Abstract
Most small nucleolar RNAs (snoRNAs) guide rRNA nucleotide modifications, some participate in pre-rRNA cleavages, and a few have both functions. These activities involve direct base-pairing of the snoRNA with pre-rRNA using different domains. It is not known if the modification and processing functions occur independently or in a coordinated manner. We address this question by mutational analysis of a yeast box H/ACA snoRNA that mediates both processing and modification. This snoRNA (snR10) contains canonical 5′- and 3′-hairpin structures with a guide domain for pseudouridylation in the 3′ hairpin. Our functional mapping results show that: (i) processing requires the 5′ hairpin exclusively, in particular a 7-nt element; (ii) loss of the 3′ hairpin or pseudouridine does not affect rRNA processing; (iii) a single nucleotide insertion in the guide domain shifts modification to an adjacent uridine in rRNA, and severely impairs both processing and cell growth; and (iv) the deleterious effects of the insertion mutation depend on the presence of the processing element in the 5′ hairpin, but not modification of the novel site. Together, the results suggest that the snoRNA hairpins function in a coordinated manner and that their interactions with pre-rRNA could be coupled.
INTRODUCTION
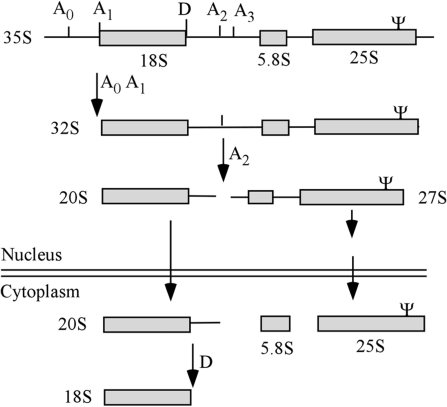
In eukaryotes, three of the four cytoplasmic rRNAs are transcribed from polycistronic coding units. The nascent transcripts contain 18S, 5.8S and 25S/28S rRNAs flanked by 5′ and 3′ external transcribed spacers (ETS) and two internal transcribed spacers (ITS1 and ITS2). This precursor undergoes nucleotide modification at scores of sites and cleavage and trimming reactions (processing) to form the mature rRNAs (Figure 1) (1–4). Both modification and processing of rRNA require small nucleolar RNAs (snoRNAs) that act through base-pairing with pre-rRNA (5–9). Most snoRNAs, functioning as snoRNPs, guide pseudouridine (Ψ) or 2′-O-methylation (Nm) modifications; only a few are required for the cleavage reactions (9). For the 76 known yeast snoRNAs, 45 have one modification guide domain, 25 have two different guide domains and two mediate both processing and modification reactions (10). These functions are targeted by complementarities to the various, specific rRNA regions.
Figure 1.
Yeast pre-rRNA processing pathway. The major rRNA precursors and final products are shown. Cleavage sites relevant to the present study are indicated, as well as the approximate location of the Ψ guided by snR10 in 25S rRNA.
While it is generally accepted that the snoRNA modification and processing reactions do not occur randomly, the basis and extent of coordination of these functions remain unknown. Binding of snoRNPs in a coordinated manner has implications for possible chaperone-like functions for snoRNAs in ribosome assembly, and for coupling modification and cleavage events. To gain insight into these important questions, we have carried out functional interference mapping of a yeast snoRNA (snR10) that is known to be required for both rRNA modification and normal processing. Our aim was to determine if the different interactions with pre-rRNA occur independently or are coordinated. Loss of this snoRNA causes slow growth and processing defects and the Ψ modification has been shown to affect ribosome translation activity in vivo (11,12). This knowledge facilitated our functional analysis of the snoRNA by mutagenesis. The results reveal a cooperative interference effect between the two functional domains that is consistent with coordinated binding to pre-rRNA.
Most nucleotide modifications in rRNA appear to occur before processing is completed; among these alterations the Ψ and Nm modifications are dominant. In eukaryotes, box H/ACA snoRNPs carry out pseudouridylation and box C/D snoRNPs mediate Nm modification (6,9,13–17). Each snoRNP contains a single snoRNA that selects the rRNA site to be modified, and a set of four family-specific core proteins, one of which catalyzes the modification reaction (5,18). The consensus structure of the H/ACA snoRNAs, the type featured here, contains two hairpin regions, linked by an H box (hinge region) and an ACA box at the 3′-end (19,20) (Figure 2A). In Saccharomyces cerevisiae, 29 H/ACA snoRNAs have been identified and all but one have been assigned to specific Ψs in rRNA. For 14 of these snoRNAs both hairpin structures target Ψ, to sites in the same or different rRNA species (10). The dual-function snoRNA featured in the present study has both Ψ guide and processing functions, with Ψ formation targeted to a single site.
Figure 2.
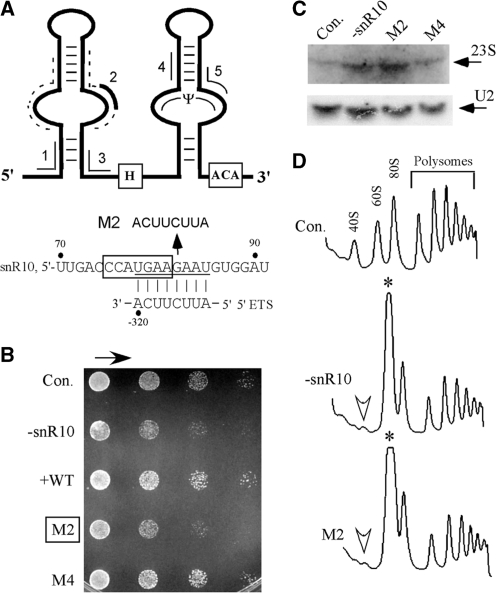
A 7-nt sequence element in snR10 is required for normal cell growth and pre-rRNA processing. (A) Schematic structure of the snR10 snoRNA. Key features are identified, including the H and ACA boxes, the Ψ guide region and five segments complementary to pre-rRNA. The complementary segments are numbered and highlighted with lines outside the snoRNA structure. Segment 2, which is complementary to the 5′ETS (thick line), is revealed in the present study to be essential for the processing function of snR10. Sites within and neighboring this segment were subjected to mutational analysis; the flanking sites are identified with dashed lines (Supplementary Table S1). A lower panel shows complementarity between segment 2 and the 5′ETS of pre-rRNA. The 7-nt element required for processing is boxed. (B) Mutation of segment 2 impairs cell growth. Test cells expressing snR10 with mutations were serially diluted (1:5), dotted on solid medium lacking Trp, and incubated at 30°C for 30 h. Control cells include: a wild-type strain (YS602) with an empty vector (Con.), and a parental test strain depleted of endogenous snR10 that contains either an empty vector (−snR10) or a plasmid that expresses wild-type snR10 (+WT). Growth properties of cells expressing snR10 variants with mutations in segment 2 or 4, (M2 or M4), are shown. The arrow above the colony patterns indicates decreasing numbers of cells. (C) Mutation of segment 2 causes accumulation of a 23S precursor to 18S rRNA: this product is defined by the 5′-end of the transcript and the A3 cleavage. Total RNA was analyzed by northern hybridization. U2 snRNA is shown as a loading control. (D) Polysome patterns are altered by mutation of segment 2. Whole-cell extracts containing similar amounts of RNA were separated by sucrose gradient fractionation and ribosomal complexes detected by UV absorbance. The identities of ribosomal complexes are indicated for control cells (Con.). Reductions in the level of free 40S subunits are marked by open arrowheads and increases in the level of free 60S subunits by asterisks.
More than 70% of the 45 Ψ and 54 Nm modifications in yeast rRNA are conserved in humans, and the functionally important regions of the ribosome are especially rich in these modifications, arguing that they have important roles in ribosome production or function. Structural studies with small model RNAs containing Ψ and Nm nucleotides have shown that these modifications stabilize RNA structure in different ways (21–23). Considerable genetic and biochemical evidence is now available showing that these modifications are indeed important, based on examining effects of blocking modification formation by deleting specific guide snoRNAs or mutating guide sequences. These results, many from our laboratory, have revealed that depleting small subsets of modifications from different rRNA regions can significantly impair ribosome biogenesis and activity (12,24–26). Modification is accepted to be the major function of the snoRNPs, however, since all snoRNPs appear to act through base-pairing with pre-rRNA, it is also possible that some modifying snoRNPs could affect pre-rRNA folding, distinct from the effect of modification itself (27). In the case of the processing snoRNPs, remodeling pre-rRNA seems highly likely (see below) (28–34).
In the yeast rRNA processing pathway, the longest detectable pre-rRNA—a 35S species, is cleaved sequentially to generate the immediate precursor of 18S rRNA (20S) and a 27S precursor that contains the 5.8S and 25S rRNAs (Figure 1). The 20S pre-rRNA, which is pertinent to the current study, is exported to the cytoplasm where it is further cleaved (at site D) to create 18S rRNA (35). A few snoRNPs are also required for specific early cleavages. In yeast, the processing snoRNPs, i.e. U3, U14, U17/snR30 and snR10, are required for 18S rRNA maturation. The first three are thought to be common to all eukaryotes, whereas snR10 is thus far yeast-specific. U3 and U14 are C/D snoRNAs and snR30 and snR10 are H/ACA species. The universal processing snoRNAs contain sequence elements that base-pair with pre-rRNA without guiding modification, however, the roles of these elements and, indeed, the snoRNPs themselves remain to be defined. Speculation includes remodeling of pre-rRNA for cleavage by cis- or trans-acting components (28,31,34,36,37).
The U3 C/D snoRNP is the first to associate with full-length 35S pre-rRNA (38). In mediating the cleavages at sites A0, A1 and A2, a 10-nt element of U3 base-pairs with a 5′ ETS segment (39–44). In addition, a 7-nt segment in U3 base-pairs with the 5′ region of 18S rRNA, which otherwise forms a pseudo-knot structure (31), inferring the U3 snoRNP has a role in pre-rRNA folding. The U14 C/D snoRNA contains two sequence elements (13 and 14 nucleotides) complementary to pre-rRNA. One base-pairs with the 5′-end of 18S rRNA and is essential for pre-rRNA cleavages (28,45). The other is a guide sequence for a methylation site in 18S rRNA (Cm414), and is dispensable for processing (28,46). Finally, production of 18S rRNA has been shown to also depend on two short complementary elements in the H/ACA snoRNA snR30/U17, which base-pair with 18S rRNA (34,47). Determining how these processing snoRNAs affect pre-rRNA cleavages remains an important challenge.
The yeast U3, U14 and snR30 snoRNAs are all essential for cell growth, whereas deletion of snR10, the snoRNA examined here, has a less severe effect, resulting in a moderately reduced growth rate and a cold-sensitive phenotype (11). Although not severe, this phenotype can be used to screen for functional requirements of the snoRNA sequence elements. In the absence of snR10, processing of 18S rRNA is moderately impaired as evidenced by accumulation of full-length pre-rRNA (35S) and an intermediate that is normally of very low-abundance (23S); the latter species is generated by cleavage at site A3 prior to removal of the 5′ ETS segment (Figure 1) (48,49). The snR10 snoRNA also directs Ψ modification at site U2923 in the A-loop of the peptidyl transferase center (PTC) region in 25S rRNA (Fig 1, Ψ) (14,15). This Ψ is not required for pre-rRNA processing, as revealed by a point deletion in the guide domain of snR10 that disrupts Ψ formation, yet has no effect on cell growth or pre-rRNA cleavages (12). This observation suggests that the processing function depends on structural elements outside the guide domain. Thus far, no processing elements have been identified in snR10, and it is not known if the modification guide domain communicates with its processing determinant(s). In this study, we identify a sequence element that is necessary for normal pre-rRNA processing. Functional mapping also revealed that the processing and modification domains are interconnected functionally.
MATERIALS AND METHODS
Plasmids
Mutations in snoRNA coding units were created using a PCR approach (24,50). All constructs are listed in Supplementary Table S1.
Strains and cell growth
A yeast strain (YS602) with an empty vector was used as a positive control throughout the study. The parental test strain lacks the snR10 snoRNA gene, which was replaced with a marker gene using targeted gene disruption. The strain was transformed with plasmids that encode wild-type or mutant snR10 snoRNAs. All experimental strains are listed in Supplementary Table S2. For growth comparisons, yeast cultures prepared in synthetic medium lacking Trp were adjusted to 2 OD600/ml, diluted 1:5 serially, dotted on plates with synthetic medium lacking Trp, and incubated at 30°C for 2–3 days (24).
RNA preparation and analysis
Total RNA was isolated from cells using the Tri-reagent procedure (Sigma) following the manufacturer’s instructions. The presence and abundance of snoRNAs and rRNA species were determined by northern blotting (51). To examine the rRNA components in an abnormal 50S pre-rRNP complex detected in a C-insertion mutant of snR10 (M + C), RNA was prepared from sucrose gradient fractions containing this complex and subjected to northern analysis (52). The Ψ and Nm modifications were detected by primer extension analysis (53,54).
Analysis of polysome patterns
Whole-cell extract was fractionated by sucrose gradient (7–47%) centrifugation and examined as described previously (24).
In vivo pulse-chase labeling of rRNA
Test cells were grown in synthetic medium lacking methionine to an OD600 of 0.8–1.0, and ∼3 OD600 units of cells were diluted to 3 ml with the same, prewarmed medium. Next, 0.1 mCi of [methyl-3H]methionine (1 mCi/ml, PerkinElmer) was added and the cells pulse-labeled at room temperature for 3 min. Labeling was terminated by addition of 200 μl of 60 mM unlabeled (d, l)-methionine. Subsequently, 800 μl samples were taken at time points 0, 2, 5 and 15 min after the chase was initiated, and immediately frozen in liquid nitrogen. Total RNA was prepared and dissolved in 10 μl water. Three microliters of RNA was fractionated on 1.2% agarose gels, transferred to a membrane and visualized with a Fuji PhosphorImager (24).
In vivo chemical probing of RNA structure
Chemical modification with dimethyl sulfate (DMS) and primer extension probing of snoRNA and rRNA structure were carried out as described earlier (24).
RESULTS
A 7-nt sequence element in the 5′ hairpin of snR10 is required for pre-rRNA processing
Our functional study of snR10 proceeded in three stages: (i) identifying and testing the importance of elements complementary to pre-rRNA for rRNA processing activity; (ii) examining effects of guide domain mutations on Ψ formation and rRNA processing; and (iii) evaluating the relationship between the processing and modification domains.
Because other processing snoRNAs act through base-pairing with pre-rRNA, we reasoned this is likely the case for snR10. Screening of the snR10 sequence with bioinformatics tools LALIGN Server (http://www.ch.embnet.org/software/LALIGN_form.html) identified five segments of at least 8 nt that are complementary to different regions of pre-rRNA (Figure 2A). The importance of these sequences was tested by replacement with complementary nucleotides and screening for effects on growth and pre-rRNA processing. We anticipated that functional defects in processing should impair growth rate, as seen earlier for deletion of snR10 (11). The mutant snoRNAs were characterized in a test strain depleted of wild-type snR10, and determined to be produced at levels comparable to wild-type snR10 expressed in the same way (data not shown). As expected, mutating the Ψ guide domain disrupted modification at U2923 in 25S rRNA (segment 5), as shown by primer extension analysis (data not shown).
The growth data revealed that only one substitution mutation caused slower growth—in particular, mutation of segment 2 in the 5′ half of snR10 (U78GAAGAAU85). Based on growth on solid medium, the mutant strain has a growth rate that is ∼20% of normal, essentially the same as the effect of snR10 depletion (M2, Figure 2B, and data not shown). This observation suggests that segment 2, which is complementary to the 5′ ETS of pre-rRNA (−327 to −320), is involved in processing. Indeed, the mutation caused significant accumulation of the normally low-abundance 23S pre-rRNA (Figure 2C). Subunit levels were also affected, with strong accumulation of free 60S subunit (∼5-fold) and dramatic reduction in the level of free 40S subunit (∼4-fold), as determined by analysis of polysome profiles by density gradient fractionation (Figure 2D). These defects mimic exactly those that accompany loss of wild-type snR10, indicating that the complementary segment is required for its processing activity.
To better define the sequence and folding requirement of this particular snoRNA region, we analyzed 10 additional substitution mutations of 1–8 nt (Supplementary Table S1). These included changes in the flanking sequences and sequences predicted to be single and double stranded (Figure 2A, indicated as dashed lines). Together, the new data defined a 7-nt sequence required for normal processing function (C75CAUGAA81). Two mutants, C75CAU78→GGUA and U78GAA81→ACUU, exhibited growth defects similar to that caused by loss of snR10 itself (Figure 2A, lower panel, and data not shown). The important 7-nt sequence partially overlaps the complementary sequence examined initially, with the 3′ 4 nt located in the complementary region (Figure 2A, lower panel). In addition, mutations designed to disrupt the predicted secondary structure of the upper stem had no effect on cell growth (data not shown). These results show the 7-nt element required for processing likely functions in a sequence-dependent manner.
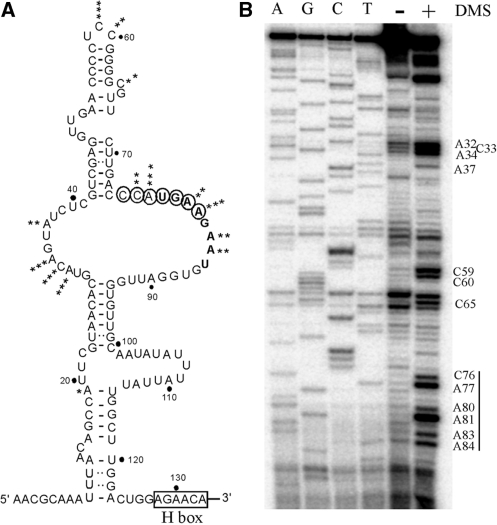
Next, we carried out an in vivo chemical probing analysis to determine if the 7-nt processing element is single-stranded and available for base-pairing. The structure probe was DMS, which methylates accessible A and C nucleotides. Following DMS treatment, total cell RNA was prepared and the relevant snoRNA region was examined by primer extension analysis. The results show that the 7-nt region is clearly modified by DMS (C76 to A84), indicating that this segment is single-stranded as predicted in the folding model (Figure 3A and B). The results also suggest that the novel processing element is not covered by protein.
Figure 3.
The 7-nt processing element in snR10 is single-stranded. (A) Secondary structure of the 5′-hairpin region in snR10 that contains the 7-nt processing element. The structure was predicted with the folding program Mfold, confirmed and refined using the in vivo chemical probing data in (B) below. The relative extent of DMS modification is indicated by the number of asterisks. The seven nucleotides required for pre-rRNA processing are circled. The potential H box is indicated. (B) The 7-nt element is present in an internal loop of the 5′ hairpin. Following DMS modification in vivo, the structure of snR10 was probed by primer extension analysis with extension products separated on an 8% sequencing gel, next to sequencing products created with the same primer. Nucleotides accessible to DMS are identified.
The snoRNA information required for processing is located exclusively in the 5′ hairpin
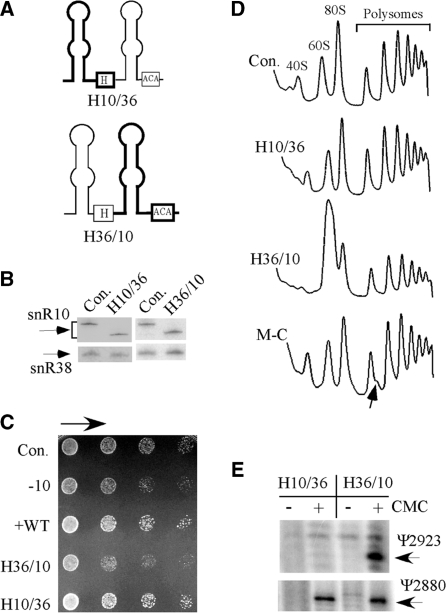
The mutational results available for the processing and modification functions of snR10 suggest that the information for processing of 18S rRNA is located in the 5′ hairpin of the snoRNA (Figure 2), and that for modification of Ψ2923 in 25S rRNA occurs solely in the 3′ hairpin (12). To test this model directly, we attempted without success to separate these functions by expressing the individual hairpins alone. While single hairpin snoRNAs occur in trypanosomes and euglena (55,56), to our knowledge, none have yet been stably expressed in S. cerevisiae. Thus, we turned to the reliable strategy of expressing hybrid snoRNAs created by combining the 5′ or 3′ hairpin of snR10 with the appropriate hairpin from another yeast H/ACA snoRNA. In these experiments, the second snoRNA was snR36, which guides Ψ modification in 18S rRNA. Depletion or over-expression of snR36 does not affect cell growth or pre-rRNA processing (our unpublished data). The origins of the hairpins are indicated in the hybrid names, for example, a hybrid (H) containing the 5′ hairpin of snR10 and 3′ hairpin of snR36 is designated H10/36, and the alternate combination is H36/10 (Figure 4A).
Figure 4.
The processing function of snR10 is mediated by its 5′ hairpin. (A) The roles of the 5′ and 3′ hairpins in snR10 function were examined with hybrid snoRNAs containing hairpins from snR10 and snR36 in test cells lacking snR10. The structures of the hybrid snoRNAs are depicted schematically, with the snR10 portion shown with thick lines and the snR36 portion with thin lines. (B) The hybrid snoRNAs are expressed at normal levels as evident in northern hybridization results. (C) Normal growth occurred with the hybrid containing the 5′ hairpin of snR10 (H10/36), but not the hybrid containing the 3′ hairpin of snR10 (H36/10). The snoRNA snR38 was used as a control for loading. (D) A normal ribosome–polysome pattern was restored by the hybrid snoRNA with the 5′ hairpin from snR10 (H10/36). Half-mer polysomes are evident in test cells that express a snR10 mutant lacking a C nucleotide (M-C) in the guide domain; this deletion blocks Ψ2923 formation (12). Accumulation of half-mer polysomes is indicated by an arrow. (E) The Ψ2923 modification is formed with a hybrid snoRNA containing the 3′ hairpin of snR10 (H36/10). Total RNA was treated with CMC and Ψ detected by primer extension analysis. The extension stops representing Ψ2923 and Ψ2880 (guided by snR10 and snR34, respectively) are identified.
The hybrid snoRNAs were expressed at normal levels in the test strain lacking snR10, and as anticipated, the hybrid containing the 5′ hairpin of snR10, but not the 3′ hairpin, restored normal growth for cells depleted of snR10 (Figure 4B and C). The inference that the 5′ hairpin of snR10 is sufficient for its processing function was borne out by the finding that this hybrid also restored a normal polysome profile (Figure 4D). Also, as expected, the hybrid containing the 3′ hairpin from snR10 restored the Ψ2923 modification (Figure 4E). Together, these data demonstrate that the information required for normal rRNA processing is exclusively located in the 5′ hairpin of snR10, and that the processing and modification functions are mediated by the 5′ and 3′ hairpins, respectively.
Earlier, we showed that the rate of rRNA processing is normal for a snR10 mutant with only a point deletion in the guide domain that disrupts its modification function (M−C) (Figure 5A) (12). The same cells also accumulate half-mer polysomes, whereas cells expressing the present hybrid with the 5′ hairpin of snR10 do not (Figure 4D); Ψ2923 formation was blocked in both strains. The new data argue that loss of the Ψ modification is not the basis of the half-mer polysome effect. A noteworthy difference between the two mutant snoRNAs is that the snoRNA with the point deletion has the potential to base pair with its normal target sequence in pre-rRNA, whereas the hybrid snoRNA with the 3′ hairpin of snR36 (H10/36) does not. These observations infer that altering the base-pairing pattern between the guide domain of snR10 and pre-rRNA may affect pre-rRNA folding or pre-rRNP assembly (see below).
Figure 5.
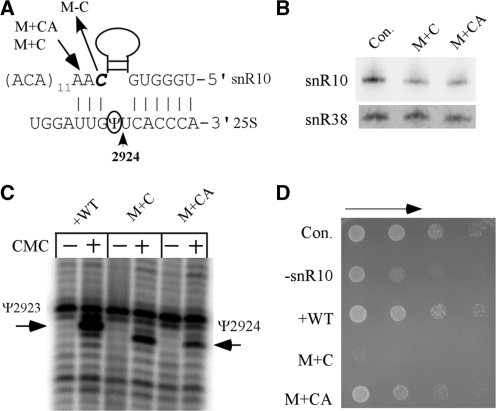
A 1-nt insertion mutation in the Ψ guide domain severely impairs cell growth. (A) Sequences of three mutant variants of snR10 snoRNA. The uridine normally targeted in 25S rRNA (U2923) is circled and an adjacent uridine that is modified with the two insertion variants of snR10 is marked with an arrow (U2924). In one mutant, a single C (italicized) has been deleted (M−C). Two other mutants contain C and CA insertions at the same position, as indicated by an arrow (M + C, M + CA). (B) Northern hybridization results show that the two insertion mutant snoRNAs are expressed at comparable levels. (C) The shift in Ψ modification site for the insertion mutations is shown by primer extension data, as in Figure 4, and the positions of the natural and novel Ψs are indicated. (D) The C-insertion mutation severely impairs cell growth (M + C). Growth was examined in a serial dilution assay as in Figure 2B.
A mutation in the guide domain that shifts Ψ modification to an adjacent site severely impairs growth
Our functional mapping included mutating the guide domain of snR10 to identify potential effects on rRNA modification and processing. We were especially interested in the possibility of shifting Ψ formation to an adjacent uridine and determining if this shift influences ribosome biogenesis or function. With the knowledge that the distance between the ACA box and the target uridine is an important determinant in pseudouridylation (14,15), we created two small insertion mutations predicted to alter modification specificity. The mutations include C and CA insertions in the 3′ element of the guide motif (M + C and M + CA) (Figure 5A). Both mutant snoRNAs were expressed at nearly normal levels in the test strain, and each caused Ψ modification to be shifted from U2923 to U2924 (Figure 5B, C). No modification appears to occur at the natural site. The modification level with the C-insertion mutation is similar to that observed with wild-type snR10, whereas that with the CA-insertion is somewhat weaker than normal (Figure 5C). These results confirm the importance and relationships of the snoRNA–substrate base-pairing interactions in modification specificity and efficiency.
Surprisingly, the C-insertion mutation causes a near-lethal phenotype in the snR10 depleted cells. Only moderate effects on growth rate occur with deletion of snR10 or with expression of snR10 bearing the C-insertion mutation in control cells (data not shown). However, expression of the C-insertion variant of snR10 in cells depleted of wild-type snR10 impairs cell growth severely, to a much greater extent than simple loss of the snR10 snoRNA (Figure 5D). The CA-insertion also causes slower growth; however, the effect is similar to that caused by snR10 depletion. These results suggest that the deleterious effects of the insertion mutations on cell growth are caused by either the shift in modification location, or the altered base-pairing between the mutant snoRNAs and pre-rRNA, or a combination of these effects.
The steady-state levels of rRNA are dramatically reduced by the C-insertion mutation
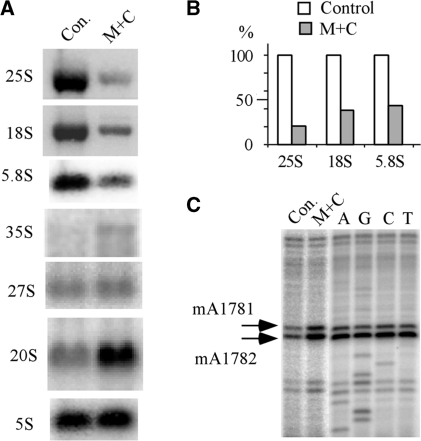
With a view to determining the basis of the severe growth defect caused by the C-insertion, we examined the rRNA levels in the mutant cells, which reflect the level of mature ribosomes. In cells expressing the C-insertion variant of snR10, the levels of mature 18S, 5.8S and 25S rRNAs were reduced by ∼60–80%, whereas the levels of the 35S full-length precursor and pre-18S rRNA (20S) were increased by ∼2-fold, as compared with control cells (Figure 6A). No significant difference was found for the 27S pre-rRNA, a precursor of the large subunit rRNAs, or for 5S rRNA, which is transcribed by RNA pol III. The levels of two control RNAs, tRNALeu and U2 snRNA, remained unchanged in the C-insertion mutant cells (not shown). Notably, for the CA-insertion mutation, no significant reduction in the levels of 18S and 25S rRNA was observed (data not shown). These results indicate that the C-insertion mutation causes a strong reduction in the level of RNAs generated by RNA pol I, presumably due to impaired processing since 35S and 20S pre-rRNAs accumulate in the mutant cells.
Figure 6.
The single nucleotide insertion in the guide domain causes a strong decrease in rRNA levels. (A) Northern hybridization probing of rRNA species reveals sharp reductions of ∼60–80% in the level of mature rRNAs, and significant accumulation of 35S and 20S pre-rRNAs. (B) The steady-state levels of 18S, 5.8S and 25S rRNAs are dramatically reduced as indicated by measuring the hybridization signals in (A), and normalizing to the amount of 5S rRNA. (C) The 20S pre-rRNA with Ψ at the novel site accumulates normally in the cytoplasm, as indicated by the presence of dimethylation modifications, which are formed only in cytoplasmic 20S pre-rRNA. The methylation modifications were detected by primer extension analysis.
In yeast, the 20S precursor to 18S rRNA is formed in the nucleolus and is then exported to the cytoplasm where it is cleaved to form mature 18S rRNA (4). To determine if the C-insertion mutation affects export of 20S pre-rRNA, we examined its cellular distribution. This was done by assaying the presence of a unique pair of dimethylation modifications (m62A1781m62A1782) that are catalyzed by the enzyme Dim1p after 20S pre-rRNA has been exported to the cytoplasm (57,58). The presence of the dimethyl modifications was probed by primer extension analysis and is reflected by strong extension stops (Figure 6C). Dimethylations were detected in both mutant and control cells showing that 20S pre-rRNA containing the mis-targeted Ψ is exported to the cytoplasm.
The C-insertion mutation severely impairs rRNA processing and causes accumulation of an abnormal rRNP complex
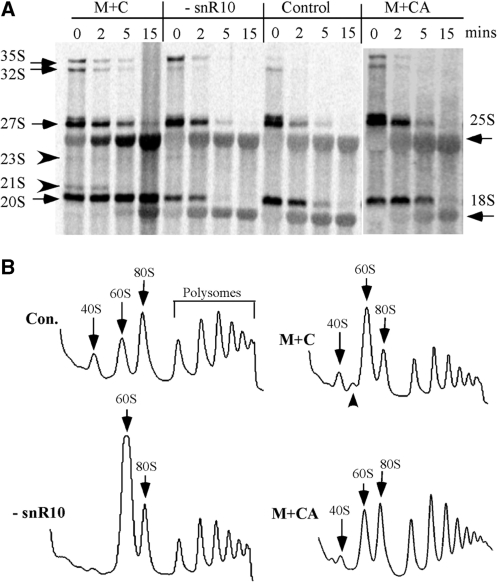
We determined that pre-rRNA processing is strongly impaired by the C-insertion mutation in the guide domain, as examined by in vivo pulse-chase labeling with [3H]-methionine and gel electrophoresis (Figure 7A; strain M + C). Production of 18S rRNA was severely inhibited, as reflected by accumulation of 20S precursor at a high level after 15 min into the chase and a strong delay in the appearance of mature 18S rRNA. In addition, two low abundance species often undetected, 23S and 21S pre-rRNAs, also accumulated. The 35S and 32S pre-rRNAs accumulated and persisted at least 5 min into the chase, whereas in control cells, these species were quickly processed and disappeared by 2 min after the chase was initiated. Thus, as a consequence of the C-insertion mutation, cleavage is delayed at the site that separates precursors for the small and large subunit rRNAs (site A2). Cleavages at sites that generate the 32S pre-rRNA (A0 and A1) and conversion of 27S precursor to 25S rRNA were also moderately delayed. Interestingly, the CA insertion mutation causes only minor processing defects, as evidenced by slightly slower maturation rates. Taken together, the results indicate that although the guide domain of snR10 and the Ψ that it normally targets are not required for pre-rRNA processing, a point mutation in the guide sequence can lead to severe impairment of rRNA processing, especially for production of 18S rRNA.
Figure 7.
rRNA processing is severely impaired by the single nucleotide insertion in the Ψ guide domain. (A) rRNA processing was examined by in vivo pulse-chase labeling. Test cells were labeled with [methyl-3H]methionine, and total RNA was prepared during a chase with unlabeled methionine. RNA was separated in a 1.2% agarose gel, transferred to a membrane, and the radioactive bands visualized with a phosphorimager. Normal rRNA species are identified by arrows and unusual pre-rRNAs are marked with arrowheads. (B) The C-insertion mutation causes an abnormal polysome pattern (M + C). Polysome profiles were analyzed as in Figure 2. The abnormal 50S complex in the mutant cells is indicated by an arrowhead.
To determine if the C-insertion mutation affects production of ribosome particles, the patterns of these complexes were analyzed for equivalent amounts of rRNA (Figure 7B). Interestingly, in the mutant cells, the level of free 40S complexes was 2- to 3-fold higher than for cells depleted of snR10, although the C-insertion mutation causes much stronger processing defects in 18S rRNA production than simple loss of snR10. The C-insertion mutant also exhibits an increase in the level of free 60S subunits (1.5-fold higher), but to a lower extent than the snR10 depletion condition, where an increase of >3-fold was seen (Figure 7B). In the C-insertion mutant, the levels of 80S ribosomes and polysomes are reduced by ∼50%, compared with control cells expressing wild type snR10. An abnormal complex of ∼50S was observed in the C-insertion mutant cells. This complex is most likely an intermediate of the small subunit, since northern probing of gradient fractions containing the complex revealed increased accumulation of 20S pre-rRNA, but not 27S pre-rRNA (data not shown). In the CA insertion mutant (M + CA), the polysome pattern is also altered, but to a much smaller extent. Here, the level of free 40S complex was reduced by ∼50% with a concomitant increase of ∼80% in free 60S complexes; levels of 80S ribosomes and polysomes were nearly normal. Thus, the data for this mutant suggest that subunit association and ribosome function are impaired moderately. Together, the results show that the C-insertion mutation has a severe negative impact on pre-rRNA processing and ribosome formation.
The snoRNA insertion mutations cause changes in ribosome structure
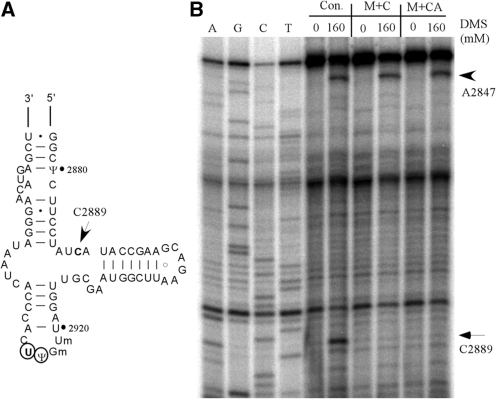
Both the C and CA insertion mutations shift the targeted Ψ to an adjacent site. The patterns of complementarity between the two mutant snoRNAs and pre-rRNA are different from each other, and also different from that of wild-type snR10 (Figure 5A). Because altered base-pairing of snoRNA with pre-rRNA could, in principle, influence pre-rRNA folding, and because Ψ modifications are already known to stabilize RNA duplexes, we next asked if the two mutant snoRNAs cause detectible changes in ribosome structure. Screening was by chemical probing of rRNA in vivo with DMS, followed by primer extension analysis of the PTC region in 25S rRNA that includes the Ψ sites of interest. One nucleotide in this region showed a qualitative change in DMS reactivity (C2889): it is strongly modified by DMS in control cells, but not in cells expressing either mutant snoRNA (Figure 8A and B). This result indicates that ribosome structure is indeed changed by the mutant snoRNAs, due either to the altered interaction between the mutant snoRNAs and pre-rRNA or the altered Ψ modification pattern.
Figure 8.
A change in rRNA structure accompanies the shift in Ψ modification. (A) Secondary structure of the A-loop region of the PTC region in 25S rRNA. The arrow identifies a nucleotide (C2889) with reduced DMS reactivity, as shown in (B). The normally modified Ψ2923 site and new site of modification (U2294) are circled. (B) Primer extension probing of rRNA structure in the A-loop region. Total RNA was prepared from cells treated with DMS and subjected to primer extension mapping. A nucleotide (C2889) showing altered DMS accessibility in test cells is marked with an arrow. A control nucleotide (A2847) sensitive to DMS treatment in all test strains is marked with an arrowhead.
The deleterious effect of the C-insertion mutation in the guide region depends on the presence of the processing element in the 5′ hairpin
While both insertion mutations cause the same shift in Ψ targeting, the C-insertion mutation results in a substantially higher modification yield (full versus ∼60%) and much stronger defects in growth and processing. These observations suggest that the much stronger detrimental effects of the C-insertion mutation are due to the different base-pairing patterns between the two insertion mutant snoRNAs with pre-rRNA, or seemingly less likely, the different levels of Ψ modification at the same novel site. Since the 3′ hairpin of snR10 can be deleted without affecting processing, an additional interesting possibility is suggested as the basis of the strong processing defects caused by the C-insertion mutation. In particular, the defects could result from a cooperative negative interference effect between the processing and modification domains of snR10. Such an effect could occur directly or indirectly. For example, an altered sequence in the guide domain could cause a change in the structure of the snoRNP that affects its interactions with pre-rRNA.
We reasoned that the importance of the modification level at the novel sites could be evaluated using new hybrid guide snoRNAs that guide modification at the novel site with the same level as the C-insertion mutation, but should not cause the detrimental effects seen for the C-insertion mutation. In this context, we examined the effects of expressing a hybrid snoRNA that contains the 3′ hairpin of snR10 with the C-insertion mutation joined with the 5′ half of snR36 (H36/10 + C) (Figure 9A). This hybrid snoRNA was expected to give the same modification specificity and yield as the C-insertion mutation. If this is indeed the case, and growth and processing are not affected or only moderately affected, those results would be strong evidence that the severe interference effects are not due to the change in modification specificity or yield.
Figure 9.
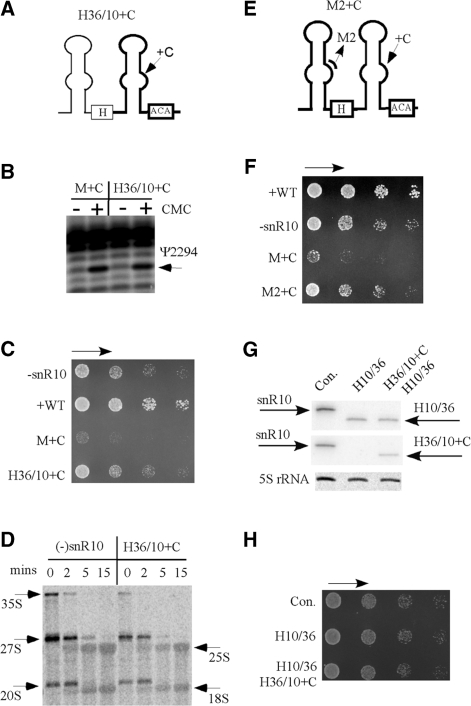
The severe growth defect caused by the C-insertion mutation is not due to mis-targeting of the Ψ modification. (A) Construction of a hybrid snoRNA with the mis-targeting guide domain from snR10 (H36/10 + C). The mutant snoRNA includes the 5′ hairpin of snR36 (thin line) and 3′ hairpin of snR10 (thick line) with the C-insertion mutation. (B) The levels of Ψ modification at the novel site are similar for both the novel hybrid and the initial snR10 variant with the C-insertion mutation (H36/10 + C and M + C), as shown by primer extension data. (C) The hybrid snoRNA with the C-insertion in the guide domain causes moderate growth defects similar to those seen for snR10 depletion. (D) Similar moderate defects in processing are also apparent for the hybrid snoRNA with the C-insertion mutation and for the snR10-depletion condition. (E) Structure of a double mutant of snR10 that includes changes in both the processing and modification hairpin domains (M2 + C). The changes are: substitution mutations in segment 2 of the 5′ hairpin, and the C-insertion in the guide region in the 3′ hairpin. (F) A 4-nt substitution in the 7-nt processing element suppresses the growth defect caused by the 3′ C-insertion mutation. (G) Co-expression of hybrid snoRNAs H10/36 and H36/10 + C in snR10-depleted cells. Total RNA was analyzed by northern hybridization using probes specific to the 5′ half (upper panel), or the 3′ half (middle panel). 5S rRNA was used as a control for loading. (H) No growth defect resulted from co-expression of the H10/36 and H36/10 + C snoRNAs.
The new hybrid snoRNA accumulated at a level comparable to the original C-insertion mutant snoRNA (data not shown), and as expected, Ψ modification was also shifted to U2924 and at a similar level (Figure 9B). Cell growth rate with this hybrid was moderately impaired (20% of the normal), but to a very much lower extent than the snR10 mutant with same 3′ domain (20% versus 1% of normal growth rate) (Figure 9C). Notably, the effect of the hybrid snoRNA on growth was comparable to loss of snR10 itself. In cells containing this hybrid snoRNA, pre-rRNA processing and ribosome fractionation patterns were also similar to those seen for snR10 depletion (Figure 9D and data not shown). The moderate defect in processing is most likely due to loss of the 5′ hairpin from snR10, which is required for normal processing. Thus, the defects on cell growth and pre-rRNA processing observed for the new hybrid snoRNA mimic those of snR10 depletion. Importantly, the defects are much less severe than those seen for the C-insertion mutation, which reduced the growth rate by ∼99% and strongly impaired processing (Figure 7). Together, these results show that the strong deleterious effects of the C-insertion mutation are not due to the change in Ψ location or level of modification. Rather these severe effects require the presence of the 5′ hairpin of snR10.
Our prediction that the deleterious effects of the C-insertion mutation require the presence of the 7-nt processing element in the 5′ half was tested by creating another snR10 double mutant. This variant contains both the C-insertion in the 3′ domain and the four nucleotide substitution mutation in the 7-nt element described above (U78GAA81) (Figure 9E). This mutant snoRNA (M2 + C) accumulated normally (data not shown), and caused a decrease in growth rate similar to that seen for snR10 depletion (20% of normal), but not the very severe growth defect seen for the C-insertion mutation alone (Figure 9F). Thus, the strong detrimental effects of the C-insertion mutation in the 3′ hairpin of snR10 is suppressed by introducing a mutation into the 7-nt processing element in the 5′ hairpin that blocks the normal processing function of snR10. These results indicate that the strong deleterious effects on growth and processing caused by the C-insertion mutation depend on the presence of the wild type processing element in the 5′ hairpin. The conclusion that the processing element and modification guide motif act in cis is further supported by the finding that co-expression of H10/36 and H36/10 + C mutants in snR10 depleted cells did not cause growth defects (Figure 9G and H). The nature of the interference effects indicates cooperative interactions between the processing and modification domains of this snoRNA.
DISCUSSION
The novel processing element
The 7-nt element in the 5′ hairpin required for normal rRNA processing is present in a single-stranded region and processing depends on the sequence of the element, but not the nearby secondary structure features. This element partially overlaps (4 nt) an 8-nt sequence complementary to pre-rRNA, suggesting that this portion and the adjoining non-essential complementary nucleotides interact with pre-rRNA. Although not all 7 nt are complementary to pre-rRNA, the element could mediate processing through partial binding to rRNA or other means, such as interacting with a protein or another RNA sequence required for activity. The DMS chemical probing data seem to argue against stable binding of the 7-nt element with a partner molecule(s), since it is accessible to DMS modification. A transient interaction for this region of the snoRNP is also possible.
Presently, we favor the possibility that the 7-nt element mediates processing by direct contact with pre-rRNA, like other processing snoRNAs. Among the snoRNAs with this function, three others bind to rRNA through sequence elements required for processing activity. These include: (i) the H/ACA processing snoRNA snR30/U17 with two essential conserved motifs of 9 and 7 nt (34,47), (ii) the C/D snoRNA U3 with two essential sequences (10 and (iii) 7 nt) (3,31,40) the C/D snoRNA U14 with an essential 13-nt element (59). Mutational results for U14 strongly infer that this element and a 14 nt methylation guide element function together in rRNA folding (see below) (28). Any one of these snoRNAs, including snR10, could mediate processing through base-pairing with pre-rRNA, for example, by assisting in an autocatalytic cleavage reaction, or recruiting a trans-acting nuclease.
The present mutational analysis also shows that loss of the 3′ hairpin or the Ψ modification it guides does not affect rRNA processing. However, a C-insertion mutation in the guide domain that causes Ψ formation to be targeted to an adjacent rRNA site strongly impairs processing, especially for 18S rRNA. This same mutation also causes accumulation of an abnormal, 50S pre-rRNP complex that is likely a precursor of the 40S subunit. These findings suggest the structure of the snoRNP–pre-rRNP complex is altered by the C-insertion mutation and that these changes are the likely basis of the interference in processing (see below).
The processing defect caused by the C-insertion mutation does not result from the modification shift
The results argue that the shift in Ψ position caused by the C-insertion is not the basis of the strong processing defects. In particular, two hybrid snoRNAs that cause the same shift do not have strong processing and growth defects (M36/10 + C and M2 + C). Notably, the levels of modification at the novel site are comparable for the three mutant snoRNAs with the same C-insertion. This situation argues that the processing defect stems from altered interaction of the dual-function snR10 snoRNP with pre-rRNA, rather than the change in modification site or yield. For example, altering the sequence in the guide domain could alter (or create) binding of a protein or interacting RNA (rRNA or other). Such an effect could influence the processing activity of the snoRNP, perhaps by interfering with the function of the 5′ processing domain. It seems unlikely, however, that the interference in processing is due solely to altered binding properties of the snoRNA guide region. Arguing against that possibility is the finding that two mutant snoRNAs with the C-insertion and a second mutation do not exhibit the strong processing defects seen for the C-insertion mutation alone. More likely, we propose, is that the severe defect is caused by a strong, negative cooperative effect involving interactions of the mutated modification region and the 5′ processing region with pre-rRNA, directly or indirectly.
Functional links between the processing and modification domains
Our functional mapping shows that the processing and modification functions of snR10 reside in distinct structural domains that can be separated genetically. However, we also show that a point insertion mutation in the 3′ hairpin severely impairs processing and that this effect depends on the presence of the 7-nt processing element in the 5′ hairpin. Mutating the processing element suppresses the severe growth defects caused by the C-insertion mutation in the guide domain, revealing that the modification and processing domains have a cooperative relationship. In addition, co-expression of hybrids H10/36 and H36/10 + C did not cause growth defects, suggesting that the two domains act in cis. An especially interesting possibility is that the two domains contact pre-rRNA simultaneously or in a tightly coupled fashion. In this context, snR10 could serve as a bridging factor to bring together or stabilize precursor forms of the small and large ribosomal subunits in a common complex. Also attractive is the possibility that such interactions could coordinate the modification and processing reactions to some benefit.
A cooperative relationship between snoRNA processing and modification elements has been observed for the universal U14 snoRNA. This snoRNA (a C/D species) contains two long sequences (13 and 14 nt) complementary to 18S rRNA; one required for rRNA processing and the other for 2′-O-methylation (46,59). Our laboratory has shown that a point mutation either in the processing element or in the modification guide domain has no effect on cell growth, but when combined cell growth is severely impaired (28). This situation suggests that the functions of the two snoRNA domains are coupled. That interpretation is supported by the fact that the complementary sequences in rRNA are in close proximity in an internal loop in the folded secondary structure, but well separated in the linear sequence. Simultaneous or tightly coupled binding to these sites was suggested as a possible basis for the cooperativity between these binding sites, although other interpretations are also possible.
Coordination of snoRNA functions at different rRNA sites also seems to be formally possible for snoRNAs with two modification guide domains and no known involvement in processing. In S. cerevisiae, 76 snoRNAs that act on rRNA have been identified and this is thought to be the full complement of such snoRNAs. About 30% of these snoRNAs contain two guide domains that target modifications at more than one site, in the same or different rRNAs (10). While the guide regions in these snoRNAs may act independently, it is also possible that the modification events are linked in some fashion and that such coordination could be an important feature in ribosome synthesis. The strong interference effects described here for the snR10 snoRNA suggest that this species may have special promise in dissecting the mechanism of action for a snoRNA that mediates both processing and modification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM19351 to M.J.F.). Funding for open access charge: National Institutes of Health (GM19351).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Ben Liu for help with early experiments, Wayne Decatur for help with the article and all laboratory members for stimulating discussions.
REFERENCES
- 1.Nazar RN. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life. 2004;56:457–465. doi: 10.1080/15216540400010867. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR. Nascent ribosomes. Cell. 2001;107:133–136. doi: 10.1016/s0092-8674(01)00531-1. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Woolford J.L., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 6.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis PP, Omer A, Lowe T. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 8.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand E, Fournier MJ. In: The Nucleolus. Olson M.OJ, editor. Georgetown, TX: Landes Bioscience Publishing; 2004. pp. 225–261. [Google Scholar]
- 10.Piekna-Przybylska D, Decatur WA, Fournier MJ. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tollervey D, Guthrie C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985;4:3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 14.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 15.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 16.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu YT, Terns RM, Terns MP. In: Fine-tuning of RNA Functions by Modification and Editing. Grosjean H, editor. Vol. 12. Berlin: Springer; 2005. pp. 223–262. [Google Scholar]
- 18.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 19.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 20.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 21.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 22.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 24.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 28.Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- 29.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 30.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 31.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA GAC-Box A′ and Box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 2001;21:6210–6221. doi: 10.1128/MCB.21.18.6210-6221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrissey JP, Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem. Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 34.Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009;28:1260–1270. doi: 10.1038/emboj.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 36.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 39.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2553. doi: 10.1093/nar/26.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10:942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borovjagin AV, Gerbi SA. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrissey JP, Tollervey D. U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma. 1997;105:515–522. doi: 10.1007/BF02510488. [DOI] [PubMed] [Google Scholar]
- 46.Dunbar DA, Baserga SJ. The U14 snoRNA is required for 2′-O-methylation of the pre-18S rRNA in Xenopus oocytes. RNA. 1998;4:195–204. [PMC free article] [PubMed] [Google Scholar]
- 47.Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, Liang XH, Piekna-Przybylska D, Liu Q, Fournier MJ. Mis-targeted methylation in rRNA can severely impair ribosome synthesis and activity. RNA Biol. 2008;5:249–254. doi: 10.4161/rna.6916. [DOI] [PubMed] [Google Scholar]
- 51.Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ. Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol. 2007;425:283–316. doi: 10.1016/S0076-6879(07)25013-X. [DOI] [PubMed] [Google Scholar]
- 52.Liang XH, Fournier MJ. The Helicase Has1p Is Required for snoRNA Release from Pre-rRNA. Mol. Cell. Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 54.Maden BE. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 55.Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11:619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell AG, Schnare MN, Gray MW. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA. 2004;10:1034–1046. doi: 10.1261/rna.7300804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brand RC, Klootwijk J, Van Steenbergen TJ, De Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 58.Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 59.Jarmolowski A, Zagorski J, Li HV, Fournier MJ. Identification of essential elements in U14 RNA of Saccharomyces cerevisiae. EMBO J. 1990;9:4503–4509. doi: 10.1002/j.1460-2075.1990.tb07901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.