Abstract
We employed the Tag-seq technique to generate global transcription profiles for different strains and life stages of the nematode C. elegans. Tag-seq generates cDNA tags as does Serial Analysis of Gene Expression (SAGE), but the method yields a much larger number of tags, generating much larger data sets than SAGE. We examined differences in the performance of SAGE and Tag-seq by comparing gene expression data for 13 pairs of libraries. We identified genes for which expression was consistently changed in long-lived worms. Additional genes emerged in the deeper Tag-seq profiles, including several ‘signature’ genes found among those zup-regulated in long-lived dauer larvae (cki-1, aak-2 and daf-16). Fifty to sixty percent of the genes differentially expressed in daf-2(−) versus daf-2(+) adults had fragmentary or no functional annotation, suggesting the involvement of as yet unstudied pathways in aging. We were able to distinguish between changes in gene expression associated with altered genotype or altered growth conditions. We found 62 cases of possible mRNA isoform switching in the 13 Tag-seq libraries, whereas the 13 SAGE libraries allowed detection of only 15 such occurrences. We observed strong expression of anti-sense transcripts for several mitochondrial genes, but nuclear anti-sense transcripts were neither abundant nor consistently expressed among the libraries.
INTRODUCTION
Genome-wide expression profiling is routinely used for genome annotation and for identifying transcripts that are differentially expressed under specific environmental conditions or in specific tissues at different times in development. DNA microarrays (1,2) or Serial Analysis of Gene Expression, SAGE (3) measure the expression levels of thousands of genes in a single experiment. However, detection and quantitation of rare transcripts has been a particular challenge (4–6). To the advantage of the SAGE platform, DNA microarrays analyze pools of mRNAs with a fixed number of oligonucleotide probes. SAGE is capable of detecting virtually any polyadenylated mRNA that contains a site for an anchoring enzyme, and even mRNAs lacking a polyA-tail if they have oligo-A stretches in their sequences.
Tag-seq (7) combines generation of short tag signatures for cellular transcripts with massively parallel sequencing. Transcript-derived tags are sequenced directly in an Illumina flow cell, rather than concatenated and cloned for serial sequencing (7). This provides a much more sensitive, cost-efficient alternative to conventional SAGE. Here, we present Caenorhabditis elegans Tag-seq data compared with previously generated SAGE data. Deeper sequencing not only allowed us to enhance the detection and quantitation of less abundant messages, but also reduced the costs by two-thirds. Lower cost allowed us to conduct replicate experiments to assess the reproducibility of our data. Hence, a major disadvantage of SAGE has been largely overcome.
In a similar study describing Polony Multiplex Analysis of Gene Expression (PMAGE), large-scale sequencing by ligation was used to enhance the SAGE protocol (8). Although PMAGE boosts the number of tags (up to 5 million cDNA sequences per run), Tag-seq surpasses it, allowing generation of up to 9-million tags per library (a SAGE library normally is sequenced to a depth of 100 000–200 000 tags). Recently, whole transcriptome shotgun sequencing, or WTSS, has been developed using random priming of cDNA to generate tags for Illumina sequencing (9). This approach provides better tag-to-gene mapping because of longer reads, and allows superior detection of alternatively spliced mRNAs because the tags are not constrained by cleavage at an NlaIII (CATG) restriction site. However, the WTSS data are not so readily comparable with existing SAGE data. The ease of such a comparison is a major benefit for the expression analysis reported here.
To directly compare Tag-seq and SAGE datasets, 13 of our Tag-seq libraries were prepared from the same RNA samples used earlier for preparation of SAGE libraries. We also compared the previous SAGE data for long-lived mutants with Tag-seq data generated from new RNA preparations from the same long-lived and wild-type strains. Finally, we analyzed Tag-seq data for negative-strand transcripts and alternatively spliced variants. Our results show that Tag-seq is a much more sensitive replacement for conventional SAGE.
MATERIALS AND METHODS
Strains, RNA preparation and library construction
For construction of Tag-seq libraries we extracted RNA from the following strains of C. elegans: the daf-2(+) strain, fer-15(b26ts), a temperature-sensitive sterile mutant of C. elegans, the long-lived daf-2(−) mutant strain, fer-15; daf-2(m41), wild-type N2 L1 larvae and N2 dauer larvae. The daf-2 gene encodes the insulin/IGF-1 receptor, and reduction of insulin-like signaling extends mean and maximum life span (10,11). The fer-15 mutation was introduced to eliminate embryonically expressed transcripts from the expression profile of synchronously aging adults.
To collect starved L1 larvae, hypochlorite-purified eggs were hatched into M9 buffer and harvested after 48 h. Well-fed L1 larvae were harvested 8 h after hatching at 20°C. All animals were grown on agar plates (except for N2 starved L1 larvae). Synchronized adult populations were grown at 25°C, then harvested at times described previously (12,13). Adults with normal life span (carrying the fer-15 mutation) were harvested on the first and sixth days of adulthood. Long-lived daf-2(−) adults were harvested on the first, sixth and 10th day of adulthood.
We also constructed 13 Tag-seq libraries from the RNA preparations used earlier for SAGE libraries (12,13). The young (2-day-old) dauer Tag-seq library was made from mRNA extracted from pheromone-induced dauer larvae. Pheromone extraction was performed as previously described (14).
Tag-seq libraries were constructed and sequenced according to the Standard Operating Procedures developed at the Michael Smith Genome Sciences Centre (7). The sequences of linkers and tags found only once (singletons) were removed. SAGE libraries were constructed from polyA+ RNA as previously described (12,15,16). Tags were mapped to C. elegans transcripts using the previously described virtual transcriptome (17,18). The virtual transcriptome pipeline uses coordinate information of all annotated exons, introns and UTRs to assemble transcript sequences, which are later used for tag alignment. Mapping tags to the virtual transcriptome was used to analyze gene expression. The number of unmapped tags was determined by their failure to perfectly align to the C. elegans genome and transcriptome.
We used a method described by others (19) to test for potential microbial contamination of our Tag-seq libraries. A set of virtual SAGE tags extracted from several bacterial transcriptomes including E. coli was checked against the sequences of detected singletons and the matches were counted.
To analyze the distribution of frequencies for tags not matching the virtual C. elegans transcriptome, we isolated the fraction of highly abundant mismatched tags (>1000 counts in a library). Using permutation analysis, we identified the tags that were single- or multiple-base mismatched derivatives of highly abundant ‘parent’ tags unambiguously matched to genes. Next, using permutation of parent sequences, we generated a virtual set of one- to five-base mismatched derivatives and scanned our data sets for the presence of such sequences. Sets of putative derivatives were filtered using Pearson correlation statistics, and the tags with expression profiles similar to that of ‘parent’ tags were used to build the distribution of the mismatch frequencies. We performed a similar analysis for linker sequences. Supplementary Table S6 lists all SAGE and Tag-Seq libraries used in our study along with the number of tag species and singletons for each library. All the data used in this study are available via the multi-sage web interface at http://elegans.bcgsc.bc.ca.
Tools for gene annotation
We used the Gene Ontology (GO) database release of January 2008 for the analysis of expression data for genes related to functional categories. To annotate genes with GO terms we used Functional Annotation Clustering with DAVID (20). With classification stringency ‘High’ and Enrichment Score ‘2.0’ was used as a threshold for significant enrichment. SAGE expression information for corresponding genes was stored locally in a mySQL database and accessed with Perl scripts using a DBI module. We also used other publicly available online tools, such as Genome Browser and Batch Genes on the Wormbase web site. All of our analyses (except human Taq-seq analysis) were done using data from Wormbase release 180, WS 180 (www.wormbase.org). For human Tag-seq analysis we used gene annotations from Ensembl database (21). We used statistical package R (version 2.6.2) for statistical analysis.
Statistical filtering
Six SAGE libraries and 12 matching Tag-seq libraries were used to extract the genes with expression pattern changes correlated with the long-lived phenotype. Prior to the analysis we normalized the total tags in all libraries to 100 000, then subjected the data to statistical analysis of the difference between expression values in the two longevity classes (long-lived daf-2 and dauer versus normal life span). As before, we employed the Audic-Claverie test for direct comparisons of tag frequencies between libraries. This test was used to identify genes with significantly altered expression levels by estimation of the P-value (P in the text) for the comparison. The same statistical test was used to compare expression profiles in triplicate sets of libraries prepared from L1 and starved L1 larvae. We also used the Audic-Claverie test for comparison of replicates prepared either in liquid culture or on agar.
RESULTS
Principal differences between SAGE and Tag-seq datasets
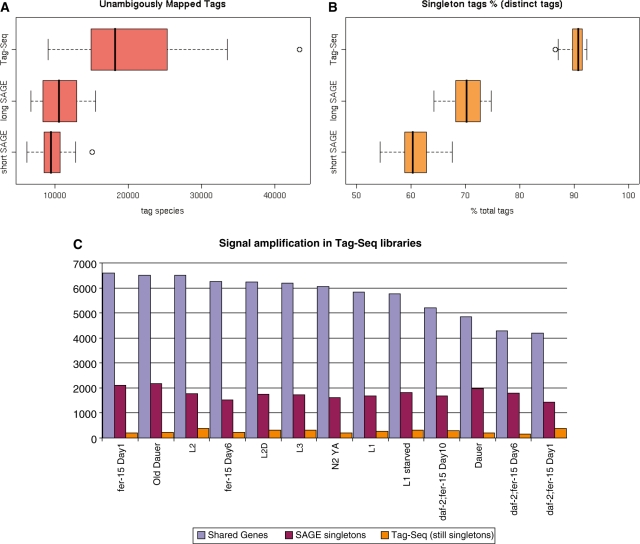
Tag-seq improves the overall sensitivity of gene expression analysis, as compared to short SAGE (producing 10-base tags) or long SAGE (producing 17-base tags). Many more tag species are detected, and consequently a larger number of unambiguously mapped tags (Figure 1A). Relative to SAGE, Tag-seq libraries had a significant increase in the number of tag species occurring just once (singletons, Figure 1B). In fact, singletons comprised ∼90% of the tag species that could not be mapped to the transcriptome. Only ∼5% of them may be interpreted as single-base mismatched tags (present in a dataset because of sequencing errors) or tags arising from bacterial contamination (data not shown). We concluded that unmapped singletons are most probably multiple-base mismatched derivatives of tags corresponding to highly expressed transcripts (see below). We removed all singletons from our analyzed datasets.
Figure 1.
General features of Tag-Seq libraries. (A) Number of unambiguously mapped tags in libraries of three types (short SAGE, long SAGE and Tag-seq). (B) percentage of singletons in the same libraries. (C) Tag-seq data show a significant increase of tag abundance for a large fraction of genes having just one SAGE tag. Expression data were compared for genes identified in matching SAGE and Tag-seq libraries. The data were visualized using the R graphical function ‘boxplot’. Shown are the median (dark line) and two nearest quartiles (area within boxes). All data points within the 1.5 inter-quartile range from the boxed area are located within the whiskers.
Tag-seq replicates of our previously generated SAGE libraries (same RNA preparation) allowed us to compare signal intensities for genes expressed in the same conditions but analyzed with different protocols. We mapped tags to genes using the C. elegans virtual transcriptome pipeline (17,18) and compared expression profiles of matching SAGE and Tag-seq libraries (Figure 1C). Analysis of 13 matching pairs of libraries showed that in each case ∼85% of the genes with a single SAGE tag had more than one tag in the matching Tag-seq library. The increased tag number is accompanied by higher tag counts for individual genes, as illustrated in Figure 1C and Supplementary Figure S1. However, signal amplification was the greatest for strongly expressed genes such as ribosomal proteins, and was more modest for tags mapped to weakly expressed genes, such as transcription regulators (Supplementary Figure S1).
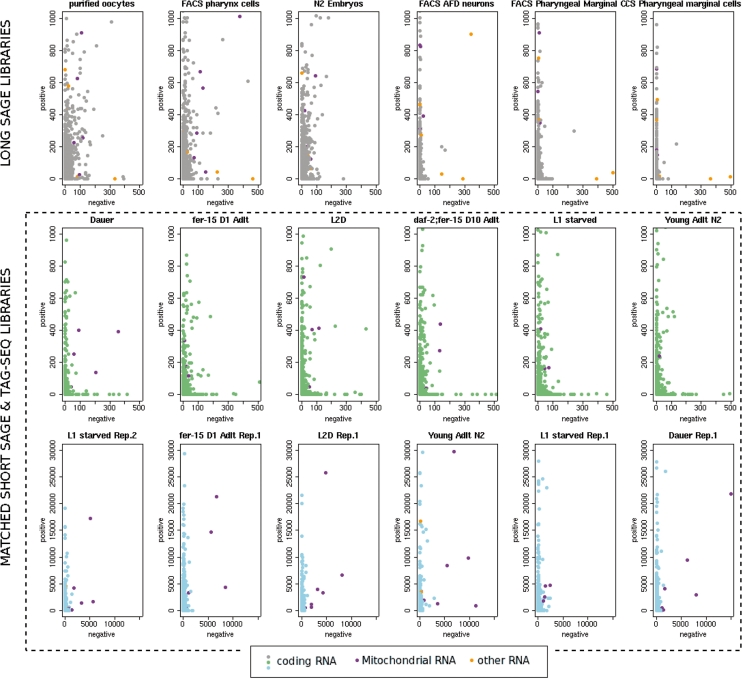
Some of the abundant tags were unambiguously aligned to the negative strand of predicted transcripts, indicating the presence of anti-sense RNA. Strong expression of anti-sense mRNAs encoded by mitochondrial genes (Figure 2, Supplementary Figure S2) corroborated previous SAGE experiments (16). Although 95% of nuclear genes have CATG sites, seven of the 12 protein-encoding genes in the mitochondria lack such a site. For the five genes with tags, the Tag-seq data showed a positive correlation between the number of sense and anti-sense tags. In total, 40–80% of the total tags (on average) for the five mitochondrial genes were anti-sense. The adjacent genes MTCE.23 and MTCE.25 had the highest antisense/sense ratio (0.9 and 3.0, respectively, averaged over the 13 pairs of libraries), but there was no obvious correlation of anti-sense transcription with chromosomal position. We observed a similar ratio of anti-sense/sense tags for two of three RNA-coding mitochondrial genes, MTCE.7 (12s rRNA) and MTCE.8 (serine tRNA) with CATG sites. Surprisingly, anti-sense tags for MTCE.33 (16S rRNA) were much less abundant, comprising only ∼1% of total tags for this gene in the 13 pairs of libraries examined. We also observed a similar pattern of transcription for mitochondria in human Tag-seq libraries (Supplementary Figure S3) using Tag-seq data from (7).
Figure 2.
Comparison of abundance for positive- and negative-strand tags originating from the same Nla III site for tags mapped to three classes of transcripts: Coding RNA, Mitochondrial Genes and Other RNA (non-coding, miRNA, etc.). Data shown for three types of experiment: SAGE, longSAGE and Tag-seq. Matching SAGE and Tag-seq libraries are framed with a dashed line. Abundances for negative and positive-strand tags shown on x and y-axes, correspondingly.
The subset of nuclear transcripts exhibiting anti-sense tags was not significantly enriched for any of the functional categories of genes described by gene ontology (GO) terms. Different libraries exhibited anti-sense RNAs for different nuclear-encoded genes, and in much smaller numbers than mitochondrial genes (a few hundred versus several thousand).
Reproducibility of Tag-seq data
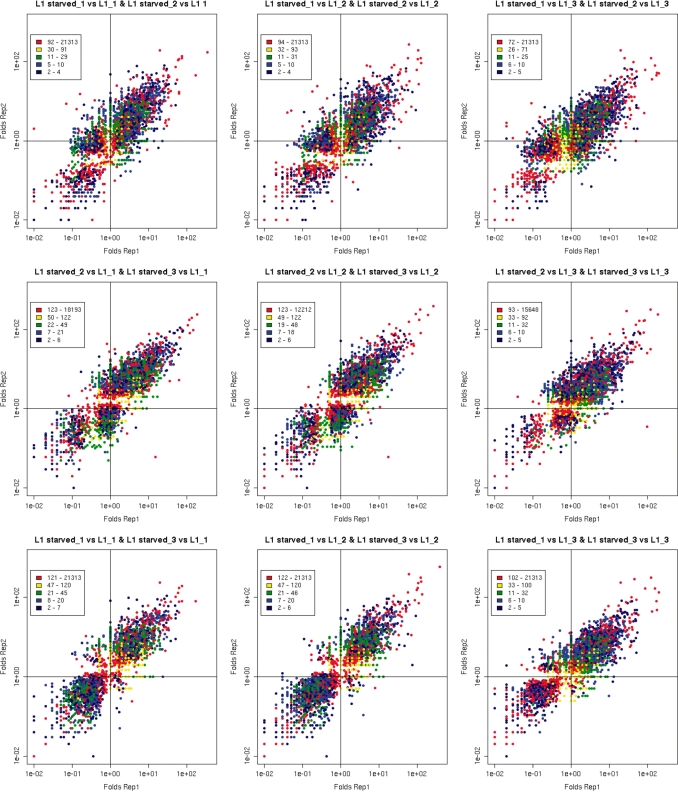
The majority of our Tag-seq libraries were prepared in duplicate using two independent RNA samples, whereas the libraries generated from feeding first-stage (L1) larvae and starved L1 larvae were prepared in triplicate. To assess the similarity between the L1 datasets, we applied Pearson statistics routinely used to measure the correlation between two independent variables. Tags were mapped to transcripts using only unambiguous mapping data. Replicate libraries showed a strong similarity both for L1 and starved L1 sets (Pearson correlation coefficient 0.8–0.9). In addition, we analyzed genes expressed differentially in fed versus starved L1 larvae using Audic–Claverie statistics. Filtered lists of genes with at least 2-fold changes in expression (P ≤ 0.05) were used to compare the differences between replicates. We plotted differences of tag abundance comparing starved L1 larvae and fed L1s using different combinations of replicate libraries. The analysis showed that most of the genes had similar differences in replicate comparisons (Figure 3). In this analysis, we observed that 75–80% of the genes analyzed showed consistent changes in expression between replicates. We conclude that the Tag-seq protocol generated highly reproducible data.
Figure 3.
Comparison of fold differences between starved and fed L1 larvae for three replicate sets of Tag-seq libraries performed for different combinations of the libraries. Levels of gene expression are color-coded (blue color shows weakly expressed genes, green shows moderately expressed genes and yellow with red show strongly expressed genes). Fold-differences were calculated for replicate pairs of starved L1 and fed L1 larvae and plotted against each other.
Highly abundant unmapped tags
Unmapped tags that match neither the C. elegans transcriptome nor the genome comprised the majority of Tag-seq species. Some of these unmapped tags were highly abundant (more than 1000 tags per library). In four Tag-seq libraries, the majority of them were linker-derived tags (including linker sequence and various single- or multiple-base mismatch derivatives). In the Tag-seq protocol, tags ligated inside a linker cassette were isolated by cutting the 85-bp band from a polyacrylamide gel. Linkers ligated to each other were found in faster migrating material (∼66 bp). Contamination of the analyzed cDNA fraction with DNA trailing from the empty linker band may explain the presence of linker-derived sequences in the dataset. Although linker sequences were often detected at some level (typically, 0.01–0.3% of all unmapped tags), several of our older RNA samples produced Tag-seq libraries with a large number of linker-derived tags (up to 20% of all tags in the most extreme case). Such contamination can be avoided by taking precautions to control mRNA quality, or by repeating the gel purification of the DNA tags inserted into the linker cassette.
Neither linker- nor gene-derived tags (data not shown) showed any distinct pattern of mismatches to the transcriptome. The mismatch frequencies for linker-derived sequences were similar to those reported in a study that used Illumina flow-cell technology for re-sequencing the C. elegans genome (23). Hence, we conclude that a major source of mismatched tags is most likely sequencing errors. However, it is possible that some of the linker-derived mismatch tags emerged from erroneous nucleotide incorporation at the step of linker synthesis. In the case of gene-derived sequences, the frequency of mismatches was higher than that for linker mismatches (Supplementary Figures S4A and S4B). Some of these mismatches might result from reverse transcriptase errors at the step of cDNA synthesis.
Alternatively spliced variants
Almost all SAGE tags arise from the 3′-most restriction site for the tagging enzyme (usually it is NlaIII, which recognizes CATG). If two alternatively spliced or polyadenylated mRNA variants have different 3′-most CATG sites, it should be possible to estimate their relative expression from Tag-seq or SAGE data. To test this, we used tag-to-gene mapping data based on Wormbase release WS180 to identify 513 genes with at least one pair of alternative mRNA isoforms distinguishable by their 3′-most SAGE tags. We then examined tag abundance for the 513 genes using data from our 13 matching pairs of SAGE and Tag-seq libraries. Of 1108 predicted 10-base tag species mapped to these 513 genes, 696 were present (had at least one tag) in the SAGE subset, and 1050 of a total of 1535 predicted 17-base mapped tag species were present in the Tag-seq subset.
We quantified differences in abundance between shorter and longer isoforms for 68 genes using SAGE data and 415 genes using Tag-seq data (Supplementary Figure S5A). These cases included only those genes with at least five tags or more in one of the analyzed libraries. There was preferential expression of one of the isoforms in 60% (SAGE) to 80% (Tag-seq) of analyzed cases (Supplementary Figure S5B), and for many of the remaining genes there was a possible transcriptional switch in abundance from one isoform to the other in some libraries. We identified 15 of the latter cases in SAGE and 62 cases in the Tag-seq libraries, which demonstrates the greater sensitivity of Tag-seq (Supplementary Figure S5C). Genes displaying such an expression pattern detected only by Tag-seq included cye-11, aak-2, gei-17 and dpy-31, all required for normal development of C. elegans. (Full lists of genes showing this pattern of tag abundance is in Supplementary Table S1).
Transcript profiling of adult longevity with Tag-seq
Construction of multiple replicate libraries enables a much deeper analysis of transcription profiles for N2 dauer larvae and adult daf-2(−) long-lived mutants than we reported previously (13). Dauer larvae are specialized, long-lived third-stage larvae that are arrested in development. Mutations in the daf-2 insulin/IGF1 receptor result in constitutive formation of dauer larvae. They also double adult life span. The evidence suggests that daf-2(−) mutant adults express longevity promoting genes that are normally expressed in the dauer stage. If longevity genes are controlled by similar pathways in adults and larvae, similarities in the expression profiles of the biologically distinct but long-lived larvae and adults should identify candidate genes for longevity (13,22).
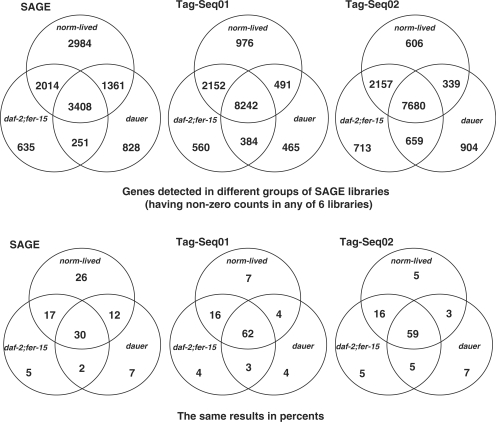
Similar to our previous study, we compared the expression data for synchronously aging daf-2(−) adults, daf-2(+) adults and N2 dauer larvae. We examined the overlap between identified genes in three groups of libraries, which were adults with normal life span, long-lived daf-2(−) adults and long-lived dauer larvae. These groups of libraries were assembled for (i) short, 10-base-tag SAGE libraries, (ii) matching Tag-seq replicates for which we used the same RNA samples used for short SAGE and (iii) a second replicate set of Tag-seq libraries (17-base tags) generated from new RNA samples. There was roughly a 2-fold increase in the total number of genes detected in both Tag-seq replicate sets, as compared to the original short SAGE libraries. Some of the daf and ins (insulin) genes are examples of genes previously missed in SAGE (Supplementary Tables S2 and S3). The number of the genes shared by all three groups of libraries was also ∼2-fold higher in Tag-seq replicates (Figure 4). As more genes were seen in each case, there was better overlap between experiments.
Figure 4.
Overlap between genes detected in three comparisons: original SAGE and two Tag-seq replicates (adults grown in liquid culture, Tag-seq01 and adults grown on plates, Tag-seq02). In each case, the normal subset consists of two daf-2(+) libraries, the daf-2(−) subset consists of three libraries and the dauer subset is represented by one Dauer library. The lower set of diagrams show the same data as percentages of total genes.
Our second set of replicate Tag-seq libraries (see ‘Materials and methods’ section) provided an opportunity to compare the transcription profiles of animals with the same genotype but grown in different conditions (liquid versus agar plate culture). We identified the genes for which the expression showed a statistical difference in either daf-2(−) adults or dauer larvae (when compared to the control with normal life span) in one of our replicate sets but not in the other. These genes may be primarily involved in adjustments to growth conditions unrelated to life span extension. Thus, the data allow us to distinguish between changes in gene expression associated with altered genotype or with altered environment.
We assembled lists of genes, the expression of which was consistently changed in long-lived worms according to the data from both replicate sets of Tag-seq libraries. The lists appear to show a strong biological relevance as we observed several ‘signature’ genes among those up-regulated in dauer larva replicates [cki-1, aak-2, daf-16 (24)] and sod-3 [a well-studied target of the daf-16 transcription factor (25)] among the genes up-regulated in daf-2(−) adult replicates. Previously generated SAGE data did not provide enough sensitivity to detect all of these genes. Several daf genes (daf-16 in particular) were not detectable when using SAGE protocol. About 50–60% of the genes in these lists had unknown or predicted function, opening the possibility that longevity may be affected by as yet uncharacterized pathways (Supplementary Tables S4 and S5).
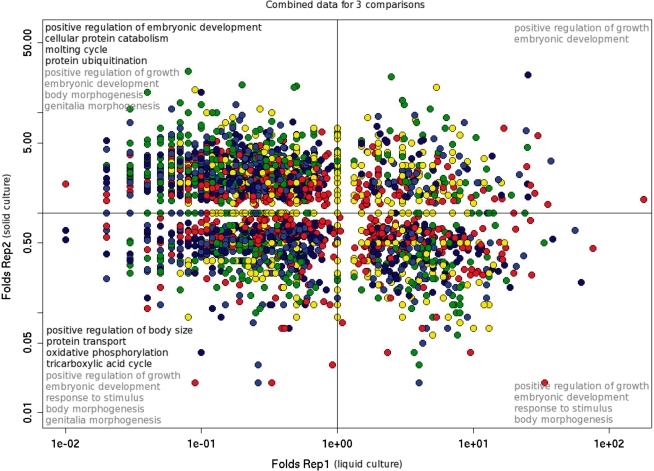
Using less stringent filtering, we analyzed enrichment in functional categories among the genes differentially expressed in long-lived C. elegans. We assembled the sets of transcripts that showed a significant (at least 2-fold, P ≤ 0.05) difference in tag abundance between normal and long-lived animals (Figure 5, Supplementary Figure S6). These sets were tested using the publicly available web program DAVID (20) for enrichment in functional categories described by GO terms (26). For the combined set of messages with reduced abundance in long-lived worms (grown either in liquid culture or on agar), the results showed enrichment of functions related to oxidative phosphorylation, TCA cycle-related genes, protein transport and positive regulation of body size. Among the genes for which transcription was down-regulated in long-lived adults grown in liquid culture but up-regulated in long-lived adults grown on plates, there was a significant enrichment of genes involved in protein ubiquitination, cellular protein catabolism, positive regulation of embryonic development and molting cycle (Figure 5). The complementary gene sets [genes up-regulated in daf-2(−) animals grown in both media and genes up-regulated in liquid culture but down-regulated on agar plates] were much smaller.
Figure 5.
Media-biased differential expression in the long-lived daf-2(−) strain. Scatter plot shows global expression in adults testing different genotypes and growth conditions. Animals were grown in liquid culture (Rep1) or on agar plates (Rep2). Data (Folds Rep1 and Folds Rep2) are signal ratios from daf-2(−) versus daf-2(+) comparisons plotted for biological replicate experiments. Levels of gene expression are color-coded (blue color shows weakly expressed genes, green shows moderately expressed genes and yellow with red show strongly expressed genes). Also shown are the results of GO term enrichment analysis for genes up- or down-regulated in long-lived daf-2(−) adults (GO terms specifically enriched in only one of the four quadrant-defined groups of genes are shown in black font, terms enriched in more than one group are shown in gray font). The quadrants of the scatter plot correspond to (clockwise from upper left quadrant): (i) genes down-regulated in liquid culture but up-regulated in daf-2(−) animals grown on agar plates, (ii) genes up-regulated in daf-2(−) animals grown in both types of media, (iii) genes down-regulated in daf-2(−) animals grown on agar but up-regulated in liquid culture, (iv) genes down-regulated in daf-2(−) animals grown in both types of media.
DISCUSSION
Gene expression profiling based on DNA sequencing greatly benefits from new technologies that provide deeper sequencing at lower cost. Tag-seq has increased sensitivity (number of detected genes) and accuracy (stronger, more reliable signal) as compared to SAGE. With Tag-seq data, we could assess the expression of genes previously undetected by SAGE. One example is daf-16, which was not detected in our SAGE libraries. Both 2-day-old dauer and 2-week old dauer Tag-seq libraries had 200+ tags for this gene, and data from other libraries also indicated strong expression for this and other daf genes. Abundance of the tags corresponding to some insulin genes also increased in Tag-seq libraries. These examples show that Tag-seq data significantly improves detection of rare transcripts that could not be reliably detected with SAGE. Strongly expressed genes may be detected with even greater confidence, as our analysis showed that relative to SAGE, amplification of tag abundance in Tag-seq is more profound for such functional categories as metabolic enzymes or structural proteins.
About 90% of all tag species in a Tag-seq library are singletons (tags seen only once in a data set), whereas our SAGE libraries had ∼60% singletons. As we found, only ∼5% of the singletons may be explained by single-base sequencing errors or by contamination with bacterial mRNA sequences (see ‘Materials and Methods’ section). The large number of Tag-seq tags allows the detection of both single- and multiple-base mismatch tags resulting from sequencing errors or spurious nucleotide incorporation at different stages of the experiment. Tag sequences derived from highly abundant ‘parent’ tags may contribute to the population of unmapped singletons. Analysis of a sample set of 69 highly expressed genes (which had >1000 tags per library) revealed thousands of apparent derivatives, mostly singletons with two or three mismatched bases. Considering the fact that in some extreme cases of linker-contaminated libraries up to 20% of all tag species were linker-derived mismatches, we suggest that highly abundant tags are the main source of unmapped singletons, including their multiple-base mismatched derivatives. On average, our Tag-Seq libraries had a few hundreds tag species with >1000 counts. We presume that this number of highly expressed parent tags may have enough derivatives to explain a large population of unmapped singletons.
We removed all singletons from our analysis, but if needed, it would be possible to reassign many of them to genes, by finding their highly abundant ‘parent’ tags. Assuming that the frequency of sequencing errors is constant, the frequency of low-abundance derivatives could be predicted and accounted for. As sequencing methods improve, the frequency of errors is declining, so we expect the number of unmapped singletons in future experiments to markedly decrease.
In a few libraries, contamination by linker sequences resulted in a large number of tags (including mismatched derivatives) that could not be mapped to genes. However, most of our libraries were almost free of the linker tag and its derivatives. Tag-seq libraries were generated using an RT polymerase lacking proofreading activity (7), so errors in reverse transcription may be a source of transcriptome mismatches. Our results indicate that errors during sequencing and cDNA synthesis are the main causes of erroneous data. Frequencies calculated for mismatched tags were higher for gene-derived tags than linker-derived tags, with the latter matching the previously reported numbers from a C. elegans genome re-sequencing study (23).
Tests of replicate well-fed L1 and starved L1 libraries using Pearson correlation statistics showed high similarities between replicate libraries (Pearson correlation coefficient 0.8–0.9). We used Audic–Claverie statistics to compose the lists of genes with significant changes in expression. The lists were used to produce a series of graphs visualizing similarities of fold differences obtained in comparisons of replicate L1 and starved L1 libraries (Figure 3). The majority of the genes had similar fold differences, indicating a strong correlation of expression in replicate experiments. A small fraction (on average, 14%) of all plotted genes had different expression in parallel comparisons. They had lower fold differences and there was little overlap between the libraries compared. It is possible that many of these outliers may represent natural fluctuations in gene expression.
Our data show that with a few exceptions nuclear transcripts have their negative-strand messages expressed at a much lower level than their positive-strand counterparts. However, it appears that mitochondria produce roughly equal amounts of negative- and positive-strand transcripts (with the apparent exception of 16s rRNA). Highly abundant tags corresponding to the negative strand of several mitochondrial transcripts were observed in SAGE data (16), but the difference in abundance between mitochondrial and nuclear anti-sense tags was even larger in Tag-seq experiments versus SAGE.
The differences in levels of anti-sense transcription between nuclear and mitochondrial RNAs might be attributed to the specific composition of the transcriptional machinery, or to differences in chromosomal structure. Biochemical studies are needed to investigate this phenomenon, which has been seen elsewhere, including our own analysis of human Tag-seq data. Studies in malarial plasmodium showed that anti-sense transcripts identified by SAGE were also detectable by other methods in follow-up experiments (27,28). Indeed, the large number of mitochondrial anti-sense transcripts may be very common. Identification of the factors that influence this expression pattern is beyond the scope of our analysis, but should be done as separate studies.
Tag-seq, like SAGE, generates short oligonucleotide sequences that may be used for analysis of alternatively spliced transcripts (29,30). Alternative splice variants may have a shared 3′-most CATG, but when the transcript has shorter isoforms there may be different 3′-most tags. Tag-seq enabled detection of ∼50% more species of alternative splice variants than SAGE. Comparing Tag-seq and SAGE data derived from the same RNA preparations (13 pairs of matching libraries), the majority of the analyzed genes had one of their isoforms preferentially expressed. For some genes there was an indication of a shift from one alternatively spliced isoform to another. Analysis of Tag-seq data provided evidence for 62 cases of putative switching in the 13 libraries, whereas SAGE data allowed detection of only 15 such cases.
Replicate libraries prepared from RNA samples of animals grown on plates versus liquid culture allowed us to estimate how strongly the environment contributes to the gene expression profile. Despite the apparent similarity in extended life span between liquid and agar cultures, the gene expression profiles showed substantial differences between these two replicate sets. We analyzed enrichment of GO terms among the genes showing statistically significant changes in expression using Tag-seq data for long-lived daf-2(−) adults and normal daf-2(+) adults grown either in liquid culture or on agar plates. To prevent contamination of aging adults with their progeny, both the long-lived daf-2(−) and the daf-2(+) strains carried a mutation in fer-15 rendering them unable to make sperm at 25°C.
Regardless of growth conditions, transcripts for a large number of genes were reduced in daf-2(−) adults. Down-regulated genes belong to the TCA cycle and oxidative phosphorylation pathways, which were previously associated with regulation of lifespan (12,13,15,22,31). Transcripts involved in the molting cycle, protein ubiquitination, positive regulation of embryonic development and cellular protein catabolism were down-regulated relative to normal adults in long-lived daf-2(−) animals grown in liquid, but up-regulated in animals with the same genotype when grown on plates.
Re-visiting our transcript profiling of dauer and adult longevity (13,30) with Tag-seq data, we analyzed two sets of replicate Tag-seq libraries constructed using RNA samples from animals grown either in liquid culture (the same RNA samples used to generate previously described SAGE libraries) or on agar plates (fresh RNA preparations). There was less similarity between our liquid and solid culture replicates than between replicates for L1 and L1 starved larvae grown in same conditions (compare Supplementary Figure S6 with Figure 3). Based on SAGE data, we suggested that daf-2(−) animals may extend their life span by modifying their oxidative phosphorylation so that production of reactive oxygen species in mitochondria was reduced. However, mitochondrial transcripts, strongly up-regulated in liquid culture daf-2(−) adults, did not show coordinated up-regulation in animals of the same genotype grown on plates. Therefore, we conclude that if reducing production of reactive oxygen species is a means to extend life span in liquid culture, it is not required for life span extension on plates. It is noteworthy, that the importance of mitochondria-induced oxidative damage in aging has been questioned by several recent studies. Knockout mice with various deleted antioxidant enzymes did not show any significant shortening of lifespan and transgeneic mice overexpressing anti-oxidant enzymes did not display increased longevity (32). In an analysis more relevant to our case, C. elegans mutant worms with deleted sod genes did not have shortened lifespan and knockout of sod-2 even made worms live longer (33). With these findings taken in account, our observations suggest that changes in expression of the genes involved in oxidative phosphorylation may be unimportant in daf-2(−) aging.
The improved sensitivity of Tag-seq allowed us to perform a stringent analysis focused on consistently up- or down-regulated genes in long-lived dauer larvae and daf-2(−) adults. For this analysis, we used both liquid and solid culture replicates for daf-2(−) adults and three dauer libraries (2-day-old dauer replicates and one 2-week-old dauer library). The resulting very compact lists contained several genes that were previously described as important elements in adult life span control and regulation of dauer formation. Our results also suggested an involvement of some as yet unstudied pathways in aging, since our lists contained several genes with fragmentary or no functional annotation. Tag-seq data may prove to be a useful resource for identifying new pathways for longevity.
Tag-seq produces high-quality data in the same format as SAGE. This is of special interest to those who have already accumulated extensive SAGE data and seek to increase the depth of rare mRNA detection. Tag-seq not only simplifies the SAGE protocol by eliminating the tag concatenation and cloning steps, it also provides much more data at reduced cost (7). The data are reproducible, as demonstrated by our comparison of library triplicates. Finally, our lists of transcripts showing changes in expression associated with longevity provide an expanded platform to pursue candidate genes and pathways for longevity using genetic and molecular methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: US National Institute on Aging, the Canadian Institutes of Health Research (MOP 79458) and a Natural Sciences and Engineering Discovery grant to D.L.R.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Marco Marra and Sorana Morrissy for helpful discussions, Martin Hirst and Stephen Jones for insights and guidance, and Allen Delaney for providing bacterial transcriptome data used to test for potential contamination of Tag-seq libraries.
REFERENCES
- 1.Kulesh DA, Clive DR, Zarlenga DS, Greene JJ. Identification of interferon-modulated proliferation-related cDNA sequences. Proc. Natl Acad. Sci. USA. 1987;84:8453–8457. doi: 10.1073/pnas.84.23.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 4.Ibrahim AF, Hedley PE, Cardle L, Kruger W, Marshall DF, Muehlbauer GJ, Waugh R. A comparative analysis of transcript abundance using SAGE and Affymetrix arrays. Funct. Integr. Genomics. 2005;5:163–174. doi: 10.1007/s10142-005-0135-4. [DOI] [PubMed] [Google Scholar]
- 5.Sun M, Zhou G, Lee S, Chen J, Shi RZ, Wang SM. SAGE is far more sensitive than EST for detecting low-abundance transcripts. BMC Genomics. 2004;5:1. doi: 10.1186/1471-2164-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilain C, Libert F, Venet D, Costagliola S, Vassart G. Small amplified RNA-SAGE: an alternative approach to study transcriptome from limiting amount of mRNA. Nucleic Acids Res. 2003;31:e24. doi: 10.1093/nar/gng024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissy A, Morin R, Delaney A, Zeng T, McDonald H, Zhao Y, Jones S, Hirst M, Marra M. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 9.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 10.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C.elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 11.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 12.Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruzanov P, Riddle DL, Marra MA, McKay SJ, Jones SM. Genes that may modulate longevity in C. elegans in both dauer larvae and long-lived daf-2 adults. Exp. Gerontol. 2007;42:825–839. doi: 10.1016/j.exger.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 15.Holt SJ, Riddle DL. SAGE surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mech. Ageing Dev. 2003;124:779–800. doi: 10.1016/s0047-6374(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 16.Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- 17.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 18.Pleasance ED, Marra MA, Jones SJ. Assessment of SAGE in transcript identification. Genome Res. 2003;13:1203–1215. doi: 10.1101/gr.873003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khattra J, Delaney AD, Zhao Y, Siddiqui A, Asano J, McDonald H, Pandoh P, Dhalla N, Prabhu AL, Ma K, et al. Large-scale production of SAGE libraries from microdissected tissues, flow-sorted cells, and cell lines. Genome Res. 2007;17:108–116. doi: 10.1101/gr.5488207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 23.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 24.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Henderson ST, Bonafè M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 26.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 2004;136:35–42. doi: 10.1016/j.molbiopara.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell. 2001;12:3114–3125. doi: 10.1091/mbc.12.10.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo BY, Chen Y, Bohacec S, Johansson O, Wasserman WW, Simpson EM. SAGE2Splice: unmapped SAGE tags reveal novel splice junctions. PLoS Comput. Biol. 2006;2:e34. doi: 10.1371/journal.pcbi.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruzanov P, Jones SJ, Riddle DL. Discovery of novel alternatively spliced C. elegans transcripts by computational analysis of SAGE data. BMC Genomics. 2007;8:447. doi: 10.1186/1471-2164-8-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanfleteren JR, De Vreese A. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. FASEB J. 1995;9:1355–1361. doi: 10.1096/fasebj.9.13.7557026. [DOI] [PubMed] [Google Scholar]
- 32.Perez VI, Bokov A, Remmen HV, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim. Biophys. Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.