Abstract
Regulation of splicing in eukaryotes occurs through the coordinated action of multiple splicing factors. Exons and introns contain numerous putative binding sites for splicing regulatory proteins. Regulation of splicing is presumably achieved by the combinatorial output of the binding of splicing factors to the corresponding binding sites. Although putative regulatory sites often overlap, no extensive study has examined whether overlapping regulatory sequences provide yet another dimension to splicing regulation. Here we analyzed experimentally-identified splicing regulatory sequences using a computational method based on the natural distribution of nucleotides and splicing regulatory sequences. We uncovered positive and negative interplay between overlapping regulatory sequences. Examination of these overlapping motifs revealed a unique spatial distribution, especially near splice donor sites of exons with weak splice donor sites. The positively selected overlapping splicing regulatory motifs were highly conserved among different species, implying functionality. Overall, these results suggest that overlap of two splicing regulatory binding sites is an evolutionary conserved widespread mechanism of splicing regulation. Finally, over-abundant motif overlaps were experimentally tested in a reporting minigene revealing that overlaps may facilitate a mode of splicing that did not occur in the presence of only one of the two regulatory sequences that comprise it.
INTRODUCTION
Splicing is a molecular mechanism by which introns are removed from an mRNA precursor and exons are ligated to form a mature mRNA (1). Most human genes that encode proteins contain multiple introns, and intronic sequence accounts for 95% of the average 28 000 nt of a transcription unit (2,3). Introns are variable in length and sequence, and thus the splicing machinery must be remarkably flexible in order to correctly recognize and excise all introns.
Splicing, which occurs in organisms as diverse as yeast and human, takes place within the spliceosome—a large complex comprising five small nuclear RNPs (U1, U2, U4, U5 and U6 snRNP) and as many as 150 proteins (4–7). The splicing machinery recognizes exons and introns using multiple signals, which presumably results in a network of interactions across exons and/or introns (8). Four main splice signals assist the splicing machinery in recognizing the proper exon–intron boundaries. These are the 5′ and 3′ splice sites, located at the 5′ and 3′ end of introns, and the branch site and the polypyrimidine tract, which are located upstream of the 3′ splice site (1,9). In metazoans, splice sites are degenerate and are postulated to provide only half of the information required for recognition by the splicing machinery (10,11). Studies of the molecular basis of splicing revealed the existence of exonic and intronic cis-acting regulatory sequences (ESRs and ISRs, respectively) that bind trans-acting factors and influence splice-site selection. These cis-acting elements are relatively short, usually 4–12 nt, are classified as exonic or intronic splicing enhancers or silencers, and are required for the regulation of both constitutive and alternative splicing (12–17). Specific binding of splicing regulatory proteins (such as SR and hnRNP proteins) to these splicing regulatory elements assists in the placement of the spliceosome on the appropriate splice sites (18,19).
Through alternative splicing more than one mRNA transcript is generated from the same mRNA precursor, giving rise to functionally different proteins (20). This requires that exons and introns be defined differently within the same sequence context (1,9,21). Thus, alternative splicing is a fundamental aspect of post-transcriptional gene regulation with significant functional implications.
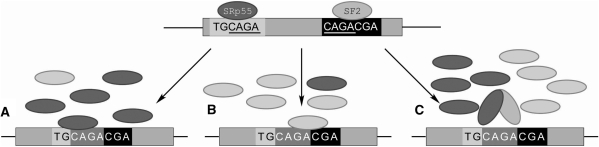
The regulation of splicing in eukaryotes is highly complex and often takes place through the coordinated action of multiple splicing regulatory factors. Smith and Valcarcel previously suggested that splicing is regulated by the combinatorial outcome of several regulatory sequences (22) and this hypothesis now has experimental support (23). A recent study examined the tendency of splicing factors to bind cooperatively (24); however, no extensive bioinformatic studies have examined whether overlapping regulatory sequences exhibit unique functions and provide another dimension to splicing regulation. For example, overlapping regulatory binding motifs may modulate exon selection by serving as a template for a competitive inhibition between two different SR proteins. The outcome of such competition may depend on the strength with which each protein binds to its specific regulatory sequence or on the expression levels of the corresponding SR proteins, their level of phosphorylation, or their spatial distribution within the cell (Figure 1). This could provide another dimension for splicing regulation. This type of mechanism is known to occur during the regulation of transcription, which is affected by the binding of different transcription factors to overlapping binding motif sequences (25–28). The use of several splicing regulatory overlapping motifs in a combinatorial fashion could allow cells to express the required amount of each splice variant at the proper time.
Figure 1.
Possible outcomes of overlapping regulatory sequences. As an example, a potential overlap between the binding sites of SRp55 (dark gray ellipse) and SF2 (light gray ellipse) is illustrated. A specific cell type or a cell at a specific time where (A) SF2 expression is down-regulated resulting in binding of SRp55 to its binding site, (B) SRp55 expression is down-regulated resulting in binding of SF2 to its binding site, (C) both SF2 and SRp55 are expressed at significant levels and compete with each other for binding to their overlapping regulatory sequences.
Here we describe a bioinformatic method, based on the natural distribution of nucleotides and splicing regulatory sequences, which indicates that there is a positive and negative interplay between overlapping splicing regulatory sequences. Specifically, we found 72 splicing regulatory motifs that overlapped more than expected and 153 splicing regulatory motif overlaps that were under negative selection. Examination of these motif overlaps revealed a unique spatial distribution; they tend to be found near the 5′ splice sites of exons, especially in exons with weak 5′ splice sites. Moreover, the over-abundant overlapping motifs are highly conserved among different species, implying functionality. As a proof of principle, we experimentally tested the effect of the significantly over-abundant motif overlaps on the splicing pattern in a reporting minigene system. The results revealed that the overlaps facilitated a mode of splicing that did not occur in the presence of only one of the two regulatory sequences that comprise it, nor did it occur in the absence of the entire overlap. Overall, the results we present suggest that overlapping splicing regulatory motifs provide another dimension of splicing regulation.
MATERIAL AND METHODS
Dataset compilation
Exonic and intronic sequences of human (hg18), mouse (mm9), rat (rn4), dog (canFam2), chicken (gal3) and zebrafish (danRer5) RefSeq genes were extracted from tables in the UCSC genome browser (http://www.genome.ucsc.edu/) (29) using GALAXY (http://main.g2.bx.psu.edu/) (30). Orthologous exons were identified using coordinates downloaded from the UCSC genome browser. In order to obtain reliable alignments, we included only orthologous exons of the same length with identity levels higher than 75%.
Construction of the SXN-derived minigenes
The overlapping regulatory motifs and its mutants were cloned into SalI and BamHI sites inside the SXN reporting exon using the following oligos by annealing: 5′-TCG ATG TTT GCG GCT GCT GGA ATG-3′ and 5′-GAT CCA TTC CAG CAG CCG CAA ACA-3′; 5′-TCG ATG TTA AGG GCT GCT GGA ATG-3′ and 5′-GAT CCA TTC CAG CAG CCC TTA ACA-3′; 5′-TCG ATG TTT GCG GCA AAA GGA ATG-3′ and 5′-GAT CCA TTC CTT TTG CCG CAA ACA-3′; 5′-TCG ATG TTA AGG GCA AAA GGA ATG-3′ and 5′-GAT CCA TTC CTT TTG CCC TTA ACA-3′; 5′-TCG ATG TTC GCG GAG GAG AAT G-3′ and 5′-GAT CCA TTC TCC TCC GCG AAC A-3′; 5′-TCG ATG TTC TCG GAG GAG AAT G-3′ and 5′-GAT CCA TTC TCC TCC GAG AAC A-3′; 5′-TCG ATG TTC GCG GAT CAG AAT G-3′ and 5′-GAT CCA TTC TGA TCC GCG AAC A-3′; 5′-TCG ATG TTC TCG GAT CAG AAT G-3′ and 5′-GAT CCA TTC TGA TCC GAG AAC A-3′; All plasmids were confirmed by DNA sequencing.
Transfection, RNA isolation and RT-PCR amplification
The 293T cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 4.5 g/ml glucose (Renium) and 10% fetal calf serum (Biological Industries). Cells were grown to 50% confluence in a 10-cm culture dish, under standard conditions, at 37°C with 5% CO2. Cells were split at a 1:8 ratio to 6-well plates 24 h prior to transfection. Transfection was performed using 3 μl of FuGENE6 (Roche) with 1 μg of plasmid DNA. Cells were harvested 48 h post-transfection. Total cytoplasmic RNA was extracted using TriReagent (Sigma), followed by treatment with 2 U of RNase–free DNase (Ambion). Reverse transcription (RT) was performed on 2 μg of total cytoplasmic RNA for 1 h at 42°C, using an oligo(dT) and 2 U of reverse transcriptase of avian myeloblastosis virus (RT-AMV, Roche). The spliced cDNA products derived from the expressed minigenes were detected by PCR, using vector-specific primers: 5′-ATC GAT CCT GCA CCT GAC TC-3′ and 5′-CAG CAT CAG GAG TGG ACA GA-3′. Amplification was performed for 30 cycles, consisting of 94°C for 30 s, 60°C for 45 s and 72°C for 1 min, using ReadyMix (Bio-Lab). The products were resolved on a 2% agarose gel. PCR products were eluted from gel and confirmed by DNA sequencing after purification (Wizard, Promega). Every result represents at least three independent experiments. The ratio of exon inclusion to exon skipping was determined using the ImageJ tool.
RESULTS
Overlapping SR protein putative binding sites may result in competitive inhibition between the two SR proteins for binding to their corresponding sequences. Such competitive inhibition might allow a delicate regulation of the splicing process. If this scenario is common, we expected functional overlapping motifs to be more abundant than expected and perhaps over-conserved. In contrast, such competitive inhibition might interfere with the proper identification of a nearby exon–intron junction. In these cases, we could expect negative selection to act against such overlapping motifs.
The putative binding sites of SR proteins tend to significantly overlap
To determine whether putative binding sites of SR proteins have a positive or negative tendency to overlap, we examined putative SR-binding sites that had been experimentally identified by SELEX (31–33); we did not analyze bioinformatically identified putative elements because were concerned they could be sequence-biased. We developed a method to examine whether two SR putative binding sites that can overlap do so. For example, the putative SRp55-binding sequence TGCAGA may overlap with the putative SF2-binding sequence CAGACGA in the context of TGCAGACGA. Briefly, we calculated the expected frequency of the unified sequence (TGCAGACGA) in the human exons dataset, given the prevalence of the first sequence (TGCAGA) and of the extension (CGA). Estimates were based on the relative frequency of the four nucleotides (A, C, G and T) in each of the possible three phases of the codon (0, 1 and 2). This was done to avoid any bias that may be introduced by protein coding constraints. The expected frequency was then compared with the observed frequency of the unified sequence and putative binding sites that were significantly over-abundant or under-represented were identified.
First, we generated a pool of all possible hexamers, heptamers and octamers previously identified as putative SR-binding sites using any of the five available matrices in the ESEfinder web server (SRp55, SRp40, SF2/ASF, SF2/ASF BRCA1 and SC35; using threshold of four to maximize credibility) (34). We then calculated all the theoretical possibilities for overlap (i.e. all possible unified sequences, 192 327 cases). Since we were interested in identifying cases of potential interaction/competitive inhibition between two different SR proteins, we discarded unifications that involved two putative binding sites of the same SR protein as such cases might actually reflect one longer binding site for a single SR protein (for a separate analysis of these sequences see Supplementary Table S2). Next, we extracted a dataset containing 202 839 human RefSeq exons. We discarded exons for which we could not extract the definite phase (e.g. exons in intronless genes, exons in the UTR, etc.) and very short or very long exons (top and bottom 0.025%); 150 941 exons remained. We examined the coding sequence of each exon and calculated the prevalence of each of the four nucleotides (A, C, T and G) in each of the three possible codon phases (0, 1 and 2).
In the next step, we calculated the expected frequency of all unified sequences. We examined the exons in the dataset to identify the prevalence of each putative binding site in each of the three phases. For example, the putative SRp55 TGCAGA sequence appeared 1086, 6503, 6046 times in phases 0, 1 and 2, respectively. Using the nucleotide frequency table in each of the phases we then calculated the expected prevalence of the unified sequence as follows:
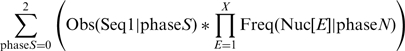 |
(1) |
where phase S is 0, 1, or 2 and Obs(Seq1|phase S) is the prevalence of Seq1 in the specific phase S; X is the number of extending nucleotides (three in the above example); E ranges from 1 to X, and represents a specific extending nucleotide (E = 1,2,3 for the extending nucleotides C,G,A, respectively); and Freq(Nuc[E]|phaseN) is the frequency of the extending nucleotide in position E, which is in phaseN.
After calculating the expected frequency for all possible unified sequences, we analyzed the dataset of exons to retrieve observed frequencies. For each of the unified sequences, the expected and observed values were subjected to a Fisher’s exact test and false discovery rate (FDR; P-value < 0.05) correction for multiple testing (35). This way, we identified potential overlapping binding sites that were significantly over-abundant or under-represented. Out of the 192 327 possible unifications of putative overlapping binding sites, we found 72 unified sequences that were significantly over-abundant and 153 that were significantly under-represented (see Supplementary Table S1 for the specific overlaps).
Several SR protein-binding sites have a significant tendency to overlap. However, this phenomenon may not be unique to splicing regulatory sequences. It may be that any similar subset of sequences would exhibit the same tendency to overlap, yielding similar numbers of over-abundant and under-represented overlaps. In order to address this issue, we generated 100 random subsets of sequences, which were not detected as splicing regulatory elements by any of the ESEfinder matrices (33,34) nor by other known bioinformatic detection methods (15,36,37). Each random subset was composed of the same number of hexamers, heptamers and octamers as the original dataset. Next, we performed the same statistical analyses to reveal significant overlaps between the random sequences in each of the 100 random subsets. We found that the number of significant over-abundant and under-represented SR-binding site overlaps was significantly higher than the one observed for all 100 random datasets containing splicing inert sequences (P < 0.01 for both the over-abundant and under-represented sequences).
We were also interested to examine whether the SR-binding sites that form the over-abundant and under-represented overlaps tend to appear in proximity, even if not in overlap. If so, their tendency to appear in overlap could be only a special case of their general tendency to be in proximity. We examined the average distance between every two SR-binding sites that formed an overlapping motif. This average distance was calculated for all SR-binding sites that formed a significantly over-abundant or under-represented overlapping motif. As control, we calculated the average distance between every two SR-binding sites that did not form over-abundant or under-represented overlapping sites. We found that the average distance between the putative SR-binding sites that form an over-abundant or an under-represented overlapping motif did not significantly differ from the control dataset (see Supplementary Figure S1). This suggests that the overlapping motif itself is important and that the results we found could not be explained by mere proximity between SR-binding sites.
Comparative analysis revealed high conservation of over-abundant motif overlaps
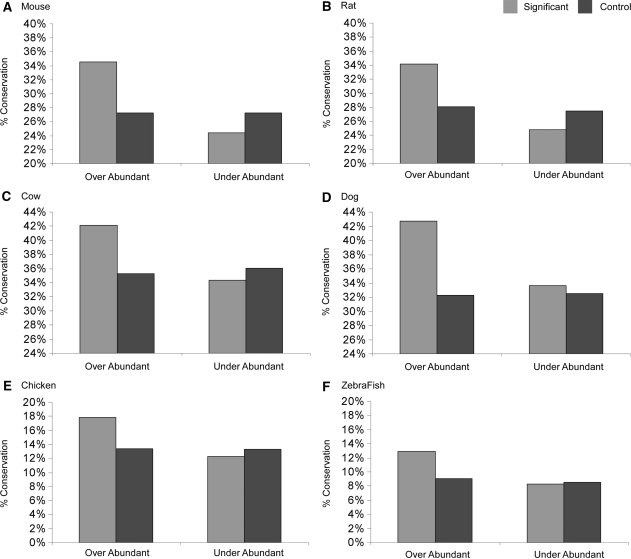
We identified 72 significantly over-abundant and 153 significantly under-represented overlapping SR protein putative binding motifs. To shed light on the functionality of these motif overlaps, we employed a comparative genomics approach. The common premise underlying comparative genomic analyses is that selective pressure dictates that functional elements evolve at a slower rate than that of non-functional sequences. Indeed, splicing regulatory elements were shown to be highly conserved, indicative of their significance in splicing regulation (38,39). To examine the conservation level of the significant overlaps we identified, we extracted a dataset of human exons and their conserved orthologs (>75% nucleotide identity) in mouse (Mus musculus), rat (Rattus norvegicus), cow (Bos taurus), dog (Canis familiaris), chicken (Gallus gallus) and zebrafish (Danio rerio). We then calculated the percentage of over-abundant and under-represented SR motif overlaps that were fully conserved (no mismatches allowed) between the human exons and their orthologous counterparts. As control for the significantly over-abundant (or under-represented) motifs, we used all overlap motifs that appeared more (or less) than expected albeit not significantly. As illustrated in Figure 2, compared to the non-significant motifs, the 72 over-abundant overlapping motifs we identified are significantly conserved between human exons and their orthologous counterparts in mouse (P = 4.77e − 60), rat (P = 1.68e − 32), cow (P = 4.72e − 18), dog (P = 1.62e − 04), chicken (P = 6.05e − 07) and zebrafish (P = 3.13e − 03, χ2 with df = 1 for all tests). Examination of the 153 under-represented motifs overlaps revealed insignificant results. We were concerned that some of the mismatches we identified do not actually change the ability of the sequence to function as a binding platform for the corresponding SR protein, i.e. the k-mer including the mismatch still belongs to the same SELEX matrix. We therefore repeated this analysis, this time considering mismatches that still preserve the ‘binding-type’ (giving ESEfinder motif score higher than the threshold) as conserved. However, this analysis yielded similar results (see Supplementary Data).
Figure 2.
Conservation of the over-abundant overlapping regulatory binding motifs. The conservation levels of the overlapping motifs were calculated between orthologous exons of human and (A) mouse, (B) rat, (C) cow, (D) dog, (E) chicken and (F) zebrafish. These calculations were performed for both significant (light-gray bars) and control (dark-gray bars) overlaps. The percent conservation level (y-axis) is indicated for both the over-abundant and the under-represented overlapping motifs (x-axis).
We next employed another statistical approach to confirm the validity of the conservation level of the over-represented motifs. Under the assumption that two adjacent N-mers are independent, the expected conservation level of each overlapping SR-binding motifs could be calculated based on three observations: (i) the conservation level of the putative SR-binding site, Cons(SRbinding); (ii) the conservation level of the corresponding extension of N-mers, Cons(extension) and (iii) the number of times the corresponding overlap was detected in the dataset, Occurrence (SRbinding + extension), as follows:
| (2) |
After calculating the expected conservation level of each overlapping sequence in the orthologous exons between human and each of the other examined species, we examined these exons to extract the actual conservation level of each of the overlapping motifs. For example, the overlap TGACTCCAG is comprised of the SRp40-binding site TGACTCC and of the extension AG, which is derived from the SR-binding site SC35 (GACTCCAG). The expected conservation level of this overlap between human and mouse, for example, was calculated by multiplying the conservation level of the SRp40-binding site (40%, for example) and the conservation level of the extension AG (77%, for example). The result was then multiplied by the number of times we observed this overlap in the data (128 times, for example) resulting in 39.22—the expected conservation level for the overlap TGACTCCAG.
Generally, the over-abundant motifs were significantly conserved between human and mouse (P = 2.90e − 04), rat (P = 3.60e − 03), cow (P = 4.88e − 03) and chicken (P = 5.14e − 03), but were not significantly conserved between human and dog (P = 0.19) or human and zebrafish (P = 0.89, χ2 with df = 1 for all tests). Examination of the 153 under-represented motif overlaps revealed significant, yet opposite results. These motifs were significantly less conserved between human and mouse (P = 2.40e − 17), rat (P = 8.34e − 08), cow (P = 1.62e − 04) and zebrafish (P = 0.043), but not between human and dog (P = 0.48) or human and chicken (P = 0.13, χ2 with df = 1 was used for all tests).
Overall, we used different methods to examine the conservation level of the over-abundant and under-represented motifs (see Supplementary Data for another method). The results imply that the over-abundant motif overlaps we identified are functionally relevant. These over-abundant motifs are significantly conserved despite millions of years of evolution since the last common ancestor of the organisms evaluated, suggesting that these motif overlaps play a functional role in the regulation of splicing. In contrast, under-represented motif overlaps were not significantly conserved compared to the random data or even exhibit a significant opposite trend. This suggests that the under-represented motifs are selected against, perhaps because they interfere with the splicing process.
Abundance of SR overlapping binding motifs correlates with splice signal strengths
We next examined whether the abundance of SR overlapping binding motifs in exons correlated with the strength of the splice sites. The splicing signals play a major role in the proper recognition of the exon/intron junctions. For example, the strength of the affinity of U1 snRNA to the 5′ splice site dictates both constitutive and alternative splicing, as well as regulates the inclusion/skipping ratio in alternative splicing (40). However, the presence of a strong ESE can compensate for a weak splice site (41). Thus, if the over-abundant SR overlapping binding sites are indeed functional in splicing regulation, we would expect them to be more abundant in exons with weak splice signals. Similarly, if the under-represented SR overlapping binding sites interfere with the splicing process, we would expect them to be selected against, especially in exons with weak splice signals.
To test these hypotheses we divided our exon dataset into quartiles according to either their 5′ splice site or their 3′ splice site strength, as computed by the MaxEntScan server (42). We defined strong or weak exons according to their presence in the upper or lower quartile, respectively. Next, for each of the exons, we determined the number of nucleotides that were part of a motif overlap and the number of nucleotides that were not (omitting the first and the last three bases as these are part of the splice signals). The over-abundant overlapping motifs were significantly more abundant in exons with weak 5′ splice site splice signals than in exons with strong ones (P = 3.77e − 20, χ2 with df = 1). Furthermore, we found that the under-represented overlapping motifs were significantly selected against in exons with weak 5′ splice site splice signals compared to exons with strong ones (P = 2.70e − 10, χ2 with df = 1). Similar results were obtained when exons were classified according to their 3′ splice site strengths. Namely, over-abundant motifs were found mainly in exons with weaker splice signals whereas under-represented motifs were selected against in this group of exons (over-abundant P =1.69e − 34; under-represented P =1.46e − 26, χ2 with df =1).
Negative selection acts against the under-represented sequence overlaps near the splice donor site
The above analyses implied that under-represented sequences interfere with the splicing process as they are selected against. Previous studies indicated that the density of exonic splicing regulatory elements is highest near splice sites and that the relative location of these elements is crucial for their function (36,38,43). Furthermore, it appears that the effect of exonic splicing regulatory sequences is more prominent near the splice donor site than the splice acceptor site (36). Thus, if the under-represented overlapping motifs we identified indeed interfere with the splicing process, we could expect they would be selected against specifically near the splice donor site where they would interfere most with splice site selection. Furthermore, we could expect such a phenomenon to be more prominent in exons with weaker splice signals, which are sub-optimally recognized to begin with, and hence depend more on auxiliary splicing factors.
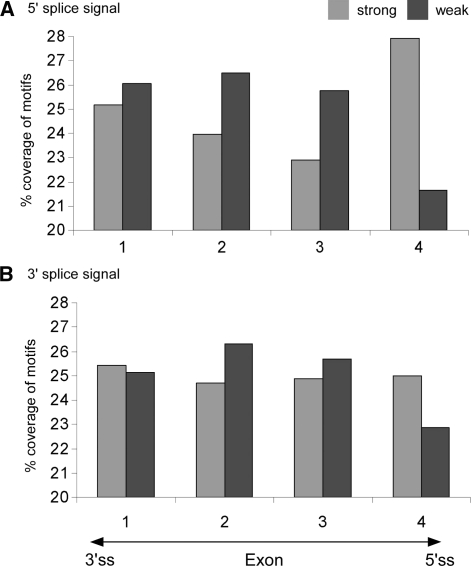
To reveal correlations between the strength of the splice signals and the distribution of the under-represented SR putative binding motif overlaps along the exons, we divided the sequence of each of the exons into quartiles. The first and last quartiles were near the splice acceptor and donor site, respectively. As above, for each of the exons, we determined the number of nucleotides that were part of a motif overlap and the number of nucleotides that were not (omitting the first and the last three bases of each exon, as these are part of the splice signals). We subdivided the exons into those with strong or weak 5′ and 3′ splice sites (see above). There was a significant difference between the distributions of under-represented overlapping motifs across the entire exon for exons with strong versus weak 5′ splice sites (P = 3.20e − 35, χ2 with df = 3; see Figure 3A). This difference in the distribution was observed mainly near the splice donor site. Similarly, there was a significant difference between the distributions of under-represented overlapping motifs across the entire exon for exons with strong versus weak 3′ splice sites (P = 2.08e − 05, χ2 with df = 3; see Figure 3B). Again, this difference was found to be more prominent near the splice donor site.
Figure 3.
Distribution of under-represented motifs along exons. The distribution of the under-represented motif overlaps along exons was calculated for exons with strong (light-gray bars) and weak (dark-gray bars) splice signals near (A) the 5′ splice signal and (B) the 3′ splice signal. Exon sequences were divided into four equally-sized quartiles (x-axis). The percentage of nucleotides which are part of under-represented motif overlaps, in each of the four quartiles, is indicated (y-axis).
Different spatial distributions of over-abundant and under-represented overlaps in exons with weak 5′ splice sites
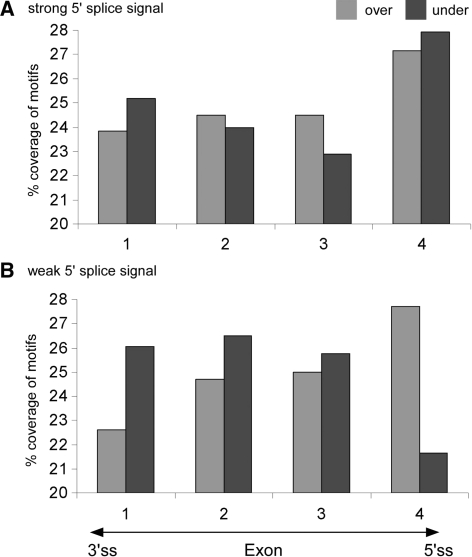
The results indicated that over-abundant overlaps are functionally important and play a regulatory role in the splicing process. If the over-abundant motifs indeed participate in splicing regulation, we would expect their distribution along exons to be different from the distribution of the under-represented motifs. Namely, we expect their prevalence to be higher near the splice donor site. Our data indicated that the relative distribution of over-abundant and under-represented motifs is significantly different. The under-represented motifs are highly selected against and over-abundant motifs are highly preferred specifically near weak 5′ splice sites (Figure 4; P = 2.56e − 51, χ2 with df = 1). In fact, in exons with a strong 5′ splice site, there was no significant preference for either type of motifs. These results suggest that exons with weak splice signals that rely on auxiliary factors for their splicing are more affected by the presence or absence of the over-abundant and under-represented motifs; in these exons, the motifs are under tight selective pressure.
Figure 4.
Comparison between the distributions of over-abundant and under-represented motifs along exons. The distribution of the over-abundant (light gray bars) and under-represented (dark gray bars) sequence overlaps along exons was calculated for exons with (A) strong 5′ splice signals and (B) weak 5′ splice signals. Exon sequences were divided into four equally sized quartiles (x-axis). The percentage of nucleotides which are part of over-abundant or under-represented motif overlaps in each of the four quartiles is indicated (y-axis).
Over-abundant motif overlaps facilitate a new mode of splicing in a reporting minigene system
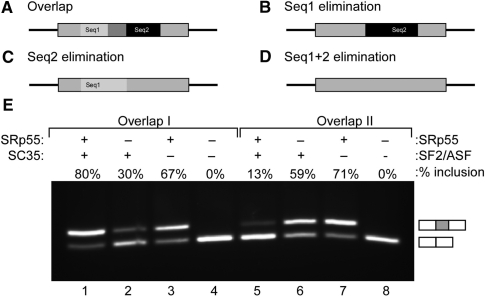
The above bioinformatic findings suggest that overlapping splicing regulatory sequences can positively or negatively interact to affect splicing. Specifically, we found 72 significantly over-abundant and conserved overlaps, suggesting they may be functional in splicing regulation. To further test the validity of the bioinformatic findings, we examined four representative over-abundant overlaps in a minigene reporting system (44). The minigene was composed of four exons, and we inserted the tested overlapping motif into the second exon. For each overlap, which is comprised of two splicing regulatory motifs (seq1 and seq2), we designed one mutation to eliminate seq1 leaving seq2 intact, and one mutation to eliminate seq2 leaving seq1 intact. We introduced each of the two mutations to the minigene independently to test the effect of the presence of seq2 or seq1 alone. We then also introduced the two mutations simultaneously to test the effect of the absence of both sequences. Two of the four overlaps we tested showed full exon skipping, a pattern that did not change regardless of the mutations we introduced (data not shown). However, the other two overlaps enabled a mode of splicing that did not occur in the presence of only one of the two regulatory sequences that comprise it, nor did it occur in the absence of the entire overlap (the overlaps’ sequence is indicated in Supplementary Table S1). The insertion of the first overlap resulted in 80% inclusion level, while mutations that eliminated each of the regulatory sequences that form the overlap, or eliminated them both, resulted in different splicing patterns (30, 67 and 0%, respectively; Figure 5, complex I). The insertion of the second overlap resulted in 13% inclusion level. Similarly, mutations that eliminated each of the regulatory sequences that form the overlap, or eliminated them both yielded different splicing patterns (59, 71 and 0%, respectively; Figure 5, complex II). These experimental results are consistent with the possibility that the overlapping splicing regulatory motifs provide regulation that is different from the effect of each regulatory sequence alone. However, it is worth noting that further experimental analysis is required to determine this matter, as it may be that the extension itself is important, rather than the relation between two different splicing factors.
Figure 5.
The effect of two over-abundant overlaps in a minigene reporting system. Each tested overlap was inserted into the second exon of a reporting minigene comprised of four exons. (A) Illustration of an overlap comprised of two spicing regulatory sequences. Seq1 is in light-gray, seq2 is in black, and the overlapping sequence is in dark-gray. For each overlap, which is comprised of two splicing regulatory motifs (seq1 and seq2), we introduced two mutations: (B) mutation to eliminate seq1 leaving seq2 intact, and (C) mutation to eliminate seq2 leaving seq1 intact. (D) Insertion of both mutations simultaneously. (E) Splicing patterns of each of the two overlaps (overlap I: lanes 1–4; and overlap II: lanes 5–8) is depicted. For each overlap, the splicing pattern in its presence, after the elimination of seq1, the elimination of seq2, or the elimination of both is depicted, respectively. The upper bands indicate exon inclusion, while the lower bands indicate exon skipping; the exact inclusion level is indicated above each lane.
DISCUSSION
To date, many layers of splicing regulation have been documented. These include the consensus splice site sequences, auxiliary signals, such as exonic and intronic enhancers and silencers [reviewed in ref. (40)], chromatin structure and transcription elongation rate (45). However, despite a vast body of knowledge on elements and sequences that affect splicing of particular genes, the splicing code in general is far from being fully understood and one cannot predict the mode of splicing of a given exon (or an intron) strictly from analysis of sequence.
In this work we provide evidence that two exonic splicing regulatory sequences that overlap might represent yet another level of splicing regulation. We describe a computational method, based on the natural distribution of nucleotides and splicing regulatory sequences, by which we identified overlapping putative splicing regulatory sequences. Dozens of such motif overlaps are favorable in exons and even more appear to be selected against, implying both positive and negative interplay between splicing regulatory motifs. Selection against overlap of regulatory sequences might not be surprising. Such overlapping motifs, which are presumably formed by two independently functional regulatory binding sites, may obstruct one another when in overlap resulting in improper splicing. On the other hand, we were surprised that some motifs have a specific and significant tendency to overlap. This finding implies that there is a layer of splicing regulation provided by a competition between factors that can either assist or obstruct the splicing machinery in proper recognition of exon/intron junctions. Such a competition could depend on the strength with which each SR protein binds to its specific regulatory sequence, the expression levels of the SR proteins, their level of phosphorylation, or their spatial distribution within the cell, which could vary in different cell types or cell stages (see also Figure 1). The outcome of such competition could result in the proper splicing mode (constitutive or alternative), or proper exon inclusion level, in each cell type or stage. It is worth noting that the complexes we identified might in fact reflect single longer motifs, however, our data provides evidence against such possibility (see Supplementary Data and Supplementary Figure S2).
By using comparative genomics we showed that the over-abundant overlapping splicing regulatory motifs are highly conserved among human and six other species. This suggests an important functional role for these overlapping motifs in splicing regulation, as they are significantly conserved despite the ∼450-million years of evolution since the last common vertebrate ancestor. The overlapping motifs we found to be under-represented were not conserved. In fact, in some species, these overlapping motifs were actually significantly less conserved than expected, suggesting that selective pressure acts against them. Therefore, such overlapping motifs have been selected against and eliminated during evolution.
We further showed that the prevalence of over-abundant SR overlapping binding motifs in exons correlates with the strength of splice signals. Over-abundant overlapping motifs were significantly more abundant in exons with weak donor/acceptor splice signals than in exons with strong ones. This suggests that this additional level of regulation introduced by the overlapping motifs is specifically important for exons with suboptimal splicing signals for which splicing regulation depends strongly on auxiliary splicing factors. The prevalence of the under-represented overlapping motifs was negatively correlated with the strength of the splice signals. Exons with suboptimal splice signals are more prone to undergo aberrant splicing than those with consensus signals and therefore may be less permissive to the presence of sequences that obstruct the splicing process.
Additional examination of the overlapping motifs revealed that over-abundant and under-represented motif overlaps exhibited different spatial distributions. In exons with weak splice donor sites but not in those with a strong donor splice site, under-represented motifs were relatively less prevalent near the splice donor site than over-abundant motif overlaps. This implies that over-abundant and under-represented motif overlaps functionally differ. We hypothesize that the over-abundant motifs we identified take part in fine tuning the delicate splicing regulation and that the under-represented motifs hinder the splicing machinery and are therefore selected against. Finally, to further support the bioinformatic findings, we experimentally tested selected over-abundant overlaps in a minigene reporting system. We revealed that the overlaps enabled a mode of splicing that did not occur when either of the splicing motifs that form the overlap was present independently, nor did it occur when the entire overlap was absent. While our analyses reveal the over-abundance and conservation of overlapping sequences, both of which are indicative of functional implications, further experimental work is needed to determine whether there is actual binding of the SR-proteins to these specific sites. Overall, the results we present are indicative of another dimension of splicing regulation that involves overlapping splicing regulatory motifs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation (ISF 61/09), Joint Germany-Israeli Research Program (ca-139), Deutsche-Israel Project (DIP MI-1317) and European Alternative Splicing Network (EURASNET). AG is supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities and EK is a fellow of the Clore Scholars Program. Funding for open access charge: EURASNET.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 8.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 9.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl Acad. Sci. USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 13.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 15.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 16.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodley L, Valcarcel J. Regulation of alternative pre-mRNA splicing. Brief Funct. Genomic Proteomic. 2002;1:266–277. doi: 10.1093/bfgp/1.3.266. [DOI] [PubMed] [Google Scholar]
- 18.Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 20.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 22.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XH, Arias MA, Ke S, Chasin LA. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10:R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackerman SL, Minden AG, Williams GT, Bobonis C, Yeung CY. Functional significance of an overlapping consensus binding motif for Sp1 and Zif268 in the murine adenosine deaminase gene promoter. Proc. Natl Acad. Sci. USA. 1991;88:7523–7527. doi: 10.1073/pnas.88.17.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollwein P. Overlapping binding sites of two different transcription factors in the promoter of the human gene for the Alzheimer amyloid precursor protein. Biochem. Biophys. Res. Commun. 1993;190:637–647. doi: 10.1006/bbrc.1993.1096. [DOI] [PubMed] [Google Scholar]
- 27.Discenza MT, Dehbi M, Pelletier J. Overlapping DNA recognition motifs between Sp1 and a novel trans-acting factor within the wt1 tumour suppressor gene promoter. Nucleic Acids Res. 1997;25:4314–4322. doi: 10.1093/nar/25.21.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrington RH, Sharma A. Transcription factors recognizing overlapping C1-A2 binding sites positively regulate insulin gene expression. J. Biol. Chem. 2001;276:104–113. doi: 10.1074/jbc.M008415200. [DOI] [PubMed] [Google Scholar]
- 29.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 34.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 36.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 40.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 41.Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol. Cell Biol. 2008;28:3513–3525. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 43.Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.