Abstract
TRAP150 has been identified as a subunit of the transcription regulatory complex TRAP/Mediator, and also a component of the spliceosome. The exact function of TRAP150, however, remains unclear. We recently identified TRAP150 by its association with the mRNA export factor TAP. TRAP150 contains an arginine/serine-rich domain and has sequence similarity with the cell death-promoting transcriptional repressor BCLAF1. We found that TRAP150 co-localizes with splicing factors in nuclear speckles, and is required for pre-mRNA splicing and activates splicing in vivo. TRAP150 remains associated with the spliced mRNA after splicing, and accordingly, it interacts with the integral exon junction complex. Unexpectedly, when tethered to a precursor mRNA, TRAP150 can trigger mRNA degradation in the nucleus. However, unlike nonsense-mediated decay, TRAP150-mediated mRNA decay is irrespective of the presence of upstream stop codons and occurs in the nucleus. Moreover, TRAP150 activates pre-mRNA splicing and induces mRNA degradation by its separable functional domains. Therefore, TRAP150 represents a multi-functional protein involved in nuclear mRNA metabolism.
INTRODUCTION
Eukaryotic mRNA biogenesis comprises several post-transcriptional events, including pre-mRNA processing, mRNA export, surveillance and turnover control. Each individual step is carried out by specialized molecular machinery, but all steps are interconnected and collectively form an intricate network, with which cells achieve faithful and efficient gene expression (1,2).
During mRNA synthesis, the transcription machinery recruits RNA processing factors for capping, splicing and polyadenylation of the growing transcripts. The recruitment of specific factors may influence regulated precursor mRNA (pre-mRNA) processing events, such as alternative splice site selection or polyadenylation site utilization (3). Pre-mRNA splicing is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex containing five small nuclear RNAs and ∼150 protein factors (4). The spliceosome assembles anew on each pre-mRNA during splicing. Upon completion of splicing, the multiprotein exon junction complex (EJC) forms ∼20 nt upstream of each splice junction of the spliced mRNA (5,6). The EJC recruits or interacts with factors that facilitate subsequent mRNA export and surveillance steps. The most well-characterized mRNA surveillance pathway is nonsense-mediated decay (NMD), which allows detection and elimination of mRNAs harboring premature termination codons (PTCs) and thereby prevents their translation (7,8).
Individual EJC components may function in pre-mRNA processing or act as an effector in subsequent mRNA maturation steps. Several EJC factors, such as UAP56, RNPS1, SRm160 and Pnn/DRS, physically associate with the pre-mRNA during splicing (5). These factors may be critical for pre-mRNA splicing or participate in different aspects of splicing regulation (9–12). Following splicing, some of the EJC-associated factors, such as the export receptor TAP and RNA-binding protein Aly/REF, act to lead mRNA export (13). Moreover, the core factors of the EJC, Y14/Magoh, recruit the NMD factor Upf3 in the nucleus. Upf3 accompanies the mRNA to the cytoplasm and sequentially recruits Upf2 and the SURF complex (consisting of UPF1, SMG1 and the release factors eRF1 and eRF3) to the mRNA (14). Upon phosphorylation of Upf1 by SMG1, the EJC inhibits translation and recruits mRNA degradation factors to destroy PTC-containing mRNAs (14,15). Therefore, the EJC not only provides a functional connection between individual steps of mRNA biogenesis but also provides a platform for cellular regulation of post-transcriptional gene expression.
We initially set out to identify protein partners of TAP, which is a critical factor for general mRNA export. One of the identified candidates was TRAP150, a potential transcription and splicing factor. In this study, we demonstrated a role for TRAP150 in pre-mRNA splicing and, unexpectedly, in nuclear mRNA degradation.
MATERIALS AND METHODS
Plasmids
The complementary DNA (cDNA) encoding full-length TRAP150 was polymerase chain reaction (PCR)-amplified from a human fetal brain cDNA library (Clontech) using specific primers. The PCR product was inserted into a pcDNA3.1 (Invitrogen)-based vector in-frame with the sequence encoding the FLAG tag to generate pcDNA-TRAP150-FLAG. Analogously, expression vectors encoding each of the truncated TRAP150 proteins (Figure 3C) were generated. The cDNAs encoding full-length or truncated TRAP150 proteins were each cloned into pCEP4 (Invitrogen), yielding the expression vectors for hemagglutinin (HA)-tagged proteins. The DNA fragments containing the HA and full-length or truncated TRAP150 sequences were subcloned into pMCP (16) to generate the vectors for expressing the MS2 coat protein (MCP)-fusion proteins. To raise an antiserum against TRAP150, the TRAP150ΔNC DNA fragment was inserted in-frame into pGEX-5X (GE Healthcare) and pET29b (Novagen) to generate the bacterial expression vector for the glutathione S-transferase (GST) and 6× histidine (His)-fused TRAP150ΔNC protein, respectively. The BCLAF1 and MLN51 cDNAs were PCR-amplified and cloned into pcDNA3.1 and pCEP4 to generate the expression vectors for FLAG- and HA-tagged proteins, respectively. The plasmids expressing FLAG-tagged Aly/REF, SRm160, Y14, TAP, Upf3B, RNPS1, hUpf1, hUpf1-R844C and the reporter plasmids used for NMD assays (including βUAA-6bs, βUAC-6bs, βG, βwt and β39) were kind gifts from Jens Lykke-Anderson (University of Colorado, Boulder, CO, USA; Ref. 17). The cDNA fragment encoding eIF4AIII protein was PCR amplified and cloned into pCEP4 vector to generate HA-tagged eIF4AIII expression vector. The βΔ1 reporter was created by substitution of the Hind III/BamH I cDNA fragment corresponding to the spliced β-globin reporter mRNA. The βΔ1-5′m reporter was generated by using PCR-based site-directed mutagenesis (Stratagene).
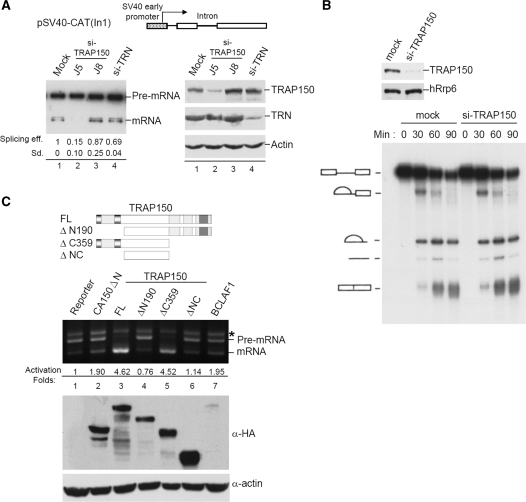
Figure 3.
TRAP150 activates pre-mRNA splicing. (A) The splicing report pSV40-CAT(In1) encodes the chloramphenicol acetyl transferase, in which human β-globin intron 1 was inserted. The pSV40-CAT(In1) reporter was co-transfected with indicated siRNA into HeLa cells. RT-PCR was performed using primers specific for the reporter exons; the PCR products were analyzed by Southern blotting using primers specific for the exons (left). Splicing efficiency in individual transfectants was calculated as the intensity of the spliced RNA over the sum of the spliced RNA and the precursor. Splicing activation fold was obtained from three independent experiments. Immunoblotting using antibodies specific to indicated protein was performed to reveal knockdown efficiency (right). (B) HeLa cells were mock-transfected or transfected with si-TRAP150 (J5). The nuclear extract was prepared from transfected cells and analyzed by immunoblotting (upper). Lower panel shows that in vitro splicing using 32P-labeled PIP85a pre-mRNA in the mock- or TRAP150-depleted nuclear extract. (C) Schematic representation of full-length and truncated TRAP150 proteins. Domains are depicted as in Figure 1. The pSV40-CAT(In1) vector was co-transfected with an expression vector encoding the indicated effector into HeLa cells. PCR was performed as in (A); asterisk denotes the precursor/spliced RNA hybrid. Splicing efficiency was also calculated as in (A) and the splicing activation fold relative to the reporter only was indicated below the gel. Immunoblotting using anti-HA shows the expression level of the effectors; actin was used as the loading control.
Antibody preparation
The GST and 6× His-TRAP150ΔNC recombinant proteins were overexpressed in Escherichia coli and purified according to the manufacturer’s instructions. 6× His-TRAP150ΔNC was used as antigen to immunize rabbits. Antibodies were affinity-purified against recombinant GST-TRAP150ΔNC protein according to the method described previously (18).
Cell culture and transfection
HEK293, HeLa, Tet-Off HeLa and NIH3T3 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin/glutamine (Invitrogen). Transfections were performed using Lipofectamine 2000 (Invitrogen) for 24 h. For immunoprecipitation experiments, HEK293 cells were transfected using calcium phosphate. At 16 h post-transfection, cells were placed in fresh medium and cultured for another 24 h before harvest.
Indirect immunofluorescence and heterokaryon assay
HeLa cells were fixed by 2% formaldehyde in phosphate-buffered saline (PBS) for 20 min and subsequently permeabilized with 100% acetone at 4°C for 3 min. The primary antibodies used included monoclonal anti-FLAG (M2; 1: 300 dilution; Sigma) and anti-SC35 (4.6 μg/ml; Sigma), and polyclonal anti-HA (1: 100 dilution; Covance Inc.) and affinity-purified polyclonal anti-TRAP150 (1: 100 dilution). The secondary antibodies used were fluorescein-conjugated anti-rabbit IgG (12 μg/ml; Cappel Laboratories) for polyclonal primary antibodies, and rhodamine-conjugated anti-mouse IgG (7.5 μg/ml, Cappel Laboratories) for monoclonal primary antibodies. The specimens were observed using a Zeiss Axiovert 200M inverted research-grade fluorescence microscope coupled with an image analysis system.
The heterokaryon assay was performed as described (17) with a minor modification. Briefly, HeLa cells were co-transfected with the vectors encoding TRAP150-FLAG and GFP-hnRNP A1 or GFP hnRNP C1. At 48 h post-transfection, HeLa cells were co-cultured with NIH3T3 cells and treated with cycloheximide (Sigma) at a concentration of 50 μg/ml for 3 h and 100 μg/ml for another 30 min. To induce cell fusion, 50% polyethylene glycol 3350 (Sigma) was added to the co-culture for 2 min. The cells were subsequently returned to fresh media containing 100 μg/ml cycloheximide for 3 or 16 h. Indirect immunofluorescence was performed as described above. To distinguish HeLa from mouse NIH 3T3 cells, the cells were counterstained with Hoechst 33258 (5 µg/ml; Sigma).
In vivo splicing and real-time-PCR
The splicing reporter pSV40-CAT(In) (18) was co-transfected with effector expression vectors (2 µg) into HeLa cells in 3.5-cm dishes at 80–90% confluency. At 24 h post-transfection, RNA samples were harvested from cells using TRIzol reagent (Invitrogen) and treated with RQ-DNase1 (Promega). Subsequently, RNAs were converted to first-strand cDNAs using SuperScript III reverse transcriptase (Invitrogen) using the 3′ primer 5′-GCAAGCTTCACTCCAGAGCGATG-3′ complementary to the CAT transcript. Subsequently, PCR and Southern blotting were performed using specific primers as described (18).
In vitro splicing and immunoprecipitation of the spliceosome
Preparation of nuclear extracts was essentially according to Hirose et al. (19), except that calcium phosphate was used for cell transfection. In general, HEK293 cells grown in 15-cm dishes at 70–80% confluency were transfected with 50 μg of expression vector encoding either FLAG-tagged full-length or truncated TRAP150. At 16 h post-transfection, medium was refreshed and cells were incubated for another 24 h. For preparation of the TRAP150-depleted nuclear extract, HeLa cells grown in 15-cm dishes at ∼60% confluency were transfected with 50 nM si-TRAP150-J5 using Lipofectamine 2000. At 48 h post-transfaction, cells were harvested for nuclear extract preparation as described (19).
In vitro splicing was performed essentially as described (20) using 32P-labeled PIP85a pre-mRNA (gift of Benjamin J. Blencowe, Toronto University) as substrate. The reaction was incubated at 30°C for 90 min, followed by subsequent incubation with anti-FLAG M2 agarose (Sigma) or anti-Sm (Y12, gift of Joan Steitz, Yale University) bound to protein A-Sepharose at 4°C for 2 h (18). The beads were washed extensively with NET-2 buffer (50 mM Tris–HCl, pH 7.4 and 150 mM NaCl) containing 0.05% NP-40. Reactions were treated with 10 mg/ml proteinase K prior to RNA collection. RNAs were analyzed by electrophoresis on 6% denaturing polyacrylamide gels.
Immunoprecipitation
For co-immunoprecipitation, HEK293 cells grown on a 10-cm dish were co-transfected with 14 μg of pCEP4-TRAP150-HA or pCEP4-eIF4AIII-HA and 7 μg of vector encoding for a Flag-tagged EJC or NMD factor or TRAP150 by calcium phosphate method. Cells were lysed in 1 ml of hypotonic lysis buffer containing 10 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, and 1× protease inhibitor cocktail (Roche) on ice for 15 min. Subsequently, additional NaCl was added to the lysate to a final concentration of 150 mM. The lysate was subjected to centrifugation at 13 400 × g at 4°C for 20 min. The supernatant was incubated with 30 μl anti-FLAG M2 agarose at 4°C for 2 h. The beads were washed with NET-2 buffer as described above. Precipitated proteins were dissociated from the beads with sample buffer containing 50 mM Tris–HCl, pH 6.8, 100 mM dithiothreitol, 2% SDS, 20% glycerol and 0.2 mg/ml bromophenol blue.
To identify TAP-interacting proteins using mass spectrometry analysis, HEK293 cells grown on a 15-cm dish at 60–70 % confluency were transfected with 60 μg of plasmid encoding Flag-tagged TAP using Lipofectamine 2000. Immunoprecipitation using anti-FLAG was carried out as described above. Precipitated proteins were separated by 10% SDS-PAGE followed by SYPR Ruby staining (Bio-Rad) and detected using Typhoon 9410 (GE Healthcare). The following procedures, including trypsinization and peptides recovery, were essentially carried out as described (18).
NMD assays and northern blotting
Tet-Off HeLa cells grown on a 3.5-cm dish were co-transfected with an MS2 site-containing NMD reporter (1 µg) and an expression vector encoding an MCP-effector (2.5 µg) as well as a control plasmid (βG; 0.5 µg). At 24 h post-transfection, total RNA was isolated using TRIzol reagent and subjected to northern blotting analysis using a 32P-labeled riboprobe complementary to exon 1 and exon 2 of β-globin. To determine mRNA half-life, 5 µg/ml actinomycin D (Sigma) was added to inhibit transcription at 10 h post-transfection. Cells were harvested at the indicated time points. For subcellular fractionation, cells were incubated with gentle lysis buffer (10 mM HEPES-KOH, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) (17) for 5 min followed by centrifugation at 2000 × g at 4°C for 8 min. The cytoplasmic and nuclear fraction was collected from the supernatant and the pellet, respectively. For knockdown experiments, cells were transfected with 100 nM siRNA for the first 48 h followed by further transfection with the effector expression vector and the reporter as above. After incubation for another 24 h, total RNA was isolated for northern blotting. siRNAs si-TRAP150-J5 and -J8 were purchased from Dharmacon and si-Luciferase (si-Luc.) and si-TRN were from Invitrogen. Their sense-strand sequences were as follows: si-TRAP150-J5, 5′-gguauaagcuccgagaugauu-3′; si-TRAP150-J8, 5′-caaaugggagggccugguauu-3′; si-Luc., 5′-ggatttcgagtcgtcttaatgtata-3′; si-TRN, 5′-acaggaaucaacuuaggaagaugcc-3′. Monoclonal anti-TRAN1 was purchased from Sigma. Protein levels were quantified using ImageQuantTL software (GE Healthcare).
RESULTS
Association of TRAP150 with TAP
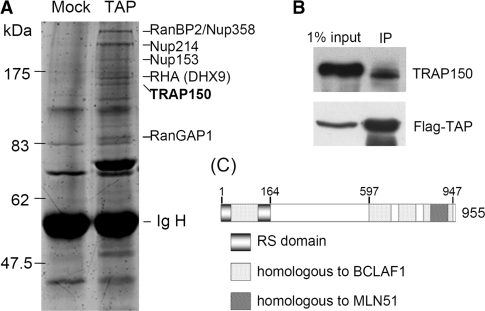
To identify TAP-associating proteins, we transiently expressed FLAG-tagged TAP in HEK293 cells and prepared the cell lysate for immunoprecipitation using an antibody against FLAG peptide. FLAG-TAP-co-precipitated proteins were detected by SDS-PAGE using SYPRO Ruby fluorescence followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis. Identified proteins included several nucleoporins, RanGAP1, RNA helicase RHA/DHX9 and TRAP150 (Figure 1A). The interaction of FLAG-TAP with endogenous TRAP150 was confirmed by immunoblotting using anti-TRAP150 (Figure 1B) and this interaction was resistant to RNase treatment (see below, for Figure 4B), suggesting that TAP may directly interact with TRAP150. TRAP150 contains an arginine/serine-rich sequence in the N-terminal region and its C-terminal region has 48% overall identity with BCLAF1/Btf (21), a cell death-promoting transcriptional repressor (Figure 1C). Within the BCLAF1 homologous domain, a ∼ 90-amino acid segment of TRAP150 shares 30% similarity with MLN51, an EJC core component (22,23). Both TRAP150 and BCLAF1 are associated with the spliceosome, indicating their potential role in pre-mRNA splicing (4,24). Moreover, the interaction of TRAP150 with TAP predicts a function for TRAP150 in post-splicing events. Therefore, in this study, we attempted to explore the potential of TRAP150 in splicing and subsequent mRNA maturation steps.
Figure 1.
TRAP150 associates with TAP. HEK 293 cells were transiently transfected with an expression vector encoding FLAG-tagged TAP or mock-transfected. Cell lysates were subjected to immunoprecipitation with anti-FLAG-conjugated resin. (A) Co-precipitated proteins were detected by SDS-PAGE using SYPRO Ruby. FLAG-TAP-associated proteins were identified by mass spectrometry. (B) Immunoblotting of FLAG-TAP-co-precipitates was performed using anti-TRAP150. (C) Schematic representation of the TRAP150 domain structure.
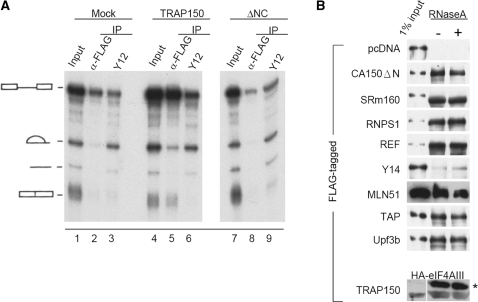
Figure 4.
TRAP150 is associated with the spliced mRNP and the EJC. (A) In vitro splicing using 32P-labeled PIP85a pre-mRNA as substrate was performed in HEK293 cell nuclear extract that contained overexpressed FLAG-TRAP150 or truncated TRAP150 (ΔNC). Immunoprecipitation was performed using anti-FLAG or anti-Sm protein (antibody Y12). The autoradiogram shows the splicing reaction and precipitated RNAs. (B) HEK293 cells were transfected with an expression vector encoding HA-TRAP150 and vector encoding the FLAG-tagged proteins as indicated. The bottom panel shows co-transfection of the vectors encoding FLAG-TRAP150 and HA-eIF4AIII. Immunoprecipitation and immunoblotting were sequentially performed using anti-FLAG and anti-HA, respectively. Asterisk denotes immunoglobulin heavy chain and below is HA-eIF4AIII.
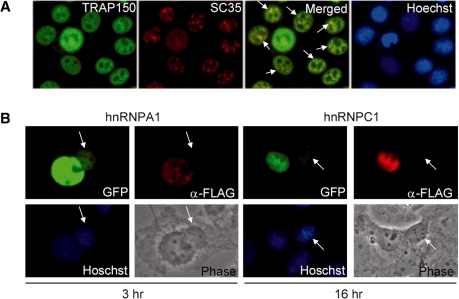
Using anti-TRAP150, we observed that TRAP150 was distributed mainly in the nucleoplasm with higher concentrations at discrete foci (Figure 2A). TRAP150 foci overlapped well with sites stained with anti-SC35 (Figure 2A), indicating that TRAP150 co-localizes with splicing factors in nuclear speckles. We also performed an interspecies heterokaryon assay to examine whether TRAP150 is exported to the cytoplasm. FLAG-tagged TRAP150 was transiently co-expressed with GFP-hnRNP A1 or GFP-hnRNP C1 in human HeLa cells. Transfected cells were subsequently fused with mouse NIH-3T3 cells. FLAG-TRAP150 was never detected in the cytoplasm of the fused cells, in contrast to the observation with hnRNP A1 and hnRNP C1 that were visible in the cytoplasm at 3 and 16 h, respectively, after cell fusion (Figure 2B). This result indicated that TRAP150 localization is very likely restricted to the nucleus.
Figure 2.
Cellular localization of human TRAP150. (A) Double immunofluorescence staining of HeLa cells was performed using anti-TRAP150 and anti-SC35. Co-localized signals are indicated by arrows. Nuclei were stained with Hoechst dye 33258. (B) HeLa cells were co-transfected with the expression vector encoding FLAG-TRAP150 and a vector encoding GFP-hnRNP A1 or GFP-hnRNP C1. Transfected HeLa cells were fused with mouse NIH3T3 cells for 3 or 16 h in the presence of cycloheximide. Immunofluorescence was performed using anti-FLAG; GFP fusion proteins were directly visualized by fluorescence microscopy. Arrows indicate mouse cell nuclei.
TRAP150 activates pre-mRNA splicing
TRAP150 has been identified in the spliceosome (24), but whether it plays any role in pre-mRNA splicing is unclear. To first examine whether TRAP150 is essential for pre-mRNA splicing, we exploited small-interfering RNA (siRNA)-mediated knockdown to deplete TRAP150 in HeLa cells. Immunoblotting showed that the protein level of TRAP150 was reduced to ∼20% of the mock by siRNA J5 (Figure 3A, lane 2). The reporter used encoded the chloramphenicol acetyl transferase (CAT) transcript, within which the first intron of human β-globin was inserted (18). TRAP150 knockdown resulted in blockage of the splicing of this reporter transcript (lane 2). However, another TRAP150 siRNA, J8, that failed to reduce TRAP150 expression had no effect on splicing (lane 3), as was the case for siRNA targeting transportin-1, (lane 4, TRN). Moreover, the splicing activity of TRAP150-knockdown cells could be restored by overexpression of a J5-resistant TRAP150, indicating the specific role of TRAP150 in pre-mRNA splicing (Supplementary Figure 1, lane 4). We also prepared the nuclear extract from TRAP150-depleted HeLa cells and performed in vitro splicing assay using a 32P-labeled PIP85a pre-mRNA as substrate. However, a significant reduction of TRAP150 protein level (Figure 3B, immunoblotting) appeared not to affect pre-mRNA splicing in vitro (Figure 3B, splicing assay). A similar result was also observed with another pre-mRNA derived from the adenovirus major late transcript (data not shown). Although the possibility that residual TRAP150 was sufficient for pre-mRNA splicing remains, our present data indicate that TRAP150 is not critical for pre-mRNA splicing in a cell-free system, but plays a role in cells. Perhaps transcription is required for TRAP150 to function in pre-mRNA splicing in vivo; this possibility remains to be examined.
We next explored whether overexpression of TRAP150 has any effect on splicing. The β-globin intron-containing CAT reporter was co-transfected with the expression vector of HA-tagged TRAP150 in HeLa cells. Overexpression of TRAP150 greatly enhanced the efficiency of reporter pre-mRNA splicing, as compared to another splicing factor, CA150 (Figure 3C, lanes 2 and 3). BCLAF1 could also promote the splicing of this reporter, but whether poor expression of BCLAF1 accounted for its lower splicing activity is unclear (lane 7). Furthermore, to examine which domain of TRAP150 contributes to splicing activation, we made truncation mutants. Deletion of the RS-rich domain severely impaired the splicing activation ability of TRAP150 (lane 4), whereas the C-terminally truncated mutant retained almost full activity (lane 5), indicating that the RS-rich domain is required for the activity of TRAP150 in pre-mRNA splicing. Together, the above assays apparently indicate that TRAP150 acts as a splicing activator and may participate in a step that is critical for pre-mRNA splicing in cells but not in the extract.
TRAP150 binds to spliced mRNA and interacts with the EJC factors
Previous report has indicated that TRAP150 is a component of the spliceosome. Our data showed that TRAP150 played an essential role in splicing of reporter pre-mRNAs in cells (Figure 3). As TRAP150 also interacts with TAP (Figure 1), we therefore explored whether TRAP150 joins the spliceosome and subsequently binds to the spliced mRNA. Nuclear extracts were prepared from transfected HEK293 cells that transiently expressed FLAG-tagged TRAP150 or its truncated non-functional fragment, ΔNC, for in vitro splicing of the PIP85a pre-mRNA. The reactions were subsequently subjected to immunoprecipitation using anti-FLAG. A substantial amount of the pre-mRNA was co-precipitated with full-length TRAP150 but not with ΔNC (Figure 4A, lanes 5 and 8). Indeed, TRAP150 became associated with the pre-mRNA even without addition of ATP in the splicing reaction, indicating that TRAP150 joins the pre-mRNP early during spliceosome assembly (Supplementary Figure 2, lane 4). In contrast with small nuclear RNP-associated splicing complexes that reacted with anti-Sm Y12, TRAP150 bound more preferentially to the spliced mRNA than the excised intron (compare lane 5 to lane 6). Therefore, TRAP150 might be a component of nuclear post-splicing mRNP complexes. Nevertheless, TRAP150 also co-precipitated the intronless PIP85a in vitro (Supplementary Figure 2, lane 12). Thus, consistent with a previous observation (24), TRAP150 could be associated with mRNPs independent of splicing.
The association of TRAP150 with the spliced mRNA promoted us to assess whether it interacts with the EJC and participates in downstream mRNA metabolic events. To test this possibility, the HA-tagged TRAP150 expression vector was co-transfected with vector encoding a FLAG-tagged splicing, EJC or NMD factor into HEK293 cells. Anti-FLAG-precipitated proteins from cell lysates were subjected to immunoblotting with anti-HA. The result showed that a substantial amount of HA-tagged TRAP150 was co-precipitated with splicing factors CA150, SRm160 and RNPS1, and that it also interacted with several components of the post-splicing EJC or NMD complex, such as REF, TAP, MLN51 and Upf3b, albeit minimally with Y14 (Figure 4B). The interactions of HA-TRAP150 with these factors were specific because it was not precipitated in the absence of bait (Figure 4B, pcDNA). Although the interaction between FLAG-eIF4AIII and HA-TRAP150 was not detected (data not shown), a significant amount of HA-eIF4AIII could be co-precipitated with FLAG-TRAP150 using anti-FLAG (Figure 4B, bottom). Therefore, our data suggested that TRAP150 remains bound to the spliced mRNA likely through its direct interaction with the EJC.
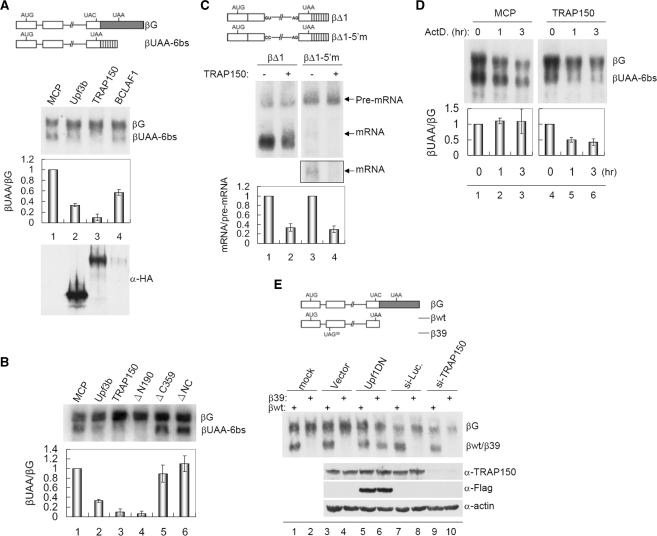
TRAP150 induces mRNA degradation but is not a canonical NMD factor
The interaction of TRAP150 with the EJC and NMD factors implied its potential role in mRNA degradation. Next, we took advantage of an NMD-tethering assay (17) to examine the activity of TRAP150 in mRNA surveillance. The reporter β-globin mRNA contained a stop codon located upstream of the repeated MS2 sites (Figure 5A, diagram, βUAA-6bs, Ref. 17); expression of the NMD factor Upf3b that was fused to the bacteriophage MS2 coat protein (MCP) triggered degradation of this reporter mRNA (northern blotting, lane 2). Tethering of TRAP150 markedly downregulated βUAA-6bs mRNA expression (lane 3). BCLAF1 could also reduce βUAA-6bs expression, although it was poorly expressed (lane 4). Next, we characterized TRAP150 domains involved in mRNA decay. Using truncated TRAP150 proteins, we found that the C-terminal domain, which contains an MLN51-like fragment, was essential for TRAP150 function in mRNA degradation, whereas the N-terminal RS domain was dispensable for this activity (Figure 5B, lanes 4 and 5). Therefore, together with the above observation, our results indicated that the activities of TRAP150 in pre-mRNA splicing and mRNA degradation involve its separated domains.
Figure 5.
TRAP150 promotes mRNA degradation in the tethering NMD assay but is not essential for NMD. (A) Schematic representation of the β-globin reporter βUAA-6bs that contains the UAA stop codon upstream of six copies of the MS2 coat protein (MCP)-binding site; βG without these sites was used as a co-transfection reference. The βUAA-6bs and βG vectors were co-transfected with an expression vector encoding the indicated effector. Northern blotting was performed using a 32P-labeled β-globin probe. In individual transfectants, the intensity of steady-state βUAA-6bs mRNA was normalized to that of βG. Bars show the relative level of the βUAA-6bs mRNA of MCP-effector transfectants to the unfused MCP. (B) The tethering NMD assay was performed by co-transfection of the reporter as in (A) and the vector encoding MCP or MCP-fused proteins as indicated. Bars shown at bottom are as in (A). (C) Schematic representation of the βΔ1 and the 5′ splice site mutant βΔ1-5′m reporters. These two reporters were each co-transfected with the MCP or MCP-TRAP150 expression vector. Northern blotting was performed as in (A); a longer exposure of the spliced product of βΔ1-5′m is shown in the outlined rectangle. The steady-state level of the spliced mRNA was normalized to that of the precursor mRNA in individual transfectants; relative levels of mRNA/pre-mRNA of the MCP-TRAP150 transfectant to the corresponding mock are indicated. (D) The βUAA-6bs and βG vectors were co-transfected with the MCP or MCP-TRAP150 expression vector for 10 h. Cells were collected after addition of actinomycin D for 0, 1 or 3 h. Northern blotting was performed as in (A). Bars show the relative level of βUAA-6bs to βG mRNA of each transfectant was normalized to that of time zero. (E) HeLa cells were transfected with the indicated siRNA for 48 h. Subsequently, the β-globin reporter containing (β39) or not containing (βwt) a translation termination codon UAG in exon 2, as depicted in the scheme, was co-transfected with the reference βG. For lanes 5 and 6, the expression vector of the dominant-negative Upf1 was co-transfected with the reporters. Northern blotting was performed as in (A). Immunoblotting of cell lysates was performed using anti-TRAP150, FLAG and anti-actin (bottom).
To reduce the arguments that MCP-TRAP150 may interfere with reporter transcription or induce degradation of primary transcripts, we had attempted to examine the expression of reporter pre-mRNAs. However, probably due to efficient splicing, the βUAA-6bs pre-mRNA was barely detected by northern blotting even in the absence of MCP-fusion proteins. Therefore, we created two reporters, which had relatively poor splicing efficiency, thereby allowing pre-mRNA to be detectable (Figure 5C, diagram). The βΔ1 reporter was created by removing the first intron from βUAA-6bs. Using this reporter, we observed that TRAP150 could reduce the level of the spliced mRNA but not the precursor (Figure 5C, lanes 1 and 2). Splicing was more severely blocked, when the 5′ splice site dinucleotides of the remaining intron 2 of βΔ1 were further mutated (Figure 5C, βΔ1-5′m). Nevertheless, the result of the TRAP150-tethering assay was similar to that observed with βΔ1, indicating that TRAP150 neither significantly impaired transcription nor degraded pre-mRNA (lanes 3 and 4). Therefore, TRAP150 may trigger mRNA degradation after splicing.
We also attempted to evaluate the kinetics of TRAP150-mediated mRNA decay. In order to detect spliced reporter mRNAs, we measured their expression level at 10 h after co-transfection with the vector of MCP-TRAP150. At 1 h after addition of actinomycin D to arrest transcription, ∼50% of spliced βUAA-6bs mRNA was reduced by TRAP150 (Figure 5D, lane 5). This result also indicated that TRAP150 is involved in post-transcriptional mRNA decay.
Next, we examined whether TRAP150 is essential for NMD. Using a reporter containing a PTC in the penultimate exon (Ref. 17; Figure 5E, diagram), we compared RNAi-mediated knockdown of TRAP150 to overexpression of the Upf1-R844C mutant that displays a dominant-negative activity in NMD. In contrast to the effect of Upf1-R844C overexpression, substantial depletion of TRAP150 did not elicit the reporter mRNA level (Figure 5E, northern blotting), indicating that TRAP150 is not critical for NMD. In the meantime, we also found that neither the translation inhibitor cycloheximide nor the Upf1-R844C mutant could block TRAP150-tethering-induced degradation of the βUAA-6bs reporter mRNA (Supplementary Figure 3). Therefore, TRAP150-induced mRNA decay was dissimilar to canonical NMD and might not be coupled to translation.
TRAP150-mediated mRNA decay is irrespective of PTC and occurs in the nucleus
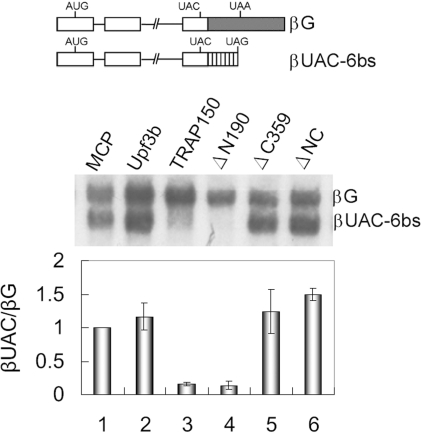
The finding that TRAP150-mediated mRNA degradation was distinct from NMD prompted us to examine whether a translation termination codon located prior to the effector tethering site is essential. Using the β-globin mRNA lacking such a stop codon (Figure 6, diagram, βUAC-6bs; Ref. 17), we observed that, unlike Upf3b, TRAP150 could induce efficient degradation of this NMD-insensitive mRNA (Figure 6, lanes 2 and 3). Therefore, TRAP150-induced mRNA decay might not necessarily depend on the termination codon upstream of its tethering site. This result also indicated that TRAP150 can induce mRNA decay no matter whether it is tethered within the 3′ untranslated region (UTR) or open reading frame. Thus, TRAP150 could bypass the requirement for a PTC to elicit degradation of the tethered mRNA, as does SMG7 (25).
Figure 6.
Premature stop codon is not required for TRAP150-mediated mRNA decay. The scheme shows the βUAC-6bs reporter, which is similar to βUAA-6bs except that the UAA codon is changed to UAC. The TRAP150-tethering assay was performed as in Figure 5B. The relative level of βUAC-6bs to βG mRNA of the transfectants was shown as in Figure 5B.
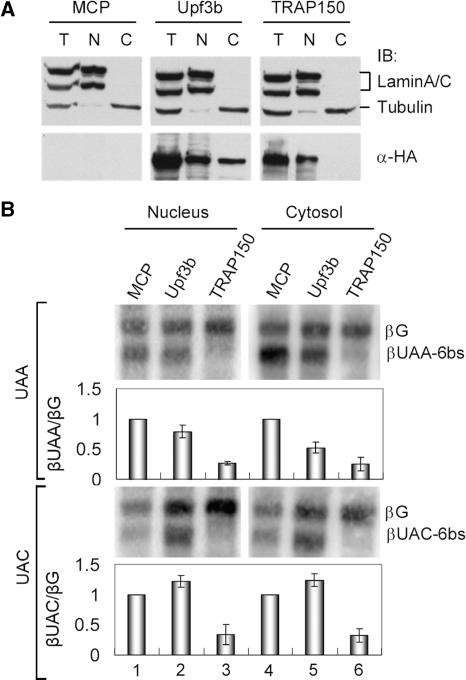
Our above data showed that TRAP150 is restricted to the nucleus (Figure 2). Thus, it is possible that TRAP150-activated mRNA decay takes place in the nucleus. To examine this possibility, we adopted the tethering assay as above but fractionated transfected Tet-Off HeLa cells for analysis. Immunoblotting of subcellular fractions showed that TRAP150 distributed only in the nucleus (Figure 7A), consistent with our immunofluorescence data (Figure 2), whereas Upf3b was detected in both the cytoplasm and the nucleus (Figure 7A). As reported (26), tethering of Upf3b led to degradation of the βUAA-6bs but not βUAC-6bs mRNA in the cytoplasm (Figure 7B, lanes 2 and 5). In contrast to that observed with Upf3b, the level of both reporter mRNAs was significantly reduced by tethering of TRAP150 in the nucleus (Figure 7B, lanes 3 and 6). Thus, as predicted, TRAP150-induced mRNA decay may occur in the nucleus, which is also substantially different from NMD.
Figure 7.
TRAP150-mediated NMD mRNA decay occurs in the nucleus. (A) Tet-Off HeLa cells were transiently transfected with an expression vector encoding MCP, MCP-Upf3b or MCP-TRAP150. Immunoblotting of total (T), nuclear (N) or cytoplasmic (C) extract of transfected HeLa cells was performed using respective antibodies against lamin, α-tubulin and HA epitope that detected MCP-fusions. (B) The βUAA/C-6bs reporter assay was performed essentially as in Figure 5A except that northern blotting was using RNAs prepared from subcellular fractions. Bars show the relative level of βUAA-6bs or βUAC-6bs to βG mRNA.
DISCUSSION
In this report, we show that the spliceosomal component TRAP150 has a dual function in post-transcriptional control. TRAP150 can activate pre-mRNA splicing when overexpressed and plays an essential role in splicing in vivo. Moreover, when tethered to mRNA transcripts, TRAP150 can induce their degradation in the nucleus. We propose that TRAP150 functions in nuclear mRNA processing and mRNA decay.
Role of TRAP150 in pre-mRNA splicing
We identified TRAP150 by its interaction with TAP (Figure 1). Previous reports have revealed that TRAP150 associates with the spliceosomal complexes, albeit in a transient and heparin-sensitive manner (24). TRAP150 may directly interact with splicing factors RNPS1, SRm160, UAP56 and SRrp86 (27). Consistently, our observation that TRAP150 is co-localized with splicing factors in nuclear speckles (Figure 2) also indicates a potential role of TRAP150 in pre-mRNA splicing.
Indeed, depletion of TRAP150 by siRNA blocked pre-mRNA splicing in vivo although it had no apparent effect on splicing in vitro (Figure 3). Previous reports indicated that TRAP150 may be recruited to emerging transcripts via its interaction with RNA polymerase II-associated splicing factors such as CA150 during transcription (18,28). Using immunoprecipitation, we observed that TRAP150 became associated with the pre-mRNA independent of ATP (Supplementary Figure 2). Therefore, participation of TRAP150 in pre-mRNA splicing in vivo may require ongoing transcription and occur during the early stage of spliceosome assembly. Moreover, TRAP150 could potently enhance splicing efficiency via its RS domain-containing N-terminus while overexpressed (Figure 3). However, our tests toward whether TRAP150 could modulate alternative 5′ splice site selection of the adenovirus E1A transcripts or alternative exon selection of CD44, a substrate of its interacting factor SRrp86 (27), did not find a role of TRAP150 in alternative splicing regulation (data not shown). Therefore, our present data indicate that TRAP150 has the splicing enhancing capacity in vivo, but may not be a general alternative splicing regulator. The questions of how TRAP150 functions in pre-mRNA splicing and whether it modulates alternative splicing in a gene-specific manner still remain to be investigated.
Role of TRAP150 in mRNA decay
We observed that TRAP150 associated with spliced mRNAs and interacted with several components of post-splicing RNP complexes (Figure 4), and it could induce degradation of spliced mRNA but not pre-mRNA (Figure 5). However, we also found that TRAP150-induced mRNA degradation has several unique features. First, tethering TRAP150 to the βUAC-6bs and βUAA-6bs reporter mRNAs could induce their degradation without discrimination (Figures 5 and 6), which indicates that TRAP150 induces mRNA decay in a PTC-independent manner. In another words, TRAP150 may degrade an mRNA, regardless of whether it is tethered within the 3′ UTR or within the open reading frame. We infer that TRAP150-mediated mRNA decay does not involve decoding of the reading frame by the ribosome. In accordance with this idea, we also showed that TRAP150-induced mRNA decay does not depend on active translation (Supplementary Figure 3). Moreover, unlike NMD, degradation of TRAP150-tethered mRNAs occurred in the nucleus (Figure 7). Therefore, it is possible that TRAP150 is not directly involved in NMD. To explore which ribonucleases are involved in TRAP150-induced mRNA decay, we examined three exonucleases Xrn1, Xrn2 and Rrp6. Depleting one or two of these nucleases by RNAi had no significant effect on TRAP150-induced RNA degradation (Supplementary Figure 4). This observation was somewhat intriguing because these three enzymes constitute the main ribonucleolytic activity of eukaryotic cells (7). Taken together, although TRAP150 likely interacts with the integral EJC (Figure 4), it confers an mRNA degradation activity that is very different from NMD. The mechanism of how TRAP150 activates mRNA degradation pathways needs further study.
TRAP150 is a dual function modular protein
Domain deletion analysis showed that truncation of the RS domain-containing N-terminus of TRAP150 caused a complete loss of its splicing activation potential but retained the RNA degradation activity, whereas deletion of the C-terminal domain had an opposite result (Figures 3, 5 and 6). This observation indicated that TRAP150 is a modular protein, in which the respective splicing activation and RNA degradation activities involve distinct and separable domains and thereby act independently. The RS-rich domain of TRAP150 very likely contributes to its splicing activation activity (Figure 3), which is similar to that observed with many SR splicing factors (29). Interestingly, the C-terminal region of TRAP150 contains a motif homologous to MLN51 and BCLAF. MLN51 is a critical factor for NMD (30), whereas TRAP150 is likely involved in nuclear mRNA decay (Figure 7). Therefore, whether this conserved motif recruits any critical factors such as ribonucleases to cause mRNA destruction remains to be studied. In conclusion, we report here that TRAP150 is a modular protein with dual post-transcriptional activities and may provide a link between pre-mRNA spicing and nuclear degradation of mRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The National Science Council of Taiwan (NSC95-2311-B-001-019); Intramural fund from Academia Sinica.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly thank Ming-Chih Lai for mass spectrometry of TAP-interacting mRNPs and identification of TRAP150, and Kang-mai Wu and Pey-Jey Peng for technical assistance. We appreciate J. Lykke-Anderson and B. J. Blencowe for reporter constructs, and J. Steitz for Y12 antisera.
REFERENCES
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Kornblihtt AR, de la Mata M, Fededa JP, Muňoz MJ, Nogués G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell. Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 9.Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–1803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 2004;24:1174–1187. doi: 10.1128/MCB.24.3.1174-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCracken S, Longman D, Johnstone IL, Cáceres JF, Blencowe BJ. An evolutionarily conserved role for SRm160 in 3'-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 2003;278:44153–44160. doi: 10.1074/jbc.M306856200. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Lin RI, Lai MC, Ouyang P, Tarn WY. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol. Cell. Biol. 2003;23:7363–7376. doi: 10.1128/MCB.23.20.7363-7376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 14.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu IW, Hsu M, Li C, Chuang TW, Lin RI, Tarn WY. Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. J. Biol. Chem. 2005;280:34507–34512. doi: 10.1074/jbc.M507658200. [DOI] [PubMed] [Google Scholar]
- 17.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 18.Lin KT, Lu RM, Tarn WY. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 2004;24:9176–9185. doi: 10.1128/MCB.24.20.9176-9185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirose T, Shu MD, Steitz JA. Splicing of U12-type introns deposits an exon junction complex competent to induce nonsense-mediated mRNA decay. Proc. Natl Acad. Sci. USA. 2004;101:17976–17981. doi: 10.1073/pnas.0408435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarn WY, Steitz JA. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 21.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 22.Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, Goetze B, Lupas A, St Johnston D, Kiebler M. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 2003;23:5778–5788. doi: 10.1523/JNEUROSCI.23-13-05778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tange TØ, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merz C, Urlaub H, Will CL, Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol. Cell. 2007;27:780–792. doi: 10.1016/j.molcel.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Hawkins IC, Harvey CD, Jennings JL, Link AJ, Patton JG. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol. Cell. Biol. 2003;23:7437–7447. doi: 10.1128/MCB.23.21.7437-7447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstrohm AC, Albrecht TR, Suñé C, Bedford MT, Garcia-Blanco MA. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 2001;21:7617–7628. doi: 10.1128/MCB.21.22.7617-7628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long JC, Cáceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 30.Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.