Abstract
In trypanosomes a 39 nucleotide exon, the spliced leader (SL) is donated to all mRNAs from a small RNA, the SL RNA, by trans-splicing. Since the discovery of trans-splicing in trypanosomes two decades ago, numerous attempts failed to reconstitute the reaction in vitro. In this study, a crude whole-cell extract utilizing the endogenous SL RNA and synthetic tubulin pre-mRNA were used to reconstitute the trans-splicing reaction. An RNase protection assay was used to detect the trans-spliced product. The reaction was optimized and shown to depend on ATP and intact U2 and U6 snRNPs. Mutations introduced at the polypyrimidine tract and the AG splice site reduced the reaction efficiency. To simplify the assay, RT–PCR and quantitative real-time PCR assays were established. The system was used to examine the structural requirements for SL RNA as a substrate in the reaction. Interestingly, synthetic SL RNA assembled poorly to its cognate particle and was not utilized in the reaction. However, SL RNA synthesized in cells lacking Sm proteins, which is defective in cap-4 modification, was active in the reaction. This study is the first step towards further elucidating the mechanism of trans-splicing, an essential reaction which determines the trypanosome transcriptome.
INTRODUCTION
mRNA maturation in trypanosomes differs from the process in most eukaryotes mainly because in trypanosomes, protein coding genes are transcribed into polycistronic RNAs. The polycistronic RNAs are dissected to individual mature mRNAs by the concerted action of trans-splicing and polyadenylation. Thus, trans-splicing functions to release mature capped mRNAs from the polycistronic RNA. In trans-splicing, a small RNA, the spliced leader RNA (SL RNA) harboring a special cap-4 and composed of a conserved exon (SL) and a more divergent intron, donates its exon to all mRNAs (1,2). Trans-splicing, like cis-splicing, proceeds through a two-step transesterification reaction, though a Y structure is formed instead of a lariat intermediate. Although first discovered in trypanosomes, trans-splicing was later found in nematodes, trematodes and even chordates (3), and more recently also in dinoflagellates (4). Cis-splicing also occurs in trypanosomes, but so far only three substrates were shown to undergo this process (5).
SL RNA is transcribed from a RNA polymerase II promoter (6,7). SL RNA undergoes modification to form the unique hypermethylated cap-4 (8). However, eliminating an individual cap modification does not interfere with the ability of the SL RNA to undergo trans-splicing (9). The SL RNA binds core Sm proteins, which are assisted by SMN and Gemin2 (10). SL RNA undergoes pseudouridylation by SLA1, which serves as a chaperone during SL RNA biogenesis. SL RNA transcription, pseudouridylation and assembly with core Sm proteins take place in a special nuclear location that we termed the SL RNP factory (11,12). The majority of SL RNA in the steady state is concentrated in this sub-domain (12,13). It is not currently known how the SL RNP is recruited to the trans-spliceosome.
Recently, much progress was made in identifying a variety of trypanosome splicing factors, and their functions were elucidated by down-regulation via RNAi (10,12,14–19). Splicing defects such as an increase in the level of SL RNA, and changes in the level of the Y structure intermediate were observed following the depletion of each splicing factor (18,19). Using this approach, the factors U2AF35, U2AF65 and SF1, which function to identify and specify the correct 3′ AG splice sites, were recently identified. The Trypanosoma brucei U2AF65 is larger than its mammalian counterpart, and surprisingly, unlike the protein’s interactions in metazoa, it interacts with SF1 but not with U2AF35 (20). An ∼45 S spliceosomal complex was identified, suggesting the existence of a single-spliceosome complex that can potentially conduct both cis- and trans-splicing (19).
In vitro splicing reactions were established in mammalian and yeast extracts >25 years ago (21–23). A very efficient in vitro trans-splicing reaction was also established in nematodes using Ascaris lumbricoides whole-cell extracts derived from developing embryos. The reaction is very efficient and is based on a synthetic pre-mRNA acceptor and SL RNA derived from the extract, or synthetic radiolabeled SL RNA (24,25). The in vitro cis-splicing systems in both mammals and yeast required different manipulations in order to optimize the reactions. Although whole-cell extracts were used in the mammalian system, nuclear extracts were later found to be more efficient (21). The in vitro reaction is more efficient using ‘tailored’ RNA molecules harboring introns which are derived from transcripts that are spliced in vivo with high efficiency. For instance, the first splicing substrate was constructed using exons and an intron from the adenovirus transcription unit (21,23) and a better substrate was subsequently obtained by shortening the exon size (26). It was demonstrated in the mammalian system that the pre-mRNA does not require either a cap at its 5′-end or a poly-(A)+ tail for its splicing, but splicing is enhanced by the addition of a cap, probably due to stabilization of the pre-mRNA (27). In yeast, a cell-free splicing system was first established using a sensitive S1 nuclease assay monitoring the production of the spliced product, and later, radiolabeled pre-mRNA was used and the spliced product and intermediates were readily detected (22). Despite overcoming many hurdles in establishing in vitro splicing systems in mammals and yeast as mentioned above, the insights obtained did not prove to be useful for creating such an in vitro trans-splicing reaction in trypanosomes.
In this study, we established an in vitro trans-splicing system using substrates whose trans-splicing was first verified in vivo in transgenic parasites expressing these constructs. The model substrate contains the splicing signals of the β-tubulin gene fused to luciferase and the upstream polyadenylation signal of the α-tubulin gene. Three sensitive assays were established to monitor the production of the trans-spliced product, RNase protection, RT–PCR and a quantitative real-time PCR (qRT–PCR) assay. Trans-splicing requires ATP, is sensitive to heat-inactivation, and is dependent on U2 and U6 snRNAs. The reaction is inhibited when the AG splice site or the polypyrimidine tract are mutated. Mutant SL RNA, lacking the cap-4 +4 modification, can be utilized in trans-splicing, and synthetic cap-4 does not inhibit the reaction. Optimization of the reaction conditions indicates that SL RNA is the limiting component in the extract, but only endogenous and not m7G capped synthetic SL RNA is active in the reaction. This system can be further used to investigate in depth the mechanism and fidelity of 3′ splice site selection.
MATERIALS AND METHODS
Cell growth, transfection and cloning
All experiments were performed with the procyclic form of T. brucei strain 29–13. Cell culture and preparation of transgenic parasites was as previously described (14).
Plasmids
pNS21 was a kind gift from George Cross (Rockefeller University, USA). This plasmid contains the T. brucei procyclin promoter, followed by a linker containing a BglII restriction site, the procyclin (EP) 3′ splice site and synthetic 5′UTR, fused to the luciferase ORF that contains an XbaI restriction site 47-nt downstream from the ATG start codon. This vector contains the phleomycin resistance gene (28). To create plasmids carrying the splicing signals of β-tubulin or EP, the corresponding sequences were amplified from the genome and were cloned between the restriction sites BglII and XbaI (28). To generate the vector pNS21-TIR carrying the β-tubulin intergenic region, a PCR product was generated using forward primer (233-Tub-luc f1) and reverse primer (234-Tub-Luc.r1) to create the correct fusion. Similarly, the pNS21-PIR was generated by amplifying the EP-2-3 (Tb927.6.520; positions 226182-227114 on chromosome 6) with forward primer (246-EP-luc f1) and reverse primer (247-EP-luc r1) to create a fusion with the luciferase gene. The plasmids were linearized with NotI for integration into the rRNA locus. RNA from transgenic parasites was used to prepare cDNA using the Reverse-iT (Thermo Scientific) kit with random primers. The cDNA was amplified using SL forward primer (220, SL RNA sense) and the luciferase reverse primer (575, T7 Luc 52-67 as). These cDNA clones were used to prepare the probes for the RNase protection assay described below. The oligonucleotide list is given in Supplementary Data 1.
Mutants of pNS21-TIR
The mutations that changed the polypyrimidine tract (PPTm), AG to AA, and the pNS21-TIRsub were generated by site directed mutagenesis using the Mega primer method. The mutants were generated using a forward primer and a reverse primer carrying the mutation, and a second pair composed of a sense primer carrying the mutation and a reverse primer containing the XbaI site (234). The two fragments carrying the mutation were amplified using the primer pair 233 and 234. The mutated fragments were cloned between the XbaI and BglII site of pNS21, and the plasmids were sequenced and used to prepare transgenic cell lines. cDNA from these cell lines were cloned and sequenced. The pairs of oligonucleotides used to generate the mutations are listed in Supplementary Data 1.
In vitro trans-splicing extracts
Extracts were prepared essentially as described (29), with minor changes. First, 2 l of T. brucei 29-13 were grown in a 5 l conical flask under constant stirring to a density of 1 × 107 cells/ml. Cells were harvested at 4°C, yielding a packed cell volume of around 1.5 ml. Cell pellets were washed twice in 10 ml of ice-cold wash solution [20 mM Tris–HCl (pH 7.4), 100 mM NaCl, 3 mM MgCl2, 1 mM EDTA] and once in 10 ml of ice-cold transcription buffer [150 mM sucrose, 20 mM potassium l-glutamate, 3 mM MgCl2, 20 mM HEPES-KOH (pH 7.7), 2 mM dithiothreitol, and leupeptin (10 µg/ml)]. Finally, pellets were suspended in 3 ml of transcription buffer and incubated for 20 min on ice. Cells were broken in a 7-ml Dounce homogenizer with a type B pestle by applying rapid strokes continuously for ∼5 min, until >75% of the cells were broken. The suspension was divided into 1 ml aliquots, frozen in liquid nitrogen and stored at −70°C. For a whole-cell extract preparation, an aliquot was thawed, 900 µl was mixed with 100 µl of transcription buffer containing 1.5 M KCl, and incubated for 20 min on ice with frequent mixing. Subsequently, the extract was centrifuged at 21 000g for 10 min at 4°C. The supernatant (∼600 µl) was transferred to a new microtube, diluted with 0.5 volumes of transcription buffer, and centrifuged again for 2 min. The supernatant was loaded on an Amicon Ultra-4, 5kDa MWCO (Millipore) to concentrate the extract. When the volume reached ∼500 µl, a second salt dilution to 40 mM KCl was performed by adding 750 µl transcription buffer, and the sample was further concentrated until a final volume of 150–200 µl was reached. The final cell extract was divided into ∼50 µl aliquots, frozen, and stored at −70°C. Protein concentration of extracts varied between 40 and 50 mg/ml.
In vitro trans-splicing
The reaction buffer was essentially as described (30) with minor changes. Each 20 µl reaction mixture contained 8 µl cell extract (300–500 µg protein) in 20 mM potassium l-glutamate, 20 mM KCl, 3 mM MgCl2, 20 mM HEPES–KOH, pH 7.7, 20 mM creatine phosphate, 0.48 mg/ml creatine kinase, 2.5% polyethylene glycol, 0.2 mM EDTA, 0.5 mM EGTA, 4 mM dithiothreitol, 10 µg/ml leupeptin, 10 µg/ml aprotinin and 25 µg/ml pre-mRNA substrate (0.5 µg). ATP (0.5 mM) was added after 15 min incubation on ice, and then the reaction was continued for 1 h at 28°C. When co-transcriptional trans-splicing was examined, 0.5 mM NTP mix was added to the reaction and 40 µg/ml of pNS21-TIR plasmid DNA was used. Total RNA was prepared and analyzed by RNase protection assay as previously described (14), except that RNA was extracted from the in vitro reaction using TriReagent (Sigma). RNase protection assay products were separated on 6% polyacrylamide/7 M urea gels and visualized by autoradiography.
RNA preparation for the in vitro reaction
Total RNA from the transgenic parasites was prepared using TriReagent (Sigma). To generate pre-mRNA for the trans-splicing reaction, PCR products were prepared using a sense primer containing the T7 promoter. The T7 in vitro transcribed pre-TIR and its derivate mutations were prepared with sense primer 259, and antisense primer 218 situated 86 nt downstream of the luciferase ATG. Synthetic SL RNA was prepared using primers 775 and 777, and plasmid coding for T. brucei SL RNA (14). RNA was synthesized using the MegaScript T7 Kit (Ambion) according to the manufacturer’s instructions. [α-32P]-UTP-labeled antisense probes for RNase protection assay were prepared by using T7 RNA polymerase (Promega).
Isolation of SL RNA transcribed in the cells
The synthesis of [α-32P]-UTP-labeled SL RNA in permeable cells was essentially as described (31). For isolation of native SL RNA, total RNA was prepared from 250 ml T. brucei parental strain (∼1 mg) and separated on a 10% acrylamide denaturing gel. SL RNA co-migrates on such gels together with the 140 nt srRNA-3. RNA was visualized by UV lamp on a TLC plate. The ∼140 nt RNA species was excised from the gel. SL RNA concentration was determined by primer extension using synthetically transcribed SL RNA as a standard (0.08 to 8 ng). Based on the primer extension calibration curve, the concentration of gel-extracted SL RNA was estimated to be 0.35 ng/µl.
RNase H digestion of U snRNAs in the splicing extracts
RNase H digestion was performed as follows: In vitro splicing extract (300–500 µg) was incubated with 200 pmol of primer, 0.4 mM ATP and 1 U RNase H (New England Biolabs, USA) in a total reaction volume of 20 µl, for 90 min on ice. The reaction was then split. Half of the treated extract was used for an in vitro reaction and the other half was subjected to primer extension. The primers used for RNase H cleavage are listed in Supplementary Data 1.
RT–PCR as a trans-splicing assay
In vitro trans-splicing reaction was performed as described above and RNA was extracted by Proteinase K treatment, followed by phenol : chloroform extraction and ethanol precipitation. The cDNA was prepared from one standard 20 µl splicing reaction using random primer and the RevertAid™ First Strand cDNA synthesis kit (Fermentas) following the manufacturer’s instructions. PCR was performed on 0.1 µl of cDNA, 1 µM primers and ReadyMix™ Taq PCR Reaction Mix with MgCl2 (Sigma). The PCR conditions were as follows: 95° for 2 min followed by 30 cycles of 95° for 30 s, 60° for 30 s and 72° for 10 s (for pNS21-TIRsub mRNA amplification) or for 30 s (for pNS21-TIRsub pre-mRNA and Tubulin amplification).
Quantitative real-time PCR
Real-time PCR was performed in a two-step reaction. First, cDNA was prepared from either 10 µg of total RNA or a standard in vitro trans-splicing reaction using random primer and the RevertAid™ First Strand cDNA synthesis kit (Fermentas) following the manufacturer’s instructions. Next, real-time PCR was performed using 1 µl of cDNA (diluted 1 : 100), 1 µM primers and Absolute Blue QPCR SYBR® Green ROX mix (Thermo Scientific). qRT–PCR was performed on a Chromo4 Real-Time PCR detection system (Bio-Rad), as follows: 95° for 2 min followed by 40 cycles of 95° for 30 s, 60° for 30 s and 72° for 10 s. A concentration curve of the trans-spliced product, purified using the QIAquick PCR purification kit (Qiagen), was determined using the Opticon Monitor3 software supplied with the Opticon4 apparatus. This concentration curve was used to determine the amount of PCR product produced in each sample. The reverse transcription could be performed using random-primers or primer specific to luciferase (216-Luc 66-68as). However, the random priming enabled simultaneous detection of the controls and the spliced product.
RESULTS
Constructing a pre-mRNA for in vitro trans-splicing
In vivo trans-splicing takes place on polycistronic pre-mRNA. A scheme depicting such a pre-mRNA is given in Figure 1A, highlighting the essential sequences required for trans-splicing including the 3′ AG splice site, polypyrimidine tract (PPT), branch point and the polyadenylation signal of the upstream gene.
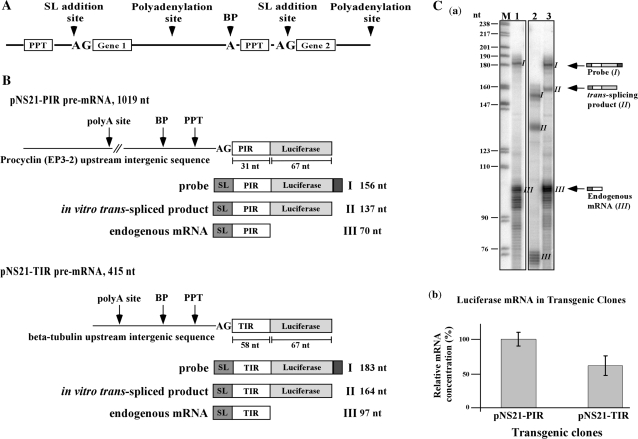
Figure 1.
(A) Schematic presentation of a trypanosome polycistronic pre-mRNA. The significant sequence elements essential for trans-splicing are indicated. (B) Pre-mRNA substrates used in this study. Schematic representation of the pre-mRNA substrates highlighting the significant sequence elements: polyadenylation site (polyA site), polypyrimidine tract (PPT) and AG splice site. The lengths of the PIR and TIR UTRs as well as the Luciferase ORF are indicated. Underneath each pre-mRNA, the respective probe, in vitro trans-spliced product and endogenous mRNA are shown, along with the expected size of the protected fragments after RNase protection. (C) RNase protection assay to monitor the expression of the TIR and PIR constructs. (a) Thirty micrograms of total RNA was hybridized with labeled antisense probe, complementary to the trans-spliced product, as described in ‘Materials and Methods’ section. Protected RNA products were separated on a 6% sequencing gel. M- DNA marker, labeled pBR322 DNA MspI digest. The size of the marker is indicated on the left. Lane contents are as follows: 1, RNA from parental strain; 2, RNA from transgenic parasites expressing the pNS21-PIR construct; 3, RNA from transgenic parasites expressing the pNS21-TIR construct. The scheme on the right hand side of the gel indicates the structure of the probe and the protected fragments. (b) Relative expression of the tubulin-luciferase or the EP-luciferase to the endogenous tubulin and EP transcripts.
Plasmid pNS-21 was used in this study; this vector was previously used to study the optimal composition and distance between the AG splice and the PPT for trans-splicing (28). The sequences in the intergenic region of the vector are partially synthetic and lack the polyadenylation signal of an upstream gene. Because of the previously shown linkage between trans-splicing and polyadenylation (28,32–34), we replaced the intergenic region with authentic sequences. The sequences between the BglII site and the XbaI site were replaced by either the 326 bp intergenic region upstream to the β-tubulin locus (pNS21-TIR) or the 630 bp EP-3-2 intergenic region (pNS21-PIR) (Supplementary Figure S2). Transgenic parasites were obtained and the expression of the luciferase gene was determined by northern analysis. The results indicated that the presence of the authentic intergenic regions of either tubulin or EP enhanced the expression of the luciferase gene compared to the parental pNS21 (data not shown). Since these constructs differ at the 5′ flanking sequences, especially in the sequences upstream to the AG splice site, the difference in expression most probably reflects differences in trans-splicing efficiency.
To establish an assay that would directly measure the production of the trans-spliced product from pre-mRNA, we prepared an antisense RNA probe (carrying a small extension at the 5′-end to distinguish the probe from the protected fragments) using a cDNA clone derived from the transgenic parasites as a template. The RNase protection assay was used to examine the production of the trans-spliced products in parasites expressing either the TIR or PIR constructs. Schematic presentation of TIR and PIR constructs and the fragments that are generated from the RNase protection are depicted in Figure 1B. Three fragments were observed: (I) the residual undigested probe; (II) the trans-spliced product; and (III) the endogenous transcript. Based on the ratio between the trans-spliced product (II) and the endogenous transcript (III), it was evident that the PIR construct was more highly expressed and trans-spliced compared to the TIR transcript (Figure 1C-b compare lanes 2 to 3). As a control for the specificity of the assay, RNA from the parental strain was used, and the only protected fragment was of the endogenous tubulin transcript (lane 1). These results suggested that we established a chimeric construct that is properly trans-spliced in vivo and can be used as a substrate for establishing the in vitro trans-splicing reaction.
In vitro reaction with the PIR and TIR pre-mRNA
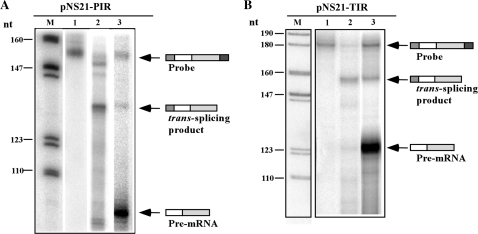
Several parameters were considered when establishing the in vitro reaction. Different protocols were used to prepare nuclear extracts (35,36) and a whole-cell extract (29). The efficiency of SL RNA transcription was examined in the extracts using an SL RNA gene plasmid harboring a 19-nt insertion mutation, and primer extension using a primer specific for the insertion (37). The results (data not shown) indicated that in our hands the most active transcription extract was the whole-cell extract prepared according to Laufer et al. (29). Note, that sucrose was used both for freezing the extract and in the reaction, rather than glycerol or sorbitol, which are used to stabilize the mammalian and yeast extracts, respectively. This change stems from the fact that the trypanosome extracts are highly enriched in glycolytic enzymes that require ATP, and hence, addition of glycerol depletes the ATP in the extract. It was previously reported that when the counter ion chloride is replaced by glutamate, the in vitro transcription in the trypanosome extracts is more efficient (35). Potassium glutamate was therefore used in the reaction. The first step was to determine if a trans-spliced product is generated in the reaction using T7 in vitro transcribed pre-TIR or pre-PIR mRNAs. The source of SL RNA was the endogenous SL RNP present in the extract. The reaction was terminated by extraction of the RNA, and the RNA was subjected to RNase protection with the corresponding probes. The RNase protection was performed in parallel on RNA extracted from transgenic parasites expressing the TIR and PIR constructs (Figure 2). Three fragments, whose identities are illustrated schematically on the right side of the gel, were observed when pre-TIR and pre-PIR were used in the reaction: (i) residual undigested probe, which migrates like the probe (Figure 2A and B, lane 1); (ii) the trans-spliced product (Figure 2A and B, lane 3), seen as the protected fragments present in the RNA from the in vitro reactions. The protected fragments were identical to the fragments detected when RNA from transgenic parasites was used (compare lanes 2 to 3); and (iii) pre-mRNA; the intensity of the protected fragment indicates the extent of pre-mRNA that existed at the end of the reaction (present only in lane 3). Although the same amount (0.5 µg) of both pre-TIR and pre-PIR were added to the reaction, massive degradation of pre-PIR was observed in the extract. The trans-spliced product (lanes 3) was excised from the gel, amplified using the SL forward primer and the luciferase reverse primer (described above) and sequenced. The sequence was identical to the product observed in the transgenic lines, suggesting that the reaction is faithful and generates a genuine trans-spliced product. The efficiency of the reaction was estimated by examining the fraction of pre-mRNA that was converted to trans-spliced product. The results were analyzed by densitometry and the intensity of the spliced product was divided by the intensity of the pre-mRNA. While this approach gives only a very rough estimation, our results indicate that between 0.5% and 2% of the pre-mRNA is trans-spliced. Note that the pre-mRNA is present in vast excess (see below).
Figure 2.
In vitro trans-splicing of synthetic pre-mRNA using T. brucei whole-cell extract. (A) A total of 0.5 µg pNS21-TIR pre-mRNA was incubated in a 20 µl reaction (∼300 µg of extract), in transcription buffer, for 1 h at 28°C, as described in ‘Materials and Methods’ section. RNA was extracted and subjected to RNase protection assay with an antisense probe complementary to the tubulin-luciferase trans-spliced transcript as described in ‘Materials and Methods’ section. Lane contents are as follows: 1, The PIR antisense probe; 2, RNase protection of RNA from transgenic pNS21-PIR; 3, RNase protection of RNA from the in vitro reaction with pre-PIR mRNA. (B) The same as in (A) but using the pNS21-TIR pre-mRNA. Lane contents are as follows: 1, The TIR antisense probe; 2, RNase protection of RNA from transgenic pNS21-TIR; 3, RNase protection of RNA from the in vitro reaction with pre-TIR mRNA. In both panels, the RNA products were separated on a 6% denaturing gel. M- DNA marker, labeled pBR322 DNA MspI digest. The size of the marker bands are indicated in nt. The identity of the different fragments is schematically illustrated.
The trans-splicing reaction is heat sensitive and requires ATP
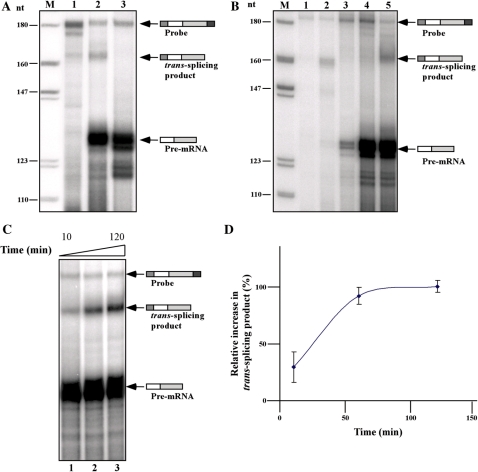
To examine if the reaction shares known properties of in vitro splicing reactions (22–24), two requirements must be met. The reaction should be inactivated by heat and should depend on the presence of ATP. The results in Figure 3A demonstrate that heat inactivation (incubating the extract for 30 min at 55°C) abolished the production of the trans-spliced product (compare lanes 2 to 3). The position of the trans-spliced product was determined by the migration of trans-spliced product generated from RNA extracted from the transgenic line expressing the TIR plasmid (lane 1). Next, the dependence of the reaction on ATP was examined. If the ATP regenerating system was omitted from the reaction, no trans-spliced product was generated (Figure 3B, compare lanes 4 and 5). If the reaction lacked pre-mRNA, and instead the TIR plasmid was added to the reaction under transcription conditions, pre-mRNA was produced but no trans-spliced product was detected, suggesting that the amount of pre-mRNA produced in vitro did not enable the detection of the trans-spliced product (Figure 3B, lane 3). Next, we examined the time dependence of the reaction; the results demonstrate that the reaction was linear up to 60 minutes (Figure 3C and D). No increase in the trans-spliced product was observed thereafter, most probably because of consumption of all the SL RNA present in the extract. The degradation of the pre-mRNA most probably takes place immediately after addition of the pre-mRNA, since no change in the level of protected pre-mRNA was observed at longer incubation times, suggesting that the pre-mRNA is protected from further degradation once it is assembled into splicing complexes (Figure 3C; compare lanes 1–3).
Figure 3.
The trans-splicing reaction is heat-inactivated, requires ATP and is time-dependent. (A) Trans-splicing in heat-inactivated extracts. For heat inactivation, the trans-splicing extract was incubated for 30 min at 55°C. The trans-splicing product was compared to that produced in untreated extract. Total RNA was prepared from the reaction and subjected to RNAse protection as described in ‘Materials and Methods’ section. Lane contents are as follows: 1, RNase protection assay with RNA (30 µg) from pNS21-TIR transgenic cells; 2, In vitro trans-splicing reaction using untreated extract (control); 3, In vitro trans-splicing reaction using heated-inactivated extract. (B) Co-transcriptional trans-splicing and ATP requirement of the trans-splicing reaction. In vitro trans-splicing was performed in the presence of 0.8 µg pNS-21-TIR plasmid DNA as described in the ‘Results’ section. For ATP depletion, the ATP regenerating system was omitted from the reaction. After incubation at 28°C for 1 h, total RNA was prepared and subjected to RNase protection as described in ‘Materials and Methods’ section. Lane contents are as follows: 1, Antisense probe (500 c.p.m.); 2, total RNA (30 µg) from the transgenic pNS21-TIR cells; 3, RNA from extracts containing plasmid pNS21-TIR; 4, RNA from trans-splicing reaction lacking the ATP regenerating system; 5, RNA from trans-splicing performed under optimal conditions (control). M- DNA marker, labeled pBR322 DNA MspI digest. (C) Kinetics of the in vitro trans-splicing reaction. In vitro trans-splicing was conducted under standard conditions but for different durations (10′, 60′ and 120′). RNA was subjected to RNase protection as described in ‘Materials and Methods’ section. (D) Quantitation of the time-dependent splicing from three independent experiments.
Optimization of the reaction conditions
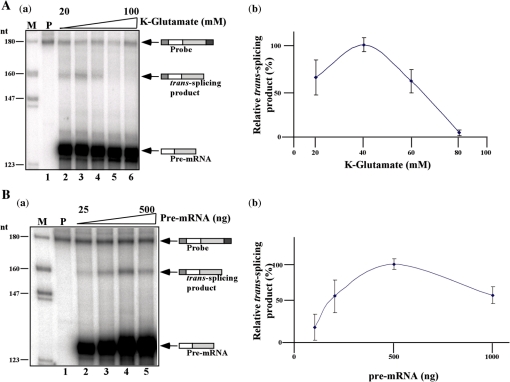
Salt concentration, the level of MgCl2, the concentration of reactants and the identity of the acceptor and donor RNAs can all dramatically change the yield of the trans-splicing reaction. Thus, it was necessary to determine the optimal concentration of each of these constituents within the reaction. Previously, the type of counter ion used was shown to affect the in vitro transcription reaction. Changing KCl to K-glutamate increases the efficiency of SL RNA transcription (35). Thus, the in vitro reaction was incubated in the presence of elevated salt concentration, and the production of the trans-spliced product was monitored. The results shown in Figure 4A indicate that 40 mM K-glutamate was the optimal salt concentration. As expected, at higher salt concentrations, above 60 mM, the reaction was completely inhibited. Next, the effect of MgCl2 concentration was examined and the results indicate that 3 mM MgCl2 was optimal (results not shown). The amount of pre-mRNA was also examined, and the results (Figure 4B) indicate that a relatively high concentration (0.5 µg per reaction) was needed to enable production of the trans-spliced product. These results demonstrate that the RNase protection assay reflects the increased amount of pre-mRNA present in the reaction. Note that concentrations higher than 0.5 µg poison the reaction. The results in the right panel present the quantification data from three independent experiments.
Figure 4.
Optimization of the in vitro trans-splicing reaction. (A) Effect of potassium-glutamate concentration on the reaction. In vitro trans-splicing reactions were carried out for 1 h at 28°C in the presence of 20, 40, 60, 80 and 100 mM K-glutamate (lanes 2–6). RNA was extracted and subjected to RNase protection. (B) Effect of increasing pre-mRNA concentration on the reaction. In vitro trans-splicing reactions were carried out in the presence of 25, 50, 125, 250 and 500 ng pNS21-TIR pre-mRNA (lanes 2–5). RNA was extracted and subjected to RNase protection. RNA samples from the different reactions were separated on a 6% sequencing gel. M- DNA marker, labeled pBR322 DNA MspI digest. P- Probe (500 c.p.m.). The identity of the fragments is presented schematically. The right panels show the quantitation of three independent experiments.
Addition of endogenous SL RNA but not in vitro transcribed SL RNA enhances the reaction efficiency
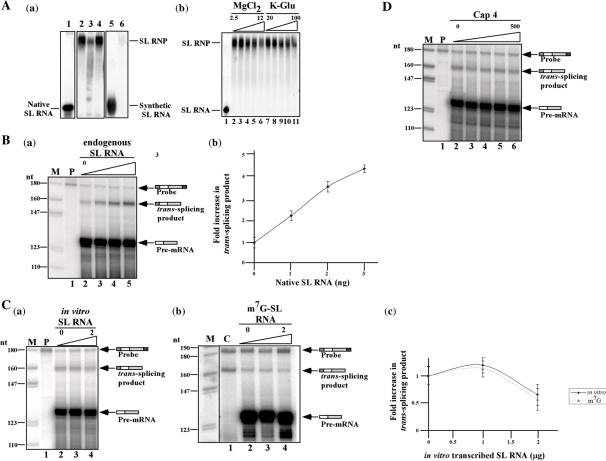
The trans-splicing reaction depends on the supply of endogenous SL RNA for the reaction to proceed. However, SL RNA might be a limiting factor in the extract. Exogenous SL RNA must first assemble into its cognate particle, the SL RNP, before it can participate in the reaction. The assembly of SL RNA into its particle was compared using in vitro transcribed SL RNA versus the assembly of native SL RNA that was synthesized in permeable cells. The results presented in Figure 5A-a, lanes 1 and 2, suggest that 100% of the native SL RNA prepared in permeable cells was incorporated into the SL RNP particle, whereas <1% of the in vitro transcribed SL RNA was assembled into the SL RNP (lanes 5 and 6). SL RNP assembly with the radiolabeled SL RNA was competed by the addition of cold-endogenous SL RNA (compare lanes 2 to 3) but not by tRNA (compare lanes 2 and 4). The assembly of SL RNA into the SL RNP was optimal when the reaction was performed with 4 mM MgCl2 and 30 mM K-glutamate (Figure 5A-b), conditions very similar to those used in the in vitro reaction (Figure 4).
Figure 5.
Assembly of authentic and synthetic SL RNA and their effect on trans-splicing. (A) (a) Assembly of the SL RNP. Nuclear extract (50 µg) was mixed with 5 × 103 c.p.m. of SL RNA synthesized in permeable cells or 2 × 105 c.p.m. of in vitro transcribed SL RNA under splicing conditions. The complexes were separated on 6% native acrylamide gel, as described (56). The position of the SL RNA and SL RNP are indicated. Lane contents are as follows: 1, Free SL RNA transcribed in permeable cells; 2, reconstitution of the particle using SL RNA transcribed in permeable cells; 3, the same as in lane 2, but 2 ng of gel-purified SL RNA was added to the reaction; 4, the same as in lane 2, but 300 ng of tRNA was added to the reaction; 5, SL RNA synthesized in vitro; 6, assembly in vitro transcribed SL RNA. (b). Assembly of SL RNP in the presence of elevated MgCl2 and K-glutamate levels. Assembly was as described in section (a), but in the presence of elevated MgCl2 (2.5, 4, 6, 8, 12 mM MgCl2) (lanes 2–6) and K-glutamate (20, 30, 40, 75, 100 mM) (lanes 7–11). The reactions were separated on a 6% native gel next to free RNA (lane 1). (B) (a) Addition of endogenous transcribed SL RNA increase trans-splicing efficiency. SL RNA was extracted from a gel, and its level was quantified by primer extension. The in vitro reaction was performed under standard conditions in the presence of elevated SL RNA (0, 1, 2, 3 ng) (lanes 2–5). The in vitro reaction was subjected to RNase protection assay. M-marker; labeled pBR322 DNA MspI digest P, antisense probe. (b) Quantitation of the increase in trans-spliced product. The data are based on three independent experiments. (C) (a). Addition of in vitro transcribed SL RNA does not increase trans-splicing efficiency. In vitro trans-splicing was performed under standard conditions in the presence of elevated levels of in vitro transcribed SL RNA (0, 1, 2 µg) (lanes 2–4). The in vitro reaction was subjected to RNase protection assay. M- marker; labeled pBR322 DNA MspI digest P, antisense probe. (b) Addition of in vitro transcribed m7G capped SL RNA does not increase trans-splicing efficiency. In vitro trans-splicing was performed under standard conditions in the presence of elevated levels of in vitro transcribed m7G capped SL RNA (0, 1, 2 µg) (lanes 2–4). The in vitro reaction was subjected to RNase protection assay. M- marker; labeled pBR322 DNA MspI digest C, total RNA (30 µg) from the transgenic pNS21-TIR cells. (c) Quantitation of the increase in trans-spliced products upon addition of in vitro transcribed SL RNA. The data are based on three independent experiments. Solid line, addition of uncapped in vitro transcribed SL RNA; dotted line, addition of m7G capped in vitro transcribed SL RNA. (D) The effect of cap-4 on trans-splicing. The in vitro trans-splicing reaction was carried out in the presence of increasing amount of synthetic cap-4 (0, 5, 50, 500, 5000 nM) (lanes 2–6). M-DNA marker, labeled pBR322 DNA MspI digest P-Probe (500 c.p.m.). The identity of the fragments is presented schematically.
Next, we examined whether addition of native SL RNA or in vitro transcribed SL RNA to the in vitro splicing reaction could increase the efficiency of producing the trans-spliced product. Addition of increasing amounts of endogenous SL RNA (up to 3 ng), increased the efficiency of the reaction, suggesting that the SL RNP is limiting in the extracts (Figure 5B). In contrast, addition of increasing amounts of synthetic SL RNA (up to 2 µg) had no effect on the production of trans-spliced product (Figure 5C-a); This result was expected based on the much more efficient assembly of endogenous SL RNA relative to the synthetic molecule. Next, it was determined if in vitro transcribed SL RNA containing an m7G cap could be used as a substrate for in vitro trans-splicing. Addition of up to 2 µg of synthetic m7G capped SL RNA did not increase the generation of the trans-spliced product (Figure 5C-b). This synthetic SL RNA also failed to efficiently assemble into SL RNP (data not shown), suggesting that the cap-4 modification might be necessary for SL RNP biogenesis. Therefore, increasing amounts of synthetic cap-4 (38) were added to the reaction. Figure 5D shows that adding cap-4 at concentrations up to 50 µM did not affect the reaction. Thus suggesting that cap-4 needs to be attached to the SL RNA in order to promote proper SL RNP assembly and that cap-4 provided in trans can not interfere with this process.
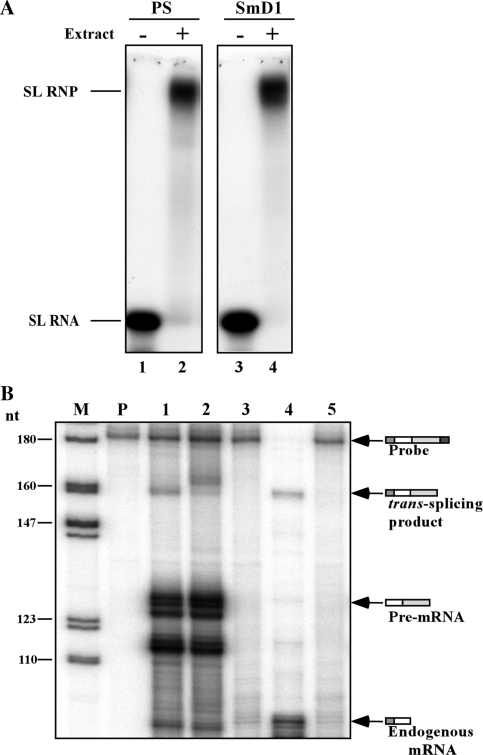
The absence of the +4 cap modification does not affect in vitro trans-splicing
To examine the dependence of trans-splicing in vitro on cap-4, we prepared an extract from SmD1-silenced cells. In these silenced cells, trans-splicing is inhibited, and SL RNA lacking modification at the +4 position of cap-4 accumulates (14). The efficiency of knockdown is variable, and silencing is not very efficient if cells are silenced in large quantities. The silenced population also contains cells that are unaffected, and thus Sm proteins are nevertheless present to some extent in the extracts. When extracts are made from such cells, the majority of SL RNA however, is ‘defective’ SL RNA. First, we examined if SL RNA synthesized from SmD1-silenced cells can assemble efficiently into SL RNP particles. SL RNA was prepared from SmD1-silenced permeable cells. The SL RNA synthesized under these conditions lacks the +4 cap-4 modification and possesses a 5-nt 3′ tail (14). Defective SL RNA synthesized in permeable SmD1-silenced cells was able to assemble properly into SL RNP particles (Figure 6A compare lanes 2 and 4). Next, we examined the capacity of extracts prepared from SmD1-silenced cells to carry out the in vitro trans-splicing reaction. Using these extracts, two distinct protected fragments were observed compared to a single protected fragment observed when wild-type splicing extract was used (Fig. 6B, compare lanes 1 and 2). In lane 2, the lower protected fragment represents splicing from the residual wild-type SL RNA present in the extract, and the upper fragment from splicing of defective SL RNA. The difference in size may reflect the fact that SL RNA lacking the cap-4 modification protects the last four hypermodified nucleotides. The results also suggest that SL RNA lacking the cap-4 +4 nucleotide and harboring a 5-nt tail can serve as a bona-fide trans-splicing substrate.
Figure 6.
Trans-splicing using extracts from SmD1-silenced cells. (A) SL RNA produced in SmD1-silenced cells can assemble into SL RNP. Nuclear extract (50 µg) was mixed with 5 × 103 c.p.m. of SL RNA synthesized in permeable parental cells or SmD1-silenced permeable cells. The complexes were separated on a 6% native acrylamide gel. Lane contents are as follows: 1, SL RNA synthesized in parental permeable cells; 2, reconstitution of SL RNP using parental cell extract; 3, free SL RNA synthesized in SmD1-silenced cells; 4, reconstitution of SL RNP in extracts prepared from SmD1-silenced cells. (B) In vitro trans-splicing reactions. The in vitro reaction was conducted with parental cell extracts or extracts prepared from the SmD1-silenced cells. SmD1 extracts were prepared from 5 × 109 cells 3 days after tetracycline addition as described (14). The RNA purified from the reactions was subjected to RNase protection. M- DNA marker, labeled pBR322 DNA MspI digest P- Probe (500 c.p.m.). Lane contents are as follows: 1, RNA from in vitro trans-splicing using parental strain extract; 2, in vitro trans-splicing using extracts from SmD1-silenced cells; 3, in vitro trans-splicing extract without pre-mRNA; 4, RNA from transgenic parasites expressing pNS21-TIR; 5, RNA from parental strain. RNA from the protection assays was separated on 6% acrylamide denaturing gels.
Inactivation of U2 or U6 snRNA reduces the splicing efficiency
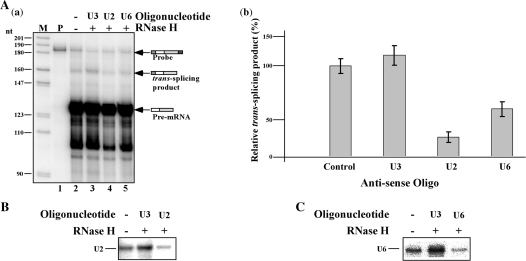
It was previously demonstrated that trans-splicing is inhibited in permeable trypanosome cells when either U2 or U6 snRNAs are inactivated by RNase H cleavage (39). To examine if trans-splicing in vitro relies on the presence of intact U2 and U6 snRNAs, extracts were incubated with U2 and U6 antisense oligonucleotides and RNase H. U3 cleavage was used as a control. The same oligonucleotides, U2SSC and U6-15, were previously used to inactivate these snRNAs in permeable cells (39). Cleavage reduced the level of U2 and U6 snRNA by 50% and 40%, respectively, as evident by primer extension that was performed on RNA extracted from the oligonucleotide-RNAse H treated extracts (Figure 7B and C). Indeed, in extracts depleted of these snRNAs, the production of the trans-spliced product was reduced by 60% and 45%, in accordance with the reduction in snRNAs (Figure 7A; lanes 4 and 5). These results suggest that, as expected, the reaction depends on the snRNAs that constitute the catalytic center of pre-mRNA splicing (40). The cleavage of U3 did not affect the level of U2 (Figure 7B, lane 2) or U6 (Figure 7C; lane 2) snRNAs and did not affect splicing (Figure 7A, lane 3).
Figure 7.
RNaseH digestion of U2 or U6 snRNAs inhibits in vitro trans-splicing. (A) (a) Whole-cell extract was incubated for 1 h on ice with oligonucleotides complementary to U3, U2 or U6 snRNAs, in the presence of 1U RNase H, as described in ‘Materials and Methods’ section. The RNA from the different reactions was analyzed by an RNase protection assay. RNA was separated on a 6% sequencing gel. Lane contents are as follows: 1, Probe (500 c.p.m.); 2, RNA from in vitro trans-splicing extract (control); 3, extract was subjected to RNase H cleavage with U3-specific oligonucleotide; 4, extract was subjected to RNase H cleavage with U2-specific oligonucleotide; 5, extract was subjected to RNase H cleavage with U6-specific oligonucleotide. M-DNA marker, labeled pBR322 DNA MspI digest. (b) Quantitation of the effect on the production of the trans-spliced product; data represent three independent experiments. (B) Primer extension to determine the amount of U2 snRNA after RNase H treatment. Primer extension was performed on RNase H-oligonucleotide cleaved extracts using U2 specific primer (listed in S-1). The identity of the antisense oligonucleotides used for cleavage is indicated. (C) Primer extension to determine the amount of U6 snRNA after RNase H treatment. Primer extension was performed on RNase H-oligonucleotide cleaved extracts using U6 specific primer (listed in S-1). The identity of the antisense oligonucleotides used for cleavage is indicated.
Mutations in the 3′ splice and polypyrimidine tract compromise the production of the trans-spliced product
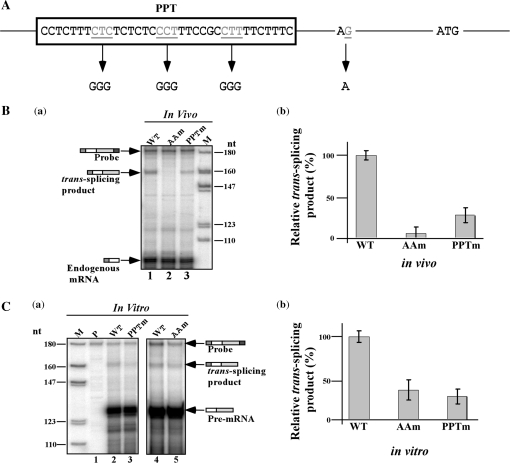
The AG splice site and the PPT are sequences essential for trans-splicing (28,32,33). It was therefore essential to demonstrate that the production of the trans-spliced product is affected if these sequences are mutated. To this end, mutations were introduced to the AG splice site and the PPT, as illustrated in Figure 8A. Mutations were constructed and the mutated genes were cloned into the pNS21 vector. Transgenic parasites were obtained, and the production of the trans-spliced protected fragment was examined by RNase protection assay using RNA isolated from the transgenic parasites. The results demonstrate (Figure 8B) that the AG to AA mutation reduced the production of the trans-spliced product by 90% (compare lanes 1 and 2), whereas the PPT mutation reduced the production of the trans-spliced product by ∼70% compared to the wild-type transcript (compare lanes 1 and 3). cDNA was prepared from the transgenic parasites and amplified with the SL forward primer and antisense luciferase primer. Although the RNase protection assay showed complete inhibition of the production of trans-spliced product using the RNase protection assay (Figure 8B), a PCR product was nevertheless obtained after PCR amplification. Sequence analysis indicated that SL was added exclusively 9-nt downstream to the authentic AG splice site, suggesting that, in the absence of a bonafide site, the splicing system ‘scans’ for an alternative AG splice site.
Figure 8.
Trans-splicing efficiency is reduced in AG and PPT mutants. (A) Schematic representation of the mutations introduced in the AG 3′ splice site (3′ss) and polypyrimidine tract (PPT) of β-tubulin upstream intergenic sequence. Generation of the mutants and preparation of the corresponding cell lines were performed as described in ‘Materials and Methods’ section. (B) (a) In vivo trans-splicing in transgenic parasites expressing the pNS21-TIR mutants. The level of the expressed trans-spliced product was measured by RNase protection using an antisense probe. RNA from the RNase protection assays was separated on a 6% acrylamide/denaturing gel. Lane contents are as follows: 1, RNA from cells expressing the wild-type transcript (pNS21-TIR); 2, RNA from cell lines expressing pNS21-TIRAAm; 3, RNA from cell lines expressing pNS21-TIRPPTm. (b) Reduction in expression of the trans-spliced product relative to the wild-type transcript. The data are derived from three independent experiments. (C) (a) In vitro trans-splicing of the AG and PPT mutants. Pre-mRNA was prepared from the mutants and the trans-splicing in vitro was compared to that of wild-type transcripts. RNA from the in vitro reaction was subjected to RNase protection and the products separated on 6% denaturing acrylamide gels. The identity of the RNA used in the reaction is indicated. M- DNA marker, labeled pBR322 DNA MspI digest. The identity of the protected fragments is indicated. (b) Quantitation of the reduction in splicing. The results are derived from three independent experiments.
To investigate whether the splicing signal mutants are trans-spliced in vitro similarly to their splicing in vivo, pre-mRNAs were prepared from the mutants and the production of the trans-spliced product was examined. The results (Figure 8C) suggest that both mutations affected the ability to form the spliced product. The reduction in the formation of the trans-spliced product for the AA and PPT mutants was 60% and 70%, respectively (Figure 8C-b). Interestingly, the PPT mutation affected the in vitro reaction more strongly than its effect in vivo. The AA mutation, however, had a less severe effect in vitro compared to in vivo (compare Figure 8C-a, lanes 3 and 5 to Figure 8B-a, lanes 2 and 3).
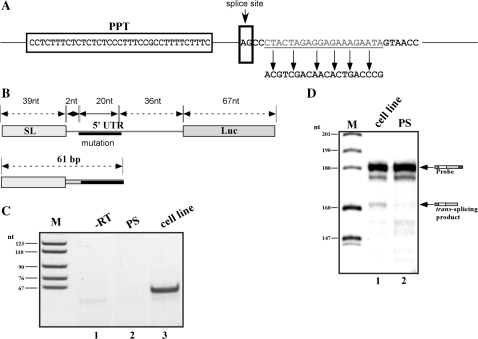
Generating a TIR mutation downstream to the AG splice site to produce a model substrate for RT–PCR or real-time PCR assays
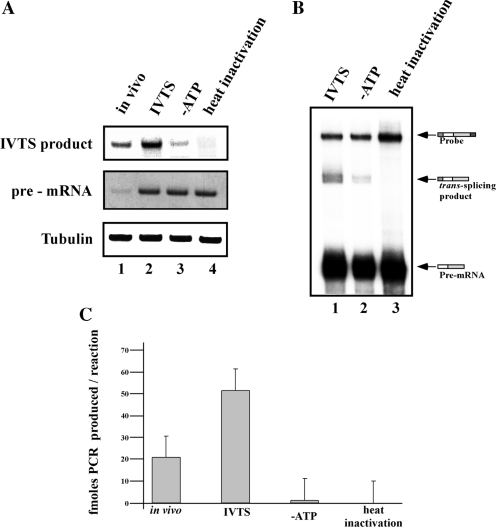
The RNase protection assay used to monitor the production of the trans-spliced product is a very laborious and time-consuming assay. However, it allows examination of the trans-spliced product without any amplification and is therefore less prone to artifacts compared to assays that involve amplification. Nevertheless, the technical difficulties in the RNase protection assay led us to develop an alternative RT–PCR assay that involves an amplification step. Performing RT–PCR on an in vitro reaction, using the pre-TIR as a substrate and the same oligonucleotides that were used to produce the cDNA in the transgenic parasites, produced a trans-spliced product whose production was insensitive to ATP depletion or to heat inactivation, suggesting that amplification can take place that is not dependent on trans-splicing. We therefore generated a 20-nt substitution mutation 2-nt downstream of the AG splice site (Figure 9A). The mutant was cloned into pNS-21, and cell lines were generated (pNS21-TIRsub). RNA was prepared from the cell line and used to amplify the cDNA using pairs of forward (SL) and antisense primers specific to the mutation (Figure 9C). No such product could be amplified using cDNA from cells expressing pNS21-TIR. The PCR product was sequenced and the results indicated that SL was added to the authentic site. Next, an antisense RNA probe was made from this clone and used in the RNase protection assay. The results in Figure 9D show that the protected fragment appeared only in RNA extracted from the mutant but not from the parental strain (compare lanes 1 and 2). Pre-mRNA was produced from pNS21-TIRsub, and the production of the in vitro trans-spliced product was examined by RNase protection assay. Production of the trans-spliced product was observed, and was dependent on ATP and inactivated by heat (Figure 10B, compare lanes 1 to 3). Next, we examined the same RNA in the RT–PCR assay. cDNA was prepared from the same reactions presented in (B), and the cDNA was amplified with three different pairs of oligonucleotides, a pair to amplify the pre-mRNA, the endogenous tubulin mRNA, and the trans-spliced product. The trans-spliced product was amplified with an antisense oligonucleotide complementary to the mutation and a forward SL oligonucleotide. The results indicate that identical amounts of pre-mRNA and extract (based on a tubulin control) were present in each reaction (Figure 10A, lanes 2 to 4), but the level of the trans-spliced product was sensitive to conditions that affected splicing. Thus, the RT–PCR results are in agreement with the RNase protection assay, and show 85% and 96% reduction in the formation of the trans-spliced product under ATP depletion or heat-inactivation, respectively (Figure 10).
Figure 9.
In vivo trans-splicing of the pNS21-TIRsub mutation by RT–PCR and RNase protection assay. (A) Schematic presentation of the substitution mutant. The PPT sequence and AG splice site are indicated, the sequence changes to create the mutation are marked. (B) The structure of the trans-spliced product. The distance between the strategic domains is given in nt. The structure and size of the RT–PCR product is shown. (C) RT–PCR assay to detect the trans-spliced product carrying the substitution mutation. cDNA was prepared from total RNA (10 µg) of parental and the pNS21-TIRsub expressing cell line, using random primers as described in ‘Materials and Methods’ section. The cDNA was amplified with the SL forward primer and reverse primer specific for the mutation (see S-1). Lane contents are as follows: 1, PCR product of RNA prepared from the cell line carrying pNS21-TIRsub; 2, cDNA prepared from parental strain; 3, cDNA was prepared from cells expressing pNS21-TIRsub. The PCR product was separated next to size markers on a 10% acrylamide gel, and the gel was stained with ethidium bromide. (D) RNase protection assay to detect the expression of pNS21-TIRsub. RNA was prepared from parental strain and from cells expressing the pNS21-TIRsub construct. After RNase protection with antisense RNA probe complementary to the mutated RNA, the RNA was separated on a 6% denaturing acrylamide gel. The identity of the fragments is indicated. M, marker, labeled pBR322 DNA MspI digest. Lane contents are as follows: 1, RNA from the cell line expressing pNS21-TIRsub; 2, RNA from parental strain.
Figure 10.
Detection of the trans-spliced product by RT-PCR and real-time PCR. (A) RT–PCR assay of the in vitro trans-splicing reaction. RNA was prepared from transgenic cells expressing pNS21-TIRsub and from in vitro trans-splicing assays using pre-mRNA of TIR-sub. cDNA was prepared from the different samples and was subjected to amplification with the different probes (pre-mRNA, tubulin and the primers specific for pNS21-TIRsub), as described in ‘Materials and Methods’ section. The primers are listed in S-1. The PCR product was separated on a 10% acrylamide gel and stained with ethidium bromide. The in vitro reaction was performed in the absence of ATP regenerating system or using heat-inactivated extract, prepared as described in Figure 3. Lane contents are as follows: 1, cDNA from cell line expressing pNS21-TIRsub; 2, cDNA prepared from in vitro reaction performed under standard conditions; 3, cDNA prepared from in vitro trans-splicing performed in the absence of ATP regenerating system; 4, cDNA prepared from in vitro trans-splicing using a heat-inactivated extract. (B) RNase protection assay. RNA prepared from the in vitro reactions performed in Panel A was subjected to RNase protection assay. The RNA was separated on a 6% denaturing gel. Lane contents are as follows: 1, RNA from in vitro trans-splicing using pre-TIRsub; 2, the same as in lane 1 but the in vitro reaction lacked ATP regenerating system; 3, the same as in lane 1 but a heat-inactivated extract was used. The identity of the fragments is indicated. (C) Quantitation of the real-time PCR reactions. Real time-PCR was performed as described in ‘Materials and Methods’ section. The concentration curve was used to determine the total amount of PCR amplified in each reaction. The results shown are the average of three independent experiments. The designation of the different columns are: in vivo, the amount of PCR product (fmol) from 10 µg total RNA extracted from the transgenic cell line; IVTS, amount of PCR product (fmol) produced from a standard in vitro trans-splicing reaction; –ATP, amount of PCR product (fmol) produced from an in vitro trans-splicing reaction in the absence of the ATP regeneration system; heat inactivation, amount of PCR product (fmol) produced from one in vitro trans-splicing reaction using heat-inactivated extract.
Real-time PCR
In order to quantify the exact amount of PCR product that is amplified from the in vitro trans-splicing reactions, and to show that the RT–PCR performed above was done while the reaction was in exponential phase, qRT–PCR was performed. A melting curve was performed to show that only a single trans-spliced product was amplified during the qRT–PCR, and the melting curve was identical for the fragment formed using the cDNA from the transgenic parasite and from the in vitro reaction (results not shown). Finally, a PCR product of known quantity was used to establish a concentration curve. This concentration curve was then used to determine the amount of PCR product amplified from an in vitro trans-splicing reaction. As can be seen in Figure 10C, a standard in vitro trans-splicing reaction, in which 4 pmol of pre-mRNA was added yielded 50 fmol of PCR product. This is comparable to the amount of PCR product amplified from 10 µg of total RNA from transgenic parasites expressing pNS21-TIRsub. Performing qRT–PCR on reactions in the absence of ATP or after heat inactivation yielded ∼1 fmol and 0 fmol, respectively. The reduction in PCR amplification in the absence of ATP or upon heat inactivation was similar to the reduction as determined by RT–PCR and RNase protection. Thus, these experiments establish three different assays that faithfully monitor the production of trans-spliced product generated in vitro from synthetic pre-mRNA and endogenous SL RNA and demonstrate some of the requirements for these assays.
DISCUSSION
In this study, we describe for the first time a system that is able to faithfully monitor the formation of a chimeric trans-spliced RNA product in vitro using a T. brucei soluble cell extract and synthetic in vitro transcribed pre-mRNA. The ability to establish such an in vitro system stems from the reaction conditions and the sensitive assays used in this study. The study demonstrates that SL RNP is a limiting factor in the reaction, and only endogenous SL RNA but not m7G capped synthetic SL RNA can be utilized. Interestingly, SL RNA lacking the +4 cap modification and possessing a 5-nt longer tail can be utilized in vitro. The assembly of synthetic and endogenous wild-type or SL RNA from SmD1-silenced cells suggests that the cap-4 modifications but not the cap-4 +4 modification are essential for SL RNP assembly. The performance of the in vitro trans-splicing reaction meets the requirements observed for other splicing systems, specifically the dependence on ATP, and on U2 and U6 snRNAs. Mutations in strategic positions such as the polypyrimidine tract and the AG splice site compromised the reaction in vivo and in vitro.
The most essential factors for establishing the in vitro system
The failure to date to establish an in vitro system in trypanosomes lies in the use of radiolabeled pre-mRNA substrates that were extensively degraded, and the deficiency in essential factor(s) in the extract. Massive degradation of pre-mRNA was also observed in our study. This degradation could not be eliminated even if the mRNA was capped at the 5′-end or at the 3′-end with pCp (our unpublished data). Our success in detecting the trans-spliced product in this study lies in three parameters: (i) the extract used is crude but contains all the factors necessary for the reaction. It is prepared very rapidly to eliminate the potential inactivation of essential factors. However, because the extract is not purified, it contains enzymes that may inactivate essential factors, thus reducing the reaction efficiency. (ii) A large amount of pre-mRNA is added to the reaction, leaving sufficient amounts of substrate (after its immediate degradation) to assemble into an active complex. The amount of pre-mRNA (4 pmol) is 50-fold higher than the amount of SL RNA in the extract (∼80 fmol). Indeed, we demonstrate that increasing the amount of pre-mRNA increases the reaction efficiency (Figure 4C). (iii) The assays used in this study. This is the most critical parameter that made it possible to detect the trans-spliced product. Three different assays were used. In all the assays, only the end-product but none of the intermediates are monitored. The reaction efficiency, roughly calculated by the RNase protection assay, is ∼1–2% of the input pre-mRNA utilizing almost 50% of the SL RNA present in the extract. Thus, the limiting factor in this system is the SL RNA.
Although the traditional assays to monitor splicing so far are based on the use of radiolabeled substrate, the real-time PCR assay represents a much desired substitute. It avoids the use of radiolabeled materials; it is quantitative, and is able to quantitatively detect the amount of trans-spliced product even at levels less than a few femtomoles. In addition, the assay is convenient relative to the RNase protection assay. Note that in all assays used in this study, the in vitro trans-spliced product was compared to the genuine trans-spliced product produced in vivo in transgenic parasites.
An additional parameter that was essential for establishing the in vitro system was the choice of pre-mRNA substrate. Our results indicate that the TIR pre-mRNA was a better substrate than the PIR pre-mRNA. The poor performance of the PIR substrate in vitro might stem from its size. The larger substrate appeared more vulnerable to degradation. In other splicing systems, it was demonstrated that shortening both the exons and the introns resulted in a substrate that is spliced more efficiently (26). In addition, microarray data of the transcriptome of cells silenced for splicing factors suggest that EP splicing requires a high level of SR proteins (Gupta and S.M., unpublished data). These factors, in the proper phosphorylation form, might be limited in the extracts used in this study. The choice of the β-tubulin gene over α-tubulin was based on the finding that efficient α-tubulin splicing in vivo requires exonic sequences present in the UTR. Such splicing might require additional factor(s) to enhance the protein binding to suboptimal sequences present in the vicinity of the AG splice site of the α-tubulin transcript (33).
The effect of pre-mRNA mutations on trans-splicing in vivo and in vitro
One of the interesting results of this study is the differential splicing of the AG and PPT mutants in vivo and in vitro. The mutation of AG to AA almost completely abolished the production of the trans-spliced product in vivo (Figure 8B). However, in vitro, only a 60% reduction was observed (Figure 8C). RT–PCR of RNA extracted from the transgenic parasites revealed the production of a trans-spliced product where an alternative AG located 6-nt downstream to the authentic site was utilized. Using this cDNA in an RNase protection assay showed that alternative selection of this splice site took place when either wild-type transcript or the mutant AG to AA were used in vitro (our unpublished data), suggesting that in vitro, as opposed to in vivo, an alternative AG splice site can be chosen.
The difference between the in vivo and the in vitro utilization of the AG to AA mutant and the choice of an alternative AG splice site may be explained by the shortage in the extract of a factor that functions in selecting the optimal AG splice site. One such candidate is the U2AF65 homologue (20). Our recent study indicates that this large protein (90 kDa) is subjected to proteolysis in the extract. In addition, the active protein is phosphorylated, and the presence of phosphatases in the extract may render this factor inactive.
The PPT mutation, like the splice site mutation, had a different effect in the in vitro reaction. This mutation affected the in vitro reaction as strongly as the AG to AA mutation. In mammals, mutation or deletion of the AG splice site completely blocks ligation (step2), while the first step proceeds, albeit at reduced efficiency. In contrast, mutation of the PPT drastically reduces the efficiency of the first step; thus, the PPT sequence seems to be more important in splicing than the AG dinucleotide (41). In yeast, the 3′ splice site mutations activate a cryptic AG splice site even if this is not preceded by a PPT (42), while in mammals, the cryptic 3′ splice site is always preceded by a PPT (43). These results suggest that PPT recognition is more essential in mammalian than in yeast splicing. The PPT seems to also be very important for trans-splicing, especially in vitro. The trypanosome PPT was shown to be essential for trans-splicing and polyadenylation of the upstream gene (44–46). Mutations introduced in the PPT of trypanosome pre-mRNA suggest that although the size of the PPT does not have a large effect on the trans-splicing efficiency, its composition does. Replacing uridines with two or three consecutive purines severely compromises in vivo trans-splicing (28). Indeed, the mutation we introduced not only reduced the level of U’s within the tract but inserted consecutive purines into the PPT site (Figure 8A).
Endogenous SL RNA but not synthetic SL RNA can enhance the in vitro reaction
In nematodes, in vitro transcribed radiolabeled SL RNA is properly capped and efficiently utilized in the in vitro trans-splicing reaction (25). This is not the case with the trypanosome SL RNA. The special cap-4 is added to the SL RNA co-transcriptionally (47), and capping requires that the nascent SL RNA adopt a special conformation dictated by its interaction with SLA1 (11). Sm assembly is coupled with the cap-4 +4 modification (14); therefore, the in vitro transcribed SL RNA can not assemble efficiently with the Sm core proteins, and hence, the SL RNA is subject to major degradation. Indeed, in the reconstitution experiments presented in Figure 5A, we could not detect unassembled SL RNA; such RNA is most probably rapidly degraded in the extract. The capping of SL RNA by m7G neither improved its stability (our unpublished data), nor its ability to be utilized in the trans-splicing reaction (Figure 5C-b). The in vitro transcribed SL RNA can not assemble properly into its cognate particle (results not shown) and therefore can not associate with the trans-spliceosome. In contrast, SL RNA derived from SmD1-silenced cells that lack the +4 cap-4 modification can properly assemble and be utilized in the reaction (Figure 6B). Although the SmD1 SL RNA is defective compared to the wild-type RNA, it contains all the cap modifications with the exception of cap +4. This may suggest that these modifications are crucial for SL RNP assembly. Indeed, our results are in agreement with the in vivo data demonstrating that an SL RNA mutant in the cap-4 +4 nucleotide has only a minor effect on its utilization compared to mutations in the +1 and +2 cap nucleotides, which severely affect trans-splicing in vivo (9).
In mammalian cells, it was demonstrated that splicing is enhanced by using capped pre-mRNA (48). In yeast, however, addition of cap analogues does not affect the reaction (22). In trypanosomes, the cap-4 may have additional biological functions during SL RNP assembly, other than just protecting the SL RNA from degradation. The main function of cap-4 might not be in the splicing reaction itself, but rather, in the assembly of functional SL RNP. The cap modification may serve as a landmark controlling the fidelity of SL RNA biogenesis; thus, only SL RNA containing the majority of modifications can properly assemble in vitro. Indeed, it was shown that effector proteins distinguish the fully methylated form of the cap-4 from the incomplete cap structure. For instance, the T. brucei nuclear cap-binding complex CBP20 binds with higher affinity to the mature cap-4 compared to uncapped SL RNA (49). In Leishmania, homologues of the cap-dependent translation initiation factor eIF4E distinguish between different SL RNA caps (50). Interestingly, although cells can resist the loss of either modification at the cap-2 (51) or at positions 3 and 4 (51,52), it is not possible to derive viable cells with complete loss of cap ribose methylation (53). Indeed, in vitro transcribed SL RNA lacking all the trypanosome-specific modifications can not assemble into SL RNP nor participate in the trans-splicing reaction in vitro. These modifications are therefore also essential for trans-splicing.
One of future challenges will be to synthesize in vitro SL RNA that can be utilized in the in vitro trans-splicing reaction. It will be interesting to use SL RNA from cells with knock-out of TbMTr1, TbMTr2, TbMTr3, or even a TbMTr-2 and 3 double knock-out, to examine the ability of undermethylated SL RNA to serve as a substrate in the in vitro trans-splicing reaction. Once the modifications that are most relevant for trans-splicing are identified, it may be possible to synthesize in vitro the properly capped SL RNA using the relevant recombinant methylating enzyme(s).
What is the significance of an in vitro trans-splicing system and how can the system be improved
In this study, we utilized the in vitro system to begin to assess the role of cap modification on the trans-splicing reaction, and this can be further extended as outlined above. However, there are still many additional open questions related to our understanding of the mechanism and the machinery of trypanosome trans-splicing. One major question is identifying the protein factor(s) that mediate the interactions between the SL RNP and the pre-mRNA machinery. In addition, relatively little is known about what protein factors and sequences are essential for determining the trans-splicing efficiency and the selection of the proper 3′ AG splice site. Transgenic parasites expressing reporter genes fused to upstream regulatory sequences were used to identify structural constraints within the 3′ AG splice site, and the PPT was shown to be highly essential for trans-splicing and its linkage to polyadenylation (28,32–34). However, such results may be influenced by mRNA stability, and the translatability of the reporter gene. An in vitro system will therefore help to determine the exact role of sequences and factors in trans-splicing. Recently, we described two polypyrimidine tract binding proteins in trypanosomes that affect both trans-splicing and mRNA stability (54). However, in our studies, it was very difficult to dissect the contribution of these RNA binding proteins to trans-splicing as compared to mRNA stability. The in vitro system should assist in defining the factors that are essential for trans-splicing of C rich polypyrimidine tract pre-mRNAs that bind PTB. Thus, in vitro trans-splicing could be used to study which factors and sequences are required for differential trans-splicing, for example of the three phosphoglyceratekinase genes present in the same polycistronic unit in T. brucei (55). However, the immediate next goal is to identify factor(s) that might be missing or inactivated in the extracts, resulting in their reduced efficiency, especially for splicing genes that require additional factors. This can be achieved by either ‘spiking’ the reaction with fractionated extracts, or adding factors such as the U2AF65 homologue as recombinant proteins.
This study is only the first step towards developing a robust cell-free system that, together with data from in vivo studies, should help to reveal the mechanism controlling the efficiency and fidelity of trypanosome trans-splicing. Trans-splicing is one of the most essential mRNA processing mechanisms in trypanosomes. This is the main biochemical process that differentiates the host from the parasite and therefore a better understanding of the mechanism may also identify novel targets for therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
United States-Israel Binational Science Foundation grant, by the Deutsche Forchshungsgemeinschaft (DFG); International Research Scholar grant from the Howard Hughes Medical Institute (to S.M., E.D.); David and Inzez Myers Chair in RNA silencing of diseases (to S.M.). Funding for open access charge: Deutsche Forchshungsgemeinschaft (DFG).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Nicolai Siegel and George Cross (Rockefeller University) for providing us with the pNS21 vector. We also thank Arthur Günzl (University of Connecticut Health Center) for valuable advice on extract preparation.
REFERENCES
- 1.Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 2.Liang XH, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in Trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenthal T. Trans-splicing and operons. WormBook. 2005:1–9. doi: 10.1895/wormbook.1.5.1. (25 June 2009, date last accessed) [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl Acad. Sci. USA. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilinger G, Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell DA, Sturm NR, Yu MC. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today. 2000;16:78–82. doi: 10.1016/s0169-4758(99)01545-8. [DOI] [PubMed] [Google Scholar]
- 8.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 9.Mandelboim M, Estraño CL, Tschudi C, Ullu E, Michaeli S. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 2002;277:35210–35218. doi: 10.1074/jbc.M201910200. [DOI] [PubMed] [Google Scholar]
- 10.Palfi Z, Jae N, Preusser C, Kaminska KH, Bujnicki JM, Lee JH, Günzl A, Kambach C, Urlaub H, Bindereif A. SMN-assisted assembly of snRNP-specific Sm cores in trypanosomes. Genes Dev. 2009;23:1650–1664. doi: 10.1101/gad.526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot. Cell. 2009;8:56–68. doi: 10.1128/EC.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tkacz ID, Lustig Y, Stern MZ, Biton M, Salmon-Divon M, Das A, Bellofatto V, Michaeli S. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA. 2007;13:30–43. doi: 10.1261/rna.174307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dossin FM, Schenkman S. Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot. Cell. 2005;4:960–970. doi: 10.1128/EC.4.5.960-970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 2003;278:51469–51478. doi: 10.1074/jbc.M308997200. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Liang XH, Uliel S, Belahcen M, Unger R, Michaeli S. Identification and functional characterization of Lsm proteins in Trypanosoma brucei. J. Biol. Chem. 2004;279:18210–18219. doi: 10.1074/jbc.M400678200. [DOI] [PubMed] [Google Scholar]
- 16.Tkacz ID, Cohen S, Salmon-Divon M, Michaeli S. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei Mol. Biochem. Parasitol. 2008;160:22–31. doi: 10.1016/j.molbiopara.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang P, Palfi Z, Preusser C, Lücke S, Lane WS, Kambach C, Bindereif A. Sm core variation in spliceosomal small nuclear ribonucleoproteins from Trypanosoma brucei. EMBO J. 2006;25:4513–4523. doi: 10.1038/sj.emboj.7601328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luz Ambrosio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Gunzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot. Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 2006;145:29–39. doi: 10.1016/j.molbiopara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Vazquez MP, Mualem D, Bercovich N, Stern MZ, Nyambega B, Barda O, Mesiga D, Kumar-Gupta S, Michaeli S, Levin MJ. Functional characterization and protein-protein interactions of trypanasome factors U2AF35, U2AF65 and SF1. Mol. Biochem. Parasitol. 2009;164:137–146. doi: 10.1016/j.molbiopara.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez N, Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983;35:89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- 22.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 23.Padgett RA, Hardy SF, Sharp PA. Splicing of adenovirus RNA in a cell-free transcription system. Proc. Natl Acad. Sci. USA. 1983;80:5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannon GJ, Maroney PA, Denker JA, Nilsen TW. Trans splicing of nematode pre-messenger RNA in vitro. Cell. 1990;61:1247–1255. doi: 10.1016/0092-8674(90)90689-c. [DOI] [PubMed] [Google Scholar]
- 25.Maroney PA, Hannon GJ, Denker JA, Nilsen TW. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J. 1990;9:3667–3673. doi: 10.1002/j.1460-2075.1990.tb07578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabowski PJ, Padgett RA, Sharp PA. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984;37:415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- 27.Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu. Rev. Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 28.Siegel TN, Tan KS, Cross GA. Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei. Mol. Cell Biol. 2005;25:9586–9594. doi: 10.1128/MCB.25.21.9586-9594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laufer G, Schaaf G, Bollgonn S, Günzl A. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 1999;19:5466–5473. doi: 10.1128/mcb.19.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimanski B, Nguyen TN, Günzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lustig Y, Sheiner L, Vagima Y, Goldshmidt H, Das A, Bellofatto V, Michaeli S. Spliced-leader RNA silencing: a novel stress-induced mechanism in Trypanosoma brucei. EMBO Rep. 2007;8:408–413. doi: 10.1038/sj.embor.7400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol. Cell. Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Estraño C, Tschudi C, Ullu E. Exonic sequences in the 5′ untranslated region of alpha-tubulin mRNA modulate trans splicing in Trypanosoma brucei. Mol. Cell Biol. 1998;18:4620–4628. doi: 10.1128/mcb.18.8.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Günzl A, Tschudi C, Nakaar V, Ullu E. Accurate transcription of the Trypanosoma brucei U2 small nuclear RNA gene in a homologous extract. J. Biol. Chem. 1995;270:17287–17291. doi: 10.1074/jbc.270.29.17287. [DOI] [PubMed] [Google Scholar]
- 36.Huie JL, He P, Bellofatto V. In vitro transcription of the Leptomonas seymouri SL RNA and U2 snRNA genes using homologous cell extracts. Mol. Biochem. Parasitol. 1997;90:183–192. doi: 10.1016/s0166-6851(97)00146-1. [DOI] [PubMed] [Google Scholar]
- 37.Günzl A, Ullu E, Dorner M, Fragoso SP, Hoffmann KF, Milner JD, Morita Y, Nguu EK, Vanacova S, Wunsch S, et al. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 1997;85:67–76. doi: 10.1016/s0166-6851(96)02816-2. [DOI] [PubMed] [Google Scholar]
- 38.Lewdorowicz M, Yoffe Y, Zuberek J, Jemielity J, Stepinski J, Kierzek R, Stolarski R, Shapira M, Darzynkiewicz E. Chemical synthesis and binding activity of the trypanosomatid cap-4 structure. RNA. 2004;10:1469–1478. doi: 10.1261/rna.7510504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschudi C, Ullu E. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 40.Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu. Rev. Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 41.Ruskin B, Green MR. Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- 42.Rymond BC, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 43.Fukumaki Y, Ghosh PK, Benz E.J., Jr, Reddy VB, Lebowitz P, Forget BG, Weissman SM. Abnormally spliced messenger RNA in erythroid cells from patients with beta+ thalassemia and monkey cells expressing a cloned beta+-thalassemic gene. Cell. 1982;28:585–593. doi: 10.1016/0092-8674(82)90213-6. [DOI] [PubMed] [Google Scholar]
- 44.Huang J, Van der Ploeg LH. Requirement of a polypyrimidine tract for trans-splicing in trypanosomes: discriminating the PARP promoter from the immediately adjacent 3′ splice acceptor site. EMBO J. 1991;10:3877–3885. doi: 10.1002/j.1460-2075.1991.tb04957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 46.Schurch N, Hehl A, Vassella E, Braun R, Roditi I. Accurate polyadenylation of procyclin mRNAs in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol. Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mair G, Ullu E, Tschudi C. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 2000;275:28994–28999. doi: 10.1074/jbc.M004193200. [DOI] [PubMed] [Google Scholar]
- 48.Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Tschudi C. Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol. Cell Biol. 2005;25:2216–2226. doi: 10.1128/MCB.25.6.2216-2226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoffe Y, Zuberek J, Lewdorowicz M, Zeira Z, Keasar C, Orr-Dahan I, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shapira M. Cap-binding activity of an eIF4E homolog from Leishmania. RNA. 2004;10:1764–1775. doi: 10.1261/rna.7520404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arhin GK, Ullu E, Tschudi C. 2′-O-methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 2006;147:137–139. doi: 10.1016/j.molbiopara.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot. Cell. 2006;5:905–915. doi: 10.1128/EC.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamudio JR, Mittra B, Campbell DA, Sturm NR. Hypermethylated cap 4 maximizes Trypanosoma brucei translation. Mol. Microbiol. 2009;72:1100–1110. doi: 10.1111/j.1365-2958.2009.06696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA. 2009;15:648–665. doi: 10.1261/rna.1230209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Blancq SM, Swinkels BW, Gibson WC, Borst P. Evidence for gene conversion between the phosphoglycerate kinase genes of Trypanosoma brucei. J. Mol. Biol. 1988;200:439–447. doi: 10.1016/0022-2836(88)90534-7. [DOI] [PubMed] [Google Scholar]
- 56.Xu YX, BenShlomo H, Michaeli S. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: The Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl Acad. Sci. USA. 1997;94:8473–8478. doi: 10.1073/pnas.94.16.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.