1.0 nm and the Van der Waals interactions were modeled by a Lennard-Jones potential with a 1.4 nm cut-off. All of the bonds were constrained for the protein using the LINCS algorithm (42). Simulations of each set-up were repeated four times with different random initial velocities. The overall root mean square fluctuations of the proteins and the theoretical B factors were calculated using the Cα atoms only.
RESULTS
Overall structures
The crystal structure of the apo sTrpRS was determined at 2.1 Å resolution (Table 1 and Figure 1A). In this structure, the asymmetric unit contains one monomer, and two monomers related by a 2-fold crystallographic symmetry form a homodimer. This structure is almost identical to that reported by Malkowski et al. (43) with a root mean square deviation (RMSD) of 0.2 Å for all Cα atoms (hereafter our structure of the apo sTrpRS will be used for further discussion). The structures of sTrpRS in complexes with Trp, TrpNH2O and TrpAMP (Figure 1B) were determined at a resolution of 2.8 Å, 3.0 Å and 2.6 Å, respectively. In all these complex structures, there are two homodimers in one asymmetric unit which are related by 2-fold noncrystallographic symmetry. Respective superposition of the two dimers of the asymmetric unit for each complex all yields an RMSD of ∼0.3 Å for all Cα atoms, indicating that the two dimers are quite similar.
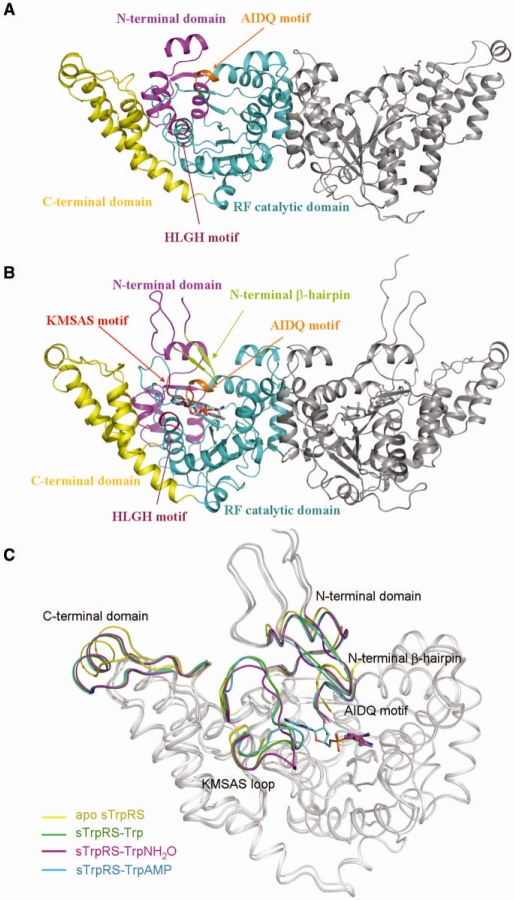
Figure 1.
Overall structures of sTrpRS in different ligand-bound forms. (A) Overall structure of sTrpRS in apo form. Two 2-fold symmetry-related monomers form a dimer. For simplicity, only one monomer is colored and labeled: the N-terminal domain in magenta, the RF catalytic domain in cyan, the C-terminal domain in yellow, and the AIDQ and HLGH motifs in orange and pink, respectively. The N-terminal 44 residues (including the β-hairpin) and a region (residues 292–299) containing the conserved KMSAS loop are disordered. (B) Overall structure of sTrpRS in complex with TrpAMP. The overall structures of sTrpRS in complexes with Trp, TrpNH2O, and TrpAMP are very similar and the sTrpRS-TrpAMP structure is presented as a representative. The bound TrpAMP molecules are shown with ball-and-stick models and colored in light grey. The color codings of the three domains and the AIDQ and HLGH motifs are the same as in (A). The N-terminal β-hairpin and the KMSAS motif are defined in all complexes and are colored in limon and red, respectively. (C) Superposition of the sTrpRS structures in different ligand-bound forms. The superposition is based on the core region of the RF catalytic domain (residues 100–254 and 264–289). Several regions undergo marked conformational changes upon ligand binding, which are differentially colored in the sTrpRS structures with the apo form in yellow and the complexes with Trp in green, with TrpNH2O in magenta and with TrpAMP in cyan. The bound ligands are shown with ball-and-stick models and colored accordingly. (D) The asymmetry between the two monomers of the homodimeric sTrpRS in the sTrpRS–Trp complex. Conformational differences of the β-hairpin, the KMSAS loop and the C-terminal domain between monomer A (green) and monomer B (purple) of the sTrpRS–Trp complex are demonstrated. At the active site, the asymmetry of the structure is accompanied by the differed interactions between the KMSAS loop and the β-hairpin. The hydrogen bonds between Thr293 and Gly34 are denoted as dashed lines. The monomers of the sTrpRS–TrpNH2O complex or the sTrpRS–TrpAMP complex are asymmetric in a similar way, and hence are not shown. (E and F) Stereoviews showing the interactions between (E) Trp or (F) the AMP moiety of TrpAMP and the surrounding residues at the active site in the sTrpRS–TrpAMP complex. The Trp binding modes are very similar in the sTrpRS–Trp and sTrpRS–TrpNH2O complexes, and hence are not shown. The involved hydrogen bonds are indicated with dashed lines.
Similar to hTrpRS, sTrpRS is composed of three domains (Figure 1A and B): an N-terminal domain (residues 1–99), an RF catalytic domain (residues 100–308 and 399–432) and a C-terminal α-helical domain (residues 309–398). As shown in Figure 1A and B, the catalytic active site of sTrpRS resides in a deep pocket of the RF domain and contains a Trp-binding subsite and an ATP-binding subsite. It is composed of several conserved structural elements including the N-terminal β-hairpin (residues 25–33), the KMSAS loop (residues 289–303), the HLGH motif (residues 117–120, helix η1), the AIDQ motif (residues 256–259, helix η5 and α15) and the linker region (residues 282–289) which connects the KMSAS loop to the core of the RF domain (the nomenclature of the secondary structure of sTrpRS is after that of hTrpRS defined by Yang et al. (25) for consistency). In the apo structure, all the residues are evident with good electron density except the N-terminal 44 residues (including the β-hairpin) and a region (residues 292–299) containing the conserved KMSAS loop. In the binary complex structures, residues 21 to 398, including the β-hairpin and the KMSAS loop are resolved, while the N-terminal 20 residues are not observed. The invisibility of the N-terminal part is probably due to the internal flexibility of this segment rather than degradation of the region as the full-length protein was present in the crystals according to the result of the SDS–PAGE analysis (data not shown). In the complex structures, at each active site the bound ligand (Trp, TrpNH2O or TrpAMP) displays high quality electron density (Supplementary Figure S1). Intriguingly, there is also a sulfate ion bound at the active site of each monomer of all three complexes (see the section ‘sulfate binding at the active site’ later).
Comparisons of the apo structure with the complexed structures show that the apo sTrpRS assumes an open conformation with the N- and C-termini relatively far from the RF domain (Figure 1C and Supplementary Table S1). Upon ligand binding, sTrpRS transforms to a compact conformation with the movement of the AIDQ motif towards the active site, followed by the rotation of the N- and C-terminal domains towards the RF domain and the closure of the KMSAS loop taking either a closed (in almost all the complexes) or a quasi-closed (in one monomer of the sTrpRS–TrpNH2O complex, see ‘Discussion’ section later) conformation. The N-terminal β-hairpin is also stabilized covering the active site (Figure 1C and Supplementary Table S1). For bTrpRS, it has been hypothesized that a covalent linkage between an AMP moiety to an occupant of either the pyrophosphate- or the Trp-binding subsite is required for a closed interdomain hinge angle and stabilization of the closed conformation, which is supported by the recent observation that the binding of tryptophan, AMP and phosphate was unable to convert bTrpRS from an open conformation of the apo form to a fully closed conformation (18). In contrast, in both sTrpRS and hTrpRS, the binding of tryptophan alone leads to a closed overall conformation (here and refs 25 and 26), indicating that such a covalent bond is not necessary for the interdomain motions in eukaryotic TrpRSs.
In the apo form, sTrpRS forms a symmetric homodimer with the two monomers related by a 2-fold crystallographic axis. As shown in Figure 1D, superposition of the monomers of the sTrpRS–Trp complex based on the RF domain (residues 100–254 and 264–289) reveals notable structural differences, consistent with an RMSD of ∼1.0 Å for about 400 Cα atoms. Compared with the apo enzyme, although the C-terminal domains of both monomers in the complex rotate towards the RF domain, the rotation angle of the C- terminal domain is slightly larger in one monomer (monomer A, 6.4°) than in the other (monomer B, 4.7°) (Supplementary Table S1), resulting in a more compact conformation of monomer A. Notably, the N-terminal β-hairpin of monomer B reaches deeper into the catalytic active site and, correspondingly, the interactions between the β-hairpin and the KMSAS loop are different. In particular, although Thr293 on the KMSAS loop forms a hydrogen-bonding interaction with the main-chain amide of Gly34 on the β-hairpin in both monomers, the interaction is stronger in monomer B (3.0 Å, via the main-chain carbonyl of Thr293) than in monomer A (3.4 Å, via the side-chain hydroxyl of Thr293) (Figure 1D). In the sTrpRS–TrpNH2O and sTrpRS–TrpAMP complexes, the two monomers exhibit a similar asymmetry (Supplementary Table S1). Asymmetry between the two monomers has also been observed in various hTrpRS complexes (20,21,25,26), indicating that such a phenomenon might be common for eukaryotic TrpRSs.
Binding of the Trp and AMP moieties
The Trp-binding subsite resides deep in the substrate-binding pocket. In the sTrpRS complexes, the bound Trp and TrpNH2O molecules and the tryptophanyl moiety of TrpAMP occupy almost identical positions (Supplementary Figure S2) and have similar interactions with the surrounding residues. Thus, we will use the structure of the sTrpRS–Trp complex as a representative to discuss the binding of Trp. To specifically recognize Trp, sTrpRS forms two hydrogen bonds with the indole nitrogen of Trp via the phenolic hydroxyl of Tyr106 (2.9 Å) and the carboxyl of Glu141 (3.2 Å) (Figure 1E). Moreover, the amino group of Trp is stabilized by the side chains of Glu146 and Gln230 via salt-bridging and hydrogen-bonding interactions (2.9 Å and 3.0 Å, respectively), while the carbonyl group of Trp is positioned by the side-chain amino of Lys147 via a salt bridge. All the residues that are involved in Trp binding are strictly conserved in hTrpRS, except Glu141 which is substituted with Gln194 in hTrpRS (Supplementary Figure S3A), indicating that the Trp recognition and binding mechanism is highly conserved between the lower and higher eukaryotes.
Upon Trp binding, sTrpRS undergoes remarkable conformational changes. With the formation of a hydrogen bond between Trp and Gln230, the Cδ atom of Gln230 is displaced by 2.4 Å and the side chain of Gln230 rotates to form a new hydrogen bond with the side-chain carbonyl group of Gln259 of the AIDQ motif, leading to ∼3 Å movement of the AIDQ motif towards the active site (Figure 1C and Supplementary Table S1). The positional displacement of the AIDQ motif is further coupled with rotations of both the N- and C-terminal domains towards the RF domain, leading to the closure of the overall structure (Figure 1C and Supplementary Table S1). Intriguingly, the N-terminal β-hairpin and the KMSAS loop of sTrpRS which are disordered in the apo structure now become stabilized. Analysis of the interactions between the β-hairpin and the AIDQ motif shows that with the movement of the AIDQ motif, helix α15 where the AIDQ motif resides is also displaced (3–4 Å) and consequently, residues Pro261 and Tyr262 (with the re-oriented side chain) of helix α15 make hydrophobic interactions with Pro29 and Trp30 of the β-hairpin. Additionally, in the vicinity residue Phe213 on helix α12 reorients its side chain towards the cluster, further reinforcing the hydrophobic microenvironment. Meanwhile, the main-chain amide of Gly34 on the β-hairpin forms a hydrogen bond with residue Thr293 on the KMSAS loop, which may attribute to the stabilization of the flexible loop. These findings indicate that upon Trp binding and the AIDQ closure, the β-hairpin plays a ‘bridging’ role for positioning the KMSAS loop at a right location for further ATP binding, which provides the structural basis for the role of the β-hairpin in ATP binding other than Trp binding as demonstrated by the previous biochemical data (25).
In the sTrpRS–TrpAMP complex, the adenine moiety of TrpAMP is covered by the side chain of Phe285, and the N1 and N6 atoms of the adenine moiety have hydrogen-bonding interactions with the main-chain amide and carbonyl of Phe286 (2.8 Å and 3.1 Å) and the main-chain carbonyl of Met296 (3.2 Å), respectively (Figure 1F). The 2′-hydroxyl of the ribose moiety has two hydrogen-bonding interactions with the main-chain amide of Ala256 (2.7 Å) and the side-chain carboxyl of Asp258 (2.6 Å), and the 3′-hydroxyl makes an interaction with the main-chain amide of Ala256 (3.5 Å). The phosphate group forms two hydrogen bonds with the side-chain Nη1 of Arg109 (2.9 Å) and the main-chain amide of Gly110 (2.7 Å) (Figure 1F). All of the interactions between sTrpRS and the AMP moiety are similar to those detected in the hTrpRS–TrpAMP complex (26) (Supplementary Figure S3A), indicating that the AMP binding mode in the product state is all conserved among eukaryotic TrpRSs.
Together, the eukaryotic TrpRSs share a similar TrpAMP binding mode with the archaeal Pyrococcus horikoshii TrpRS (44), which is different from that of the bacterial bTrpRS (Supplementary Figure S3B). Briefly, in the bTrpRS–TrpAMP complex, the indole nitrogen of the tryptophanyl moiety is recognized by Asp132 and Phe5 in contrast to Glu141 and Tyr106 of sTrpRS. In addition, the amino and carbonyl groups are bound by Tyr125 and Gln9, respectively, which take the roles of Glu146, Gln230 and Lys147 of sTrpRS. Most importantly, the α-phosphate is bound by Lys195 of the KMSKS loop which is missing in eukaryotic TrpRSs (see ‘Discussion’ section later).
Sulfate binding at the active site
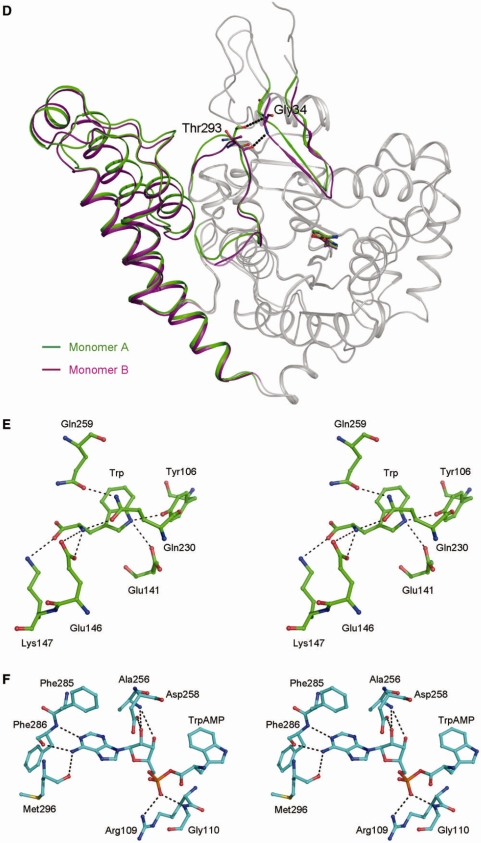
Intriguingly, in the sTrpRS–Trp and sTrpRS–TrpAMP complexes, there is a sulfate ion bound at the active site of each monomer, which is most likely introduced during the crystallization process due to the presence of ammonium sulfate as a precipitant. Binding of sulfate ions by TyrRSs has been observed in the structures of E. coli TyrRS in complexes with tyrosine and TyrAMS, and the apo A. pernix TyrRS (45,46). In the TyrRS structures the sulfate ion occupies a position similar to that of the γ-phosphate of ATP and has no interaction with the KMSKS loop. Structural comparisons of the sTrpRS–Trp and sTrpRS–TrpAMP complexes and the hTrpRS–TrpNH2O–ATP complex indicate that the sulfate ion occupies a position similar to that of the β-phosphate of ATP (Figure 2A). It imposes hydrogen bonds with the main-chain amide of Ala298 on the KMSAS loop and the side chains of His117 and His120 of the HLGH motif, and forms a salt bridge with the side-chain Nη1 of Arg109 on strand β1 (Figure 2B and C). As it has been well-documented that the KMSAS loop of hTrpRS (26) or the KMSKS loop of bTrpRS (14,16) has a high affinity for the β-phosphate rather than the γ-phosphate, the unique position of the sulfate ion and its interaction with the KMSAS loop imply that it mimics the β-phosphate of ATP and correspondingly, the equivalent phosphate of the leaving pyrophosphate group, suggesting that the HLGH motif and the KMSAS loop of sTrpRS are involved in the release of the pyrophosphate during the nucleophilic reaction.
Figure 2.
Sulfate binding in the sTrpRS complexes. (A) Structural comparison of the bound ligands among the sTrpRS complexes and the hTrpRS–TrpNH2O–ATP complex. There is a sulfate ion bound at each active site of the three sTrpRS complexes. Analyses of the sTrpRS complexes and the hTrpRS–TrpNH2O–ATP complex show that during the amino activation reaction the Trp moieties remain at similar positions, while the α-phosphate appears to approach Trp from a distant position observed in the hTrpRS–TrpNH2O–ATP complex (orange) to an intermediate position similar to that of the sulfate ion in monomer A of the sTrpRS–TrpNH2O complex (magenta), and to a final position in the sTrpRS–TrpAMP complex (cyan) when the reaction is completed. The sulfate ions in the sTrpRS–Trp (green) and sTrpRS–TrpAMP (cyan) complexes occupy a position equivalent to that of the β-phosphate of ATP in the hTrpRS–TrpNH2O–ATP complex. For clarity, all the ligands are colored in gray except the α-phosphate of ATP and the sulfate ions. (B–E) Stereoviews showing the interactions of the sulfate ions with the surrounding residues at the active site in (B) the sTrpRS-Trp complex, (C) the sTrpRS–TrpAMP complex, and (D) monomer A and (E) monomer B of the sTrpRS–TrpNH2O complex, respectively.
In the sTrpRS–TrpNH2O complex, there is also a sulfate ion bound at the active site of each monomer but in different ways compared with that in the sTrpRS–Trp and sTrpRS–TrpAMP complexes (Figure 2D and E). In monomer A of the sTrpRS–TrpNH2O complex, the sulfate ion moves towards TrpNH2O by ∼3 Å to form a hydrogen bond with the amino group of TrpNH2O (2.8 Å) (Figure 2D). Correspondingly, the sulfate ion has no interaction with Ala298 of the KMSAS loop or His117 of the HLGH motif while the interactions with His120 of the HLGH motif and Arg109 of strand β1 remain similar. Consequently, the loss of the interaction between the sulfate ion and the KMSAS loop leads to a quasi-closed conformation of the KMSAS loop. In monomer B, the sulfate ion locates at a position similar to that in the sTrpRS–Trp and sTrpRS–TrpAMP complexes and the KMSAS loop assumes a closed conformation (Figure 2E). Although there are some changes in its interactions with the surrounding residues (Figure 2E), the differences seem to be resulted from the rotation of the ion by a small angle which is not related to the Trp activation process.
The tryptophan analog TrpNH2O differs from Trp in that the carboxyl group of tryptophan is replaced by an amide group, which confers a partial positive charge to TrpNH2O for potential interaction between TrpNH2O and the sulfate ion. However, in monomer B of the sTrpRS–TrpNH2O complex, the position of the sulfate ion and the conformation of the KMSAS loop remain the same as in the sTrpRS–Trp complex, indicating that the change of the electrostatic property of the ligand alone is not sufficient to induce the changes observed in monomer A. Instead, the substantial differences in the sulfate binding mode between monomers A and B can be attributed to the aforementioned asymmetry of the monomers: the relatively stronger interactions between the KMSAS loop and the β-hairpin in monomer B probably stabilizes the closed conformation of the KMSAS loop and hence the interactions between the KMSAS loop and the sulfate ion, whereas in monomer A with the weaker interactions between the β-hairpin and the KMSAS loop, the KMSAS loop is not as stable and hence is unable to restrain the sulfate ion from the attraction of TrpNH2O. Intriguingly, the sulfate ion observed in monomer A of the sTrpRS–TrpNH2O complex is located between the α-phosphate of ATP in the hTrpRS–TrpNH2O–ATP complex and the phosphate moiety of TrpAMP in the sTrpRS–TrpAMP complex in a way that the three ions are basically in line without overlapping each other (Figure 2A), suggesting that the sulfate ion might mimic an intermediate state of the α-phosphate from the pretransition state to the transition state (see ‘Discussion’ section later).
Molecular dynamics simulation studies of the hTrpRS complexes
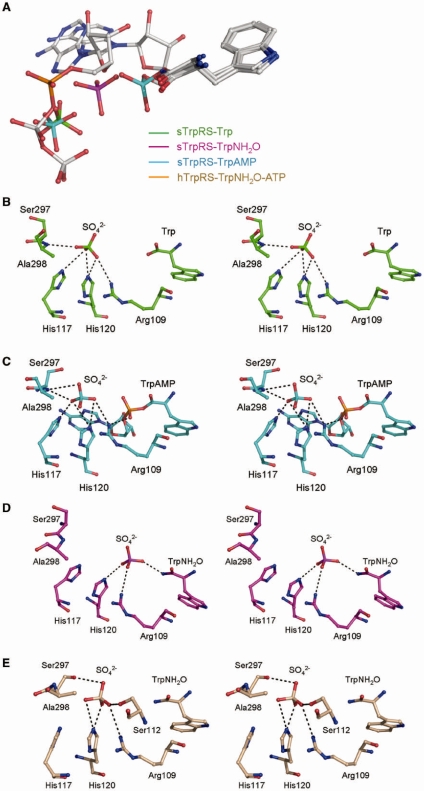
Comparison of the structure of monomer A of the sTrpRS–TrpNH2O complex with those of the hTrpRS–TrpNH2O–ATP (representing the pretransition state) and hTrpRS–TrpAMP/sTrpRS–TrpAMP (representing the product state) complexes (this study and ref. 26) shows that the positions of Trp and its binding modes are almost identical during the reaction, whereas the α-phosphate of TrpAMP shows a positional displacement of 5.5 Å in relation to that of ATP, suggesting that the α-phosphate of ATP moves towards Trp for the nucleophilic attack. This now raises questions about how the α-phosphate group moves and what are the accompanying events at the active site during the catalytic process. For bTrpRS, simulation studies of the bTrpRS–ATP–Mg2+ and bTrpRS–AQP complexes suggest that the reaction proceeds through the transition state with minimum structural and energetic changes and the coupling of the interactions between Mg2+ and ATP and those between ATP and the active-site lysine residues is necessary to sustain the high-energy state (16). Moreover, the Mg2+ functions cooperatively with the KMSKS loop during the reaction (16). Due to the unavailability of a structure of eukaryotic TrpRS in complex with AQP, we built a model of the hTrpRS–Trp–ATP complex based on the crystal structure of the hTrpRS–TrpNH2O–ATP complex and carried out simulations of the model to track the movements of the substrates and the changes of the surrounding residues and/or structural elements during the reaction, and to identify potential interactions that cannot be detected in the crystal structures. For comparison, the model of the hTrpRS–ATP complex was built and simulation studies of this model and the hTrpRS–TrpNH2O–ATP structure were also performed.
For the hTrpRS–TrpNH2O–ATP complex the theoretical B factors of the protein calculated based on the mean of squared atomic displacements from the simulation were compared with the experimental B factors of the crystal structure. As shown in Figure 3A, the theoretical and experimental B factors exhibit good correlation in general except two short regions, providing a good validation of the MD trajectories. As the simulation was performed with a monomer only, it is not surprising that one region (approximately residues 260–290) participating in the dimer formation has higher theoretical B factors. The other region (approximately residues 390–400) is involved in crystal packing, which may account for its lower B factors in the crystal structure. Consistent with the observation that the positions of the Trp moieties are almost identical in the pretransition state and the product state (26), during the 5 ns simulations Trp or TrpNH2O is generally stable showing very small deviations. Therefore, the oxygen bridging the Trp moiety and the AMP group is assumed to occupy a position close to that of the nucleophilic oxygen at the transition state. For clarity, the hTrpRS–TrpAMP complex (26) was superposed to the hTrpRS–TrpNH2O–ATP structure and the bridging oxygen atom between the Trp and AMP moieties served as a reference for the positions of the moving α-phosphate of ATP during all the simulations. For the hTrpRS–Trp–ATP complex which represents the pre-transition state, the simulation results show that the configurations of the residues at the active site remain similar except for residue Arg162 (see ‘Discussion’ section later). In addition, the presence or absence of Mg2+ does not appear to have any significant effect. However, as Mg2+ was docked to the complex based on the bTrpRS–ATP–Mg2+ structure (13), the contribution of Mg2+ in the Trp activation mechanism of eukaryotic TrpRSs needs further assessment. As shown in Figure 3B, in the presence of tryptophan, the Pα atom of ATP moves quickly towards tryptophan during the beginning 200 ps, then oscillates while continuing to approach tryptophan (200–800 ps), followed by stable behavior with modest fluctuations at positions of ∼2 Å from the reference atom (>800 ps). As the bridging oxygen in TrpAMP is assumed to take a position close to that of the attacking nucleophile during the reaction, the results suggest that the distance between the nucleophile of tryptophan and the Pα atom of ATP may be as short as 2 Å, which is close to the distance of ∼1.7 Å proposed for an associative mechanism (17). Such changes only occur when both tryptophan and ATP are present, as in the simulations of the hTrpRS–TrpNH2O–ATP and hTrpRS–ATP complexes, the respective distances between Pα of ATP and the reference position remain at 6–7 Å (Figure 3B), implying that the binding of tryptophan and ATP at the right positions and the subsequent attraction between the carboxylate of tryptophan and the positively charged Pα of ATP initiate the reaction.
Figure 3.
Molecular dynamics simulations of the hTrpRS complexes. (A) Comparison of the theoretical B factors of the Cα atoms of the TrpRS–TrpNH2O–ATP complex during the MD trajectories (black) with the crystallographic B factors (red). The calculated B factors correlate well with the experimental B factors in general. (B) Displacement of Pα of ATP during the simulation of the hTrpRS–Trp–ATP complex. The hTrpRS–TrpAMP complex was superposed with the hTrpRS–TrpNH2O–ATP complex, and the oxygen atom bridging the AMP and Trp moieties of TrpAMP (which adopts a position approximating that of the nucleophilic oxygen at the transition state) served as a reference to the moving α-phosphate of ATP for the simulation studies. During the beginning 1.5 ns of the 5 ns simulations of the hTrpRS–Trp–ATP (red), hTrpRS–TrpNH2O–ATP (black) and hTrpRS–ATP (green) complexes, the respective distances between Pα of ATP and the reference position were measured and compared. (C) Comparison of the conformations of ATP, Trp and Arg162H at 450 ps of the simulation of the hTrpRS-Trp-ATP complex (orange) with those of the sulfate ion, TrpNH2O and Arg109S in monomer A of the sTrpRS-TrpNH2O complex (magenta). The superposed hTrpRS–TrpAMP complex (cyan) is also included as a positional reference. For clarity, the ligands are colored in gray except the α-phosphate groups and the sulfate ion. Residues Arg162S and Arg109H are labeled and colored accordingly. (D) Potential interaction of Arg162H with the oxygen bridging the α-phosphate and the β,γ-pyrophosphate group of ATP at the transition state. During the simulation of the hTrpRS–Trp–ATP complex, an interaction between Arg162 and the bridging oxygen (indicated by a dashed line) is formed when the Pα atom is about 2 Å from the nucleophile. A representative scheme at 860 ps of the simulation is shown. The color codings are the same as above.
Intriguingly, the sulfate ion in monomer A of the sTrpRS-TrpNH2O complex occupies a position similar to that of the α-phosphate of ATP during 200–600 ps of the simulation of the hTrpRS–Trp–ATP complex (with a comparison of the structure with the scheme of the simulation at 450 ps being presented in Figure 3C), implying that the sulfate ion may mimic an intermediate state of the α-phosphate prior to phosphoryl transfer and thus represent a snapshot of the α-phosphate motion during the tryptophan activation reaction. Albeit in monomer A of the sTrpRS–TrpNH2O complex a salt bridge is formed between Arg109 (equivalent to Arg162 of hTrpRS) and the sulfate ion, Arg162 cannot make a salt-bridging or hydrogen-bonding interaction with the α-phosphate group during this period. Further progression of the simulation (>800 ps) shows that in states approximating an associative transition state with Pα of ATP at positions of about 2 Å from the reference oxygen, Arg162 interacts with the oxygen bridging the α-phosphate and the β,γ-pyrophosphate group (Figure 3D), suggesting an important role of Arg162 for the bond cleavage and the stabilization of the leaving group (see ‘Discussion’ later).
DISCUSSION
Mechanism of tryptophan activation by eukaryotic TrpRSs
With multiple bTrpRS structures and the related simulation studies, Retailleau et al. proposed that bTrpRS uses a dissociative mechanism in the tryptophan activation reaction (16–18). In particular, in the bTrpRS–AQP structure, the α-phosphate of AQP shows a positional displacement of 0.4 Å towards the tryptophan carboxylate nucleophile compared with the α-phosphate of ATP in the closed bTrpRS–ATP–Mg2+ complex, and the γ- and δ-phosphates mimic the pyrophosphate leaving group (17). The distance between Pα and the oxygen atom bridging the β- and γ- phosphates is 2.7 Å, which is close to the expected 3.3 Å of a dissociative transition state, hence favoring a catalytic mechanism with dissociative character for bTrpRS (17). Due to the limited structural information of eukaryotic TrpRSs, the molecular mechanism of nucleophilic substitution in the tryptophan activation remains unclear. Analysis of the crystal structures of the sTrpRS complexes and the simulation studies of the hTrpRS complexes presented here shed some light onto the mechanism of Trp activation by eukaryotic TrpRSs.
For bTrpRS, several lysine residues at the active site (Lys111B, Lys192B and Lys195B) have been shown to play important roles in the transition state (16–19) (for simplicity, hereafter the residues of bTprRS, sTrpRS and hTrpRS are denoted with superscripted letters B, S and H, respectively). Specifically, in the transition-state model constructed based on the bTrpRS-AQP structure, Lys195B contributes to charge stabilization of the oxygen atom in the scissile bond, suggesting a critical role of the residue in the nucleophilic attack (17). Consistently, mutation of the residue to Ala resulted in about 100-fold decrease in kcat (19). In eukaryotic TrpRSs, there is no residue equivalent to Lys111B (as Lys111B is located in an insertion of bTrpRS compared with eukaryotic TrpRSs) and the residue equivalent to Lys195B is an Ala residue (Ala298S or Ala352H) which is unable to interact with the oxygen atom. Instead, a sequentially distant Arg (Arg109S or Arg162H) seems to take a similar role as Lys195B. In the hTrpRS–TrpNH2O–ATP complex, Arg162H interacts solely with the γ-phosphate. However, in the hTrpRS–TrpAMP complex the α-phosphate group has a positional displacement of 5.5 Å towards the Trp moiety, and correspondingly the side chain of Arg162H reaches a position similar to Lys195B from an opposite direction and forms a salt bridge with the phosphate of TrpAMP which is equivalent to the α-phosphate of ATP (26). In the sTrpRS–TrpAMP structure, Arg109S binds to the phosphate of TrpAMP (Figure 1F) and additionally to the sulfate ion which mimics the β-phosphate of the leaving group (Figure 2D), suggesting that Arg109S plays a role in stabilization of the newly formed product. Consistently, the computational studies show that during the simulations of the hTrpRS–Trp–ATP complex, Arg162H interacts with the oxygen bridging Pα and Pβ of ATP, implying that Arg162H might facilitate to stabilize the increased negative charge on this atom during phosphoryl transfer (Figure 3D). Together, these findings imply that in eukaryotic TrpRSs, the Arg residue probably compensates for the missing Lys of the KMSKS motif by facilitating the cleavage of the bond between the α- and β-phosphates of ATP and the stabilization of both Pα and the pyrophosphate leaving group.
The proposed critical functions of this Arg residue and the requirement of displacement of the α-phosphate of ATP in the catalytic process well explain the biochemical data (25). Previously, the mutational study showed that Arg162H (equivalent to Arg109S) plays a critical role in ATP binding and/or the catalysis as the R162A mutant of hTrpRS had substantially decreased ATP affinity and catalytic activity (25). When Ala352H (equivalent to Ala298S) was further mutated to generate the R162A/A352K double mutant, the ATP binding affinity was restored, however, the catalytic activity was not rescued, providing further evidence for an indispensible role of Arg162H in the catalysis. Moreover, the R162A/A352K double mutant appeared to have an even lower catalytic activity than R162A with a decreased kcat (0.01 versus 0.05 s−1) (25). Considering that the KMSKS loop where Ala352 resides interacts with the α- and β-phosphate groups in the pre-transition state (26), conversion of Ala352 to Lys might enhance the interactions between the KMSAS loop and the triphosphate group of ATP, and consequently impede the movement of the α-phosphate towards tryptophan during the catalytic process.
Although Arg162H functions in the bond cleavage and the product stabilization, it does not act exactly the same as Lys195B. For bTrpRS, in the modeled transition state, the distance between the Trp carboxylate and Pα is about ∼3.5 Å, which is very close to the expected distance of 3.3 Å for a dissociative transition state, and Lys195B is considered to interact with the oxygen bridging Pα and Pβ of ATP to compensate for the increased negative charge on this oxygen during the reaction (17). In the pre-transition state of hTrpRS (26), Arg162H locates at the opposite side of the phosphate group relative to Lys195B and is unlikely to reach the bridging oxygen between α- and β-phosphates with the distance of 5.3 Å (26). During the simulation of the hTrpRS–Trp–ATP complex, the possibility of formation of a hydrogen bond between Arg162H and the bridging oxygen was examined when Pα of ATP is about 3.3 Å from the tryptophan carboxylate. Our analysis shows that under this circumstance the distance between Arg162H and the bridging oxygen is larger than 4.5 Å, implying that such an interaction is unlikely to form as in bTrpRS. As the interactions of Lys195B of the KMSKS loop and Asn18B of the TIGN motif (equivalent to the HVGH motif of hTrpRS) with this oxygen atom in the scissile bond are important to compensate the increased negative charge on the atom in the proposed dissociative mechanism for bTrpRS (17), the incapability of Arg162H to execute a similar function as Lys195B under an equivalent situation and the replacement of Asn18B by a weakly charged residue His173H would substantially reduce the effect and hence our results do not favor a dissociative mechanism. On the other hand, considering that at the transition state Pα of ATP may move as close as ∼2 Å to the nucleophilic oxygen (Figure 3B) and simultaneously an interaction forms between Arg162H and the oxygen bridging Pα and Pβ of ATP (Figure 3D), our data suggest an associative mechanism of Trp activation by eukaryotic TrpRSs, which is in accordance with the conclusion of the previous investigation of the phosphoryl transfer reaction of bovine TrpRS by examination of the primary and secondary oxygen kinetic isotope effects (47).
Implications for anti-fungal drug design
Due to their essential roles in protein synthesis, aaRSs have been therapeutic targets against microbial infections. To date, mupirocin (BactrobanTM), a potent inhibitor of bacterial IleRS, has been clinically applied to treat skin infections caused by Gram-positive bacteria (48,49). For fungal infections, an LeuRS inhibitor (AN-2690) and an IleRS inhibitor (icofungipen) have been shown to be effective and are currently evaluated under clinical trials (50–53). For suppression of TrpRSs, two antibiotics, namely indolmycin (54,55) and chuangxinmycin (56) which are the biosynthetic derivatives of l-tryptophan, have been developed to treat bacterial infections. However, no inhibitor specific for fungal TrpRSs is available, probably due to the high sequence similarities (>40%) among eukaryotic TrpRSs.
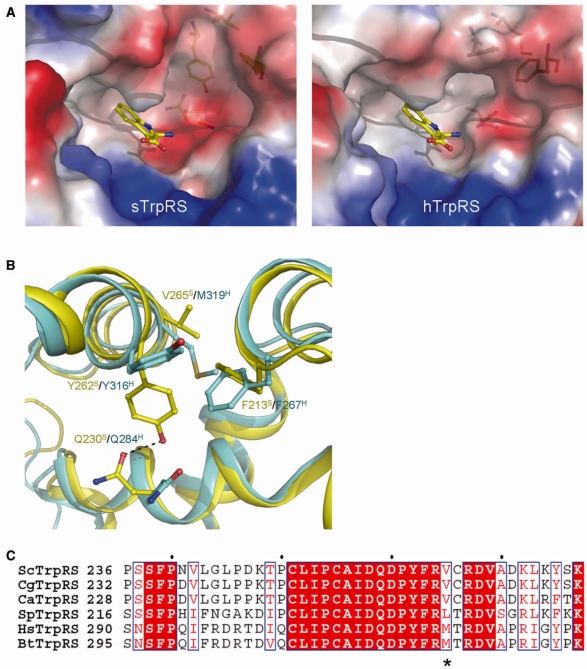
Structural information has been successfully employed in drug design and development by discovering meticulous differences between homologous proteins and revealing the details of interactions between drugs and their target proteins. Recently, crystal structures of human and fungal cytosolic LeuRS editing domains were determined, providing hints for optimization of available drugs such as AN-2690 and rational design of new drugs to improve the potency or enhance the specificity towards particular pathogens (57). Similarly, a detailed comparison between the apo form structures of sTrpRS and hTrpRS (this study and ref. 26) shows that although their overall structures and the structures of the active site are quite similar, local differences exist in the Trp-binding subsite. In particular, compared with hTrpRS, sTrpRS has an additional small cavity at the Trp-binding subsite (Figure 4A). Such a feature is also observed in the previously reported structure of the apo sTrpRS grown from a different crystallization condition (43), indicating that the observed differences are caused by intrinsic characters.
Figure 4.
Conformational differences of the Trp-binding subsite between the apo sTrpRS and hTrpRS structures. (A) Electrostatic surfaces of the Trp-binding pockets in the apo sTrpRS (left panel) and hTrpRS (right panel) structures. A tryptophan molecule is docked to show the Trp-binding subsite. (B) Structural comparison of the Trp-binding subsite of the apo sTrpRS (yellow) with that of the apo hTrpRS (cyan). The residues responsible for the structural differences are labeled and shown with stick-and-ball models. The sequence variance at position Val265 S/Met319H is a key for the structural differences of the Trp-binding subsite. (C) Sequence alignment of sTrpRS with TrpRSs from other yeasts including C. glabrata, C. albicans and S. pombe, and with those from higher eukaryotes including H. sapiens and B. taurus. The position equivalent to that of Val265S/Met319H is marked by an asterisk. The sequence alignment is generated by ESPript (58).
Analysis of the residues comprising the Trp-binding subsite reveals conformational differences of the residues at two positions (corresponding to Tyr262S/Tyr316H and Gln230S/Gln284H, respectively). In sTrpRS, the aromatic ring of Tyr262S points towards the active site interacting with Gln230S via a hydrogen bond, whereas in hTrpRS, the side chain of Tyr316H flips toward outside without a detectable interaction with Gln284H, leaving the side chain of Gln284H in a distinct orientation (Figure 4B). Analysis of the residues in the vicinity shows that in hTrpRS, a neighboring residue Phe267H takes a conformation different from that of the equivalent Phe213S, which does not allow Tyr316H to take a conformation similar to that of Tyr262S. Further analysis shows that the conformational difference between Phe267H and Phe213S is resulted from a sequence difference at a position corresponding to Met319H or Val265S. In the superposed structures, Met319H occupies a similar position as Val265S; however, with a significantly larger side chain, the distance between Met319H and Phe213S is about 1.8 Å, indicating a potential steric conflict. Thus, the side chain of Phe267H takes a conformation different from that of Phe213S, which establishes an acceptable distance of 3.3 Å between Phe267H and Met319H. Therefore, residues Met319H and Val265S appear to play a fundamental role for the observed conformational differences of the Trp-binding subsites between sTrpRS and hTrpRS. Inspiringly, sequence alignment indicates that the Val residue (equivalent to Val265S) is conserved among many yeast genera including Candida, while Met is conserved among higher eukaryotes (Figure 4C). Thus, compounds with selectivity for the Trp-binding subsite of the apo fungal enzymes over that of the mammalian enzymes and further with ability to lock the fungal TrpRSs in an open conformation would be beneficial for specific suppression of fungal TrpRSs in mammalians. Therefore, our findings open a possibility to discriminate between TrpRSs from the pathogenic fungal genera and the mammalian TrpRSs, and hence will allow de novo rationalized drug design targeting fungal TrpRSs.
PROTEIN DATA BANK ACCESSION CODES
The structures of S. cerevisiae TrpRS in apo form and in complexes with Trp, TrpNH2O and TrpAMP have been deposited with the RCSB Protein Data Bank under accession codes 3KT0, 3KT6, 3KT8 and 3KT3, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China (2006AA02A313, 2007CB914302); National Natural Science Foundation of China (30730028, 30770480); Chinese Academy of Sciences (KSCX2-YW-R-107, SIBS2008002); the Science and Technology Commission of Shanghai Municipality (07XD14032, 07ZR14131). Funding for open access charge: the National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members at Photon Factory, Japan for technical support in diffraction data collection and other members of our group for helpful discussion.
REFERENCES
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 3.Burbaum JJ, Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J. Biol. Chem. 1991;266:16965–16968. [PubMed] [Google Scholar]
- 4.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 5.Cusack S. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1997;7:881–889. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 6.Cavarelli J, Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993;7:79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- 7.Cusack S. Eleven down and nine to go. Nat. Struct. Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 8.Admiraal SJ, Herschlag D. Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. Chem. Biol. 1995;2:729–739. doi: 10.1016/1074-5521(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 9.Aqvist J, Kolmodin K, Florian J, Warshel A. Mechanistic alternatives in phosphate monoester hydrolysis: what conclusions can be drawn from available experimental data? Chem. Biol. 1999;6:R71–R80. doi: 10.1016/S1074-5521(99)89003-6. [DOI] [PubMed] [Google Scholar]
- 10.Florian J, Warshel A. Phosphate ester hydrolysis in aqueous solution: associative versus dissociative mechanisms. J. Phys. Chem. B. 1998;102:719–734. [Google Scholar]
- 11.Doublie S, Bricogne G, Gilmore C, Carter C.W., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 12.Ilyin VA, Temple B, Hu M, Li G, Yin Y, Vachette P, Carter C.W., Jr 2.9 Å crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci. 2000;9:218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Retailleau P, Yin Y, Hu M, Roach J, Bricogne G, Vonrhein C, Roversi P, Blanc E, Sweet RM, Carter C.W., Jr High-resolution experimental phases for tryptophanyl-tRNA synthetase (TrpRS) complexed with tryptophanyl-5′AMP. Acta Crystallogr. 2001;D57:1595–1608. doi: 10.1107/s090744490101215x. [DOI] [PubMed] [Google Scholar]
- 14.Retailleau P, Huang X, Yin Y, Hu M, Weinreb V, Vachette P, Vonrhein C, Bricogne G, Roversi P, Ilyin V, et al. Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations. J. Mol. Biol. 2003;325:39–63. doi: 10.1016/s0022-2836(02)01156-7. [DOI] [PubMed] [Google Scholar]
- 15.Kapustina M, Carter C.W., Jr Computational studies of tryptophanyl-tRNA synthetase: activation of ATP by induced-fit. J. Mol. Biol. 2006;362:1159–1180. doi: 10.1016/j.jmb.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 16.Kapustina M, Weinreb V, Li L, Kuhlman B, Carter C.W., Jr A conformational transition state accompanies tryptophan activation by B. stearothermophilus tryptophanyl-tRNA synthetase. Structure. 2007;15:1272–1284. doi: 10.1016/j.str.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Retailleau P, Weinreb V, Hu M, Carter C.W., Jr Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5′ tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases. J. Mol. Biol. 2007;369:108–128. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laowanapiban P, Kapustina M, Vonrhein C, Delarue M, Koehl P, Carter C.W., Jr Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations. Proc. Natl Acad. Sci. USA. 2009;106:1790–1795. doi: 10.1073/pnas.0812752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinreb V, Li L, Campbell CL, Kaguni LS, Carter C.W., Jr Mg2+-assisted catalysis by B. stearothermophilus TrpRS is promoted by allosteric effects. Structure. 2009;17:952–964. doi: 10.1016/j.str.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R, Yokoyama S, Kim S, Nureki O. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment: insights into substrate recognition, tRNA binding, and angiogenesis activity. J. Biol. Chem. 2004;279:8378–8388. doi: 10.1074/jbc.M311284200. [DOI] [PubMed] [Google Scholar]
- 23.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XL, Guo M, Kapoor M, Ewalt KL, Otero FJ, Skene RJ, McRee DE, Schimmel P. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen N, Zhou M, Yang B, Yu Y, Dong X, Ding J. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Res. 2008;36:1288–1299. doi: 10.1093/nar/gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. 2007;D63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 35.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 36.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 37.Schuettelkopf AW, van Aalten D.MF. PRODRG – a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 38.Hermans J, Berendsen H.JC, Van Gunsteren WF, Postma J.PM. A consistent empirical potential for water-protein interactions. Biopolymers. 1984;23:1513–1518. [Google Scholar]
- 39.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 40.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 41.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 42.Hess B, Bekker H, Berendsen H.JC, Fraaije J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 43.Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, Said M, Luft JR, Phizicky EM, DeTitta GT, et al. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc. Natl Acad. Sci. USA. 2007;104:6678–6683. doi: 10.1073/pnas.0610337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong X, Zhou M, Zhong C, Yang B, Shen N, Ding J. Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1053. doi:10.1093/nar/gkp1053 [Epub ahead of print 26 November 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi T, Takimura T, Sekine R, Kelly VP, Kamata K, Sakamoto K, Nishimura S, Yokoyama S. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase. J. Mol. Biol. 2005;346:105–117. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 46.Kuratani M, Sakai H, Takahashi M, Yanagisawa T, Kobayashi T, Murayama K, Chen L, Liu ZJ, Wang BC, Kuroishi C, et al. Crystal structures of tyrosyl-tRNA synthetases from Archaea. J. Mol. Biol. 2006;355:395–408. doi: 10.1016/j.jmb.2005.10.073. [DOI] [PubMed] [Google Scholar]
- 47.Lowe G, Tansley G. An investigation of the mechanism of activation of tryptophan by tryptophanyl-tRNA synthetase from beef pancreas. Eur. J. Biochem. 1984;138:597–602. doi: 10.1111/j.1432-1033.1984.tb07956.x. [DOI] [PubMed] [Google Scholar]
- 48.Fuller AT, Mellows G, Woolford M, Banks GT, Barrow KD, Chain EB. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature. 1971;234:416–417. doi: 10.1038/234416a0. [DOI] [PubMed] [Google Scholar]
- 49.Nakama T, Nureki O, Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 2001;276:47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 50.Petraitis V, Petraitiene R, Kelaher AM, Sarafandi AA, Sein T, Mickiene D, Bacher J, Groll AH, Walsh TJ. Efficacy of PLD-118, a novel inhibitor of candida isoleucyl-tRNA synthetase, against experimental oropharyngeal and esophageal candidiasis caused by fluconazole-resistant C. albicans. Antimicrob. Agents Chemother. 2004;48:3959–3967. doi: 10.1128/AAC.48.10.3959-3967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeates C. Icofungipen (PLIVA) Curr. Opin. Investig. Drugs. 2005;6:838–844. [PubMed] [Google Scholar]
- 52.Ferrer E. Spotlight on targeting aminoacyl-tRNA synthetases for the treatment of fungal infections. Drug News Perspect. 2006;19:347–348. [PubMed] [Google Scholar]
- 53.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 54.Werner RG, Thorpe LF, Reuter W, Nierhaus KH. Indolmycin inhibits prokaryotic tryptophanyl-tRNA ligase. Eur. J. Biochem. 1976;68:1–3. doi: 10.1111/j.1432-1033.1976.tb10758.x. [DOI] [PubMed] [Google Scholar]
- 55.Werner RG, Reuter W. Interaction of indolmycin in the metabolism of tryptophan in rat liver. Arzneimittelforschung. 1979;29:59–63. [PubMed] [Google Scholar]
- 56.Brown MJ, Carter PS, Fenwick AS, Fosberry AP, Hamprecht DW, Hibbs MJ, Jarvest RL, Mensah L, Milner PH, O'H;anlon PJ, et al. The antimicrobial natural product chuangxinmycin and some synthetic analogues are potent and selective inhibitors of bacterial tryptophanyl tRNA synthetase. Bioorg. Med. Chem. Lett. 2002;12:3171–3174. doi: 10.1016/s0960-894x(02)00604-2. [DOI] [PubMed] [Google Scholar]
- 57.Seiradake E, Mao W, Hernandez V, Baker SJ, Plattner JJ, Alley MR, Cusack S. Crystal structures of the human and fungal cytosolic Leucyl-tRNA synthetase editing domains: a structural basis for the rational design of antifungal benzoxaboroles. J. Mol. Biol. 2009;390:196–207. doi: 10.1016/j.jmb.2009.04.073. [DOI] [PubMed] [Google Scholar]
- 58.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.