Abstract
Up to 450 000 non-coding RNAs (ncRNAs) have been predicted to be transcribed from the human genome. However, it still has to be elucidated which of these transcripts represent functional ncRNAs. Since all functional ncRNAs in Eukarya form ribonucleo-protein particles (RNPs), we generated specialized cDNA libraries from size-fractionated RNPs and validated the presence of selected ncRNAs within RNPs by glycerol gradient centrifugation. As a proof of concept, we applied the RNP method to human Hela cells or total mouse brain, and subjected cDNA libraries, generated from the two model systems, to deep-sequencing. Bioinformatical analysis of cDNA sequences revealed several hundred ncRNP candidates. Thereby, ncRNAs candidates were mainly located in intergenic as well as intronic regions of the genome, with a significant overrepresentation of intron-derived ncRNA sequences. Additionally, a number of ncRNAs mapped to repetitive sequences. Thus, our RNP approach provides an efficient way to identify new functional small ncRNA candidates, involved in RNP formation.
INTRODUCTION
Two major classes of RNA species have been identified in cells of all organisms: protein-coding RNAs or messenger RNAs (mRNAs), which serve as templates for protein synthesis, and non-protein-coding RNAs (ncRNAs), which are not translated into proteins, but instead, function at the level of the RNA itself. Interestingly, recent reports by the ENCODE project, focusing in high resolution on the analysis of ∼1% of the human genome, have shown that up to 90% of the genome is being transcribed (1), with only a minor portion of RNA transcripts (1.5%) encoding for protein open reading frames. Hence, it was suggested that the remaining 88.5% of RNA transcripts might serve as a source for regulatory ncRNAs. These findings implied the presence of a, so far, ‘hidden layer’ of regulatory elements within the human and other eukaryal genomes, represented by ncRNAs (2), with more than 450.000 ncRNAs genes predicted to be encoded by the human genome (3). However, it has been argued that many of the ncRNA transcripts from the human or other higher eukaryal genomes merely represent spurious non-functional transcription products (4,5). Therefore, identification of the full set of functional ncRNAs, either by in silico or by experimental approaches (or a combination of these), is of fundamental importance, until all functional ncRNAs have been identified within the transcribed, but not translated portions of eukaryal genomes.
In Eukarya, most if not all known ncRNAs are associated with RNA binding proteins thus forming ribonucleo-protein particles or RNPs (6). Numerous ncRNAs serve as so-called ‘guide RNAs’ for these proteins, guiding them to nucleic acids targets (i.e. DNA or RNA), where the proteins subsequently exert their enzymatic activity (7). Prominent examples of these guide RNAs are represented by the classes of miRNAs or snoRNAs (8–10). Therefore, identification of functional, and thus biologically relevant, ncRNAs can be achieved by isolation of ncRNAs binding to proteins, thereby forming so-called ncRNPs.
For ncRNA identification, in the past, isolation of phenol extracted, protein-devoid ncRNA species was followed by size-separation on denaturing gels and cDNA cloning (11–13). Generally, however, this lead to the repeated identification of cDNA clones encoding ribosomal RNAs or other known ncRNA species (14–17). In contrast, co-immunoprecipitation based cDNA library generation, employing an antibody targeting an RNA-binding protein of interest, only allowed identification of ncRNAs associated to this protein (18,19).
By employing a novel cDNA library generation approach from human or mouse cells, based on the size-selection of RNPs on glycerol gradients, we have identified new candidates for functional ncRNAs in Eukarya. Bioinformatical analysis mapped ∼95% of the deep-sequencing reads and identified ∼40% of the clusters as known ncRNAs in both libraries. The remaining 60% of the clusters, corresponding to new unannotated ncRNA candidates were found in intronic, and to a smaller extent, in intergenic regions of the respective genomes, and some of these ncRNA candidates were derived from repetitive elements. We confirmed the presence of selected candidates within RNPs, demonstrating that our RNP selection approach is a powerful tool to identify novel functional ncRNA genes in eukaryal genomes.
MATERIALS AND METHODS
Preparation of protein extracts
HeLa cells were harvested from cell culture media by centrifugation at 700 g for 5 min at 4°C. Pelleted cells were suspended in five volumes of ice-cold Dulbecco’s Phosphate Buffered Saline (PBS) medium (PAA Laboratories, Pasching, Austria) and collected twice by centrifugation as described above. All following steps were performed at 4°C according to the previously described protocol (20). Briefly, cells were suspended in five packed cell pellet volumes of buffer A (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF) and were incubated on ice for 10 min. Subsequently, cells were collected by centrifugation as described above, re-suspended in two packed cell pellet volumes of buffer A and lysed by 10 strokes of a Teflon-glass Dounce homogenizer (Fisher Scientific, Vienna, Austria) or until 80% of the cells were lysed, which was microscopically verified employing Trypan blue. The homogenate was centrifuged for 15 min at 700g to pellet nuclei. The supernatant was mixed with 0.11 volumes of buffer B (300 mM Hepes pH 7.9, 30 mM MgCl2, 1.4 M KCl), and centrifuged for 60 min at 100 000g. The high-speed supernatant was dialysed two times for 2 h against 20 volumes of buffer D (20 mM Hepes pH 7.9, 100 mM KCL, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol) and was assigned as the S100 or cytoplasmatic fraction.
The nuclear extract was prepared as follows: the pellet obtained from low speed centrifugation of the homogenate (see above) was resuspended in 0.5 ml of low salt buffer (20 mM Hepes pH 7.9, 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 25% glycerol). The resulting suspension was stirred gently with a magnetic stirring bar and mixed for 30 min with 0.5 ml of high salt buffer (20 mM Hepes pH 7.9, 1.2 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 25% glycerol), and then centrifuged for 30 min at 9000g at 4°C. The supernatant was dialysed against 50 volumes of buffer D containing additionally 0.2 mM PMSF two times for 2 h. The dialysate was centrifuged at 9000g for 25 min at 4°C and the supernatant was designated as nuclear extract. Aliquots from both S100 and nuclear extracts were snap frozen in liquid nitrogen and stored at −80°C.
Mouse brains were washed three times in buffer A [with 0.5 mM PMSF instead of 0.2 mM and one Complete, Mini, EDTA-free, Protease Inhibitor Cocktail Tablet (Roche, Vienna, Austria) for 7 ml of buffer], and suspended in 2 ml/g of tissue of the same buffer, containing a 10-fold increase in protease inhibitor (Buffer A with 5 mM PMSF and one Complete, Mini, EDTA-free, Protease Inhibitor Cocktail Tablet [Roche, Vienna, Austria) for 1 ml of buffer]. Brains were minced on ice with a scalpel and cells were lysed by five strokes of a Teflon-glass Dounce homogenizer at 4°C. Following steps for isolation of nuclear or cytoplasmatic extracts were identical to the preparation of protein extracts from HeLa cells, except for the composition of buffer B (100 mM Hepes pH 7.9, 30 mM MgCl2, 250 mM KCl).
Sedimentation of RNP extracts on glycerol gradient
Cytoplasmatic, nuclear extracts, or a mixture of both, were mixed (1:1) with gradient dilution buffer (20 mM Hepes, pH 7.9; 100 mM KCl, 1 mM MgCl2) and layered onto a 10–30% glycerol gradient containing 20 mM HEPES pH 7.9, 100 mM KCI and 1 mM MgCl2. Gradients were spun at 100 000g (34 000 rpm) for 18 h at 4°C in a Beckman SW41 rotor and fractionated into 28 samples.
RNA library generation
RNA libraries were generated according to the previously described protocol (13). Briefly, after ultracentrifugation of the gradient, 28 fractions were collected, and pooled four by four, but always excluding the two first (bottom) and the two last (top of the gradient) fractions, which were expected to contain rRNA contaminations and various RNA degradation products, respectively. Samples were twice extracted with phenol–chloroform, ethanol precipitated, and poly(C)-tailed employing poly(A) polymerase from yeast (Epicentre, Madison, WI, USA). C-tailed RNAs were ligated to a 19-nt long 5′ linker (5′-GTC AGC AAT CCC TAA C GAG, with bold representing ribonucleotides) by T4 RNA ligase. RNAs were subsequently converted into cDNAs by reverse transcription (RT) using an anchor primer (5′-AGG AGC CAT CGT ATG TCG GGG GGG GH), amplified by PCR as described, employing complementary primers to 5′ linkers and the poly(C) tail (forward, 5′-GTC AGC AAT CCC TAA CGA G; reverse, 5′-AGG AGC CAT CGT ATG TCG), and cloned into pGEM-T vector (Promega, Mannheim, Germany) for diagnostic Sanger sequencing. Alternatively, samples were directly submitted to pyrosequencing (GS-FLX system, Roche, GATC company, Konstanz, Germany), which required utilization of an alternative pair of primers (forward, 5′-GCC TCC CTC GCG CCA TCA GGT CAG CAA TCC CTAA CGA G; reverse, 5′-GCC TTG CCA GCC CGC TCA GAG GAG CCA TCG TAT GTC G).
Northern blotting
RNA was size fractionated on denaturing polyacrylamide gels (PAGE). For PAGE, 1–30 µg of RNA isolated from tissue, cells or glycerol gradient fractions was denatured for 1 min at 95°C, separated on a 8% denaturing polyacrylamide gel (7 M urea, 1× TBE buffer (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA) and transferred onto a nylon membrane (Hybond N+, Amersham, GE Healthcare, Little Chalfont, UK) using the Bio-Rad semi-dry blotting apparatus (Trans-blot SD; Bio-Rad, Vienna, Austria). Immobilizing of RNAs on membranes was performed using the STRATAGENE UV crosslinker (Stratagene, La Jolla, CA, USA) (120 mJ/cm2). Oligonucleotides (Microsynth, Balgach, Swiss) from 18 to 35 nt in size, complementary to potentially new RNA species, were end-labeled employing [γ-32P]-ATP (Hartmann Analytic, Vienna, Austria) and T4 polynucleotide kinase (Promega, Mannheim, Germany). Depending on the Tm of the respective oligonucleotides, hybridization was carried out from 42 to 58°C in hybridization buffer (178 mM Na2HPO4, NaH2PO4, pH 6.2, 7% SDS) for 12 h. Blots were washed twice, i.e. once at room temperature in 2× SSC buffer, 0.1% SDS for 10 min and subsequently at the respective hybridization temperature in 0.1× SSC, 0.1% SDS for 1–10 min when more stringent washing was required. Membranes were exposed to Kodak MS-1 film from 15 min to 5 days (Kodak, Bagnolet, France).
Bioinformatics
In the first step, the 5′ and 3′ adaptors were removed from the 454 reads. While the 5′ adaptors could be efficiently identified, identification of the 3′ adaptors is complicated by the decreasing quality of the sequences towards the end of the 454 reads. Therefore, we used a customized Gotoh (21) alignment algorithm to identify adaptor sequences. After the adaptor sequences were removed, only reads with a minimum length of 15-nt were used for further computations; All shorter reads were discarded. The sequences were mapped onto the genomes using the program Segemehl (22). Segemehl is a suffix array based program that is especially suited for the mapping of 454 reads, as it takes mismatches and insertions/deletions, which are the most frequent errors observed with 454 sequencing, into account. Because of Segemehl’s memory requirements, genomes were split in four or five parts, containing at least four chromosomes each. The 454 reads were matched onto the respective genomes, and the results were joined so that all mappings covering <90% of the read or with an e-value >5 were discarded. For the remaining reads, we retained the best mapping as well as all suboptimal mappings with at most two additional errors. For comparison with existing annotations, we used data from Ensembl, tRNAdb, UCSC and the ncRNA.org genome browser databases (23–26). To take into account the difference in reliability between the annotation sources, we ranked the annotations, in the order Ensembl ncRNA entries (including entries from miRbase), tRNAdb entries, UCSC entries and ncRNA.org annotations. A read was considered to be annotated if any of the suboptimal mappings could be annotated. Only the annotation from the highest ranked source was kept for each read. However, annotations of the same level were kept, as it is, for example, not possible to distinguish between different loci of the same miRNA (e.g. in miR-9-1, miR-9-2, miR-9-3). Finally, we classified unannotated reads into intergenic, exonic, intronic and intron/exon junction. Protein annotations for human (hg18, NCBI Build 36.1, March 2006) and mouse (mm9, NCBI Build 37, July 2007) were retrieved from the UCSC Genome Browser (http://www.genome.ucsc.edu). We used Ensembl and Refseq gene collections as well as the RepeatMasker tracks. Analysis were in part performed using the Galaxy Browser (http://main.g2.bx.psu.edu/).
We used Blast (27) to find sequence similarities with known ncRNAs (Rfam, ncRNA.org databases). For all hits that remained unannotated, the genomic sequences, including 50-nt up- and down-stream, were used for further prediction using Infernal (28) to match the sequences to all structural alignments of the Rfam (29). Furthermore, RNAz (30) was used to find structurally conserved sequences, and snoReport (31) was used to find candidates for snoRNAs within the unknown hits.
RESULTS
Size-fractionation and cDNA library generation of ncRNPs from HeLa cells
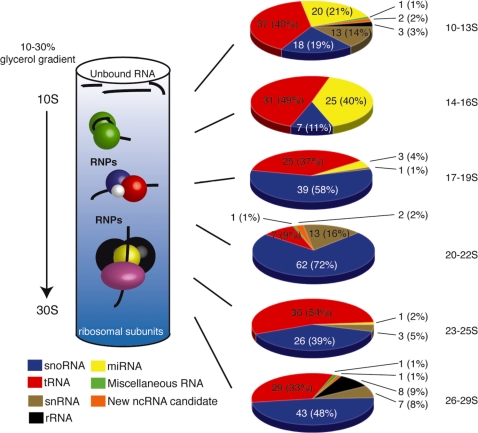
For initial analysis, we combined nuclear and cytoplasmic extracts from HeLa cells and fractionated them by a glycerol gradient in the range of 10–30S. Subsequently, six fractions representing different sedimentation values, i.e. from 10 to 13S, 14 to 16S, 17 to 19S, 20 to 22S, 23 to 25S and 26 to 29S, respectively, were collected from the gradient (Figure 1). After phenol extraction, six cDNA libraries were generated from these fractions as described, encoding ncRNAs derived from RNA–protein complexes (see ‘Materials and Methods’ section). Following cDNA library generation, 96 cDNA clones from each size-fractioned RNP pool (i.e. ∼600 cDNA sequences in total) were analysed by Sanger sequencing and further subjected to bioinformatical analysis (Figure 1).
Figure 1.
Schematic representation of RNP cDNA library generation and analysis. Left: RNP extracts were size-fractionated on glycerol gradients, with separation of RNPs in the size range from ∼10S to 30S. RNPs, consisting of ncRNAs and proteins, penetrate into the gradient, whereas short, non-functional RNA degradation products appear at the top of the gradient and are discarded; ribosomes and ribosomal subunits are pelleted at the bottom of the gradient. Each cDNA library corresponds to a distinct sedimentation range in the gradient (see text). Right: overview of sequence analysis of 96 clones from each cDNA library. From top to bottom, 94, 63, 68, 85, 66 and 89 clones out of 96 were exploitable, respectively. (Miscellaneous RNA: RNAse P RNA, 7SK RNA, Y RNA, 7SL RNA).
This analysis revealed a significant enrichment towards functional ncRNAs species (e.g. miRNAs, snoRNAs, or snRNAs), all of which had been previously reported to be involved in the formation of RNPs. We thereby noted a particularly strong representation of miRNPs at the top of the gradient (i.e. from 10S to 16S, Figure 1). In contrast, snRNAs, tRNAs were found—to a large extent—within all fractions, while snoRNAs were mainly present in the centre and bottom of the gradient (i.e. from 17S to 29S, Figure 1). This might be due to the fact that various classes of snRNAs or snoRNAs, exhibiting different sizes, protein binding partners and assembly states, migrate at different S-values (32). In the case of tRNAs, which exhibit similar sizes, various protein interaction partners, such as aminoacyl-synthetases, elongation factors or processing/modification enzymes (33) are likely responsible for their distribution throughout the gradient (Figure 1).
Notably, we observed an almost complete absence of both RNA degradation products (e.g. mRNA fragments) and highly abundant rRNA species, in contrast to previously reported RNA library generation approaches, derived from size-fractionated, phenol-extracted total RNA (14–17,34). Thereby, the absence of small ribosomal RNAs (5S and 5.8S rRNAs) is likely due to the absence of small and large ribosomal subunits, which sediment at 40S and 60S, respectively, consequently being excluded from cDNA cloning and analysis (see above), which is a highly advantageous outcome.
Importantly, a number of cDNA reads from this analysis corresponded to non-annotated intergenic or intronic regions (designated as new ncRNA candidates, Figure 1), likely representing novel RNP-forming ncRNA candidates. The presence of new and known ncRNA candidates in the library, forming RNPs, confirmed the validity of our approach, i.e. to be able to experimentally identify novel RNA species by RNP fractionation. Since new and known ncRNAs were found in all fractions ranging from 10S to 30S within the gradient, for future analysis, we pooled these fractions and generated RNP libraries from glycerol gradients within this size range.
HeLa and mouse brain cDNA libraries generated from size-fractionated RNPs
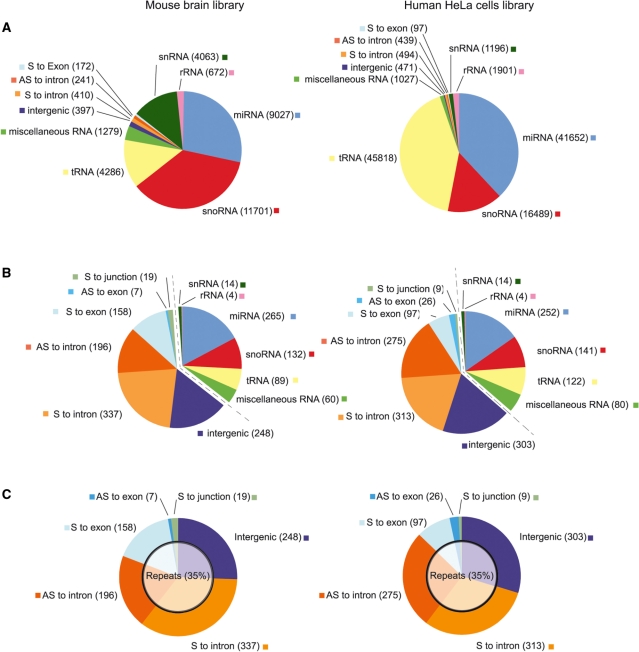
As a proof of concept, in order to compare the small ncRNP transcriptomes from two different cellular systems and organisms, we generated RNP libraries from mouse (brain) cells and human (HeLa) cells by fractionation of pooled nuclear and cytoplasmic cell extracts on 10–30S glycerol gradients, as described above. Subsequently, we subjected the two cDNA libraries to deep-sequencing (35). Raw data were deposited at the short read archive (SRA) with the accession numbers SRA009169.2 and SRA009016.3 for the HeLa and mouse brain libraries, respectively. Obtained sequence reads were bioinformatically analysed (for detailed description, see ‘Materials and Methods’ section). Briefly, after removal of the adaptors sequences, we obtained 166.184 and 56.430 cDNA reads, which were subsequently mapped (see Materials and methods for details), to give rise to two final datasets of 108.518 and 31.882 reads for the HeLa and the mouse brain RNP–cDNA libraries, respectively (Table 1, Figure 2 and Supplementary Data: ‘All mapped reads’ section). Size distribution of the reads in these datasets can be found in Supplementary Figure S1. These final datasets were used for the following statistics.
Table 1.
Distribution of total and unique sequences from human HeLa cells and mouse brain RNP libraries
|
HeLa |
Mouse brain |
|||||
|---|---|---|---|---|---|---|
| Total reads | Unique clusters | Clusters >2 | Total reads | Unique clusters | Clusters >2 | |
| Analysed sequences | 108 518 (100%) | 1636 | 398 | 31 882 (100%) | 1530 | 313 |
| snoRNA | 16 489 (15%) | 141 (9%) | 106 (27%) | 11 701 (37%) | 132 (9%) | 84 (15%) |
| tRNA | 45 818 (42%) | 122 (7%) | 88 (22%) | 4286 (13%) | 89 (6%) | 46 (15%) |
| miRNA | 41 652 (38%) | 252 (15%) | 143 (36%) | 9027 (28%) | 265 (17%) | 106 (15%) |
| rRNA | 1901 (2%) | 4 (<1%) | 4 (1%) | 672 (2%) | 4 (<1%) | 4 (15%) |
| snRNA | 1196 (1%) | 14 (<1%) | 11 (3%) | 4063 (13%) | 14 (<1%) | 14 (15%) |
| Miscellaneous RNA | 1027 (<1%) | 80 (5%) | 29 (7%) | 1279 (4%) | 60 (4%) | 25 (15%) |
| Sense to intron | 494 (<1%) | 313 (19%) | 5 (1%) | 410 (<1%) | 337 (22%) | 10 (3%) |
| Antisense to intron | 439 (<1%) | 275 (16%) | 5 (1%) | 241 (<1%) | 196 (13%) | 7 (2%) |
| Sense to exon | 97 (<1%) | 97 (6%) | 0 | 172 (<1%) | 158 (10%) | 2 (<1%) |
| Antisense to exon | 26 (<1%) | 26 (2%) | 0 | 7 (<1%) | 7 (<1%) | 0 |
| Sense to junction | 9 (<1%) | 9 (<1%) | 0 | 21 (<1%) | 19 (1%) | 0 |
| Antisense to junction | 0 | 0 | 0 | 1 (<1%) | 1 (<1%) | 0 |
| Intergenic | 471 (<1%) | 303 (19%) | 7 (2%) | 397 (1%) | 248 (16%) | 15 (5%) |
After clustering, sequences retrieved from both HeLa and mouse brain libraries were classified into a specific ncRNA family by blasting cDNA sequences against the corresponding genomic nucleotide database (for details see ‘Materials and Methods’ section). Total number of sequences, as well as number of unique sequence clusters, for each ncRNA class, is indicated. The third column in each library indicates the unique sequences represented by at least three cDNA clones. S stands for sense, AS for antisense. Miscellaneous RNA: RNA 7SL, RNA 7SK, piRNAs, vault RNA, RNA Y, RNAse P RNA, RNAse MRP RNA.
Figure 2.
Distribution of cDNA sequences generated from deep sequencing of RNP libraries raised from human HeLa or mouse brain cells. Two RNP libraries, from human HeLa cells and mouse brain, respectively, were subjected to high throughput sequencing. (A) The distribution of the total sequence reads of each ncRNA family (tRNA, miRNA, miscellaneous RNA, rRNA, snRNA, snoRNA and putatively new RNA candidates) is indicated. The numbers indicated correspond to the numbers of total reads retrieved in the respective libraries (see also Table 1). (B) The distribution of unique clusters from each ncRNA class (tRNA, miRNA, miscellaneous RNA, rRNA, snRNA, snoRNA and putatively new RNA candidates) is indicated (see also Table 1). ncRNA candidates are represented to the left of the dashed line. (Miscellaneous RNA: RNAse P RNA, 7SK RNA, Y RNA, 7SL RNA, piRNAs, scRNA, telomerase, RNAse MRP RNA). (C) Distribution of the candidate clusters only. One thousand and twenty-three and 966 candidates were identified in the human and mouse libraries, respectively, and classified with respect to their genomic localization. The inner circle represents the proportion, ∼35%, of the candidate clusters mapping additionally to repeats. AS: antisense; S: sense.
Analysis and comparison of total cDNA reads, i.e. before assembly, showed many similarities, but also several discrepancies between the HeLa and mouse libraries (Figure 2A). From the known ncRNA species, miRNAs and tRNAs were found to be over-represented in the HeLa library compared to the mouse brain library (see below), while snoRNAs and snRNA were found in higher numbers in the mouse brain library (Figure 2A). We subsequently clustered all overlapping cDNA reads into unique sequences, designated as contigs or clusters. Interestingly, distribution of clusters was nearly identical in both libraries (Figure 2B).
Coverage of cDNA libraries and quality assessment
To estimate the ncRNA coverage of the two libraries, we compared all known miRNA to the miRNA clusters identified by the RNP approach. Thereby, we recovered 265 and 252 different miRNAs from the mouse and human genome, respectively, corresponding to 47 and 38% of all known miRNAs in these organisms (Figure 2B). These yields are comparable to the number of miRNAs obtained using the miRDeep program, specifically dedicated to miRNA identification from deep sequencing data (36). Though the number of total miRNA reads was higher in the HeLa than in the mouse brain library (Figure 2A), the overall miRNA transcriptome distribution was rather similar between the two libraries (Figure 2B). We still noticed—as expected—the presence of several brain-specific miRNAs (e.g. miR-9, -124, -127, -128, -132, -153) (37), in the mouse brain library, only. Interestingly, we also observed the presence of few piRNAs (PIWI interacting RNAs), previously only reported to be expressed in germ cells (38). The obtained rare piRNAs clusters were classified with other ncRNA species, i.e. 7SL, 7SK, in the miscellaneous RNA class (Figure 2 and Table 1).
In addition, we recovered 122 different human cellular tRNAs clusters (i.e. 28% of all cellular tRNAs) as well as 89 mouse tRNAs clusters (i.e. 20% of all mouse tRNAs) (Figure 2B). Considering that tRNAs, because of their stable tertiary structure as well as due to nucleotide modifications, are generally highly refractory to RT and hence cDNA analysis (34,39,40), these numbers point to efficient cloning of ncRNAs, in general. The higher number of total tRNA reads retrieved from the HeLa library (42%) versus the mouse brain library (13%, Figure 2A) might be due to the higher rate of cell divisions of HeLa cells, requiring a very efficient translation machinery and therefore, a higher number of tRNAs. Interestingly, among tRNAs from both libraries, we observed numerous partial sequences representing tRNA halves (see Supplementary Data: ‘All mapped reads’ section). These tRNA fragments have previously been reported to be involved in translation regulation in eukaryal organisms such as fungi and humans (14,41–43).
SnRNAs or snoRNA clusters retrieved from each cDNA library show a rather similar distribution with ∼140 different snoRNAs recovered in both libraries (40% of all known snoRNAs; Figure 2B). As a notable difference between HeLa and mouse brain libraries, we observed the presence of two previously identified mouse brain-specific orphan snoRNAs (44), MBII-52 (12% of all mapped reads in the library) and MBII-85 (5% of all mapped reads in the library), respectively. Thereby, MBII-52 and MBII-85 snoRNAs were highly represented in the mouse brain library, corresponding to 33 and 13%, respectively, of all identified snoRNA reads.
A general disadvantage of cDNA expression libraries is that very low abundant ncRNAs or ncRNAs which are expressed in other tissues, during a different developmental stage or upon particular conditions of stress might be missed. Their identification would require the generation of additional libraries for each condition in order to be detected. In addition, some ncRNAs might be involved in RNP complexes larger than 30S or might dissociate from their protein partners during library generation and thus would be missed. Therefore, taking these pitfalls into account, it is clear that the RNP approach, as observed for other ncRNA cloning methods, will likely not reveal all ncRNAs forming RNPs in a tissue or an entire organism. At this point, it is therefore difficult to estimate the complete number of ncRNPs in human HeLa cells or mouse brain cells.
Identification of novel ncRNAs candidates in human and mouse RNP libraries
Any cluster that could not be annotated as a known ncRNA and neither was sequentially or structurally related to known ncRNA families is considered to be a new ncRNA candidate. We identified 1023 such candidates in human and 966 in mouse (Figure 2C).
Intergenic ncRNA candidates
In addition to all known classes of ncRNPs observed in both libraries, ∼25% of the candidate clusters corresponded to non-annotated intergenic regions of the human or mouse genome (Table 1, Figure 2, Supplementary Data: ‘Candidates’ section). In the HeLa library we retrieved a total of 303 intergenic clusters, corresponding to 29% of all candidate clusters (Figure 2C). In the mouse brain library, we retrieved 248 clusters from intergenic regions, corresponding to 25% of the candidate clusters (Figure 2C).
Interestingly, intergenic regions have previously been shown to contain numerous functional ncRNA species within eukaryal genomes (45,46). This strongly suggested that many genes encoding novel functional ncRNAs, forming RNPs, will be located within these intergenic regions as well. By northern blotting, we validated the expression of a selected subset of intergenic ncRNA candidates (Figure 3A). We also analysed the distance between each intergenic ncRNA candidate gene and the closest protein-coding genes (Supplementary Figure S2). In both libraries, the number of intergenic candidates increases towards the proximity of coding genes, with the highest number of clusters found within a 10-kb distance from a coding gene. A higher resolution analysis revealed that a large number of candidates are even found within 1 kb of the closest coding gene (Supplementary Figure S2, see inserts), implying a possible functional relationship. Thereby, some intergenic ncRNA transcripts located in the immediate vicinity of a protein coding region, might be processed from the 5′- or 3′-UTR of the corresponding mRNA.
Figure 3.
Analysis of selected novel ncRNA candidates by northern-blotting. (A) Expression and sizes of a subset of selected candidates was analysed by northern-blotting. Sizes of ncRNAs, as deduced by northern blotting, appeared in several cases larger than the corresponding cDNAs sizes (see Supplementary Table S1), reflecting that some sequenced clones might correspond to fragments of larger new ncRNA transcripts. hs: human candidate; mm: mouse candidate; AS: antisense; S: sense; SR: sense to repeats; ASR: antisense to repeat. (B) Predicted secondary-structures of putatively novel ncRNA candidates. Structures were predicted using the program RNAfold from the Vienna package (69). When the reads were too short to be folded, 50-nt up- and down-stream of the read were added to the sequence and locally stable structures containing the read were looked for. The corresponding structures were then folded, the grey backbone representing the read itself. The 0–1 color scale represents the base-pair probabilities.
Intronic ncRNA candidates: sense and antisense orientation
In both libraries, ∼55% of cDNA clusters mapped to intronic regions, in either sense or antisense orientation (Table 1, Figure 2, Supplementary Data: ‘Candidates’ and ‘Antisense to intron’ sections). In the HeLa library we retrieved a total of 588 intronic clusters, corresponding to 56% of the candidate clusters (Figure 2C). In the mouse brain library, we retrieved 533 clusters from intronic regions, corresponding to 54% of the candidate clusters (Figure 2C). Notably, intronic reads were significantly over-represented compared to what would be expected from random genome transcription (47) of the total genome. This is consistent with the observation that previously identified functional ncRNAs, such as miRNAs or snoRNAs, have been reported to be encoded by intronic portions of the genome (2,48).
Interestingly, in both HeLa and mouse brain cells libraries, about half of the intronic candidates mapped in antisense orientation to their ‘host’ pre-mRNAs (Table 1, Figure 2C, Supplementary Data: ‘Antisense to intron’ section). In contrast this was observed only for few exon-encoded candidates (see below), and only one exon/intron junction-encoded new ncRNA candidate (Table 1, Figure 2, Supplementary Data: ‘Candidates’ and ‘Antisense to exon or junction’ sections). At this point, we cannot completely exclude the possibility that some of the intron-derived ncRNA candidates reflect RNA degradation products or splicing intermediates. However, expression and distinct sizes of a subset of intronic ncRNA candidates could be verified by northern blotting (Figure 3A).
Alternatively to being splicing intermediates, at least some of these intronic ncRNA candidates, especially those in antisense orientation, might be involved in splicing regulation of their complementary host genes by an antisense-like mechanism, as previously reported for artificial intronic antisense RNAs (49). In that respect it is noteworthy that one ncRNA candidate mapped exactly to an intron/exon junction in antisense orientation in the mouse library (Figure 3A, Supplementary Data: ‘Antisense to exon or junction’ section).
Therefore, we analysed the distance between the intronic candidates and the closest splice site of the host gene (Supplementary Figure S3A). In both HeLa and mouse brain datasets, the number of intronic candidates mapping in antisense orientation increased towards the vicinity of splice sites (Supplementary Figure S3B). Thereby, the highest number of these candidates was found within 1 kb of a splice site. By a higher resolution analysis, the proportion of antisense candidates, located in very close proximity to a splice site (i.e. within 250 nt) is more pronounced in the HeLa library compared to the mouse brain library (Supplementary Figure S3B, see inserts). Although most of the host mRNAs correspond to alternatively spliced transcripts, so far no intronic antisense ncRNA could be identified that mapped in the vicinity of a known alternative splice site in the EBI database (50). Future experimental analysis will have to reveal which of the intron-derived antisense ncRNA candidates are directly involved in splicing regulation of ‘host genes’ and which represent independent ncRNA genes.
ncRNAs located in exonic regions of mRNAs
To a significantly lesser extent, some ncRNA clusters could also be mapped to exonic regions of protein-coding genes (Table 1, Figure 2, Supplementary Data: ‘Candidates’ and ‘Antisense to exon or junction’ section). We retrieved a total of 97 clusters, corresponding to 9% of the candidate clusters in the HeLa library and 158 clusters corresponding to 16% of the clusters in the mouse brain library, mapped in sense orientation to their ‘host’ pre-mRNAs, suggesting that they might derive from mRNA degradation (Figure 2C); alternatively, they might represent new ncRNA species processed from exons, as previously reported for some viral exons (51,52).
Additionally, 26 (2%) and 7 (0.7%) of the clusters mapped in antisense orientation to exonic regions in the HeLa and mouse libraries, respectively (Figure 2C, Supplementary Data: ‘Antisense to exon or junction’ section). Some of these candidates are expressed and exhibit a defined size (Figure 3A), arguing against mRNA degradation products. Thus, exon-derived antisense ncRNAs, might be involved in regulation of gene expression, as previously observed for several antisense transcripts in Eukarya and Bacteria (53–57).
ncRNAs located in repetitive elements
A few master genes provide the RNA templates for repetitive elements (58) and mobile elements can serve as sources for ncRNAs, often by co-transcription if fortuitously located in transcription units in either orientation. Alus, for instance, are derived from 7SL RNA (59), whereas several miRNAs were predicted to originate from retroposons as LINEs and SINEs (60). In both our libraries, ∼35% of the candidate clusters mapped to repetitive element regions of the genome (Figure 2C and Supplementary Data: ‘Candidates’ section). Among those, 25% of the candidate clusters reside in retrotransposons, which exhibit RNA intermediates during retro-transposition; about half of these sequence clusters are transcribed in sense orientation to their respective retrotransposons. Taken into consideration that repeats are indeed actively transcribed, this would explain the transcription of a fraction of the total set of ncRNA candidates derived from repeats. Besides, we also find a small fraction of intronic reads that are antisense to both introns and repeats as well as few intergenic reads that are antisense to repeats (Supplementary Data: ‘Candidates’ and ‘Antisense to introns’ sections).
Next, we further characterized the identified repeats by comparing them to their consensus sequence. The similarity between repeat and consensus was used to estimate the activity of the repeats in transposition. We found that the majority of repeats are truncated and had accumulated numerous mutations. As a consequence, 75% of repeat associated reads, map less than six times to the genome, and 50% even map only once (data not shown). Most of these fragmented repeats map to the terminal regions of the full-length consensus sequences. We also analyzed the frequencies of the individual repeat classes and families, and compared them to the genomic background (intergenic or introns). Intergenic reads are dominated by LTRs and LINEs, intronic ones by SINEs. This constitutes an interesting observation, as LTRs and LINEs carry their own promoters, potentially enabling their autonomous transcription, while SINEs require an external promoter. However, reads that match LTRs are rather transcribed in antisense orientation relative to the repeat, whereas those in SINEs and LINEs are rather found in the same orientation. Therefore, the fraction of reads in sense orientation to LINEs is probably amongst the most likely candidates for novel ncRNAs. The terminal (5′) regions of the repeats where these reads reside contain promoters and thus provide sites of transcriptional regulation. However, in general, the repeat sequences themselves diverged from consensus sequences such that they most likely lost their original identity and function. It is tempting to speculate that these reads might be evolving new ncRNAs putting ‘evolution in progress’ on display.
Novel ncRNA classes and conservation of ncRNAs
Thus far, we have only identified few human ncRNA candidates having homologs in the mouse brain library and vice versa. Blasting human reads against mouse reads with an e-value of 1e−3 resulted in 17 human clusters matching 12 mouse clusters for the intergenic and 16 human clusters matching to 27 mouse clusters for intronic hits (Supplementary Data: ‘Candidates’ section). Moreover, except for three pairs, reads in mouse and human did not map to loci that are annotated as syntenic in the human.net track of the UCSC genome browser for the mouse genome. The lack of homologous human and mouse sequences suggests that the majority of new ncRNA candidates might correspond to tissue- or cell-type-specifically expressed RNA species.
Nevertheless, all ncRNA candidates tend to map to more highly conserved regions of the genome. To that aim, we looked at the vertebrate PhastCons (61) conservation track of the UCSC genome browser (44 vertebrates for human, 30 for mouse track), and compared the mean base-wise conservation of all sequence reads which mapped to introns, exons and intergenic regions with the mean of the total genomes. In human, exonic ncRNA candidates appear to be more highly conserved, with a mean PhastCons score of 0.539 compared to the genomic background of 0.428, the increase for intergenic (0.097 versus 0.090) and intronic (0.116 versus 0.084) ncRNA candidates was much smaller, however. For mouse ncRNA candidates, the effect is even stronger, with 0.602 versus 0.468 for exonic ncRNAs, 0.167 versus 0.113 for introns and 0.165 versus 0.102 for intergenic ncRNAs.
Nevertheless, we were unable yet to group these new ncRNA candidates into any novel RNA families, based on primary- or secondary-structure conservation. However, this might be a challenging task at this point, considering that small degenerate sequence motifs combined with conserved secondary structures are sufficient to define a novel class of ncRNA species, as exemplified by snoRNAs (32).
Selected ncRNA candidates from our analysis form RNPs
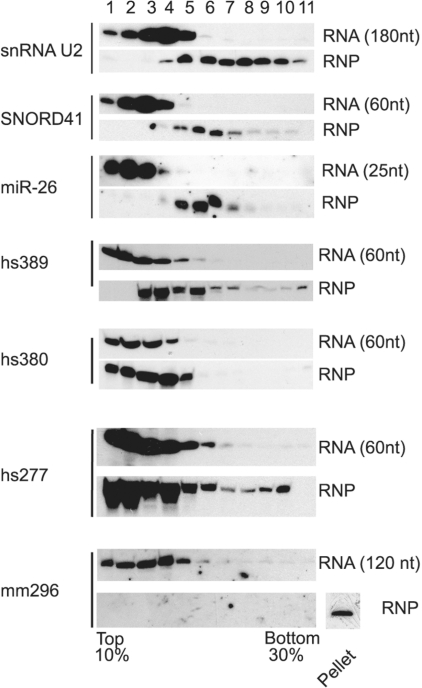
By northern blotting, we verified expression and sizes of selected candidates (Figure 3A). Sizes of ncRNA candidates, as estimated by northern blotting, appeared in several cases larger than sizes deduced from cDNA sequencing (see Supplementary Table S1), reflecting that some sequence reads might correspond to fragments of larger new ncRNA transcripts. Though all ncRNA molecules present in the libraries are likely derived from RNP particles, in agreement with our RNP selection procedure (see above), we also verified interaction of some selected ncRNAs candidates with proteins. To that end, total cellular protein extract or phenol-extracted total RNA were fractionated on 10–30S glycerol gradients, and the sedimentation profile of ncRNA was analyzed by northern-blotting employing specific oligonucleotide probes directed against ncRNA candidates (Figure 4). For the ncRNA candidates tested, we observed that phenol extracted RNA samples did not penetrate into the gradient (as deduced by northern blotting), while the sedimentation profile was shifted to higher S-values when a cellular RNA/protein extract was used. These observations are consistent with novel ncRNA candidates being involved in RNP formation and thus likely exerting biologically relevant functions, as observed for other known ncRNAs forming functional RNPs. In addition, in silico predictions revealed that novel ncRNA candidates could fold into stable secondary structures, supporting that they correspond to stable and functional RNA species (Figure 3B). Surprisingly, for ncRNAs mapping to repetitive elements we could also detect their presence within protein complexes of higher molecular weight (Figure 4, hs277 and mm296). In particular, clone mm296, a mouse candidate mapping to a LINE element, might be associated with an RNP particle of considerable size, as it was predominantly observed in the gradient pellet, comprised of RNPs larger than 30S. Since clone mm296 was originally isolated from the 10–30S fraction, it might be enriched in the pellet by binding to a larger RNP or protein complex. A potential function of mm296 in regulation of translation initiation, e.g. by binding to the ribosome or ribosomal subunits could be envisioned, as previously described for the BC1/BC200 ncRNAs, which are acting through interaction with poly(A)-binding protein, PABP (62).
Figure 4.
Selected novel ncRNA candidates are present as RNPs in cell extracts. From four novel ncRNA candidates, sedimentation values were determined by glycerol gradient centrifugation either employing phenol extracted RNA (upper panels) or total cellular extracts, containing RNPs (lower panels). Sedimentation profiles of the candidates were analyzed by northern blotting of respective fractions and hybridization employing radioactively labeled DNA oligonucleotide complementary to the ncRNA candidate. As controls, the sedimentation profiles of three ncRNAs known to be part of RNPs were also analysed. The pellet fraction was tested when there was one.
DISCUSSION
The total number of ncRNAs in the human or mouse genome remains controversial, with estimates ranging from a few hundred to hundreds of thousand ncRNA species (2,3). Hence, to separate the ‘transcriptional noise’ from biologically functional ncRNAs, novel methods for their identification have to be applied. In the past, deep-sequencing of cDNA libraries, generated from phenol extracted and size separated ncRNA species, resulted in the identification of novel ncRNA species, but also in a large number of ncRNA transcripts of unknown biological significance. Thus, to identify new functional ncRNAs in human and mouse genomes, we have established a novel selection procedure based on the isolation of ncRNAs forming RNP complexes. Through this approach, we were able to (i) identify several hundred new ncRNAs that are likely components of ribonucleo-protein complexes and (ii) identify the majority of all known ncRNAs forming RNPs, validating the RNP method.
Thus, by RNP selection we significantly increased the probability of the presence of biologically relevant ncRNA species in our data set. This is extremely important since the functional analysis even of single ncRNA species is a highly time-consuming and laborious effort (63). It is important to note, however, that in this study we have only focused on the small ncRNA/ncRNP transcriptome in two defined systems, human HeLa cells and mouse brain cells. In order to obtain a complete picture of both human and mouse RNP transcriptomes, additional tissues and conditions (such as different developmental conditions or stress) will have to be investigated. Moreover, by selecting RNPs in the size range of 10–30S, we might have missed ncRNAs forming larger RNP complexes or ncRNAs dissociating from proteins during the isolation procedure. Furthermore, longer ncRNA species (e.g. in antisense orientation to protein coding genes), excluded from our study, which might function even in the absence of protein binding partners, could represent yet another large class of unidentified ncRNA species (64).
Still, by our RNP selection approach, we have identified numerous new ncRNAs candidates, encoded in intronic or intergenic regions with a significant number of repeats-derived ncRNA candidates within the mouse or human genomes. Thus far, we were unable to computationally identify new families or classes of ncRNAs, based on shared sequence or structural motifs. However, taken into account the limited conserved features of some existing classes of ncRNAs, such as miRNAs or snoRNAs (32), the establishment of novel ncRNA classes based on our sequences and other known RNA candidates will warrant further efforts. Alternatively to forming novel ncRNA classes, ncRNA species identified in our study might themselves correspond to tissue- or cell-type-specific RNA species, as was shown for antisense or mRNA-like ncRNA transcripts (65–67). To unequivocally identify functions of all newly identified ncRNAs, high-throughput knock-down strategies, determination of ncRNA targets or protein binding partners will be required for future analysis (68).
ACCESSION NUMBERS
Raw data were deposited at the SRA with the Accession Numbers SRA009169.2 and SRA009016.3 for the HeLa and mouse brain libraries, respectively. Annotations for human (hg18, NCBI Build 36.1, March 2006) and mouse (mm9, NCBI Build 37, July 2007) were retrieved from the UCSC Genome Browser (http://www.genome.ucsc.edu). Complete annotation of the data can be found as Supplementary Data on our website under the link http://www.tbi.univie.ac.at/papers/SUPPLEMENTS/454/Identification.html.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical University of Innsbruck and the Austrian Genome Research (GEN-AU, http://www.gen-au.at/) funding program (Grants D-110420-011-013 and D-11420-011-015 to A.H.). Funding for open access charge: GEN-AU grant D-11420-011-015.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jürgen Brosius, Bénédicte Sohm and Andreas Ploner for critically reading the manuscript and Jonathan Vosper for careful proofreading.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14(Spec No 1):R121–132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 3.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J. Waste not, want not–transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Eddy SR. Non-coding RNA genes and the modern RNA world. Nature Rev. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 7.Huttenhofer A, Schattner P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nature Rev. 2006;7:475–482. doi: 10.1038/nrg1855. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 11.Huttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 12.Huttenhofer A, Cavaille J, Bachellerie JP. Experimental RNomics: a global approach to identifying small nuclear RNAs and their targets in different model organisms. Methods Mol. Biol. 2004;265:409–428. doi: 10.1385/1-59259-775-0:409. [DOI] [PubMed] [Google Scholar]
- 13.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madej MJ, Alfonzo JD, Huttenhofer A. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 2007;35:1544–1554. doi: 10.1093/nar/gkm004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genetics. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–D92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 32.Reichow SL, Hamma T, Ferre-D'A;mare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryckelynck M, Giege R, Frugier M. tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations. Biochimie. 2005;87:835–845. doi: 10.1016/j.biochi.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Huttenhofer A, Kiefmann M, Meier-Ewert S, O'B;rien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 37.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 38.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 39.Verma IM, Baltimore D. Purification of the RNA-directed DNA polymerase from avian myeloblastosis virus and its assay with polynucleotide templates. Methods Enzymol. 1974;29:125–130. doi: 10.1016/0076-6879(74)29015-3. [DOI] [PubMed] [Google Scholar]
- 40.Wittig B, Wittig S. Reverse transcription of tRNA. Nucleic Acids Res. 1978;5:1165–1178. doi: 10.1093/nar/5.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 42.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Liu M, Zhang L, Zhang W, Gao G, Zhu Z, Wei L, Fan Q, Long M. Detection of intergenic non-coding RNAs expressed in the main developmental stages in Drosophila melanogaster. Nucleic Acids Res. 2009;37:4308–4314. doi: 10.1093/nar/gkp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 47.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 48.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 49.Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum. Mol. Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 50.Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullen BR. Viral RNAs: lessons from the enemy. Cell. 2009;136:592–597. doi: 10.1016/j.cell.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 54.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, Kenny P.J., 3rd, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's; disease and drives rapid feed-forward regulation of beta-secretase. Nature Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheele C, Petrovic N, Faghihi MA, Lassmann T, Fredriksson K, Rooyackers O, Wahlestedt C, Good L, Timmons JA. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics. 2007;8:74. doi: 10.1186/1471-2164-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 58.Kim J, Martignetti JA, Shen MR, Brosius J, Deininger P. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc. Natl Acad. Sci. USA. 1994;91:3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res. 1992;20:3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 61.Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Huttenhofer A, Tiedge H, et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 63.Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genetics. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Rev. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 65.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 66.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 67.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature Rev. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 68.Ploner A, Ploner C, Lukasser M, Niederegger H, Huttenhofer A. Methodological obstacles in knocking down small noncoding RNAs. RNA. 2009;15:1797–1804. doi: 10.1261/rna.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.