Abstract
The majority of human miRNA genes is transcribed by polymerase II and can be classified as class II genes similar to protein-coding genes. Whereas current research on miRNAs has focused on the physiological and pathological functions, the molecular mechanisms underlying their transcriptional regulation are largely unknown. We recently reported that lipopolysaccharide (LPS) alters mature miRNA expression profile in human biliary epithelial cells. In this study, we tested the role of transcription factor NF-κB in LPS-induced transcription of select miRNA genes. Of the majority of LPS-up-regulated mature miRNAs in cultured human biliary epithelial cells, potential NF-κB binding sites were identified in the putative promoter elements of their corresponding genes. Inhibition of NF-κB activation by SC-514, an IKK2 inhibitor, blocked LPS-induced up-regulation of a subset of pri-miRNAs, including pri-miR-17-92, pri-miR-125b-1, pri-miR-21, pri-miR-23b-27b-24-1, pri-miR-30b, pri-miR-130a and pri-miR-29a. Moreover, direct binding of NF-κB p65 subunit to the promoter elements of mir-17-92, mir-125b-1, mir-21, mir-23b-27b-24-1, mir-30b and mir-130a genes was identified by chromatin immunoprecipitation analysis and confirmed by the luciferase reporter assay. Thus, a subset of miRNA genes is regulated in human biliary epithelial cells through NF-κB activation induced by LPS, suggesting a role of the NF-κB pathway in the transcriptional regulation of miRNA genes.
INTRODUCTION
MicroRNAs (miRNAs) are initially transcribed as primary transcripts known as pri-miRNAs and cropped into 70–100-nt-long hairpin precursors (termed pre-miRNAs) by the RNAse III Drosha in the nucleus (1,2). Pre-miRNAs are actively transported to the cytoplasm where they are cleaved by the enzyme Dicer to form mature miRNAs. They eventually become incorporated as single-stranded RNAs into a ribonucleoprotein complex, known as the RNA-induced silencing complex (RISC) (3). The RISC identifies target mRNA based on complementarity resulting in mRNA cleavage and/or translational suppression (4,5).
Whereas current research on miRNAs has focused on their physiological and pathological functions, the molecular mechanisms of transcriptional regulation of miRNA genes remain largely unknown. MicroRNAs exhibit tissue-specific or developmental-stage-specific expression and respond to extracellular stimuli, indicating that their expression is tightly controlled (6,7). The human genome encodes ≈700 known miRNA genes and ∼50% of them are expressed from non-protein-coding transcripts (8). The majority of miRNA genes are located in intergenic regions or in antisense orientation to annotated genes, indicating that they form independent transcription units (2). Other miRNAs are found in intronic regions, which may be transcribed as part of the annotated genes (9). Most of human miRNA genes are transcribed by polymerase II (pol II) and can be classified as class II genes along with all protein-coding genes (1,2). Therefore, like other pol II-dependent genes, miRNA genes can be elaborately controlled through various regulatory mechanisms including transactivation and transrepression by nuclear transcription factors. Up-regulation of miR-146a/b in response to Toll-like receptor (TLR) signaling is NF-κB-dependent (10). The TLR4 ligand lipopolysaccharide (LPS) has been shown to affect expression of miR-132, miR-146 and miR-155 in human THP-1 monocytes (10,11). Other transcription factors, such as c-Myc, STAT3 and C/EBPα, also appear to be involved in the expression of miRNAs (12–14).
Epithelial cells of the skin and mucosa represent the host’s first line of defense against microbial infection. Beyond the role of epithelial cells in creating a physical barrier to infection, epithelial cells are critical in the initiation, regulation and resolution of both innate and adaptive immune responses (15). Emerging studies implicate specific miRNAs in controlling epithelial cell processes such as regulation of cellular differentiation, determination of epithelial cell fate (cell death and proliferation), initiation and regulation of anti-microbial immunity, fine-control of inflammatory responses and activation of intracellular signaling pathways (15–17). Transcription of miRNA genes in epithelial cells is finely controlled in response to extracellular stimuli (17). Infection of cultured human biliary epithelial cells with Cryptosporidium parvum, a parasite that causes intestinal and biliary disease, activates host cell NF-κB signaling resulting in transactivation of a subset of miRNA genes (18). Transcriptional suppression of let-7i following C. parvum infection involves the activation and aggregation of the NF-κB p50 and C/EBPβ (19).
Our previous studies revealed similar alterations in miRNA expression profiles in cultured human biliary epithelial cells following C. parvum infection and LPS stimulation (18). In this report, we further analyzed the role of NF-κB signaling in LPS-up-regulated miRNA expression. We found that inhibition of NF-κB signaling blocked LPS-induced transactivation of a subset of miRNA genes. Direct binding of NF-κB p65 subunit to the promoter elements of select miRNA genes was confirmed by chromatin immunoprecipitation (ChIP) and promoter luciferase analysis. Thus, a subset of miRNA genes is transactivated by LPS through promoter binding of NF-κB subunit p65 in human biliary epithelial cells.
MATERIALS AND METHODS
Human cholangiocyte cell line and reagents
H69 cells are SV40 transformed human biliary epithelial cells originally derived from normal liver harvested for transplant. This cell line continues to express biliary epithelial cell markers and TLRs consistent with biliary function and has been extensively characterized (20). SC-514 (100 µM, Calbiochem), a potent IKK-2 inhibitor was used to inhibit NF-κB activation (21). LPS (E. coli strain K12) was purchased from Invivogen (San Diego, CA). SC-514 and LPS at the utilized concentrations showed no cytotoxic effects on H69 cells.
Real-time PCR
For real-time PCR analysis of mature miRNAs, total RNAs were extracted using the mirVana™ miRNA Isolation Kit (Ambion). An amount of 0.05 µg total RNAs was reverse-transcribed by using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). Comparative real-time PCR was performed in triplicate with the use of the Taqman Universal PCR Master Mix (Applied Biosystems) on the Applied Biosystems 7500 FAST real-time PCR System. Mature miRNA-specific primers and probes were obtained from Applied Biosystems. Normalization was performed with the RNU6B primers and relative expression was calculated employing the comparative CT method.
For analysis of pri-miRNAs, total RNA was isolated from cells with Trizol reagent (Ambion). RNAs were treated with DNA-freeTM Kit (Ambion) to remove any remaining DNA. Comparative real-time PCR was performed by using the SYBR Green PCR Master Mix (Applied Biosystems). Specific primers for pri-miRNAs were listed in Supplementary Table S1. All reactions were run in triplicate. The cycle threshold (Ct) values were analyzed by using the comparative Ct (ΔΔCt) method. The amount of target was obtained by normalizing to the endogenous reference (GAPDH) and shown as the relative to the control (non-treated cells).
Northern blot
Total RNAs harvested with Trizol reagent were run on a 15% Tris/Borate/EDTA (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3)–urea gel (Invitrogen) and transferred to a Nytran nylon transfer membrane (Ambion). LNA DIG-probes for selected miRNAs (Exiqon) were hybridized using UltraHyb reagents (Ambion) according to the manufacturer’s instructions with snRNA RNU6B blotted as a control.
ChIP analysis
ChIP analysis was performed with a commercially available ChIP Assay Kit (Upstate Biotechnologies) according to the manufacturer’s instructions. Briefly, 1 × 106 H69 cells were cultured in 15-cm culture dishes and stimulated with LPS for 2 h in the presence or absence of SC-514. The chromatin fraction was immunoprecipitated with an anti-NF-κB p65 antibody (Upstate Biotechnologies) overnight at 4°C. PCR amplification was performed in a total volume of 25 μl with specific primers listed in Supplementary Table S1. The PCR procedure for each target was optimized to ensure that measurements were taken in the linear range of amplification. Expression level of a target DNA sequence was determined relative to its abundance in the input chromatin and represented as fold enrichment compared with the non-LPS treated control.
Luciferase reporter constructs and luciferase assay
Promoters of miRNAs were amplified by PCR from human genomic DNA. PCR primers were listed in Supplementary Table S1. The PCR products were separated by agarose gel electrophoresis, and the DNA fragments then isolated and cloned into the restriction enzyme digested pGL3 Basic Vector (Promega) using T4 DNA ligase (Fisher scientific). All constructs were confirmed by sequencing. Mutations were introduced into the NF-κB binding sites using the QuikChange Site-directed Mutagenesis Kit (Stratagene). H69 cells were transfected with each reporter construct for 24 h and then exposed to LPS for 4 h in the presence or absence of SC-514 followed by assessment of luciferase activity. Luciferase activities were then measured and normalized to the control β-gal level. The luciferase activity of each construct was compared with that of the promoterless pGL3 basic vector.
RESULTS
Kinetics of up-regulation of mature miRNAs in cultured human biliary epithelial cells following LPS stimulation
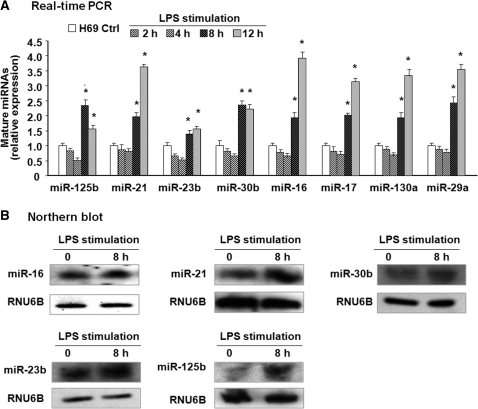
We previously described the altered expression profile of mature miRNAs in human biliary epithelial cells following LPS stimulation (18). Of these miRNAs expressed in H69 cells, expression of miR-125b, miR-146b, miR-26a, miR-18a, miR-483, miR-484, miR-486 and miR-519e* was significantly (p ≤ 0.05) increased after exposure to LPS (1 µg/ml) for 8 h as revealed by the miRCURY™ LNA human microRNAs assays. Additional 13 miRNAs (miR-29a, miR-15b, miR-106b, miR-27b, miR-16 and miR-21, etc.) showed a tendency to increase (0.05 < P ≤ 0.20). These data are shown again in Supplementary Table S2 for the purpose of exploring the molecular mechanisms underlying LPS-up-regulated miRNA expression in epithelial cells in the current study. Based on our array data, we assessed the kinetics of expression of selected miRNAs up-regulated in H69 cells following LPS stimulation by real-time PCR. Increased expression of miR-125b, miR-21, miR-23b, miR-30b, miR-16, miR-17, miR-130a and miR-29a was detected in cells after exposure to LPS for 8 and 12 h, with a 1.5–4.0-fold increase compared with non-LPS treated control (Figure 1A). Increased expression of miR-16, miR-125b, miR-21, miR-23b and miR-30b was also confirmed by northern blot (Figure 1B).
Figure 1.
Altered expression of selected miRNAs identified by real-time PCR and northern blot following LPS stimulation in H69 cells. (A) Alterations of selected miRNA expression in H69 cells after exposure to LPS for various periods of time as assessed by real-time PCR. The amount of mature miRNAs was obtained by normalizing to the level of snRNA RNU6B in the samples. Data are expressed as the amount of mature miRNAs in LPS-stimulated samples relative to the control non-stimulated samples and representative of three independent experiments. (B) Alterations of selected miRNA expression in cells after exposure to LPS for 8 h as determined by northern blot. snRNA RNU6B was used as a control to ensure equal loading. Representative northern blots (LPS-stimulated cells versus non-stimulated control) from three independent experiments are shown. *P < 0.05 t-test versus non-stimulated cells.
Database analysis of up-regulated miRNAs in biliary epithelial cells following LPS stimulating reveals potential NF-κB binding sites in their promoter elements
Binding of transcription factors, such as NF-κB subunits, to the promoter elements has been implicated in the transcriptional regulation of miRNA genes in response to microbe infection (10,11,18,22). The NF-κB pathway is activated by LPS in biliary epithelial cells and thus, may be involved in the transcriptional regulation of miRNA genes induced by LPS. To test this possibility, we performed a general characteristic analysis of these LPS-responsive miRNAs that showed a significant increase in H69 cells following LPS stimulation. First, several sets of LPS-up-regulated miRNAs were identified as cluster miRNAs as implicated by previous studies (18) (Table 1). miR-23b and miR-27b have previously reported as cluster miRNAs (9) and all of them showed an increased expression in LPS-treated cells. miR-17, miR-18a and miR-20a are also cluster miRNAs (23) and were up-regulated in cells following LPS stimulation. Second, many of LPS-responsive miRNAs are within introns of either protein-coding or non-coding transcriptional units, including miR-17-92, miR-23b-27b-24-1 and miR-125b-2 (Table 1). Those miRNA genes are usually co-transcribed with their host genes and then processed to form the mature miRNAs (9). Other LPS-stimulated miRNAs genes are located in the intergenic regions (so-called intergenic miRNAs), such as miR-21, miR-30b and miR-130a, indicating that they form independent transcription units (Table 1). Thirdly, most of those LPS-responsive miRNAs have potential NF-κB binding sites in their promoter elements of the corresponding genes (Table 1) based on TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and MOTIF (http://motif.genome.jp/) database search (24,25). The putative NF-κB binding sites are usually within 3000 bp upstream of those known miRNA genes (Table 1), consistent with the results from a recent report (8). Transactivation of most NF-κB-dependent genes requires NF-κB p65 binding to the promoter (26). We then focused on determining whether p65 binds to the promoters and transactivates the miRNA genes following LPS stimulation.
Table 1.
Analysis of LPS-up-regulated miRNAs in cholangiocytes reveals potential transactivation of their genes by NF-κB
| Mature miRNAs | miRNA genes (or cluster) | Chromosome (strand) | Host gene | Predicted NF-kB binding sites (from miRNA TSS) | References |
|---|---|---|---|---|---|
| miR-125b | miR-125b-1 | 11 (−) | None | GGGGCTTTCC (−1059 to −1050) | (18) |
| GGGAATTTCA (−2455 to −2446) | |||||
| miR-125b-2 | 21 (+) | C21orf34 | GAGAATTTCC (−893 to −884) | (9) | |
| miR-21 | miR-21 | 17 (+) | None | GGGAATTTTC (+1167 to +1176) | (1,13) |
| GGGAATTCTC (+1395 to +1404) | |||||
| miR-23b | miR-23b-27b-24-1 | 9 (+) | C9orf3 | GGGACTCTCC (−1263 to −1254) | (9,18) |
| miR-27b | |||||
| miR-24-1 | |||||
| miR-30b | miR-30b | 8 (−) | None | AGGAATTTAC (−347 to −338) | (18) |
| miR-30c | miR-30c-1 | 1 (+) | NFYC | TGGAATTACC (−689 to −680) | (9,12) |
| miR-30a-30c-2 | 6 (−) | C6orf155 | GGAAAGCCCT (−208 to −199) | (9,12) | |
| miR-16 | miR-15a-16-1 | 13 (−) | DLEU2 | None | (9,12) |
| miR-15b | (9,12) | ||||
| miR-26a | miR-26a-1 | 3 (+) | CTDSPL | GGGGATTCGC (−700 to −691) | (9,12) |
| miR-26a-2 | 12 (−) | CTDSP2 | TGGGCTTTCC (−890 to −881) | (9,12) | |
| miR-146b | miR-146b | 10 (+) | None | GGGGAGTCCC (−769 to −760) | (10) |
| GGGGAGTCCC (−800 to −791) | |||||
| miR-106b | miR-106b-25 | 7 (−) | MCM7 | None | (28) |
| miR-17 | miR-17-92 | 13 (+) | C13orf25 | TGGAATTTCC (−1698 to −1689) | (29) |
| miR-18a | TGGGATTTCC (−1442 to −1433) | ||||
| miR-20a | CGGAATTTCC (−827 to −818) | ||||
| miR-130a | miR-130a | 11 (+) | None | GGGAATTTGC (−2977 to −2968) | (30) |
| miR-29a | miR-29b-1-29a | 7 (−) | None | Promoter element unknown | (12) |
| miR-483 | miR-483 | 11 (−) | IGF2 | GGGACTTTCC (−1305 to −1296) | |
| miR-484 | miR-484 | 16 (+) | NDE1 | None | (9) |
| miR-486 | miR-486 | 8 (−) | ANK1 | None | (9) |
| miR-519e* | miR-1323-1383-1 | 19 (+) | None | Promoter element unknown | (9) |
MicroRNA genes related to LPS-up-regulated mature miRNAs, their chromosomal location and co-expressed host genes were identified by the miRBase (http://microrna.sanger.ac.uk/) database search and confirmed by previous studies as referred. Potential promoter element for each miRNA was based on the referred studies and potential NF-κB binding sites were identified by the TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and MOTIF (http://motif.genome.jp/) search.
Differential expression of primary transcripts (pri-miRNAs) of LPS-up-regulated mature miRNAs in H69 cells
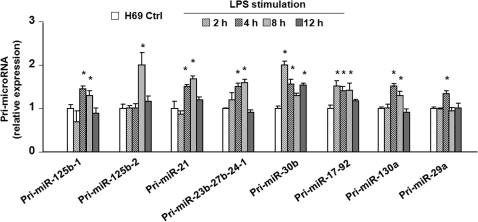
We analyzed the kinetics of alterations in the pri-miRNAs of selected mature miRNAs which were up-regulated following LPS stimulation as listed in Supplementary Table S2. H69 cells were exposed to LPS for various periods of time and pri-miRNAs of select were quantified by real-time PCR (primers listed in Supplementary Table S1). Expression of pri-miR-125b-1, pri-miR-125b-2, pri-miR-21, pri-miR-23b-27b-24-1, pri-miR-30b, pri-miR-17-92, pri-miR-130a and pri-miR-29a showed a time-dependent increase in cells following LPS stimulation (Figure 2). Expression of most pri-miRNAs was increased at 4 h after LPS challenge and declined to the basal level at 12 h following LPS stimulation. The expression of pri-miR-17-92 and pri-miR-30b began to increase 2 h after exposed to LPS. Most of those pri-miRNAs showed a 1.5–2.0-fold increase compared with non-LPS treated control (Figure 2).
Figure 2.
Differential expression of primary transcripts of selected LPS-up-regulated mature miRNAs in H69 cells. H69 cells were exposed to LPS for 2–12 h and primary transcripts (pri-miRNAs) of select miRNAs were quantified by real-time PCR. The amount of pri-miRNAs was obtained by normalizing to the level of GAPDH in the samples. Data are expressed as the amount of pri-miRNAs in LPS-stimulated samples relative to the control non-stimulated samples and representative of three independent experiments. *P < 0.05 t-test versus the non-stimulated cells.
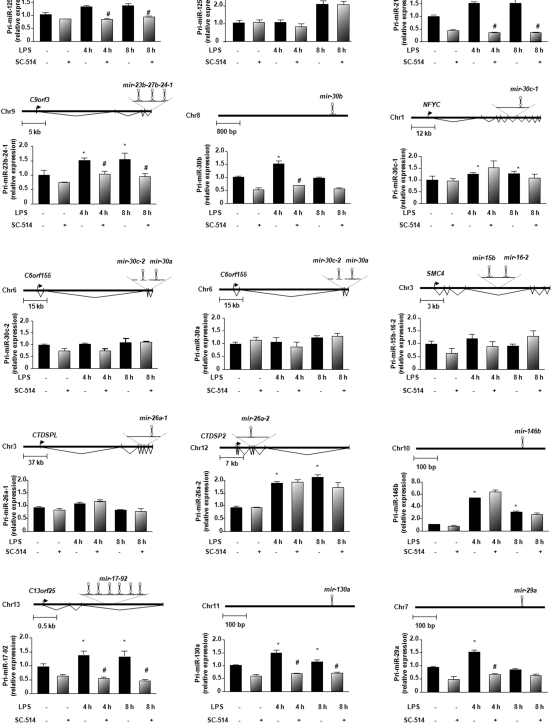
Up-regulation of a subset of pri-miRNAs in biliary epithelial cells following LPS stimulation is NF-κB dependent
To test the role of NF-κB in LPS-altered miRNA expression, we measured the levels of pri-miRNAs of select miRNA genes in H69 cells in response to LPS in the presence or absence of a specific IKK2 inhibitor, SC-514, which inhibits p65-associated transcriptional activation of the NF-κB pathway (21). We focused on those LPS-up-regulated miRNAs which have the putative NF-κB binding sites in their promoter elements as listed in Table 1. Using IL-8 as a positive control of NF-κB dependent gene (27), we detected a significant increase of IL-8 mRNA in cells exposed to LPS for 4 and 8 h (Supplementary Figure S1). Accordingly, a significant inhibition of LPS-induced IL-8 mRNA increase was observed in cells simultaneously treated with SC-514 (Supplementary Figure S1). By real-time PCR analysis using primers designed for select pri-miRNAs as listed in Supplementary Table S1, we detected that pri-miR-125b-1, pri-miR-21, pri-miR-23b-27b-24-1, pri-miR-30b, pri-miR-17-92, pri-miR-130a and pri-miR-29a were significantly increased in cells exposed to LPS and SC-514 blocked LPS-induced increase of those pri-miRNAs in the treated cells (Figure 3). Increased expression of pri-miR-125b-2, pri-miR-26a-2 and pri-miR-146b was detected in cells following LPS stimulation but SC-514 showed no inhibitory effect on their expression (Figure 3).
Figure 3.
NF-κB dependent miRNAs identified among a subset of LPS-up-regulated mature miRNAs in biliary epithelial cells. Data are presented as the relative expression level of each pri-miRNA in H69 cells following LPS stimulation for 4 and 8 h in the presence or absence of SC-514 as assessed by real-time PCR. A schematic diagram shows the structure of each miRNA gene. The amount of pri-miRNAs was obtained by normalizing to the level of GAPDH in the samples. Data are expressed as the amount of pri-miRNAs in the stimulated samples relative to the control non-stimulated samples and representative of three independent experiments. *P < 0.05 t-test versus the non-stimulated cells; #P < 0.05 t-test versus LPS-stimulated cells.
Individual mature miRNA can be derived from different pri-miRNA transcripts. Pri-miR-125b-1 and pri-miR-125b-2 encode the same mature miRNA, miR-125b, and both pri-miR-26a-1 and pri-miR-26a-2 encode miR-26a. LPS increased expression of both pri-miR-125b-1 and pri-miR-125b-2, but SC-514 only blocked LPS-induced increase the expression of pri-miR-125b-1 (Figure 3). The expression of pri-miR-26a-2 was increased after LPS stimulation and SC-514 showed no inhibitory effects. There were no changes in pri-miR-26a-1 expression in LPS-treated cells (Figure 3). These data suggest that transactivation of miRNA genes that produce the same mature miRNA can be differentially controlled. Although microarray analysis showed a significant increase of mature miR-16 in H69 cells in response to LPS, pri-miR-15b-16-2 showed no change in its cellular expression (Figure 3). miR-483 and miR-519e*, both with putative NF-κB binding sites in their promoter elements, were increased in H69 cells exposed to LPS (Supplementary Table S2). However, the expression level of pri-miR-483 and pri-miR-519e* was not detectable in H69 cells using real-time PCR (data not shown).
Promoter binding of NF-κB p65 subunit is required for the transcription of selected NF-κB-dependent miRNA genes upon LPS stimulation in H69 cells
Transcriptional inhibition of selected miRNA genes by SC-514 in LPS-stimulated cells suggests the involvement of transcriptional regulation by NF-κB components, in particular, the p65 component. To test this possibility, we performed the ChIP analysis to identify the binding of p65 components of NF-κB to the putative binding sites in the promoter elements of selected miRNA genes. We focused on those miRNA genes that are induced in a NF-κB-dependent manner in response to LPS as revealed in Figure 3. The binding of p65 to the putative binding sites in the promoter elements was demonstrated by ChIP analysis. Specific primers were designed and synthesized to cover the potential NF-κB binding sites for each miRNA gene listed in Supplementary Table S1. We also generated luciferase reporter constructs covering the potential elements, as well as various deletions and mutations, of the putative promoters of select miRNA genes to test the transcriptional regulation by p65.
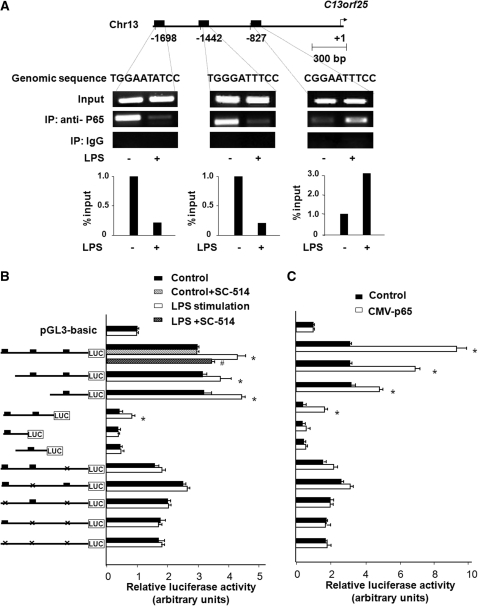
The mir-17-92 cluster is comprised of miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92-1, housed in the expressed sequence tag (EST) C13orf25 (23,28). Expression of miR-17, miR-18a and miR-20a was significantly increased in LPS-treated cells (Supplementary Table S2). Database analysis revealed three potential p65 binding sites in the promoter element within 2000-bp of the upstream from the transcription start site (TSS) of C13orf25 (29) (Figure 4A). NF-κB p65 binding was identified to all the three potential binding sites in non-stimulated H69 cells. LPS stimulation increased p65 binding to the putative binding site at −827, but slightly decreased p65 binding to the sites at −1698 and −1442 (Figure 4A) as demonstrated by ChIP analysis with the use of specific primers for each putative binding site listed in Supplementary Table S1. To further test the potential transactivation of the mir-17-92 cluster by the p65 subunit, we cloned the upstream of the C13orf25 transcript and generated a series of luciferase reporter constructs that contain different nucleotide genomic regions with the putative p65 binding sites. As shown in Figure 4B, LPS stimulation increased luciferase activity in cells transfected with the luciferase constructs that encompass the putative NF-κB binding site at −827. SC-514 significantly inhibited LPS-induced increase of luciferase activity in cells transfected with the construct that encodes all three putative NF-κB binding sites. Deletion of the binding site at −827 blocked LPS-induced luciferase activity. No transcriptional activities was identified for the luciferase constructs that encompass either one or both of the putative binding sites at −1698 and −1442, compared with the PGL3-basic construct (Figure 4B). Moreover, p65-associated transactivation of the C13orf25 promoter was also confirmed by the up-regulation of luciferase activity in cells overexpressing NF-κB p65 (Figure 4C). As an additional control, we analyzed IL-8 transactivation, a p65-dependent process induced by LPS in epithelial cells. Binding of p65 to the promoter of IL-8 gene in cells exposed to LPS were confirmed (Supplementary Figure S1). Taken together, the above data suggest that p65 binding to the promoter element of the C13orf25 mediates pri-miR-17-92 up-regulation in H69 cells in response to LPS stimulation.
Figure 4.
Promoter binding of p65 transactivates mir-17-92 gene in H69 cells following LPS stimulation. (A) LPS induced the promoter element binding of p65 to C13orf25 gene. The schematic diagram shows three potential binding sites in the putative promoter element of to C13orf25 gene. ChIP analysis revealed increased binding of p65 to the binding site at −827, but not at −1442 and −1698, of C13orf25 promoter element in cells following LPS stimulation. Representative ChIP gels are shown in the upper panel and densitometry analysis in the lower panel. (B) H69 cells were transfected with various luciferase reporter constructs spanning the potential p65 binding sites of the C13orf25 promoter. The transfected cells were exposed to LPS in the presence or absence of SC-514. Luciferase activity was measured and presented as the ratio of the activity of the test construct with the control luciferase reporter construct. Reporter constructs containing different nucleotide genomic regions with the putative p65 binding sites were also utilized for the analysis as indicated. H69 cells were co-transfected with the pCMV-p65 to overexpress p65 and the luciferase reporter constructs for 24 h followed by measurement of luciferase activity. *P < 0.05 t-test versus the non-stimulated cells or empty pCMV vector control; #P < 0.05 t-test versus LPS-stimulated cells.
Using the same approaches, we analyzed the role of p65 promoter element binding in LPS-induced transcription of pri-miR-125b-1, pri-miR-21, pri-miR-23b-27b-24-1, pri-miR-30b and pri-miR-130a. Our data were summarized in Table 2 and presented in detail in Supplementary Figures S2–S6. Specifically, binding of p65 to the promoter element of mir-130a (at −2968) (30) is involved in LPS-induced miR-130a expression (Supplementary Figure S2). p65 binding to the promoter element around −1059 of the mir-125b-1 gene mediates miR-125b up-regulation in H69 cells in response to LPS treated (Supplementary Figure S3). p65 binding to the putative p65 binding site around +1395 of the mir-21 gene appears to be associated with LPS-induced transcription of pri-miR-21 (Supplementary Figure S4). LPS increases transcription of pri-miR-23b-27b-24-1 cluster, as well as the host gene transcript, C9orf3, via promoter binding of p65 to a binding site at −1254 of the immediately upstream of the gene (Supplementary Figure S5). Increased transcription of pri-miR-30b induced by LPS is p65-dependent and may involve with p65 binding to the site at −338 downstream of the potential transcription start site (Supplementary Figure S6). We failed to identify a potential binding of p65 to the promoter element of mir-29a gene (12) (data not shown).
Table 2.
Promoter binding of NF-κB p65 subunit in LPS-stimulation transactivation of miRNA genes in H69 cells
| LPS-up-regulated mature miRNAs | Corresponding pri-miRNAs | Potential NF-κB binding site(s) within the promoter region | Promoter p65 binding by ChIP | Conformed by promoter reporter assay |
|---|---|---|---|---|
| miR-17 | pri-miR-17-92 | TGGAATTTCC (−1698 to −1689) | − | − |
| miR-18a | TGGGATTTCC (−1442 to −1433) | − | − | |
| miR-20a | CGGAATTTCC (−827 to −818) | + | + | |
| miR-125b | pri-miR-125b-1 | GGGAATTTCA (−2455 to −2446) | − | − |
| GGGGCTTTCC (−1059 to −1050) | + | + | ||
| miR-21 | pri-miR-21 | GGGAATTTTC (+1167 to +1176) | − | − |
| GGGAATTCTC (+1395 to +1404) | + | + | ||
| miR-23b | pri-miR-23b-27b-24-1 | GGGACTCTCC (−1263 to −1254) | + | + |
| miR-27b | ||||
| miR-24-1 | ||||
| miR-30b | pri-miR-30b | AGGAATTTAC (−347 to −338) | + | + |
| miR-130a | pri-miR-130a | GGGAATTTGC (−2977 to −2968) | + | + |
| miR-29a | pri-miR-29b-1-29a | AGGATTTTCC (−89 to −80)* | NS | NS |
Putative promoter element for each miRNA gene was based on previous studies as referred in Table 1. Potential NF-κB binding site(s) in the promoter region was identified by the TFSEARCH (http://www.cbrc.ip/research/db/ TFSEARCH.html) and MOTIF (http://motif.genome.jp/). LPS-induced promoter binding of NF-κB p65 subunit to the predicted binding site was confirmed by ChIP analysis and marked as ‘+’; otherwise marked as ‘−’ if p65 binding was not enhanced after LPS stimulation. LPS-induced transactivation of miRNA gene by p65 was confirmed by using luciferase reporter gene constructs that spanned the promoter region of each individual gene. If LPS stimulation increased luciferase activity in cells transfected with the luciferase constructs containing the binding site for p65 and this induction was blocked by SC-514, it was presented as ‘+’; otherwise presented as ‘−’. The putative NF-κB binding site of miR-29a was identified from the start site of pre-miR-29a, which was indicated by asterisk. NS, not selected for further ChIP analysis or luciferase reporter assay in this study. Refer to Supplementary Figures S2–S6 for details.
DISCUSSION
Similar to other RNA molecules, most miRNAs are initially transcribed as primary transcripts by Poly II in the nucleus and can be regulated by transcriptional factors (1,2). In this report, we demonstrate that LPS-induced transactivation of miRNA genes is regulated by promoter binding of NF-κB p65 subunit. Specifically, promoter binding of NF-κB p65 subunit is required for the transactivation of mir-17-92, mir-125b-1, mir-23b-27b-24-1, mir-21, mir-30b and mir-130a genes in cultured human biliary epithelial cells following LPS stimulation. Inhibition of NF-κB activation by SC-514 blocked LPS-induced up-regulation of the pri-miRNAs for those miRNA genes. Each of those NF-κB-associated miRNAs has at least one potential NF-κB binding site in its putative promoter element. Importantly, binding of NF-κB p65 subunit to the promoter element of each selected miRNA gene was confirmed by ChIP analysis. Moreover, NF-κB-driven transcriptional activity of associated miRNA genes was further demonstrated using the luciferase reporter assay. Thus, a subset of miRNA genes is regulated through NF-κB activation in human biliary epithelial cells following LPS stimulation.
Approximately 40% of human miRNA loci are located in <3 kb from the adjacent miRNA locus (8,31). Some miRNA genes are organized in clusters in the genome. It has been speculated that such clustered miRNA genes may be transcribed from a common promoter to generate a polycistronic primary transcript, proceeding to produce multiple mature miRNAs (9). Our analysis on LPS-induced miRNA expression in human biliary epithelial cells partially supports this hypothesis. Several miRNA cluster genes were transactivated and some of their corresponding mature miRNAs were coordinately up-regulated in epithelial cells following LPS stimulation. Of note, miR-23b, miR-27b and miR-24 are from the mir-23b-27b-24-1 gene cluster (9). Pri-miR-23b-27b-24-1 and two corresponding mature miRNAs, miR-23b and miR-27b, were up-regulated in biliary epithelial cells following LPS stimulation. No up-regulation of miR-24 was detected in LPS-treated cells. The mir-17-92 gene can be transcribed to generate pri-miR-17-92, which encodes six miRNAs: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92-1 (23,28,29). Pri-miR-17-92, as well as miR-17, miR-18a and miR-20a, were up-regulated in LPS-stimulated cells. Nevertheless, no up-regulation of miR-19a, miR-19b-1 and miR-92-1 was detected in LPS-treated cells. Moreover, miR-125b-2 may be clustered with let-7c and miR-99a in the c21orf34 transcript (32). Whereas miR-125b was up-regulated by LPS, array analysis failed to detect up-regulation of let-7c and miR-99a in LPS-treated cells. The inconsistence between above pri-miRNAs and their selective corresponding mature miRNAs in LPS-treated cells may be due to the differential posttranscriptional regulation of miRNA molecules or cell type specificity. Indeed, the fold changes for these selected pri-miRNAs were also not consistent with the fold changes for their responsive mature miRNAs at each time point following LPS stimulation. Recent studies suggest that posttranscriptional mechanisms play an important role in the regulation of miRNA expression in various cell types. Several RNA-binding proteins, such as the KH-type splicing regulatory protein (KSRP) and TAR RNA binding protein (TRBP), can interact with Dicer resulting in modulation of miRNA maturation (33,34). The MERK signaling pathway has recently been reported to regulate miRNA biogenesis at the posttranscriptional level through phosphorylation of the human microRNA-generating complex (35).
NF-κB regulation of genes is through binding to κB sites at the promoter elements. p65, c-Rel, and RelB of NF-κB family members contain the transcription activation domain (TAD) and usually activate gene transcription. In contrast, binding of those NF-κB subunits without TAD (i.e. p50 and p52) may actually suppress gene transcription unless they associate with a TAD-containing NF-κB family member or another protein capable of coactivator recruitment (36). Indeed, our ChIP analysis revealed binding of p65 to the promoter elements of those miRNAs that were up-regulated upon LPS stimulation. Regulation of those miRNA genes by NF-κB was experimentally confirmed by promoter reporter assay with a subset of selected miRNAs. Overexpression of p65 also significantly increased the reporter activities in cells transfected with the reporter construct containing the promoter elements of associated miRNA genes. Whether NF-κB, in particular, for those components without the TAD, is involved in LPS-induced down-regulation of these miRNAs is currently under investigation. Transcription of let-7i gene has been reported to be suppressed through promoter binding by NF-κB subunit p50 along with another transcription factor, C/EBPβ (19). In addition, up-regulation of miR-146a and miR-155 appears to be NF-κB-dependent in human THP-1 monocytes in response to LPS stimulation (10,11). Alterations in miR-146a and miR-155 expression were not detected in H69 cells following LPS stimulation, suggesting cell-type specificity of NF-κB-associated miR-146a and miR-155 expression.
Other transcription factors, including AP-1, c-myc and C/EBPα, may be involved in the transcriptional regulation of miRNA genes in epithelial cells following LPS stimulation. Previous studies suggest that c-myc is involved in the transcriptional regulation of mir-17-92 gene in fibroblasts cells (28). How these transcription factors are coordinated to regulate miRNA genes under physiologic and pathological conditions is still unclear. We recently identified a subset of miRNA genes that are transactivated by NF-κB p65 promoter binding in biliary epithelial cells following C. parvum infection (18). Both C. parvum and LPS can activate TLR4/NF-κB signaling in biliary epithelial cells (37). Interestingly, these NF-κB-responsive miRNA genes identified in cells following C. parvum infection, including mir-125b, mir-21, mir-30b and mir-23b-27b-24-1, were confirmed in cells following LPS stimulation. Nevertheless, transactivation of mir-17-92 gene cluster by promoter p65 binding was identified in LPS-treated cells, but not in cells following C. parvum infection (18). One explanation could be the activation of various intracellular signaling pathways resulting in differentially coordinated regulation of miRNA genes by multiple transcription factors in cells following C. parvum infection and LPS stimulation. Transcription of miRNA genes should be a dynamic process in response to the constant changes of intracellular signals. Moreover, the outcome of miRNA expression should reflect a final integrated result of all those related signals on miRNA biogenesis at both transcriptional and posttranscriptional levels.
The TLR/NF-κB signaling is critical to the fine-control of epithelial cell reactions to extracellular stimuli including epithelial innate defense response against microbial infection. The role for miRNAs in TLR/NF-κB signaling-mediated epithelial responses is largely unclear. Our data indicate that a subset of miRNAs is regulated through NF-κB activation and involved in LPS-induced miRNA expression in human biliary epithelial cells. Further studies should test the possibility that transactivation of miRNA genes through promoter binding of NF-κB subunits may be involved in the regulation of TLR/NF-κB signaling-mediated epithelial responses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Supported by National Institutes of Health Grants AI071321 and by the Nebraska Tobacco Settlement Biomedical Research Program (LB692) (to X.-M.C). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jun Liu and Xiaoqing Li for stimulated discussions.
REFERENCES
- 1.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO. J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 4.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol. Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome. Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'C;onnell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 14.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Drescher KM, Chen XM. MicroRNAs and Epithelial Immunity. Int. Rev. Immunol. 2009;28:139–154. doi: 10.1080/08830180902943058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, Chen XM. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 2009;183:1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong AY, Zhou R, Hu G, Li X, Splinter PL, O'H;ara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in interferon-gamma-induced B7-H1 expression in cholangiocytes. J. Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent Transactivation of miRNA Genes Following Cryptosporidium parvum Infection Stimulates Epithelial Cell Immune Responses. PLoS. Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Hara SP, Splinter PS, Gajdos GB, Christy K, Trussoni CK, Chen XM, LaRusso NF. NF&[kappa]B p50-CCAAT-enhancer binding protein beta (C/EBP&[beta])- mediated transcriptional repression of microRNA let-7i following microbial infection. J. Biol. Chem. 2010;285:216–225. doi: 10.1074/jbc.M109.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am. J. Physiol. 1994;266(6 Pt 1):G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 21.Kishore N, Sommers C, Mathialagan S, Guzova J, Yao M, Hauser S, Huynh K, Bonar S, Mielke C, Albee L, et al. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J. Biol. Chem. 2003;278:32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 22.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:8052–8058. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 23.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer. Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 24.Kast C, Wang M, Whiteway M. The ERK/MAPK pathway regulates the activity of the human tissue factor pathway inhibitor-2 promoter. J. Biol. Chem. 2003;278:6787–6794. doi: 10.1074/jbc.M210935200. [DOI] [PubMed] [Google Scholar]
- 25.Musikacharoen T, Matsuguchi T, Kikuchi T, Yoshikai Y. NF-kappa B and STAT5 play important roles in the regulation of mouse Toll-like receptor 2 gene expression. J. Immunol. 2001;166:4516–4524. doi: 10.4049/jimmunol.166.7.4516. [DOI] [PubMed] [Google Scholar]
- 26.Chan C, Li L, McCall CE, Yoza BK. Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB transactivation at the IL-1beta promoter. J. Immunol. 2005;175:461–468. doi: 10.4049/jimmunol.175.1.461. [DOI] [PubMed] [Google Scholar]
- 27.Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-kappa B p65 (RelA) and NF-IL-6. J. Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- 28.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic microRNA cluster by E2F transcription factors. J. Biol. Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 30.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gefen N, Binder V, Zaliova M, Linka Y, Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia. 2010;24:89–96. doi: 10.1038/leu.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Chen XM, O'H;ara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, Limper AH, LaRusso NF. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial responses to Cryptoaporidium parvum via activation of NF-kappaB. J. Immunol. 2005;175:7447–7456. doi: 10.4049/jimmunol.175.11.7447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.