Abstract
A method is proposed to measure global bending in DNA and RNA structures. It relies on a properly defined averaging of base-fixed coordinate frames, computes mean frames of suitably chosen groups of bases and uses these mean frames to evaluate bending. The method is applied to DNA A-tracts, known to induce considerable bend to the double helix. We performed atomistic molecular dynamics simulations of sequences containing the A4T4 and T4A4 tracts, in a single copy and in two copies phased with the helical repeat. Various temperature and salt conditions were investigated. Our simulations indicate bending by roughly 10° per A4T4 tract into the minor groove, and an essentially straight structure containing T4A4, in agreement with electrophoretic mobility data. In contrast, we show that the published NMR structures of analogous sequences containing A4T4 and T4A4 tracts are significantly bent into the minor groove for both sequences, although bending is less pronounced for the T4A4 containing sequence. The bending magnitudes obtained by frame averaging are confirmed by the analysis of superhelices composed of repeated tract monomers.
INTRODUCTION
Since the first single-crystal structure of a DNA oligomer was resolved 30 years ago (1), it has become clear that the conformation of double-stranded DNA departs from a straight, uniform double helix characteristic for idealized B or A forms. DNA conformation is commonly characterized by intra-base pair local conformational parameters (buckle, propeller, opening, shear, stretch and stagger) a by inter-base pair, or step parameters (tilt, roll, twist, shift, slide and rise) (2). One of the most important structural irregularities is DNA bending. At the level of base pair steps, bending has two components, roll and tilt. Of considerable interest, however, is also global bending, that is, the bending magnitude and direction of a longer DNA fragment as a whole.
Many authors define global bending magnitude as the angle between normal vectors of base pairs at the two ends of the DNA fragment. Dickerson and co-workers (3) proposed to plot the projections of base pair normals onto a reference plane perpendicular to a suitably chosen helical axis, yielding the normal vector plot. Bending magnitude and direction are deduced from vectors connecting the projected normals. While the method provides a quick and easy-to-understand picture of the oligomer bending, its precise quantitative evaluation is made more difficult by various approximations involved in the scheme.
If the base pair normals are not available directly, they can be inferred from the local conformational parameters. The 3DNA code by Lu and Olson (4) includes a rebuilding functionality which enables one to reconstruct the whole oligomer from the values of inter- and intra-base pair parameters. The subsequent analysis by 3DNA of such a rebuilt structure then yields, among other things, all the base pair normals.
A related approach has been adopted in the Madbend code by Strahs and Schlick (5). Here, local roll, tilt and twist are used to compute quantities called global roll and global tilt, which describe both the bending magnitude and direction. The precise meaning of these quantities depends on the definition of local roll, tilt and twist used to calculate them.
While the use of base pair normals provides a rather straightforward relationship between local and global bending, it also has disadvantages. In particular, it emphasizes the influence of local bends, especially at the ends of the oligomer. Thus, methods to smooth out local bending variations have been proposed.
The program Curves developed by Lavery and Sklenar (6) yields a global curvilinear helical axis computed by a least squares optimization scheme. In the recently released Curves+ (7), the global curvilinear axis is obtained from the average local screw axes by polynomial smoothing. The curvilinear axis as computed by Curves has been widely used, not only to evaluate shape but also to estimate global elastic properties of DNA (8,9). The disadvantage of such approaches is a rather complicated relationship between the local structure and global bending defined in this way.
Other approaches have been published. Goodsell and Dickerson (10) proposed a simple smoothing method based on computing arithmetic mean of base pair normals over a window of varying length. Lankas et al. (11) used two segments of a straight helical axis, each associated with a short fragment of the DNA double helix, to characterize the magnitude and direction of kinks in simulated minicircles. Curuksu et al. (12) calculated arithmetic mean of a set of screw axes, each connecting intra-strand neighbouring bases.
The most prominent base sequences that cause the DNA double helix to bend globally are A-tracts, stretches of at least four A·T base pairs without a TpA step (13–15). The bend occurs when A-tracts are inserted into a non-A-tract sequence. If more A-tracts are phased with DNA helical repeat, the individual bends add constructively. The precise magnitude of bending and details of how it is achieved at the local level differ somehow from one structure to another (16,17).
Gel electrophoretic mobility experiments performed by Hagerman (18) and Crothers and co-workers (19,20) showed that sequences containing A5–6 (19) and A4T4 (20) tracts are bent towards the minor groove at the tract centre; in contrast, T4A4 sequences were found to induce no appreciable bending (18). Koo et al. (21) found that an A6 tract bends the DNA helix by 17–21°. The value was deduced by fitting Monte Carlo simulations of a DNA model to cyclization data. Lutter and co-workers (22) proposed a topological method to measure A-tract bending. The A6 tract bend angle they found is in agreement with the previous estimate (21).
Several studies investigated the dependence of the A-tract induced bending on temperature and ionic environment. Koo et al. (23) found a decrease in bending magnitude when passing from 4°C to room temperature, and Diekmann (24) revealed that the decrease with temperature is monotonic (exceptional cases could be explained by a loss of planarity of the construct). The decrease of bending at elevated temperatures has been attributed to a premelting transition of the A-tract, well described by a two-state model (25). Diekmann (24) also found that bending decreases with increasing concentration of NaCl.
At the atomic resolution level, A-tract DNA has been extensively studied by X-ray crystallography (14,17). The observed global bending direction of A-tract sequences differs from the direction suggested by solution experiments, which may be due to crystal packing effects (26). The analysis of crystal packing effects, on the other hand, can provide valuable information on DNA structure and deformability (27).
NMR spectroscopy including residual dipolar couplings (RDC) represents a unique tool to study atomistic structure of DNA oligomers in solution. In particular, the use of RDC is essential to capture global bending (28,29). Several groups used the NMR technique with RDC to study A-tracts (16,28–30). The structure of a DNA dodecamer containing the A6 tract obtained by MacDonald et al. (28) shows a global bend of 19° in a geometry consistent with earlier experiments. Barbic et al. (16) investigated a decamer containing an A4 tract and found agreement with electrophoretic data and with a cyclization assay. Stefl et al. (30) obtained NMR structures of sequences containing the A4T4 or T4A4 tracts. They are discussed in detail below.
A-tract sequences have also been investigated by atomic resolution computer modelling. An early study based on energy minimization (31) was followed by extensive investigation using explicit-solvent molecular dynamics (MD) simulations. Beveridge and co-workers (32–36) conducted a series of pioneering studies aimed at investigating various A-tract properties. Their results on A-tract induced bending, as well as those by Strahs and Schlick (5) suggest general agreement with solution experiments. The work of Lankas et al. (37) indicated that the N2 amino group in the minor groove has a decisive effect on structure and mechanical properties of polypurine tracts. Other studies focused on revealing the origins of A-tract induced bending by investigating the effect of single nucleotide substitutions within an A-tract (38) and the role of ions (39).
These studies reflect the state of the art in DNA modelling of their time. The length of the simulated trajectories was in the nanosecond range, which in some cases limited the statistical quality of the collected data. The force fields used in these studies [parm94 (40) or parm99 (41)] were later shown to exhibit serious defects in longer B-DNA simulations, which were largely eliminated in the newly developed parmbsc0 force field (42).
Recently, Curuksu et al. investigated restrained bending of A-tract containing sequences by energy minimization and by MD with umbrella sampling, using the parm94 (12) and parmbsc0 (43) force fields. They computed energy profiles as a function of the bending angle and found a minimum corresponding roughly to the experimental bending magnitude.
The present study develops the subject in two directions. First, we propose a new method for global bending measurement, which uses a properly defined averaging of two or more base-fixed coordinate frames. Second, we use the method to study sequences containing either A4T4 or T4A4 tracts. For this purpose, we carried out an extended set of new reference MD simulations. The length of the simulated trajectories is increased by almost two orders of magnitude compared with the early studies. We use the parmbsc0 force field and a range of simulation conditions. We also analyse published experimental data, in particular the NMR and electrophoretic mobility data of closely related sequences obtained by Stefl et al. (30), and compare the structural information provided by the different methods.
METHODS
We performed explicit solvent, atomic resolution MD simulations of a set of DNA oligomers containing either A4T4 or T4A4 tracts using various temperature and ionic conditions. The simulations are summarized in Table 1. Our oligomers contain the sequences GCA4T4GC and CGT4A4CG studied by NMR by Stefl et al. (30) (PDB codes 1rvh and 1rvi, respectively).
Table 1.
Overview of the simulations
| Sequence | Code | tsim (ns) | T (K) | Ions |
|---|---|---|---|---|
| GGCA4T4GCC | AT_short | 150 | 300 | 150 mM KCl |
| AT_short_neut | 50 | 300 | K+ | |
| AT_short_lowT | 50 | 283 | K+ | |
| AT_short_Na | 50 | 300 | 150 mM NaCl | |
| CCGT4A4CGG | TA_short | 150 | 300 | 150 mM KCl |
| TA_short_neut | 50 | 300 | K+ | |
| TA_short_lowT | 50 | 283 | K+ | |
| TA_short_Na | 50 | 300 | 150 mM NaCl | |
| G2CA4T4GCA4T4GC2 | AT_long | 150 | 300 | 150 mM KCl |
| C2GT4A4CGT4A4CG2 | TA_long | 150 | 300 | 150 mM KCl |
Trajectory length is denoted by tsim, T stands for simulation temperature. Simulations were performed either with net-neutralizing K+ or Na+ ions and 150 mM of the corresponding added salt (KCl or NaCl), or with net-neutralizing K+ ions only.
The simulations were carried out using the AMBER 9 program package with the parmbsc0 force field (42). The ions were parametrized according to Dang (44) and the SPC/E water model was used. A truncated octahedral periodic box, particle mesh Ewald technique, a 2 fs time step and the Berendsen thermostat and barostat with time constants of 5 ps were applied. The system preparation and equilibration protocol closely follows the one used in a recent extensive DNA simulation study (45), where more details can be found.
Each MD snapshot was analysed by the 3DNA program (4). The snapshots were then filtered using two criteria. First, we checked the hydrogen bonds (H-bonds) defining a Watson–Crick base pair. We considered an H-bond broken if the donor–acceptor distance was >4 Å. Notice that any cutoff distance is artificial, since the free energy profile is in fact continuous. We found that the 4 Å cutoff allows for enough H-bond length fluctuations while capturing all the important breaking events. Second, we monitored the torsion angle γ in the backbone fragments connecting adjacent bases and considered a backbone fragment non-canonical if the value of γ was >110°. We found that excluding non-canonical γ also eliminates non-canonical torsion angles α and β. From each trajectory, only snapshots with no broken H-bond and no non-canonical base-connecting backbone fragment anywhere in the oligomer (including terminal base pairs and steps) were further analysed. Additionally, the first nanosecond of each trajectory was removed. We refer to the resulting data sets as filtered trajectories.
For each filtered trajectory, arithmetic means of the intra- and inter-base pair conformational parameters were computed. Based on these mean parameters, the average structure was rebuilt using 3DNA. We also analysed the NMR structures of AT and TA tracts reported by Stefl et al. (30) (see above), denoted, respectively, by AT_nmr and TA_nmr. The average structures were obtained analogously to those from MD, using the ensemble of NMR models. All the NMR models exhibit intact H-bonds and canonical γ torsions, so that no filtering was performed.
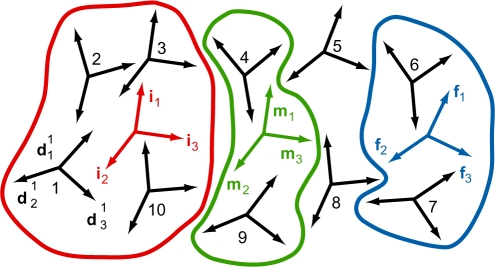
The average structures were re-analysed by 3DNA to obtain base-fixed frames (46), which were then used to measure global bending. We propose the following method for global bending measurement. We choose three groups of bases (called initial, middle and final group) and compute the average of the base-fixed frames in each group. To properly define the average frame, we employ the theory of averaging in the group of rotations developed by Moakher (47). The bending magnitude and direction are defined using these average frames. The method can be applied to a broad range of DNA and RNA structures. It is described in more detail in Section S1 of the Supplementary Data, and illustrated in Figure 1.
Figure 1.
Illustration of the proposed bending measurement method. A hypothetical nucleic acid structure is considered, involving 10 bases. First, a coordinate frame is attached to each base, using a standard procedure (black frames). Three groups of bases are then chosen: the initial (bases 1–3 and 10, red), middle (bases 4 and 9, green) and final (bases 6 and 7, blue) group. Bases 5 and 8 do not belong to any group. For each group, the average frame is computed. The global bending magnitude is defined as the angle between the average frame vector i3 of the initial group and the average frame vector f3 of the final group. The bending direction is evaluated with respect to the average frame of the middle group. A detailed description of the method is given in Section S1 of the Supplementary Data.
The reported minor groove widths are averages of the widths from the snapshots of the filtered trajectories or NMR models as measured by 3DNA (refined values). To assess convergence of the MD results, the filtering and analysis were repeated on the two halves of each trajectory separately.
To further validate the results on global bending, we also used another, independent method. For each of the average structures, we took the central 10-bp fragment and built a 15-mer by chaining the individual fragment copies one after another in space, using 3DNA. For the junction steps between monomers, we took the conformational parameters averaged over the two steps flanking the 10-bp fragment in the average structure. The shape of the 15-mer is close to a superhelix in all cases. Chaining identical DNA fragments was already used by Stefl et al. (30). Here, we refine the procedure as follows. We fit an optimal superhelix to the DNA centerline with the program Helfit (48) which, in contrast with previously published algorithms, optimizes all the helix parameters simultaneously. For each 15-mer we have 14 junction points between successive monomers, which we project onto the superhelix axis. The averaged distance between the projected points defines a fragment of the optimal superhelix. The angle between the tangent vectors at its two ends provides another estimate of the global bending magnitude of the monomer unit.
Finally, we measured the bending magnitude by the bending angle of the global, curvilinear axis assigned to the average structures by the programs Curves (6) and Curves+ (7). Terminal base pairs of AT_short and TA_short structures were discarded to keep the oligomer length consistent with the other bending measurements.
RESULTS AND DISCUSSION
Global bending
Table 2 compares bending magnitudes and directions for the simulated and NMR structures. Underscore marks the bases whose frames, together with those of the paired bases in the complementary strand, were used to measure bending. They correspond from left to right to the initial, middle and final group of frames (see Section 2, Figure 1, and Section S1 of the Supplementary Data). Thus, each group includes four base-fixed frames to average. The middle group represents the oligomer centre. It follows from the definition of the base-fixed frames, and from the frame averaging procedure, that the x-axis of the middle average frame (m1 in Figure 1) points towards the major groove.
Table 2.
Global bending magnitude and direction
| Sequence | Code | Bending magnitude (°) | Direction (°) |
|---|---|---|---|
| GGCAAAATTTTGCC | AT_short | 9.9 (10.3, 10.1) | 178 (190, 162) |
| AT_short_neut | 8.4 (8.0, 8.7) | 174 (164, 182) | |
| AT_short_lowT | 10.1 (10.6, 9.9) | 182 (177, 188) | |
| AT_short_Na | 8.7 (8.2, 9.3) | 173 (180, 162) | |
| GCAAAATTTTGC | AT_nmr | 32.0 (24.5, 35.5) | 180 (167, 194) |
| CCGTTTTAAAACGG | TA_short | 1.9 (0.7, 4.0) | 202 (129, 211) |
| TA_short_neut | 2.6 | 63 | |
| TA_short_lowT | 0.9 (0.9, 1.2) | 27 (337, 67) | |
| TA_short_Na | 1.4 (2.8, 4.5) | 222 (324, 180) | |
| CGTTTTAAAACG | TA_nmr | 23.5 (20.2, 27.2) | 176 (164, 182) |
| GGCA4T4GCA4T4GCC | AT_long | 17.4 (18.6, 16.6) | 357 (351, 4) |
| CCGT4A4CGT4A4CGG | TA_long | 4.4 (4.3, 5.6) | 30 (36, 16) |
The first values are for the entire filtered MD trajectories, or for the ensemble of NMR models, values in parentheses are for the two halves of each trajectory (MD data) or the maximum and minimum value for the NMR models. Base frames at underlined locations, in the reference and the complementary strands, are used to measure bending as described in Section 2, Figure 1, and Section S1 of the Supplementary Data. Directions around 0° or 360° correspond to bending into the major groove in the oligomer centre, directions around 180° imply bending towards the minor groove. For TA_short_neut, only the first 18 ns have been analysed since the rest of the trajectory is contaminated by a flip of the backbone torsion angle γ.
The bending direction close to 0° (or 360°) then implies bending into the major groove in the oligomer centre, whereas the direction ∼180° indicates bending into the minor groove.
All the simulated AT oligomers are bent into the minor groove in the oligomer centre, in line with experiments on A-tracts in solution. The magnitude of bending is roughly 10° The values of bending magnitude and direction computed from the two halves of each trajectory agree with the values for the whole trajectory, which indicates good convergence of the results. The values are only weakly dependent on the simulated temperature and salt conditions, with a slight reduction of bending in the neutral solution (no added salt) and in NaCl compared with KCl solution. The TA oligomers are essentially straight, as indicated by very small bending magnitude and poorly defined bending direction.
Experiments on A-tracts show a premelting transition which changes bent structures at lower temperatures to nearly straight structures at elevated temperatures, with the transition curve centred around 30–40°C (14). Since our standard simulation temperature (300 K, or 27°C) is at the edge of this interval, we performed additional simulations at 283 K (or 10°C) to make sure that the 300 K simulations are not influenced by premelting. Moreover, 283 K is close to the temperatures at which the NMR and electrophoretic mobility experiments of Stefl et al. (30) were done. We observe essentially no change in bending when comparing the 283 K and 300 K simulations (Table 2), suggesting that our results are not influenced by the premelting transition.
The results for simulated AT or TA tandem-tract structures (AT_long and TA_long) are entirely consistent with the results for the single-tract structures. In fact, the first AT tract in the AT tandem is bent by 9.1°, the second by 8.9°, both into the minor groove in their respective centres, so that the tandem as a whole is bent by roughly twice that amount but into the major groove in its centre. The TA single tracts are nearly straight, and so is the TA tandem. Thus, we observe no cooperativity in bending when passing from a single tract to a tandem.
The simulation results can be compared with the electrophoretic mobility data of Stefl et al. (30). In agreement with the findings of Koo and Crothers (20), we assume that the bending magnitude is proportional to the square root of the quantity RL−1, where RL = Lapp/Lreal and Lapp, Lreal are, respectively, the apparent length of the bent sequence and its real, contour length. We take N=15 in Figure 5 of Stefl et al. (i.e. we consider their measured sequence of 15-tract monomers). We assume a 20° bend for the A6 tract, in agreement with cyclization experiments (21), topological measurements (22) and NMR data (28). From this value and from the quadratic relationship between RL−1 and the bending magnitude, we obtain a 13° bend for an AT tract and 5° for a TA tract, both in NH4Cl and KCl, and 10° for AT and 3° for TA in NaCl. The bending magnitudes of our simulated structures (Table 2) are consistent with these electrophoresis data.
In contrast, the NMR data of Stefl et al. (30) differ from the simulation and the electrophoretic mobility results. Data in Table 2 show that the NMR structures of both the AT and the TA tracts are significantly bent into the minor groove, although the bending is weaker for the TA tract. The range of values for the individual NMR models confirms this finding.
In order to check that our measured bending magnitudes are not unduly influenced by the bending measurement method used, we also employed the helix fitting procedure described in Section 2, which is independent of the frame averaging method. The results are shown in Table 3. We see that the simulated structures yield slim, elongated superhelices with monomer units bent by 12.5° for AT tract and 3.3° for TA tract. The bending angle is consistent with the one computed for single tracts using the frame averaging method, as reported in Table 2. Moreover, the bending angle is very close to the one we deduced from the electrophoretic mobility experiments.
Table 3.
Parameters of superhelices fitted to 15-mers of the average structure fragments and bending magnitudes of a monomer fragment inferred from the fitted helices
| Code | Superhelix radius (Å) | Pitch (Å) | Bending angle (°) |
|---|---|---|---|
| AT_short | 22.7 | 333.9 | 12.5 |
| TA_short | 7.1 | 396.5 | 3.3 |
| AT_nmr | 55.8 | 117.6 | 30.8 |
| TA_nmr | 53.7 | 212.4 | 25.3 |
Finally, we measured the bending magnitude of the average structures using the global curvilinear axis computed by Curves and Curves+. The results are shown in Table 4, together with the bending magnitudes by frame averaging and helix fitting reproduced from Tables 2 and 3, respectively. Although the Curves and Curves+ values somehow differ from one another and from the values obtained by frame averaging and helix fitting, they confirm the overall trend, namely moderately bent AT tract and nearly straight TA tract from simulations, and a significant bend for AT tract and to a lesser extent also for TA tract from NMR.
Table 4.
Summary of bending magnitudes computed using different methods
| Code | Bending magnitude (°) |
|||
|---|---|---|---|---|
| Frame averaging | Helix fitting | Curves (UU) | Curves+ | |
| AT_short | 9.9 | 12.5 | 7.1 | 8.8 |
| TA_short | 1.9 | 3.3 | 4.2 | 1.0 |
| AT_nmr | 32.0 | 30.8 | 33.4 | 26.1 |
| TA_nmr | 23.5 | 25.3 | 16.0 | 14.4 |
The values from frame averaging and helix fitting discussed above (see Tables 2 and 3) are supplemented with the bending angles of the global curvilinear axis provided by Curves (the angle of the axis vectors, or UU) and Curves+. The values are to be compared with those inferred from electrophoretic mobility experiments, i.e. 13° for an AT sequence and 5° for a TA sequence (see text). The bending of the simulated oligomers (AT_short and TA_short) is consistent with the electrophoresis data. In contrast, bending of the NMR structures is much more pronounced, both for AT and for TA sequences.
Thus, the various bending measurement methods yield consistent results both for the simulated and NMR data, and confirm the contrast between the simulated structures and electrophoretic mobility data on one side and the NMR structures on the other side.
Local structure
The global bending magnitude and direction result from the local structure of the DNA oligomers. Besides that, the local structure of A-tracts is also biologically important in itself (15,49).
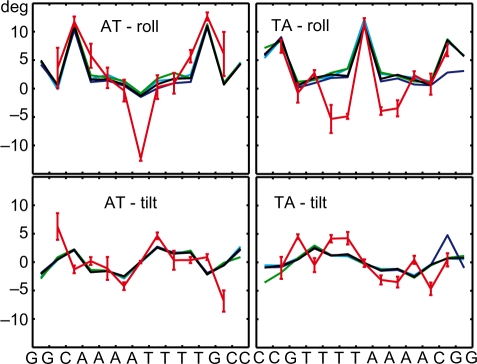
The profiles of average roll and tilt are shown in Figure 2. The simulated values, despite the fact that they were obtained in a range of simulation conditions, are very close to each other. The only visible anomaly appears in the last two steps at the 3′-end in the TA_short_lowT structure (blue line in the right part of the figure). It is probably related to the permanently broken 3′-end pair in that structure that we allowed to pass through the filtering (see Section S2 of the Supplementary Data). The NMR and the simulated values are quite similar, but with two notable exceptions: roll in the central AT step of the AT tract (–12° from NMR and –2° from MD) and roll in the two innermost TT/AA steps in the TA tract (roughly –4° from NMR and close to 0 from MD). A detailed examination of the available X-ray and other NMR structures (29,50–54) found no conclusive evidence supporting such an extreme negative roll in the AT step within AT tracts, nor the moderately negative roll in the AA/TT steps within TA tracts.
Figure 2.
Profiles of average roll and tilt in the simulated and NMR structures. The lines connect vertices marking values for the individual steps, placed in the middle of each indicated step. Values for AT_small (black), AT_short_lowT (blue), AT_short_Na (green), AT_short_neut (cyan) simulations and the NMR data AT_nmr (red) are shown. The colour coding for TA tracts is analogous. Error bars represent 1 SD for the ensemble of NMR models.
Figures analogous to Figure 2 for twist and slide are in the Supplementary Data. There the agreement is quite good for AT tract but qualitative differences are seen for TA tract. The complete sets of intra- and inter-base pair parameters for the AT_short and TA_short simulated structures are also provided in the Supplementary Data.
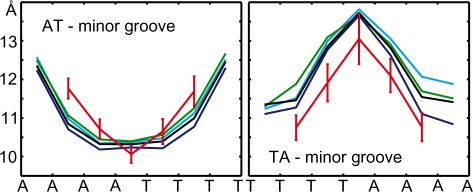
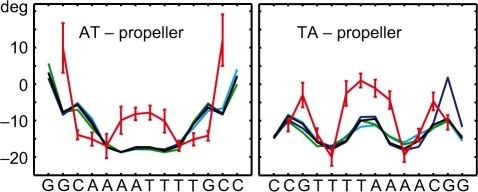
Other characteristic features of A-tracts include a progressive narrowing of the minor groove in the 5′ to 3′ direction of the adenine strand (55), and high negative propeller twist (14,17). The minor groove narrowing in MD structures (Figure 3) is saturated at the third step from the 5′-end of the adenine strand. Similar saturation has been observed in X-ray (56) and NMR (28) structures of the A6 tract. The simulated propeller twist profiles (shown in Figure 4) exhibit analogous saturation at the third base pair. Although A-tracts are commonly defined as having four or more AċT pairs in a row, early experiments by Koo et al. (23) revealed slight electrophoretic anomaly also for A3 tracts. Thus, A-tracts as short as A3 may show unique local structural features. This has also been suggested in a recent study on the role of DNA shape in protein–DNA recognition (49).
Figure 3.
Profiles of average minor groove width. Colour coding and error bars are as in Figure 2.
Figure 4.
Profiles of average propeller twist. Colour coding and error bars are as in Figure 2.
The profiles of conformational parameters for the simulated AT and TA tract tandems (AT_long and TA_long) almost precisely repeat the profiles for the simulated monomers twice in a row. We demonstrate this by the minor groove width profiles shown in the Supplementary Data.
We also checked for the presence of bifurcated hydrogen bonds in the major groove of the AA steps, involving the N6 atom of adenine and the O4 atom of thymine in the next base pair along the 5′ to 3′ direction of the adenine strand. They were observed in some A-tract structures but not in others, and are not a prerequisite for A-tract induced bending (14,17). A careful examination of the N6–O4 distances in the MD and NMR structures studied here provides no evidence for the presence of such bonds.
We finally examined the N6–N6 distances in the AT step, extensively studied by Sponer and Hobza (57) using crystal structures and quantum chemical calculations. The frequent close N6–N6 major groove contacts in AT steps were explained by activated partial sp3 pyramidalization of the adenine amino groups (57,58). Although the force field does not account for this effect, our simulated N6–N6 distances are roughly midway between the optimal (3.26 Å) and repulsive values (3.52 Å) found by Sponer and Hobza. This indicates the overall tendency of the AT step to bring the two N6 amino groups into close contact. The NMR distances in the structures examined here (4.2±0.1 Å) are too large for any contact to take place and do not agree with X-ray data.
In our analysis, we filter out snapshots with broken intra-base pair H-bonds and non-canonical backbone states (γ-flips). In this way, we limit our sampling to the B-DNA structural family. The H-bond breaks we observe are different from the base pair breathing events detected by imino proton exchange, which take place at much longer time scales. For the A4T4 tract, the experimental base pair lifetimes are roughly 100 ms, whereas the open state lifetimes are of the order of 10 ns (59). The fast opening events in our simulations may be related to a subtle underestimation of base pair stability by the force field, for instance due to the lack of an explicit polarization term. Nevertheless, the H-bond breaks we observe are by and large limited to terminal pairs. Indeed, the maximum population of a broken H-bond outside the terminal pairs is only 0.8%. A detailed check of the simulated data shows that, except for terminal pairs and steps (and next to terminal ones in two cases), the influence of H-bond breaks on the averaged intra- and inter- base pair parameters is minor. This is in line with a recent simulation study (60) that, however, revealed a dramatic influence of broken H-bonds on the standard deviations of the parameters.
The situation is different in the case of γ-flips. In the crystal structures of naked B-DNA, the flips are present only in exceptional circumstances (61–63). Thus, simulations of naked B-DNA should exhibit a very low population of γ-flips. Longer simulations using the earlier parm94 or parm99 force fields resulted in accumulation of irreversible γ-flips that had a detrimental effect on the helical structure. Irreversible flips have been eliminated in parmbsc0 (42), but random, infrequent reversible flips do occur. In fact, it is not possible to block the flips completely. Any realistic force field should make the γ/trans state easily accessible, since it is common in protein–DNA complexes and in RNA. However, the flips seem to be overpopulated at the time scale of our simulations. Indeed, the longest flip we observe (in TA_long) lasts for >70 ns, roughly half of the total simulation time. Once formed, a flip significantly affects local conformation: we observe, for instance, a decrease of twist and roll by 4° and of slide by 0.5 Å. Moreover, the effect is not limited to the flipped step itself but is non-local (see Supplementary Data for additional information). As a solution, we propose to filter out any shapshsot with a γ-flip, which yields a conformation in the limit of negligible population of γ-flips. Since no flip >80 ns has been observed even in the 1.2 μs B-DNA simulation (64), it is well possible that the population of γ-flips indeed becomes negligible at very long simulation times. However, for simulations of the order of 100 ns, we rather suggest to filter out the flips, since the occasional long-living flips may bias the values of structural parameters.
CONCLUSIONS
In this article, we propose a method to measure global bending magnitude and direction in nucleic acids structures. The method relies on a properly defined average of the base-fixed coordinate frames within suitably chosen groups of DNA or RNA bases. The bending magnitude and direction is then computed using the average frames of the groups.
The proposed method is applied to AT and TA tracts of double-helical DNA, which we study using our own atomistic MD simulations as well as published NMR and electrophoretic mobility data. We show that the simulated structures exhibit characteristic local conformational features and their global bending magnitudes agree with the electrophoresis data, whereas the NMR structures show much more pronounced bends. We expose similarities and differences among the results obtained from MD, NMR and electrophoresis, hoping that such a comparison may stimulate further research in the field.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding to complete this work: Academy of Sciences of the Czech Republic (J.E. Purkyně Fellowship and Z40550506 to F.L.); Ministry of Education, Youth and Sports of the Czech Republic (LC512 to F.L., AV0Z50040507, AV0Z50040702 and LC06030 to N.S. and J.S.). Funding for open access charge: The Wellcome Trust to J.S.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 2.Dickerson RE, Bansal M, Calladine CR, Diekmann S, Hunter WN, Kennard O, Lavery R, Nelson H.CM, Olson WK, Saenger W, et al. Definitions and nomenclature of nucleic acid structure parameters. J. Mol. Biol. 1989;205:787–791. doi: 10.1016/0022-2836(89)90324-0. [DOI] [PubMed] [Google Scholar]
- 3.Dickerson RE, Kopka ML, Pjura P. A random-walk model for helix bending in B-DNA. Proc. Natl Acad. Sci. USA. 1983;80:7099–7103. doi: 10.1073/pnas.80.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strahs D, Schlick T. A-tract bending: insights into experimental structures by computational models. J. Mol. Biol. 2000;301:643–663. doi: 10.1006/jmbi.2000.3863. [DOI] [PubMed] [Google Scholar]
- 6.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 7.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lankas F, Sponer J, Hobza P, Langowski J. Sequence-dependent elastic properties of DNA. J. Mol. Biol. 2000;299:695–709. doi: 10.1006/jmbi.2000.3781. [DOI] [PubMed] [Google Scholar]
- 9.Mazur AK. Evaluation of elastic properties of atomistic DNA models. Biophys. J. 2006;91:4507–4518. doi: 10.1529/biophysj.106.091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodsell DS, Dickerson RE. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lankas F, Lavery R, Maddocks JH. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 2006;14:1527–1534. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Curuksu J, Zakrzewska K, Zacharias M. Magnitude and direction of DNA bending induced by screw-axis orientation: influence of sequence, mismatches and abasic sites. Nucleic Acids Res. 2008;36:2268–2283. doi: 10.1093/nar/gkm1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagerman PJ. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 14.Crothers DM, Shakked Z. Oxford Handbook of Nucleic Acid Structure. Oxford: Oxford University Press; 1999. DNA bending by adenine-thymine tracts; pp. 455–470. [Google Scholar]
- 15.Haran TE, Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 16.Barbic A, Zimmer DP, Crothers DM. Structural origins of adenine-tract bending. Proc. Natl Acad. Sci. USA. 2003;100:2369–2373. doi: 10.1073/pnas.0437877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidle S. Principles of Nucleic Acid Structure. London: 1st edn. Elsevier; 2008. [Google Scholar]
- 18.Hagerman PJ. Sequence-directed curvature of DNA. Nature. 1986;321:449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- 19.Zinkel SS, Crothers DM. DNA bend direction by phase sensitive detection. Nature. 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 20.Koo HS, Crothers DM. Calibration of DNA curvature and unified description of sequence-directed bending. Proc. Natl Acad. Sci. USA. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo HS, Drak J, Rice JA, Crothers DM. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990;29:4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- 22.Tchernaenko V, Radlinska M, Drabik C, Bujnicki J, Halvorson HR, Lutter LC. Topological measurement of an A-tract bend angle: comparison of the bent and straightened states. J. Mol. Biol. 2003;326:737–749. doi: 10.1016/s0022-2836(02)01468-7. [DOI] [PubMed] [Google Scholar]
- 23.Koo HS, Wu HM, Crothers DM. DNA bending at adenine thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 24.Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987;15:247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan SS, Breslauer KJ, Austin RH, Hogan ME. Thermodynamics and premelting conformational changes of phased (dA)5 tracts. Biochemistry. 1993;32:11776–11784. doi: 10.1021/bi00095a005. [DOI] [PubMed] [Google Scholar]
- 26.DiGabriele AD, Sanderson MR, Steitz TA. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc. Natl Acad. Sci. USA. 1989;86:1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickerson RE, Goodsell DS, Neidle S. “…the tyranny of the lattice”. Proc. Natl Acad. Sci. USA. 1994;91:3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald D, Herbert K, Zhang X, Polgruto T, Lu P. Solution structure of an A-tract DNA bend. J. Mol. Biol. 2001;306:1081–1098. doi: 10.1006/jmbi.2001.4447. [DOI] [PubMed] [Google Scholar]
- 29.McAteer K, Aceves-Gaona A, Michalczyk R, Buchko GW, Isern NG, Silks LA, Miller JH, Kennedy MA. Compensating bends in a 16-base-pair DNA oligomer containing a T3A3 segment: a NMR study of global DNA curvature. Biopolymers. 2004;75:497–511. doi: 10.1002/bip.20168. [DOI] [PubMed] [Google Scholar]
- 30.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs dT4A4 conundrum. Proc. Natl Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanghani SR, Zakrzewska K, Harvey SC, Lavery R. Molecular modelling of (A4T4NN)n and (T4A4NN)n: sequence elements responsible for curvature. Nucleic Acids Res. 1996;24:1632–1637. doi: 10.1093/nar/24.9.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young MA, Beveridge DL. Molecular dynamics simulations of an oligonucleotide duplex with adenine tracts phased by a full helix turn. J. Mol. Biol. 1998;281:675–687. doi: 10.1006/jmbi.1998.1962. [DOI] [PubMed] [Google Scholar]
- 33.Sprous D, Young MA, Beveridge DL. Molecular dynamics studies of axis bending in d(G5-(GA4T4C)2-C5 ) and d(G5-(GT4A4C)2-C5): effects of sequence polarity on dna curvature. J. Mol. Biol. 1999;285:1623–1632. doi: 10.1006/jmbi.1998.2241. [DOI] [PubMed] [Google Scholar]
- 34.McConnell KJ, Beveridge DL. Molecular dynamics simulations of B'-DNA: sequence effects on A-tract-induced bending and flexibility. J. Mol. Biol. 2001;314:23–40. doi: 10.1006/jmbi.2001.4926. [DOI] [PubMed] [Google Scholar]
- 35.Beveridge DL, Dixit SB, Barreiro G, Thayer KM. Molecular dynamics simulations of DNA curvature and flexibility: helix phasing and premelting. Biopolymers. 2004;73:380–403. doi: 10.1002/bip.20019. [DOI] [PubMed] [Google Scholar]
- 36.Dixit SB, Pitici F, Beveridge DL. Structure and axis curvature in two dA6 · dT6 DNA oligonucleotides: comparison of molecular dynamics simulations with results from crystallography and NMR spectroscopy. Biopolymers. 2004;75:468–479. doi: 10.1002/bip.20157. [DOI] [PubMed] [Google Scholar]
- 37.Lankas F, Cheatham T.E., III, Spackova N, Hobza P, Langowski J, Sponer J. Critical effect of the N2 amino group on structure, dynamics and elasticity of DNA polypurine tracts. Biophys. J. 2002;82:2592–2609. doi: 10.1016/s0006-3495(02)75601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherer EC, Harris SA, Soliva R, Orozco M, Laughton C. Molecular dynamics studies of DNA A-tract structure and flexibility. J. Am. Chem. Soc. 1999;121:5981–5991. [Google Scholar]
- 39.Mocci F, Saba G. Molecular dynamics simulations of AċT-rich oligomers: sequence-specific binding of Na+ in the minor groove of B-DNA. Biopolymers. 2003;68:471–475. doi: 10.1002/bip.10334. [DOI] [PubMed] [Google Scholar]
- 40.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz Jr KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 41.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 42.Perez A, Marchan I, Svozil D, Sponer J, Cheatham T.E., III, Laughton CE, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curuksu J, Zacharias M, Lavery R, Zakrzewska K. Local and global effects of strong DNA bending induced during molecular dynamics simulations. Nucleic Acids Res. 2009;37:3766–3773. doi: 10.1093/nar/gkp234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang LX. Mechanism and thermodynamics of ion selectivity in aqueous solutions of 18-crown-6 ether: a molecular dynamics study. J. Am. Chem. Soc. 1995;117:6954–6960. [Google Scholar]
- 45.Lavery R, Zakrzewska K, Beveridge D, Bishop TC, Case DA, Cheatham T.E., III, Dixit S, Jayaram B, Lankas F, Laughton C, et al. A systematic molecular dynamics study of nearest-neighbor effects on base pair and base pair step conformations and fluctuations in B-DNA. Nucleic Acids Res. 2009;38:299–313. doi: 10.1093/nar/gkp834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu X.-J, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 47.Moakher M. Means and averaging in the group of rotations. SIAM J. Matrix Anal. Appl. 2002;24:1–16. [Google Scholar]
- 48.Enkhbayar P, Damdinsuren S, Osaki M, Matsushima N. HELFIT: helix fitting by a total least squares method. Comput. Biol. Chem. 2008;32:307–310. doi: 10.1016/j.compbiolchem.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: a comparative analysis of T-A and A-T steps within A-tracts. J. Mol. Biol. 2001;312:1037–1049. doi: 10.1006/jmbi.2001.4994. [DOI] [PubMed] [Google Scholar]
- 51.Edwards KJ, Brown DG, Spink N, Skelly JV, Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2: an examination of propeller twist and minor-groove water structure at 2.2 Å resolution. J. Mol. Biol. 1992;226:1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- 52.Woods KK, Maehigashi T, Howerton SB, Sines CC, Tannenbaum S, Williams LD. High-resolution structure of an extended A-tract: [dCGCAAATTTGCG)]2. J. Am. Chem. Soc. 2004;126:15330–15331. doi: 10.1021/ja045207x. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Dalaglio F, Tjandra N, Zhurkin VB, Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and 31P chemical shift anisotropy. J. Biomol. NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- 54.Morozov AV, Fortney K, Gaykalova DA, Studitsky VM, Widom J, Siggia ED. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burkhoff AM, Tullius TD. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 56.Nelson HCM, Finch JT, Luisi BF, Klug A. The structure of and oligo(dA) · oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 57.Sponer J, Hobza P. Bifurcated hydrogen bonds in DNA crystal structures. An ab initio quantum chemical study. J. Am. Chem. Soc. 1994;116:709–714. [Google Scholar]
- 58.Yildirim I, Stern HA, Sponer J, Spackova N, Turner DH. Effects of restrained sampling space and nonplanar amino groups on free-energy predictions for RNA with imino and sheared tandem GA base pairs flanked by GC, CG, iGiC or iCiG base pairs. J. Chem. Theor. Comput. 2009;5:2088–2100. doi: 10.1021/ct800540c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warmländer S, Sponer JE, Sponer J, Leijon M. The influence of the thymine C5 methyl group on spontaneous base pair breathing in DNA. J. Biol. Chem. 2002;32:28491–28497. doi: 10.1074/jbc.M202989200. [DOI] [PubMed] [Google Scholar]
- 60.Lankas F, Gonzalez O, Heffler LM, Stoll G, Moakher M, Maddocks JH. On the parameterization of rigid base and basepair models of DNA from molecular dynamics simulations. Phys. Chem. Chem. Phys. 2009;11:10565–10588. doi: 10.1039/b919565n. [DOI] [PubMed] [Google Scholar]
- 61.Varnai P, Djuranovic D, Lavery R, Hartmann B. α/γ transitions in the B-DNA backbone. Nucleic Acids Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Djuranovic D, Hartmann B. DNA fine structure and dynamics in crystals and in solution: the impact of BI/BII backbone conformations. Biopolymers. 2004;73:356–368. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 63.Svozil D, Kalina J, Omelka M, Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–3706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez A, Luque FJ, Orozco M. Dynamics of B-DNA on the microsecond time scale. J. Am. Chem. Soc. 2007;129:14739–14745. doi: 10.1021/ja0753546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.