Abstract
In bacteria, gene regulation is one of the fundamental characteristics of survival, colonization and pathogenesis. Operons play a key role in regulating expression of diverse genes involved in metabolism and virulence. However, operon structures in pathogenic bacteria have been determined only by in silico approaches that are dependent on factors such as intergenic distances and terminator/promoter sequences. Knowledge of operon structures is crucial to fully understand the pathophysiology of infections. Presently, transcriptome data obtained from growth curves in a defined medium were used to predict operons in Staphylococcus aureus. This unbiased approach and the use of five highly reproducible biological replicates resulted in 93.5% significantly regulated genes. These data, combined with Pearson’s correlation coefficients of the transcriptional profiles, enabled us to accurately compile 93% of the genome in operon structures. A total of 1640 genes of different functional classes were identified in operons. Interestingly, we found several operons containing virulence genes and showed synergistic effects for two complement convertase inhibitors transcribed in one operon. This is the first experimental approach to fully identify operon structures in S. aureus. It forms the basis for further in vitro regulation studies that will profoundly advance the understanding of bacterial pathophysiology in vivo.
INTRODUCTION
Staphylococcus aureus is the major cause of intravascular and systemic infections such as bacteremia, endocarditis and sepsis (1,2). Nonetheless, knowledge of the regulation of responses of S. aureus to growth in vitro and upon interaction with the human host in vivo is limited (3). Prokaryotic gene expression is tightly regulated under different conditions, depending on cell density (quorum sensing), energy availability and environmental signals (4,5).
Microbial growth under laboratory conditions can be divided in three phases: (i) lag phase, when nutrients are abundant and cell density is low; (ii) log phase, when cells grow exponentially; and (iii) stationary phase, when nutrients are scarce or absent and cell density is high. In general, in log phase many ribosomal proteins are abundantly expressed, while in stationary phase stress response genes and quorum sensing genes are up-regulated (6–9). The expression of genes related to virulence is of special interest in the interaction with the host. Virulence gene expression in many pathogens including S. aureus and Group A Streptococci is required to evade the innate immune system and establish microbial survival in the host (10,11). Usually, virulence genes encoding surface proteins are up-regulated during log phase, while toxins are up-regulated during stationary phase (12).
An operon is a series of genes transcribed as a single mRNA, mostly identified by short intergenic distances and the presence of a single promoter in front of the first gene and a terminator at the end, but more complex structures have been described (13–18). Several theories have been postulated to explain the formation of operons. Firstly, genes transcribed in an operon are usually functionally related and are often involved in the same metabolic pathway (16). Secondly, operons ensure cotransfer of genes to other genomes via horizontal transfer, thereby increasing fitness and preservation of constituent genes (19). Operons have an important role in regulated gene expression and an estimated 50% of the genes in prokaryotes are part of an operon (16). However, hardly any operon structures have been experimentally identified for important pathogenic Gram-positive bacteria. In addition, the role of operons in the regulation of virulence genes is hardly known. Operon predictions have mainly been based on the Escherichia coli genome and, to lesser extent, the Bacillus subtilis genome. These predictions take into account intergenic distances, conservation of gene clusters, functional relations and the limited available experimental evidence (13,20). For S. aureus, mainly in silico operon predictions are available based on the intergenic distances, conserved gene clusters and, to a lesser extent, rho-independent terminators and the few experimentally validated operons (13,18). Co-expression patterns from microarray experiments and high-density oligonucleotide probe arrays in combination with in silico predictions have already been successfully used as an operon prediction tool in E. coli and are considered as accurate (21,22).
Another important feature for understanding gene regulation is the presence or absence of 5′- and 3′-untranslated regions (UTRs). Prokaryotic 5′-UTRs can have important regulatory functions, since riboswitches, which are known to regulate metabolic pathways, are located within the 5′-UTR and many trans-encoded small non-coding RNAs (sRNAs) bind to the 5′-UTR to regulate translation and/or stability of the mRNA (23–25). The 3′-UTRs have a stabilizing effect and prolong the half-life of the mRNA transcript (23,24).
In the present study, we performed a growth-dependent RNA expression analysis of the highly virulent, community-acquired methicillin-susceptible S. aureus strain MSSA476 to determine operon structures in the staphylococcal genome. We found 62% of the genes located within an operon. Data were compared to and combined with an online in silico prediction method, which is to our knowledge the most complete available operon prediction for S. aureus, as well as to a computational operon prediction (18).
MATERIALS AND METHODS
Bacterial strain
The sequenced, highly virulent, community-acquired methicillin-susceptible S. aureus strain MSSA476 (26) was used.
Growth conditions
MSSA476 was grown overnight in Iscove’s Modified Dulbecco’s Medium (IMDM) (Invitrogen, Carlsbad, CA, USA). These overnight cultures were diluted (1:7) in fresh prewarmed IMDM and grown twice to mid-log phase culture (A660nm ∼0.5) prior to the growth experiment. The second mid-log phase culture was diluted to an A660nm of 0.3 with prewarmed IMDM and directly transferred to fresh prewarmed IMDM to obtain an A660nm of 0.03. Samples were taken at 1, 2, 3, 4, 5, 6 and 9 h post-inoculation. The A660nm was measured and dilutions were plated on sheep blood–agar plates to determine colony forming units (CFUs). Cultures were incubated at 37°C and 180 rpm.
RNA extraction
RNA extraction was performed at room temperature unless stated otherwise. RNA was purified using the NucleoSpin RNA II total RNA isolation kit (Macherey-Nagel, Düren, Germany) according to manufacturer’s protocol with some adjustments. Bacteria were spun for 30 s at 13000 r.p.m., immediately resuspended in 350 µl RA1 buffer supplemented with 3.5 µl β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA) and vortexed vigorously. Resuspended bacteria were added to 0.5 ml of 0.1 mm silica beads (Merlin, Breda, The Netherlands) and disrupted using a mini-Beadbeater (BioSpec Products, Bartlesville, OK, USA) for 30 s at 5000 r.p.m. Resulting samples were frozen at −80°C overnight. The samples were thawed slowly and purified. Total RNA was eluted in 60 µl RNase-free MilliQ water. RNA yield was measured using a NanoDrop ND-1000 (Nanodrop Technologies, Wilmington, DE, USA) and quality was measured using a 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA). Both the RNA integrity number (RIN) and the presence of degradation products were checked.
Microarray design
A whole genome Agilent microarray (8 × 15k) was designed using the MSSA476 sequence with the Agilent Technologies eArray microarray design software (https://earray.chem.agilent.com/earray/). The complete design was performed in a two-step procedure. First, 60-mer probes were designed to target all protein-coding genes, as well as rRNA, tRNA, sRNA of MSSA476, and the naturally occurring plasmid pSAS. Probes were mainly designed at the 3′-end of the genes. One probe per target was designed and tested for cross-hybridization. Intergenic regions were defined as non-coding regions between adjacent genes irrespective of their orientation with no gene present on the opposite strand. Probes were designed to cover the complete, specified intergenic regions on both strands with at least one probe per hundred nucleotides, where possible. Secondly, probes were validated. BLAST was used to exclude from the analysis all probes which, besides the target, matched the genome over a length of 20 nt or more. Furthermore, all probes that did not give a signal in a comparative genomic hybridization experiment using the same array design were excluded from the analysis.
Labeling, hybridization and scanning
Total RNA was labeled in a one-step labeling with fluorescent dyes by direct labeling. A total of 10 µg RNA was randomly primed with Superscript II reverse transcriptase (Invitrogen) and random hexamers (12.5 ng/µl) in a total volume of 30 µl for 2 h at 42°C with the incorporation of Cy5- or Cy3-dUTP (Agilent Technologies) with a ratio dUTP/dTTP of 3/1, yielding ∼4 µg labeled cDNA. RNA template was removed by hydrolysis with 3 µl 2.5 M NaOH (Sigma-Aldrich) for 15 min at 70°C. Hydrolysis was stopped by neutralization with 15 µl 2 M MOPS (Sigma-Aldrich) and put on ice. Labeled cDNA was purified using Qia-quick polymerase chain reaction (PCR) purification kit (Qiagen, Valencia, CA, USA). Incorporation of Cy3 or Cy5 was determined using a NanoDrop ND-1000 apparatus.
The common reference was created by pooling 400 ng Cy5-labeled cDNA sample of each time point of each growth curve, 35 points in total. Labeled cDNA was hybridized according to the manufacturer’s protocol (Agilent Technologies). A total of 300 ng Cy3-labeled cDNA and 300 ng Cy5-labeled common reference was mixed and 10 × blocking agent was added to a total volume of 25 µl. The mixture was heated to 95°C for 3 min, followed by addition of 2 × hybridization buffer to a volume of 50 µl. A total of 40 µl was loaded onto an 8 × 15k array and hybridized for 18 h at 65°C and 20 r.p.m. in a dedicated hybridization oven (Agilent Technologies).
After the hybridization, the arrays were washed in buffer 1 for 1 min at room temperature, 5 min in wash buffer 1 at room temperature and, finally, 1 min in wash buffer 2 at 37°C (Agilent Technologies). Slides were spun for 3 min at 300 r.p.m. to dry and were scanned immediately. Data was extracted and processed using Feature Extraction™ version 9.5.1 software (Agilent Technologies). Median spot intensities were determined using the same software.
Data analysis and statistical analyses
Processing of the data was performed using R (version 2.7.0) and the Bioconductor MAANOVA package (version 1.10.0). All slides were subjected to a set of quality control checks, which consisted of visual inspection of the scans, examination of the consistency among the replicated samples by principal component analysis (PCA), testing against criteria for signal to noise ratios, testing for consistent performance of the labelling dyes and visual inspection of pre- and post-normalized data with box and ratio-intensity plots. When the data was checked for effects of (random) experimental factors, slide and sample effects were observed. Slide effects were detected because eight arrays were printed on one glass slide and sample effects occurred as a consequence of the repeated measure design. After log2 transformation, the data were normalized by a LOWESS smoothing procedure to correct for dye bias effects. The resulting data were analyzed using a two-stage mixed ANOVA model (27,28). The gene specific model included terms for Array, Slide and Sample effects (random), and Time and Reference (fixed). Genes that were differentially expressed between any of the time points were identified by a permutation test. Resulting P-values were corrected for multiple testing by calculating the false discovery rate (FDR) (29). The significance threshold was set at 0.05 FDR.
Operon prediction
Correlation coefficients of transcriptional profiles were determined for all adjacent probes, including the profiles of probes in intergenic regions. Pearson’s correlation coefficients of the transcriptional profiles for all adjacent probes were calculated over all time points and replicas using the following formula:
A correlation coefficient of ≥0.80 was used for the prediction. Operons were determined by correlating the time-dependent transcriptional profiles of adjacent probes with all five replicas included. Correlation was thus calculated over 35 points and visualized. The distribution of non-correlated probes was determined for probes that were either 50 probes separated from each other or for probes that were adjacent to each other but on opposite strands. Both analyses resulted in similar distributions. Based on this distribution, 6.2% of the probes would indicate false positive operon predictions at a correlation coefficient of 0.80. Predicted operons were compared to an online in silico prediction (http://bioinformatics.biol.rug.nl/websoftware/operon/operon_start.php) and to a computational analysis based on strain Mu50 (18). These predictions are based on intergenic distances and rho-independent terminators or gene orientation, intergenic distances, conserved gene clusters, terminators and the confidence score of adjacent genes to be in an operon, respectively.
PCR
A subset of predicted operons was validated using reverse transcription (RT)-PCR. Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen) was used in combination with specific primers (listed in Table 1) to reverse transcribe RNA. Complimentary DNA (cDNA) synthesis was performed with 1 μg total RNA according to manufacturer’s protocol for specific primers. The reaction was incubated at 55°C for 50 min and stopped by incubating at 85°C for 5 min. RNA was removed by adding 2 U RNase H and incubation at 37°C for 20 min. The cDNA products were subsequently detected by PCR using primers listed in Table 1. PCRs were carried out in 25 µl reactions and consisted of 1 µl cDNA, 0.25 µM of each primer, 1 × Phusion HF buffer, 0.5 mM MgCl2, 2 mM dNTPs and 0.2 U Phusion high-fidelity polymerase (Bioké, Leiden, The Netherlands). Amplification was performed with an initial denaturation of 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 20 s and extension at 72°C for 1 min 30 s, followed by a final extension at 72°C for 5 min. Resulting DNA fragments were separated on a 0.8% agarose gel in TBE buffer with a 1 kb marker (Invitrogen) and visualized with ethidium bromide staining.
Table 1.
Primers used in this study
| Primer location | Name | Sequence |
|---|---|---|
| SAS0670–SAS0673 | 0673_F | AGTTGGTGCTGTTGCCTCT |
| 0670_R | TTGTTGCGCGAGTTCATTAG | |
| SAS1172–SAS1175 | 1175_R | TTGGTGTGTGTAATGGGAATG |
| 1172_F | TTCGTTTAACACGTTTAGGTTCAA | |
| SAS1431–SAS1435 | 1435_F | ACGCAATACGAGGTAGATATTA |
| 1431_R | GTTCTGGTGCAATGCCTGTA | |
| SAS0057–SAS0058 | 0058_R | CTTCTACGTTCTTTGGCCTGA |
| 0057_F | TGGGTTGTCAACGTACAGGA | |
| 0056_F | TAGCCAAGCAAGGGCAATTA | |
| SAS1765 | 1763_R | TTTTATCTGTAAACTGACCCTTGTC |
| 1765_R | TGGTCGAATGTTCCATAATCG | |
| 1765_F | TTCATTGTTCGGATTTACATTTAG | |
| SAS1091–SAS1092 | 1091_F | CGAAGGATACGGTCCAAGAG |
| 1092_R | GCATCAGCCATTGATACGAA | |
| SAS1739–SAS1746 | 1739_RV | ACCACGAATGATCTCCAAGC |
| 1740_FW | AATCCACATCCGGTTAATGC | |
| 1740_RV | GCATTAACCGGATGTGGATT | |
| 1741_F | CAAGTTAATAAATCAAAGGAGTT | |
| 1741_RV | AACTCCTTTGATTTATTAACTTG | |
| 1742_FW | TGTGATGAAAAACCATGACGA | |
| 1742_RV | TCGTCATGGTTTTTCATCACA | |
| 1743_FW | TGTAATCGCGTCAACAAACG | |
| 1743_RV | TCAAATGAATTCCAGAACTTTATA | |
| 1744_FW | TTTGTTTGGTGGACTTTCAGG | |
| 1744_RV | TTGCGATGCTAAATCCATTG | |
| 1745_FW1 | CCGAAATCGAAATTCCAAAA | |
| 1745_RV1 | TTCGGGTCCTCGATAAGATG | |
| 1745_FW2 | AACAGGTTTCGGGACAACAA | |
| 1745_RV2 | AATTGTTGTCCCGAAACCTG | |
| 1745_RV3 | GGATTGATTCTTTCATCTGAGCA | |
| 1746_FW | TGATTCAGCAGGTGACGAAC |
Alternative pathway (AP) hemolytic assay
Preparation of recombinant extracellular fibrinogen binding protein (Efb) and staphylococcal complement inhibitor (SCIN)-B was previously described (30). AP hemolytic assays were performed as described earlier (31). Briefly, 10% human serum was pre-incubated with Efb or SCIN-B (both at 0.6 or 1 μg/ml) alone or with Efb and SCIN-B together at a total inhibitor concentration of 0.6 μg/ml (0.3 μg/ml Efb plus 0.3 μg/ml SCIN-B) or 1 μg/ml (0.5 μg/ml plus 0.5 μg/ml SCIN-B). Rabbit erythrocytes were added, incubated for 1 h at 37°C and lysis was measured. Data was analyzed by a two-tailed unpaired student’s t-test.
Northern blot
A total of 7 µg RNA was separated on a 0.8% agarose–0.66 M formaldehyde gel (Sigma-Aldrich). RNA was transferred to a Brightstar®-plus positively charged membrane (Applied Biosystems, Foster City, CA, USA) overnight by capillary transfer and fixed to the membrane at 80°C for 2 h. The probe was created by amplification of the gene of interest and purified with Qiaquick PCR purification kit (Qiagen). DNA probe was labeled with [α-32P]-dATP using the nick-translation kit (Invitrogen). Blot was hybridized overnight with the probe in ULTRAhyb hybridization buffer (Applied Biosystems) at 42°C, subsequently washed 2 × 5 min at 42°C with 2 × SSC/0.1% SDS and 2 × 15 min at 42°C with 0.1 × SSC/0.1% SDS. Blot was overnight autoradiographed on a Bio-Rad phosphoimager (Applied Biosystems).
RESULTS
High reproducibility of five independent growth curves
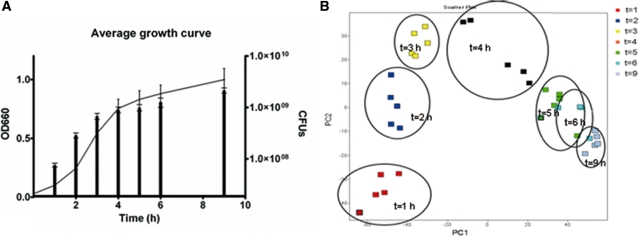
Bacterial growth in defined medium was optimally synchronized. The resulting growth curves were highly reproducible (Figure 1A). Three growth phases could be distinguished, log phase (1–3 h post inoculation; p.i.), late log phase (4 h p.i.) and stationary phase (5–9 h p.i.). A lag phase (typically in which bacteria adjust to new circumstances and start dividing in a nutrient rich environment) was not observed in our experiments. The reproducibility of the five replicates was further assessed by PCA of the normalized microarray data (Figure 1B), which showed clustering of the five biological replicas at the sampled time points.
Figure 1.
Growth of MSSA476 in IMDM and quality control. (A) Three growth phases could be identified: log, late log and stationary phase. Lines represent average of five growth curves (A660nm) and bars represent total CFUs with error bars. (B) PCA of microarray data showing all time points of five independent growth curves. Replicates cluster together indicating high reproducibility of the growth curves.
Four basic gene expression profiles during growth
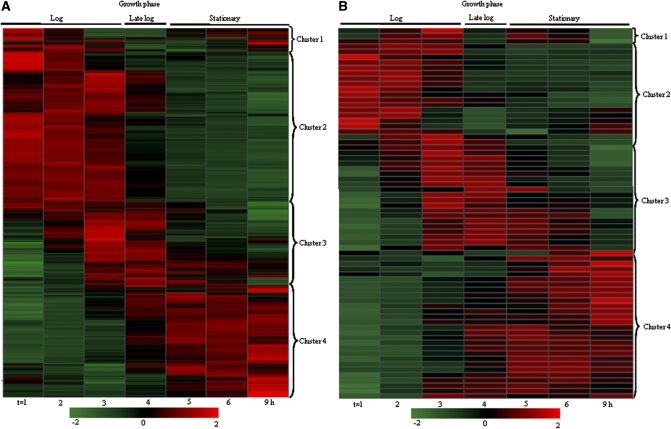
Many of the genes (2473 of 2644, corresponding to 93.5% of the genome) were significantly regulated somewhere during growth (false discovery rate < 0.05). The gene expression data were visually represented by hierarchical clustering using Ward’s method on a heatmap, where the Z-score normalized averaged signal intensities from the five independent growth curves were shown (Figure 2). Z-score normalization expresses each gene expression profile as a deviation from the mean in standard-deviation units and allows the comparison of gene expression patterns whose absolute expression levels may differ by orders of magnitude (32). The individual, significantly regulated genes were grouped with other genes based on shared expression profiles. Four basic expression profiles could be distinguished (Figure 2A). Cluster related function analyses were based on main functions, JCVI subroles and Gene Ontology (GO) functions (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) and mapped to all genes (see Supplementary Table S1 for complete lists of regulated genes). Cluster 1 consisted of genes that were down-regulated in log phase and up-regulated in stationary phase represented by 122 genes (4.9% of significantly regulated genes). These genes encoded energy and DNA metabolic functions mainly, but were also identified as virulence factors including drug transporter and drug resistance. Cluster 2 consisted of 1147 genes (46.3% of significantly regulated genes) that were down-regulated over time. This cluster mainly contained ribosomal proteins. In addition, many cell envelope genes (murC/D) and genes encoding proteins involved in cellular processes were represented. Cluster 3 consisted of 463 genes (17.8% of significantly regulated genes) that were up-regulated in log phase, then down-regulated in stationary phase. This cluster mainly consisted of genes involved in iron binding and transport, like srtB and genes encoding iron compound ABC transporters. Moreover, agrABCD genes belong to this group as well. Cluster 4 consisted of 746 genes (30.1% of significantly regulated genes) that mainly encoded proteins with metabolic functions and stress responses. In addition, RNAIII and the quorum sensing genes luxS and traP were up-regulated over time.
Figure 2.
Heatmap of significantly regulated genes divided over three growth phases. Rows represent individual gene probes and columns represent individual time points. The scale is represented by red (Z > 0), green (Z < 0) and black (Z = 0). Cluster 1: down-regulated during log phase. Cluster 2: down-regulated over time. Cluster 3: up-regulated during log phase. Cluster 4: up-regulated over time. (A) Heatmap containing 2.473 genes. (B) Heatmap of virulence genes.
Furthermore, Ward’s clustering of virulence genes showed expression profiles similar to the profiles observed for the complete gene set (Figure 2B). Remarkably, the group of up-regulated virulence genes during log phase was relatively much larger than was observed in the overall gene expression analysis; 31.9% compared to 17.8%. In this group, genes encoding immune evasion proteins like complement inhibitors, but also genes encoding proteins for eukaryotic cell lysis and bacterial transmission, were highly represented. As expected, genes encoding proteins for toxin production and resistance were mostly up-regulated during stationary phase. Genes encoding surface proteins for colonization of the host did not group together but were found in all four clusters (see Supplementary Table S2 for complete lists of regulated virulence genes). The functionally related virulence genes probably have overlapping, but not identical functions, which could explain the differences in regulation during growth. This is in accordance with the fact that bacteria tend to loose non-functional or redundant genes (33).
Operons and UTRs determined by adjacent probe correlation
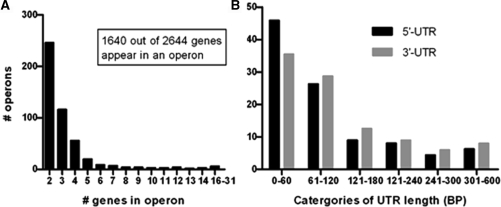
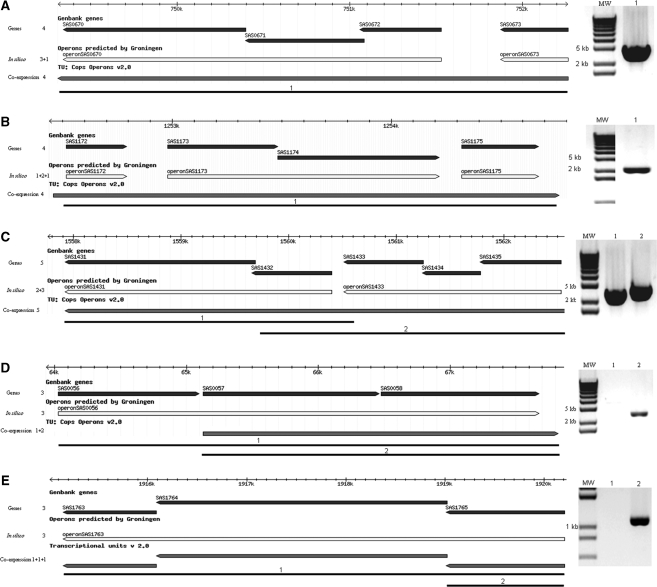
Genes transcribed in an operon or containing a 5′- or 3′-UTR were determined using correlations between adjacent probes, considering both the probes in coding regions and in intergenic regions. With a correlation coefficient cut-off set to 0.80, we found 483 operons containing two or more genes and 1004 single genes, omitting tRNAs and rRNAs (see Supplementary Table S3 for the complete list of predicted operons). In total, 1640 of 2644 genes were transcribed in an operon, corresponding to 62% of the total genome (Figure 3A ). We found no prevalence of operons on the original half or terminus half, nor on the leading or lagging strands. The data in this study and the in silico prediction were 68.8% concordant for both operons and single genes (http://bioinformatics.biol.rug.nl/websoftware/operon/operon_start.php). We predicted 139 operons to be larger, 88 operons to be smaller, 11 to be potentially differentially regulated and 52 to be completely different compared to the in silico data. Compared to the computational prediction, 60% of the operons were predicted concordantly. The in silico and computational predictions were 76% concordant. The percentages can be explained due to gene content differences between strains Mu50 and MSSA476 and based on the similar parameters for the in silico and computational predictions. We found 176 (6.6%) genes with a correlation coefficient between 0.65 and 0.80, which we assigned as uncertain. The computational prediction assigned several of these genes to an operon and the others as single genes, indicating the uncertainty in this range of correlation coefficients. The computational study referred to 36 operons described in literature as a validation for their prediction (18). We found eight differences, three of which (splABCDEF, pheST and egc) could be explained by gene content differences between MSSA476 and Mu50 (34–36). The remaining five operons (mnh, femAB, lac, sigB and sirABC) differed from both the computational prediction as well as the previously described operons (37–41). The operon containing mnh genes and the lac operon were not experimentally validated, and were described as operons based on the functional relation of the genes and promoter/terminator sequences found (37,38). The femAB operon and the sigB operon have been experimentally validated (41,42). FemAB was validated as a two gene operon (42), while we predicted two single genes with a correlation coefficient of 0.51. The sigB operon was described in S. aureus as a four gene operon (41) and in B. subtilis as an eight gene operon (43). We assigned this potentially eight gene operon as uncertain since the correlation coefficients were between 0.65 and 0.80. The sirABC operon has been described previously as a three gene operon (39,44). However, the operon was not experimentally validated and a knock-out inactivation of sirA described no downstream effect on sirB (44), while this would be expected if the genes would be transcribed in an operon. We predicted a single and a two gene operon. One operon not described previously (18) is the sae operon. This operon was predicted in both the computational and the in silico predictions as a three gene operon and one single gene. However, this operon has already been reported as a four gene operon and experimentally validated with RT-PCR and northern blotting (45). According to our prediction, the sae operon is also a four gene operon (Figure 4A).
Figure 3.
Predicted operons and UTR length. (A) Number of operons containing two or more genes. (B) The percentage of 5′- and 3′-UTR in length categories.
Figure 4.
Subset of genes validated with RT–PCR. Genes (black), in silico predictions (white) and co-expression predictions (dark grey) are visualized. Co-expression predictions were based on correlation coefficients between gene probes as well as intergenic probes. RT–PCR products were separated on a 1% TBE agarose gel with a 1 kb ladder. (A) sae operon containing four genes. (B and C) Operons predicted to be larger compared to in silico data. RT–PCR showed correct size of bands as predicted experimentally. (D and E) Operons predicted to be smaller compared to in silico data. RT-PCR indicated absence of complete operon and presence of smaller operon. Lane 1: in silico predicted operon, lane 2: co-expression based predicted operon.
Correlation coefficients were also used to predict 5′- and 3′-UTRs. Of the 1487 operons and single genes 454 (30%) contained a 5′-UTR and 487 (33%) contained a 3′-UTR; 125 (8%) of these operons or single genes contained both a 5′- and 3′-UTRs (Supplementary Table S4). Genes or operons containing UTRs had mostly 5′-UTRs smaller than 100 bp. Large UTRs (>100 bp) were more frequently found on the 3′-end (Figure 3B). UTRs were associated with genes of all different functional classes.
Operon structure of S. aureus
Supplementary Table S5 presents information on the operon structure of S. aureus. The prediction based upon the expression data in this study, in silico prediction, computational prediction based on Mu50 and the conclusion based on the combination of these three predictions is presented.
RT-PCR confirmation of operons
A subset of operons was validated with RT-PCR. A Superscript III-free reaction was added as control for absence of DNA contamination (results not shown). In Figure 4, five operons are shown, of which three operons were predicted to be larger and two operons were predicted to be smaller compared to the in silico data. Genes and the in silico predictions are visualized together with the expression based predictions. The sae operon was predicted in silico to be expressed as a three gene operon and a single gene, while we predicted a four gene operon. RT-PCR confirmed a four gene operon (Figure 4A: ∼2.9 kb), in concordance with a previous study (45). RT-PCR for the operons predicted to be larger revealed an amplification product at ∼2.3 kb and at both ∼2.1 and ∼2.5 kb for SAS1172–SAS1175 and SAS1431–SAS1435, respectively, which corresponded to the size of the complete operons (Figures 4B and C). For the operons predicted to be smaller, an amplification product was present at ∼2.6 kb for SAS0056–SAS0057 and at ∼1.1 kb for SAS1765, while no amplification product was present at the size of the in silico predicted operons SAS0056–SAS0058 and SAS1763–SAS1765. Expression profiles are visualized in Supplementary Figure S1.
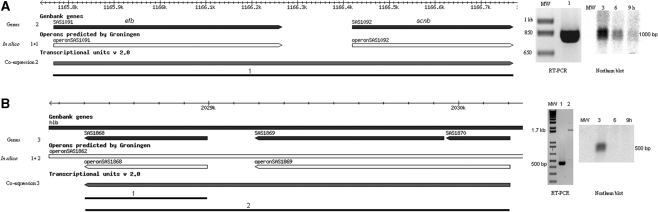
Finally, two operons consisting of virulence genes were validated. One operon consisted of two genes, efb and scnb (30). The other operon consisted of three genes, sak, an autolysin and a holin encoding gene (46) (Figure 5). Both operons were analyzed with RT-PCR and showed an amplification product of the expected sizes of ∼1 and ∼1.8 kb, respectively. Northern blot analysis of the efb-scnb operon confirmed the presence of an ∼1 kb transcript, analysis of the sak-autolysin-holin operon only showed a transcript of ∼500 bp corresponding to the length of sak, indicating that sak is probably transcribed as a single gene as well as in an operon. The operon was only transcribed at low expression levels according to the microarray data.
Figure 5.
Two operons containing virulence genes. (A) RT–PCR and northern blot showed efb located within an operon with scnb. (B) RT–PCR showed sak transcribed in an operon with autolysin and holin encoding genes. Northern blot analysis only showed a 500 bp transcript identifying sak.
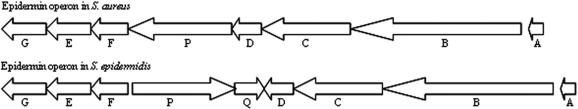
In the expression profile analysis, 11 operons were found that seemed to be differentially regulated in a growth phase-dependent manner (see Supplementary Table S6 for the complete list). For example, epiABCDPFEG, an eight gene operon containing lantibiotic genes, was previously identified in Staphylococcus epidermidis as epiABCD, epiPQ and epiFEG with a transcription start site in front of epiF (Figure 6). The epidermin operon in S. aureus was expressed as one operon in log phase, but split into two operons in the stationary phase according to the expression data (Figure 7). The complete operon was up-regulated in log-phase until 4 h p.i., then four genes were down-regulated and four genes were up-regulated in the stationary phase. RT-PCR showed the presence of the complete transcript throughout the growth curve. 5′-RACE experiments showed a potential transcription initiation site in front of epiF, in accordance with the co-expression based prediction (data not shown). Previous studies described internal promoters or terminators in operons for several bacteria. For example, in B. subtilis two operons, sigB (15) and resABCDE (17) had internal promoters. In S. aureus, three examples of differentially regulated operons have been described: cidABC and lrgAB with an internal promoter site (14,47) and srrAB with no additional transcription initiation site (48).
Figure 6.
Epidermin operon in S. aureus and S. epidermidis. In S. aureus, all genes are located on the same strand, while in S. epidermidis, epiPQ are located on the other strand of epiABCD and epiFEG.
Figure 7.
Operon epiABCDPFEG. (A) Expression profiles showing equal expression patterns for eight genes in the log phase and different patterns in the stationary phase. (B) In silico predictions compared to expression predictions. At the bottom, the tiling of the RT-PCR amplifications is indicated. (C) Tiling RT-PCR showed presence of eight genes expressed as an operon at t = 2 and 9.
Efb and SCIN-B have a synergistic effect on complement inhibition
The advantage of two genes occurring in one operon is that combined expression is a benefit. This is obvious for metabolic genes encoding enzymes that work in a cascade. For virulence genes we hypothesized that if the gene products are related and even synergistic this would be very beneficial. Therefore we did in-depth analysis of the operon containing Efb and SCIN-B, since for both gene products the molecular mode of action has been described in great detail.
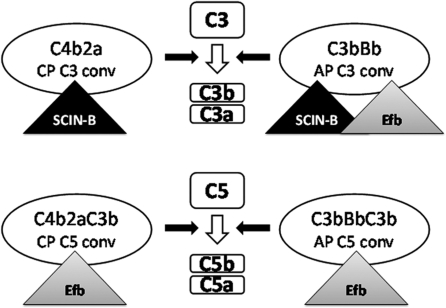
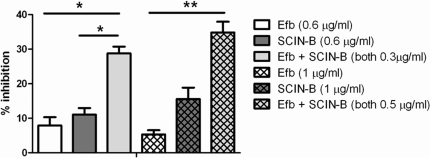
The microarray data indicated that the genes encoding Efb and SCIN-B are transcribed in one operon. Efb and SCIN-B both inhibit the complement system, but via different mechanisms (Figure 8). SCIN-B is known to target C3 convertases of the Classical, Alternative and Lectin Pathway of the complement system, thereby effectively inhibiting C3b deposition on the bacterial surface and, thus, phagocytosis and C5a generation by all pathways. Since the genes encoding for Efb and SCIN-B are in one operon and inhibit the complement system by different mechanisms (31,49–51) and interaction sites, we hypothesized that Efb and SCIN-B might have synergistic effects. Therefore, we used an AP dependent hemolytic assay, in which MAC-dependent killing of rabbit erythrocytes was used as a read-out for complement activity. Complement-mediated lysis of rabbit erythrocytes was only inhibited for 10% by Efb or SCIN-B alone (0.6 µg/ml) but addition of Efb and SCIN-B together (0.3 µg/ml Efb and 0.3 µg/ml SCIN-B) significantly increased this inhibition up to 30% (Figure 9). Similar results were obtained for Efb and SCIN-B at a concentration of 1 µg/ml.
Figure 8.
Predicted synergism from modes of action. Depiction of the two C3 convertases and two C5 convertases of the complement system and the targets of the staphylococcal complement inhibitors Efb and SCIN-B. The Classical Pathway C3 convertase (CP C3 conv) is only inhibited by SCIN-B. Alternative Pathway C3 convertase (AP C3 conv) can be inhibited by both Efb and SCIN-B. Classical Pathway C5 convertase (CP C5 conv) is only inhibited by Efb. Alternative Pathway C5 (AP C5 conv) is only inhibited by Efb. The two molecules together can inhibit all complement convertases.
Figure 9.
Synergistic effect between Efb and SCIN-B in complement inhibition. AP-dependent hemolytic assay. Rabbit erythrocytes were incubated with 10% human serum in the presence of Efb (0.6 or 1 μg/ml), SCIN-B (0.6 or 1 μg/ml) or Efb and SCIN-B together (0.3 + 0.3 or 0.5 + 0.5 μg/ml), *P < 0.05, **P < 0.01. Figure represents the mean ± SEM of four separate experiments.
DISCUSSION
Using data from our transcriptome analysis through time, we were able to identify the operon structure from the entire S. aureus genome, covering the majority of open reading frames. A reliable prediction based on the transcriptome is only possible when many genes are significantly regulated and the reproducibility is high. Therefore, we used five independent, highly reproducible growth curves with seven time points each. This resulted in discrimination of significantly regulated genes at a fold change as small as 0.28 and differential regulation of 93.5% of all genes. This is extremely sensitive as compared to other studies and is crucial to the prediction of the complete operon structure (6,7,9).
Operons were predicted by calculating the correlation coefficients of transcriptome data of all adjacent probes at all seven time points. We used a relatively high correlation cut-off value of ≥0.80 to reduce the number of false-positive operons. Validation of this cut-off was achieved by RT-PCR of several operons and showed an accurate prediction above this cut-off, regardless of the size of the intergenic region. Below a correlation coefficient of 0.65, absence of operons was predicted and validated. Even though intergenic regions were small, visual inspection of the expression patterns showed large differences in this correlation coefficient range. Therefore, we conclude that predictions based on expression data are more accurate than the in silico prediction for these correlation coefficients. Genes showing low correlation coefficients for expression were also shown to be transcribed in an operon in studies in E. coli (52,53). Between correlation coefficients 0.65 and 0.80 several operons were detected that were not predicted with the cut-off we used. The computational and in silico predictions for these operons did not give a conclusive answer either. This indicates that validation is essential for predictions with correlation coefficients between 0.65 and 0.80, representing only 6.6% of the genome.
A comparison of studies in E. coli and B. subtilis showed that among the different prediction methods, the intergenic distance was the most valuable single prediction variable (54). However, the combination of intergenic distance with functional information or gene expression data proved to be even more accurate (20,21,54–56). We compared and combined the operons predicted, using expression data with in silico predictions based on intergenic distances. We conclude that operon predictions using highly reproducible and large numbers of expression data are more accurate than predictions based on intergenic distances only.
Bacterial pathogenesis is dependent on the presence of virulence genes, but also on the expression regulation of these genes. To be able to understand the transcriptional regulation, in vitro and in vivo, knowledge of the operon structure of S. aureus is essential (16). Virulence genes are usually located on mobile elements and are exchanged regularly. Regulation of virulence genes at an operon level is, in general, not expected to exist, because of the regular exchange. Nonetheless, we found several virulence genes transcribed in an operon with other virulence genes like efb in combination with scnb and sak in combination with autolysin and holin encoding genes (Figure 5), but also virulence genes that were transcribed in an operon with genes encoding hypothetical proteins or acetyltransferases. We showed that addition of Efb and SCIN-B together enhanced complement inhibition significantly, indicating that S. aureus has evolved this operon to counterattack complement activation even more efficiently as with the single inhibitors alone.
In conclusion, the high number of significantly regulated genes in combination with the statistical power of seven time points sampled in five biological replicas used to calculate correlation coefficients enabled us to accurately predict operons in the genome of S. aureus in an unbiased approach. It identified the presence of virulence genes within an operon and the synergistic action of the translated proteins was proven. Herewith, a basis has been set for future studies on gene regulation and host-pathogen interactions both in vitro and in vivo.
ACCESSION NUMBERS
The microarray data have been submitted to GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number: GSE16488.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission grant agreement number HEALTH-F3-2008-222718 (Control of Community-acquired MRSA: Rationale and Development of Counteractions – CONCORD).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr Willem van Schaik for practical assistance with northern blots, Dr Wim de Leeuw for bioinformatical support and Dr Martijs Jonker for statistical support.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Sibbald MJJB, Ziebandt AK, Engelmann S, Hecker M, de JA, Harmsen HJM, Raangs GC, Stokroos I, Arends JP, Dubois JYF, et al. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garzoni C, Francois P, Huyghe A, Couzinet S, Tapparel C, Charbonnier Y, Renzoni A, Lucchini S, Lew DP, Vaudaux P, et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 6.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J. Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues F, Sarkar-Tyson M, Harding SV, Sim SH, Chua HH, Lin CH, Han X, Karuturi RK, Sung K, Yu K, et al. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J. Bacteriol. 2006;188:8178–8188. doi: 10.1128/JB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 9.Thompson LJ, Merrell DS, Neilan BA, Mitchell H, Lee A, Falkow S. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 2003;71:2643–2655. doi: 10.1128/IAI.71.5.2643-2655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster TJ. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 11.Rooijakkers SHM, van Kessel KPM, van Strijp JAG. Staphylococcal innate immune evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 13.Brouwer RWW, Kuipers OP, Hijum SAFT. The relative value of operon predictions. Brief. Bioinform. 2008;9:367–375. doi: 10.1093/bib/bbn019. [DOI] [PubMed] [Google Scholar]
- 14.Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann JD, Wu MF, Kobel PA, Gamo FJ, Wilson M, Morshedi MM, Navre M, Paddon C. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 2001;183:7318–7328. doi: 10.1128/JB.183.24.7318-7328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda S, Kawashima S, Kobayashi K, Ogasawara N, Kanehisa M, Goto S. Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli. BMC Genomics. 2007;8:48. doi: 10.1186/1471-2164-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan MF, Sorokin A, Pujic P, Ehrlich SD, Hulett FM. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Trawick JD, Yamamoto R, Zamudio C. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 2004;32:3689–3702. doi: 10.1093/nar/gkh694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence JG, Roth JR. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatti C, Rohlin L, Oh MK, Liao JC. Co-expression pattern from DNA microarray experiments as a tool for operon prediction. Nucleic Acids Res. 2002;30:2886–2893. doi: 10.1093/nar/gkf388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agaisse H, Lereclus D. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 1996;20:633–643. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen LH, Emory SA, Bricker AL, Bouvet P, Belasco JG. Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J. Bacteriol. 1991;173:4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 26.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl Acad. Sci. USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- 28.Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 29.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 30.Jongerius I, Kohl J, Pandey MK, Ruyken M, van Kessel KPM, van Strijp JAG, Rooijakkers SHM. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooijakkers SHM, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJB, van Kessel KPM, van Strijp JAG. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 32.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savill NJ, Higgs PG. Redundant and non-functional guide RNA genes in Trypanosoma brucei are a consequence of multiple genes per minicircle. Gene. 2000;256:245–252. doi: 10.1016/s0378-1119(00)00345-0. [DOI] [PubMed] [Google Scholar]
- 34.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 35.Reed SB, Wesson CA, Liou LE, Trumble WR, Schlievert PM, Bohach GA, Bayles KW. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 2001;69:1521–1527. doi: 10.1128/IAI.69.3.1521-1527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savopoulos JW, Hibbs M, Jones EJ, Mensah L, Richardson C, Fosberry A, Downes R, Fox SG, Brown JR, Jenkins O. Identification, cloning, and expression of a functional phenylalanyl-tRNA synthetase (pheRS) from Staphylococcus aureus. Protein Expr. Purif. 2001;21:470–484. doi: 10.1006/prep.2001.1407. [DOI] [PubMed] [Google Scholar]
- 37.Breidt F, Jr, Hengstenberg W, Finkeldei U, Stewart GC. Identification of the genes for the lactose-specific components of the phosphotransferase system in the lac operon of Staphylococcus aureus. J. Biol. Chem. 1987;262:16444–16449. [PubMed] [Google Scholar]
- 38.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 1998;180:6642–6648. doi: 10.1128/jb.180.24.6642-6648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kopp U, Roos M, Wecke J, Labischinski H. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb. Drug Resist. 1996;2:29–41. doi: 10.1089/mdr.1996.2.29. [DOI] [PubMed] [Google Scholar]
- 41.Kullik II, Giachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 42.Berger-Bachi B, Barberis-Maino L, Strassle A, Kayser FH. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 1989;219:263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- 43.Wise AA, Price CW. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J. Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dale SE, Sebulsky MT, Heinrichs DE. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 2004;186:8356–8362. doi: 10.1128/JB.186.24.8356-8362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinhuber A, Goerke C, Bayer MG, Doring G, Wolz C. Molecular architecture of the regulatory Locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 2003;185:6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rooijakkers SH, van Wamel WJ, Ruyken M, van Kessel KP, van Strijp JA. Anti-opsonic properties of staphylokinase. Microbes. Infect. 2005;7:476. doi: 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Rice KC, Patton T, Yang SJ, Dumoulin A, Bischoff M, Bayles KW. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 2004;186:3029–3037. doi: 10.1128/JB.186.10.3029-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 2004;186:2430–2438. doi: 10.1128/JB.186.8.2430-2438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. A structural basis for complement inhibition by Staphylococcus aureus. Nat. Immunol. 2007;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 50.Rooijakkers SH, van Strijp JA. Bacterial complement evasion. Mol. Immunol. 2007;44:23. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Rooijakkers SHM, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp J.AG, et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao G, Martinez-Vaz B, Pan W, Khodursky AB. Operon information improves gene expression estimation for cDNA microarrays. BMC Genomics. 2006;7:87. doi: 10.1186/1471-2164-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karimpour-Fard A, Leach SM, Gill RT, Hunter LE. Predicting protein linkages in bacteria: which method is best depends on task. BMC Bioinform. 2008;9:397. doi: 10.1186/1471-2105-9-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bockhorst J, Qiu Y, Glasner J, Liu M, Blattner F, Craven M. Predicting bacterial transcription units using sequence and expression data. Bioinformatics. 2003;19:i34–i43. doi: 10.1093/bioinformatics/btg1003. [DOI] [PubMed] [Google Scholar]
- 56.de Hoon MJL, Imoto S, Kobayashi K, Ogasawara N, Miyano S. Predicting the operon structure of Bacillus subtilis using operon length, intergene distance, and gene expression information. Pac. Symp. Biocomput. 2004;9:276–287. doi: 10.1142/9789812704856_0027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.