Abstract
RNA editing is a post-transcriptional modification of pre-mRNA that results in increased diversity in transcriptomes and proteomes. It occurs in a wide variety of eukaryotic organisms and in some viruses. One of the most common forms of pre-mRNA editing is A-to-I editing, in which adenosine is deaminated to inosine, which is read as guanosine during translation. This phenomenon has been observed in numerous transcripts, including the mammalian 5-HT2C receptor, which can be edited at five distinct sites. Methods used to date to quantify 5-HT2C receptor editing are labor-intensive, expensive and provide limited information regarding the relative abundance of 5-HT2C receptor editing variants. Here, we present a novel, ultra high-throughput method to quantify 5-HT2C receptor editing, compare it to a more conventional method, and use it to assess the effect of a range of genetic and pharmacologic manipulations on 5-HT2C editing. We conclude that this new method is powerful and economical, and we provide evidence that alterations in 5-HT2C editing appear to be a result of regional changes in brain activity, rather than a mechanism to normalize 5-HT2C signaling.
INTRODUCTION
The post-transcriptional modification of RNA, or RNA editing, was first reported in trypanosome mitochondria (1). De-amination of adenosine to inosine, the most common type of RNA editing in higher eukaryotes (2), was first demonstrated in mammals at murine glutamate receptor subunit transcripts (3) and has been reported in organisms ranging from fruit flies to rodents and humans, and a number of instances have been reported in viruses (4). A family of enzymes referred to as Adenosine Deaminases that Act on RNA (ADAR) performs the deaminations, which underlie this type of RNA editing (2). It has been shown that the genetic deletion of all ADAR activity in Drosophila severely impairs central nervous system (CNS) function and integrity (5). A-to-I editing has been shown to be critical for normal embryogenesis in mammals, since genetic deletion of ADAR1 in mice is embryonically lethal (6). Genetic deletion of ADAR2 in mice, on the other hand, increases seizure susceptibility and decreases post-natal survival (7). The inosine content of transcripts isolated from different tissues suggests that the known edited transcripts only account for a small fraction of the editing that is likely to take place, that RNA editing takes place predominantly in non-coding regions of RNA transcripts containing inverted repetitive elements of the Alu and L1 subclass, and that RNA editing is most common in the brain (8–13).
The 5-hydroxytryptamine-2C (5-HT2C; HTR2C) serotonin receptor is the only G protein-coupled receptor (GPCR), whose transcripts have been shown to undergo RNA editing. RNA editing of 5-HT2C transcripts takes place at one or more of five closely spaced adenosines in a region that codes for a portion of the second intracellular loop of the receptor (14,15). ADAR1 has been shown to be primarily responsible for editing the two 5′ sites, while ADAR2 edits the two 3′ sites (16). RNA editing has been shown to reduce the efficiency of 5-HT2C receptor-G protein-coupling (17,14). Recent in vivo studies suggest that the altered signaling properties of 5-HT2C isoforms that result from RNA editing can lead to physiological consequences, as mutant mice expressing only the fully edited VGV isoform are characterized by significant abnormalities in receptor expression, feeding, and metabolism (18).
5-HT2C RNA editing abnormalities have been reported in the brains of suicide victims with a history of major depression, whereas treatment with the antidepressant drug fluoxetine induces changes in the opposite direction in mice (19). Other studies have suggested that 5-HT2C editing is decreased in the frontal cortex of schizophrenia patients (20), and serotonin depletion by parachlorophenylalanine (pCPA) induces alterations similar to those seen after fluoxetine treatment (21). Indeed, a number of studies have suggested changes in editing after treatment with medications that target serotonin receptors or alter serotonin levels, but the results have been both inconsistent and difficult to replicate (15,22). Furthermore, it is difficult to reconcile any mechanistic hypothesis with the observed changes, given the known activities and specificities of ADAR1 and ADAR2.
Several methods have been developed to facilitate measurement of RNA editing at the 5-HT2C receptor and other edited pre-mRNAs. One commonly used and informative method is to individually sequence large numbers of transcripts (anywhere from 20 to 100 or more) (14,23,24). This individual sequencing approach is both labor intensive and inadequate for quantifying the levels of rare transcripts. Other methods include primer extension assays (25) and pyrosequencing (26), which are quantitative and provide information about editing frequencies at each site, but give insufficient or no information regarding the frequencies of the different transcripts/isoforms. We have adapted a newly developed ultra high-throughput sequencing (HTS) technology, the Illumina Genome Analyzer II, to quantify 5-HT2C pre-mRNA editing. This approach is several orders of magnitude less expensive on a per transcript basis, and provides more comprehensive and quantitative information regarding RNA editing events. We compare our HTS-based method to the most commonly used individual sequencing method and assess the effect of a range of genetic manipulations and pharmacologic treatments on 5-HT2C editing.
MATERIALS AND METHODS
Mice
A detailed description of how the Pet-1 mice were generated has been reported previously (27). C57BL/6 mice were purchased from Jackson Laboratories. C57BL/6 mice were injected with either saline or drug daily for 10, 14 or 28 days and sacrificed on the last day of injections. Different brain regions were then microdissected and frozen at −80°C until use. Treatment dose and length were based on literature reports of drug regimens that resulted in measurable effects at the biochemical and/or behavioral level (see the relevant ‘Results’ section for the individual citations). All experiments were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University or the University of North Carolina, Chapel Hill. Mice were housed under standard conditions—12 h light/dark cycle and food and water ad libitum.
Generation of cDNA from RNA
Trizol (Invitrogen) was used to extract RNA from microdissected hippocampal tissue. Ten µg of RNA was treated with DNAse (DNA-free, Ambion), and 2 µg of the DNase-treated RNA was added to a reverse transcription reaction which was performed using the SuperscriptTM III RNase H Reverse Transcriptase kit (Invitrogen) with Oligo-(dT)12–18 primers (Invitrogen). cDNA was used as template to generate a double-stranded DNA fragment by PCR for both the low-throughput sequencing (LTS) and high-throughput sequencing (HTS) experiments.
LTS
A PCR fragment (containing the edited site) that was 327 bp in length was generated for each of three Pet-1 wild-type and three Pet-1 knockout animals (Forward primer: 5′ AAA GGATCC TGT GCT ATT TTC AAC TGC GTC CAT CAT G 3′; Reverse primer: 5′ AAA GAATTC CGG CGT AGG ACG TAG ATC GTTAAG 3′) (24). The PCR DNA concentrations were determined, and the DNA from the three animals of each genotype was mixed in equal amounts and inserted into the BamHI/EcoRI sites of pcDNA3 (Rapid DNA ligation kit, Roche). The ligation product was transformed into bacteria and plated; each clone represents an individual transcript, and the clones were assumed to be evenly distributed in origin between the three animals used for each ligation since equal amounts of DNA were used from each in the ligation reaction. Clones were miniprepped using the Wizard Plus SV Miniprep kit (Promega), and they were sequenced by Sanger sequencing to assess 5-HT2C RNA editing. Seventy-eight wild-type transcripts and 88 knockout transcripts were sequenced.
Ultra HTS
5-HT2C editing in Pet-1 mice
One microliter of cDNA (of 20 µl) was used as template for a 50 µl PCR reaction (conditions: 1 cycle—95°C, 2 min; 30 cycles—95°C 30 s, 52°C 30 s, 72°C 1 min; 1 cycle—72°C 10 min) to amplify a fragment (lowercase letters for the portions of the primers complementary to 5-HT2C sequence) containing the edited region of interest using Pfu turbo polymerase (Stratagene). The primers used also contained adapter sequences (uppercase letters in bold) necessary for cluster generation:
Forward primer: AATGATACGGCGACCACCGAGATCTACACTgcgccatatcgctggaccggtat;
Reverse primer: CAAGCAGAAGACGGCATACGAGATgccacgaaggacccgatgagaacg
PCR fragments (283 bp in length) were gel purified using the QIAquick gel extraction kit, and 1 µl was used as template for another round of PCR, though a second round of PCR is typically unnecessary since <100 ng of DNA is needed from each experiment using fragments of this size.
5-HT2C editing in chronic saline and drug-treated C57BL/6 mice
Performed as described above, except the round one PCR reaction was 20 µl. Additionally, the forward primer contained a sequence corresponding to a sequencing primer optimized by Illumina for use with the Genome Analyzer II (non-bolded uppercase letters), as well as a sample identification tag, which was a random nucleotide sequence 5–10 nucleotides in length (underlined uppercase letters, with ‘AAAAA’ being the tag for sample one and the other 25 tags as follows: ATCAT, GGGGG, TTTTT, AAGGT, GGTAT, AATTG, AGTGA, TTGGA, TTAAG, GTGTA, ATATG, ATCAT ATCAT, AAAAA AAAAA, GGGGG GGGGG, TTTTT TTTTT, AAGGT AAGGT, GGTAT GGTAT, AATTG AATTG, AGTGA AGTGA, TTGGA TTGGA, TTAAG TTAAG, GTGTA GTGTA, ATATG ATATG, AAAAA GGGGG, GGGGG AAAAA). As before, the bolded, italic portions of the primers represent adapter sequence, and the lower case portions represent sequence complementary to 5-HT2C pre-mRNA sequence. Each sample used a different forward primer (26 different forward primers total) containing a different identification tag, with the rest of the primer being identical. All reactions used the same reverse primer.
Forward primer:
AATGATACGGCGACCACCGAGATCTACACTACACTCTTTCCCTACACGACGCTCTTCCGATCTAAAAAtcgctggaccggtatgtagc
Reverse primer:
CAAGCAGAAGACGGCATACGAGATcgtccctcagtccaatcacagg
One microliter of PCR reaction one was used as template for another round of PCR, the volume of these reactions being 50 µl each, and the 257-bp fragment from this second round of PCR was gel purified using the MinElute PCR Purification kit (Qiagen). A portion of the gel-purified PCR DNA from each sample was used for gel quantification, and 24–26 samples were mixed in equal parts for HTS.
Gel purified PCR DNA was diluted to a concentration of 15 nM. Two microliters was used for denaturation (total volume 20 µl). 4 µl of the denaturation mixture was diluted in 996 µl of hybridization solution. The hybridization mixture (final DNA concentration about 6 pM) was loaded into the Cluster Station for cluster generation. Primer hybridization was performed on the Cluster Station using 6.6 µl of 500 nM sequencing primer (PET-1 experiment primer sequence: gcgccatatcgctggaccggtat; multiplexed experiment primer sequence: acactctttccctacacgacgctcttccgatct) diluted in 1313 µl of hybridization buffer. Cluster generation was performed for 36 cycles (Pet-1 experiment) or 76 cycles (fluoxetine experiment), followed by base-by-base sequencing initiated by the sequencing primer on the Genome Analyzer II. The Genome Analyzer II uses two different lasers to excite the dye attached to each nucleotide. Since the emission spectra of these four dyes overlap, the four images thus obtained are not independent. As in Sanger sequencing, the frequency cross-talk is deconvolved using a frequency cross-talk matrix. Therefore, the crosstalk matrix calculation requires control lanes for samples with skewed base compositions. Thus, a control human genomic DNA sample was run in parallel on the same flow-cell concurrently with 5-HT2C editing samples. Any non-skewed DNA library can be used for this purpose. For the Pet-1 experiment, sequences that passed all three of our quality filters were sorted and counted using Textpad. MySQL 4.0, an open source and multi-platform relational database management system, was used to sort and count transcript reads that passed the first filter, which permitted comparison of ‘false’ transcript counts to theoretically real transcript counts. For the multiplexed experiment, sequences were analyzed using a Perl 5 script (available at http://pdsp-temp.pha-med.unc.edu/Download/code.php) written by us to filter the data through three quality filters and sort the data which passed the filters. Further data analysis was performed in Microsoft Excel and Graphpad Prism 5.0. All statistical analyses were performed in Graphpad Prism 5.0. N = 3 littermate pairs for the samples used to measure hippocampal 5-HT2C RNA editing in Pet-1 wildtype and knockout mice, whereas N = 4 for the multiplexed experiment. For the purposes of making statistical comparisons, all the reads generated from one animal in the HTS studies were together treated as one experiment (N = 3–4 for each genotype). For the LTS analysis, each read was treated as an experiment (N = 78 for WT, N = 88 for KO).
RESULTS
Processing transcript reads obtained by ultra HTS
In order to explore the potential of ultra HTS methods for assessing 5-HT2C transcript editing frequencies, we measured RNA editing in hippocampi from Pet-1 wild-type and knockout mice. The hippocampus was chosen because it is the region with the highest expression of neuronal 5-HT2C receptor mRNA (28). Pet-1 is an ETS domain transcription factor that is necessary for the normal development of serotonergic neurons (27), and Pet-1 knockout mice have very low levels of brain serotonin. We also compared two methods for measuring RNA editing and transcript/isoform frequencies: sequencing individual transcripts by Sanger sequencing versus ultra HTS using reversible terminator chemistry (29,30) with the Illumina Genome Analyzer II platform.
The 5-HT2C receptor is A-to-I edited at varying frequencies at one or more of five sites: A, B, C, D and E (Figure 1A). Adenosines deaminated to inosine are read as guanosines when 5-HT2C RNA is reverse-transcribed, PCR-amplified and sequenced. Since the Genome Analyzer II-based ultra HTS read length is 36 bp, the sequencing primer was designed such that it provided reads beginning at the −5 position relative to the first edited site (Table 1). We analyzed three samples from each genotype, resulting in a total of 22 652 442 raw sequence reads for the six mice. All sequences were subsequently processed through three sets of ‘quality’ filters intended to retain only those sequences which appeared to provide error-free, reliable reads (Figure 1B). All quality filters are an application of a stringent binary DNA sequence quality criterion: reads would be divided into two groups, those containing one or more detectable sequencing errors, and those containing no detectable sequencing errors. Only those with no detectable errors would be used for transcript quantification, though it should be noted that our analysis indicated that less stringent filtering (for example, by using a shorter initial proofreading filter) did not change results significantly (data not shown). For the first quality filter, we used the five non-edited positions preceding the first edited (‘A’) site and the 17 non-edited positions downstream from the last edited (‘D’) site to filter the initial pool of sequences. All reads containing errors in any of the aforementioned 22 sites were removed from the analysis (13 790 980 sequences). The remaining reads (8 861 462 sequences) were then passed through two additional filters, which removed all transcripts containing a sequencing error in any of the eight non-edited sites within the region of interest spanning the A to D sites and/or contained a cytosine or a thymine at one of the edited sites (213 997 sequences). Sequences that failed these last two filters were binned by read and the number of reads in each bin was counted. Interestingly, some of these ‘false transcripts’ occurred at higher frequencies than a number of the rarer transcripts that passed all the filters. Notably, the same false transcripts recurred at high frequency in all the samples examined, and they typically differed from one of the most common transcripts by one base, suggesting that they resulted from misreads of common transcripts. The remaining sequences that passed all three ‘quality filters’ were sorted into 32 bins, one for each theoretically possible transcript (Table 1), and the number of sequence reads in each of these bins was then counted.
Figure 1.
Schematic of the ultra HTS strategy used to measure RNA editing. (A) A representative full-sequence read with the five edited sites labeled. Guanosines at edited sites correspond to inosines in the original RNA transcript from which the sequenced DNA is derived. (B) Ultra HTS sequencing produced 22 652 442/122 828 564 sequences. Non-edited region 1 and non-edited region 2 were used to filter sequences with misreads in those regions (13 790 980 sequences). Non-edited regions 3, 4 and 5 were then used to filter the remaining 8 861 462/92 958 400 sequences in a similar fashion to remove the sequences containing misreads in those regions, and theoretically impossible transcripts (in other words, those with a C or T at an edited site) were also filtered (213 997/21 770 405 sequences failed after applying the last two filters), leaving 8 647 465/71 187 995 sequences used for subsequent analysis.
Table 1.
Complete transcript reads
| Isoform | Transcript | Complete read |
|---|---|---|
| INI | AAAAA | TAGCAATACGTAATCCTATTGAGCATAGCCGGTTC |
| INV | AAAAG | TAGCAATACGTAATCCTGTTGAGCATAGCCGGTTC |
| ISI | AAAGA | TAGCAATACGTAGTCCTATTGAGCATAGCCGGTTC |
| ISV | AAAGG | TAGCAATACGTAGTCCTGTTGAGCATAGCCGGTTC |
| IDI | AAGAA | TAGCAATACGTGATCCTATTGAGCATAGCCGGTTC |
| IDV | AAGAG | TAGCAATACGTGATCCTGTTGAGCATAGCCGGTTC |
| IGI | AAGGA | TAGCAATACGTGGTCCTATTGAGCATAGCCGGTTC |
| IGV | AAGGG | TAGCAATACGTGGTCCTGTTGAGCATAGCCGGTTC |
| MNI | AGAAA | TAGCAATGCGTAATCCTATTGAGCATAGCCGGTTC |
| MNV | AGAAG | TAGCAATGCGTAATCCTGTTGAGCATAGCCGGTTC |
| MSI | AGAGA | TAGCAATGCGTAGTCCTATTGAGCATAGCCGGTTC |
| MSV | AGAGG | TAGCAATGCGTAGTCCTGTTGAGCATAGCCGGTTC |
| MDI | AGGAA | TAGCAATGCGTGATCCTATTGAGCATAGCCGGTTC |
| MDV | AGGAG | TAGCAATGCGTGATCCTGTTGAGCATAGCCGGTTC |
| MGI | AGGGA | TAGCAATGCGTGGTCCTATTGAGCATAGCCGGTTC |
| MGV | AGGGG | TAGCAATGCGTGGTCCTGTTGAGCATAGCCGGTTC |
| VDI | GGGAA | TAGCAGTGCGTGATCCTATTGAGCATAGCCGGTTC |
| VDV | GGGAG | TAGCAGTGCGTGATCCTGTTGAGCATAGCCGGTTC |
| VGI | GGGGA | TAGCAGTGCGTGGTCCTATTGAGCATAGCCGGTTC |
| VGV | GGGGG | TAGCAGTGCGTGGTCCTGTTGAGCATAGCCGGTTC |
| VNI | GGAAA | TAGCAGTGCGTAATCCTATTGAGCATAGCCGGTTC |
| VNV | GGAAG | TAGCAGTGCGTAATCCTGTTGAGCATAGCCGGTTC |
| VSI | GGAGA | TAGCAGTGCGTAGTCCTATTGAGCATAGCCGGTTC |
| VSV | GGAGG | TAGCAGTGCGTAGTCCTGTTGAGCATAGCCGGTTC |
| VDI | GAGAA | TAGCAGTACGTGATCCTATTGAGCATAGCCGGTTC |
| VDV | GAGAG | TAGCAGTACGTGATCCTGTTGAGCATAGCCGGTTC |
| VGI | GAGGA | TAGCAGTACGTGGTCCTATTGAGCATAGCCGGTTC |
| VGV | GAGGG | TAGCAGTACGTGGTCCTGTTGAGCATAGCCGGTTC |
| VNI | GAAAA | TAGCAGTACGTAATCCTATTGAGCATAGCCGGTTC |
| VNV | GAAAG | TAGCAGTACGTAATCCTGTTGAGCATAGCCGGTTC |
| VSI | GAAGA | TAGCAGTACGTAGTCCTATTGAGCATAGCCGGTTC |
| VSV | GAAGG | TAGCAGTACGTAGTCCTGTTGAGCATAGCCGGTTC |
Calculating transcript frequency thresholds
The recurrence of the same false transcripts in each sample, some at frequencies higher than theoretically possible transcripts, suggested the possibility that the reads of some of the rarer transcripts may have arisen largely due to A-to-G or G-to-A misreads of more common transcripts. Since it was impossible to determine a priori whether or not this was the case, we first undertook a detailed analysis of the error rates at the first three adenines and guanines in non-edited region 2 (Figure 1B). Noting that adenines and guanines were most commonly misread as cytosine, and least commonly as thymine, we calculated the A-to-G and G-to-A error rates at each site for each animal and averaged all three sites, since the rates at the different sites were not significantly different from each other, and they did not depend on the genotype of the samples (Table 2 and data not shown). We reasoned that each transcript read could have arisen from an A-to-G or G-to-A misread at any one of the five edited sites, meaning, in other words, that each transcript is separated from five other transcripts by a single base pair difference. By multiplying our calculated error rates by the frequencies of the occurrence of the five closely related transcripts and summing the five resulting values, we were able to estimate the proportion of the reads of a given transcript that were likely to have arisen due to misreads of other transcripts (Table 3). Our calculations indicated that the majority of transcript reads, in every instance, do not result from sequencing error, even in the cases of the rarest transcripts, which were seen at lower frequency than the most common false transcripts. In fact, our calculated estimates of the expected frequencies of occurrence of the most common false transcripts were consistent with what was seen experimentally (data not shown), suggesting that our calculated thresholds were reasonable. Thus, we considered all of the transcripts to have occurred at rates above our detection threshold, though some appear to be extremely rare.
Table 2.
Estimate of A-to-G and G-to-A Error Rates (%)
| Site | WT1 | WT2 | WT3 | KO1 | KO2 | KO3 | AVG | |
|---|---|---|---|---|---|---|---|---|
| Average A-to-G error rate | ||||||||
| Site 1 | TTGAGCATAGCCGGTT | 0.08 | 0.07 | 0.08 | 0.07 | 0.07 | 0.05 | |
| Site 2 | TTGAGCATAGCCGGTT | 0.05 | 0.06 | 0.05 | 0.05 | 0.05 | 0.04 | 0.05 |
| Site 3 | TTGAGCATAGCCGGTT | 0.04 | 0.05 | 0.04 | 0.05 | 0.04 | 0.04 | |
| Average G-to-A error rate | ||||||||
| Site 1 | TTGAGCATAGCCGGTT | 0.05 | 0.04 | 0.04 | 0.04 | 0.05 | 0.05 | |
| Site 2 | TTGAGCATAGCCGGTT | 0.06 | 0.05 | 0.04 | 0.05 | 0.05 | 0.05 | 0.04 |
| Site 3 | TTGAGCATAGCCGGTT | 0.03 | 0.04 | 0.04 | 0.03 | 0.05 | 0.04 |
Table 3.
Percentage of each transcript arising from sequencing error
| Isoform | Transcript | % Arising from error—Method 1 | % Arising from error—Method 2 |
|---|---|---|---|
| INI | AAAAA | 0.19 ± 0.01 | 0.06 ± 0.01 |
| INV | AAAAG | 0.10 ± 0.01 | 0.03 ± 0.01 |
| ISI | AAAGA | 0.34 ± 0.05 | 0.11 ± 0.02 |
| ISV | AAAGG | 0.14 ± 0.02 | 0.04 ± 0.01 |
| IDI | AAGAA | 1.08 ± 0.33 | 0.34 ± 0.05 |
| IDV | AAGAG | 1.81 ± 0.83 | 0.65 ± 0.08 |
| IGI | AAGGA | 1.09 ± 0.55 | 0.34 ± 0.05 |
| IGV | AAGGG | 2.92 ± 0.92 | 0.91 ± 0.13 |
| MNI | AGAAA | 2.03 ± 0.82 | 0.63 ± 0.09 |
| MNV | AGAAG | 4.51 ± 1.08 | 1.40 ± 0.20 |
| MSI | AGAGA | 32.66 ± 9.96 | 10.14 ± 1.48 |
| MSV | AGAGG | 19.55 ± 9.48 | 6.07 ± 0.89 |
| MDI | AGGAA | 48.26 ± 16.30 | 14.99 ± 2.19 |
| MDV | AGGAG | 16.12 ± 9.47 | 5.01 ± 0.73 |
| MGI | AGGGA | 0.37 ± 0.24 | 0.11 ± 0.02 |
| MGV | AGGGG | 13.44 ± 12.88 | 4.17 ± 0.61 |
| VDI | GGGAA | 1.76 ± 0.50 | 0.55 ± 0.08 |
| VDV | GGGAG | 1.82 ± 0.58 | 0.57 ± 0.08 |
| VGI | GGGGA | 3.44 ± 1.18 | 1.07 ± 0.16 |
| VGV | GGGGG | 3.39 ± 0.53 | 1.05 ± 0.15 |
| VNI | GGAAA | 0.28 ± 0.02 | 0.09 ± 0.01 |
| VNV | GGAAG | 0.06 ± 0.01 | 0.02 ± 0.01 |
| VSI | GGAGA | 0.17 ± 0.01 | 0.05 ± 0.01 |
| VSV | GGAGG | 0.09 ± 0.01 | 0.03 ± 0.01 |
| VDI | GAGAA | 0.33 ± 0.07 | 0.10 ± 0.01 |
| VDV | GAGAG | 0.88 ± 0.14 | 0.27 ± 0.04 |
| VGI | GAGGA | 0.16 ± 0.03 | 0.05 ± 0.01 |
| VGV | GAGGG | 0.14 ± 0.05 | 0.04 ± 0.01 |
| VNI | GAAAA | 0.25 ± 0.03 | 0.08 ± 0.01 |
| VNV | GAAAG | 0.51 ± 0.09 | 0.16 ± 0.02 |
| VSI | GAAGA | 2.14 ± 0.32 | 0.66 ± 0.10 |
| VSV | GAAGG | 1.69 ± 0.44 | 0.52 ± 0.08 |
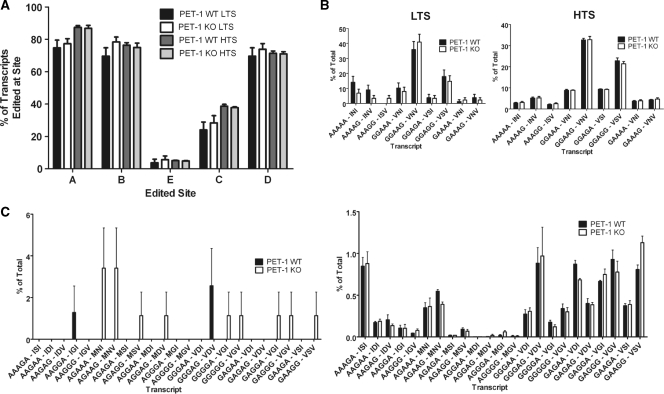
Comparison of results obtained by LTS and HTS
In parallel with our ultra HTS experiment, we also measured 5-HT2C RNA editing by the most common LTS approach to provide a realistic comparison of findings (24). First, we compared both methods with respect to their ability to detect all 32 possible transcripts (Table 4). Not surprisingly, HTS was able to detect all transcripts in both Pet-1 wild-type and knockout mice, whereas LTS detected only 20 of 32 possible transcripts, with 12 of those 20 detected in only one or the other genotype. Our LTS results are consistent with many previous studies in being able to detect only a subset of all theoretically possible transcripts (15). Thus, HTS appears to be superior to LTS with respect to the ability to comprehensively detect all transcripts.
Table 4.
Comparison of transcripts detected using the LTS and HTS methods
| WT LTS | KO LTS | WT HTS | KO HTS | |||||
|---|---|---|---|---|---|---|---|---|
| AAAAA | 14.10 ± 3.94 | AAAAA | 6.82 ± 2.69 | AAAAA | 2.870 ± 0.263 | AAAAA | 3.086 ± 0.468 | |
| AAAAG | 8.97 ± 3.24 | AAAAG | 3.41 ± 1.93 | AAAAG | 5.158 ± 0.477 | AAAAG | 5.305 ± 0.665 | |
| AAAGA | 0.847 ± 0.106 | AAAGA | 0.879 ± 0.142 | |||||
| AAAGG | 3.41 ± 1.93 | AAAGG | 2.158 ± 0.216 | AAAGG | 2.524 ± 0.388 | |||
| AAGAA | 0.172 ± 0.015 | AAGAA | 0.188 ± 0.025 | |||||
| AAGAG | 0.206 ± 0.059 | AAGAG | 0.136 ± 0.020 | |||||
| AAGGA | 1.28 ± 1.27 | AAGGA | 0.106 ± 0.035 | AAGGA | 0.105 ± 0.045 | |||
| AAGGG | 0.044 ± 0.003 | AAGGG | 0.078 ± 0.019 | |||||
| AGAAA | 3.41 ± 1.93 | AGAAA | 0.349 ± 0.062 | AGAAA | 0.364 ± 0.101 | |||
| AGAAG | 3.41 ± 1.93 | AGAAG | 0.546 ± 0.023 | AGAAG | 0.390 ± 0.029 | |||
| AGAGA | 0.019 ± 0.005 | AGAGA | 0.019 ± 0.002 | |||||
| AGAGG | 1.14 ± 1.13 | AGAGG | 0.094 ± 0.017 | AGAGG | 0.068 ± 0.023 | |||
| AGGAA | 0.001 ± 0.001 | AGGAA | 0.001 ± 0.001 | |||||
| AGGAG | 1.14 ± 1.13 | AGGAG | 0.005 ± 0.001 | AGGAG | 0.018 ± 0.014 | |||
| AGGGA | 0.019 ± 0.011 | AGGGA | 0.060 ± 0.018 | |||||
| AGGGG | 0.009 ± 0.009 | AGGGG | 0.009 ± 0.007 | |||||
| GGGAA | 0.274 ± 0.053 | GGGAA | 0.307 ± 0.041 | |||||
| GGGAG | 2.56 ± 1.79 | GGGAG | 0.883 ± 0.186 | GGGAG | 0.970 ± 0.345 | |||
| GGGGA | 1.14 ± 1.13 | GGGGA | 0.177 ± 0.024 | GGGGA | 0.123 ± 0.024 | |||
| GGGGG | 1.14 ± 1.13 | GGGGG | 0.340 ± 0.054 | GGGGG | 0.301 ± 0.049 | |||
| GGAAA | 10.25 ± 3.43 | GGAAA | 7.95 ± 2.88 | GGAAA | 8.852 ± 0.472 | GGAAA | 8.867 ± 0.183 | |
| GGAAG | 35.90 ± 5.43 | GGAAG | 40.91 ± 5.24 | GGAAG | 32.644 ± 0.793 | GGAAG | 32.799 ± 1.559 | |
| GGAGA | 3.85 ± 2.18 | GGAGA | 3.41 ± 1.93 | GGAGA | 9.363 ± 0.144 | GGAGA | 9.262 ± 0.151 | |
| GGAGG | 17.95 ± 4.35 | GGAGG | 14.77 ± 3.78 | GGAGG | 22.795 ± 1.395 | GGAGG | 21.382 ± 1.022 | |
| GAGAA | 0.871 ± 0.046 | GAGAA | 0.685 ± 0.016 | |||||
| GAGAG | 0.402 ± 0.057 | GAGAG | 0.386 ± 0.026 | |||||
| GAGGA | 1.14 ± 1.13 | GAGGA | 0.664 ± 0.013 | GAGGA | 0.749 ± 0.064 | |||
| GAGGG | 1.14 ± 1.13 | GAGGG | 0.928 ± 0.112 | GAGGG | 0.777 ± 0.130 | |||
| GAAAA | 1.28 ± 1.27 | GAAAA | 2.27 ± 1.59 | GAAAA | 3.679 ± 0.365 | GAAAA | 3.908 ± 0.478 | |
| GAAAG | 3.85 ± 2.18 | GAAAG | 2.27 ± 1.59 | GAAAG | 4.342 ± 0.232 | GAAAG | 4.737 ± 0.579 | |
| GAAGA | 0.374 ± 0.025 | GAAGA | 0.388 ± 0.046 | |||||
| GAAGG | 1.14 ± 1.13 | GAAGG | 0.807 ± 0.055 | GAAGG | 1.129 ± 0.079 | |||
Comparison of the overall editing frequencies at each of the five sites indicates that HTS and LTS report similar editing frequencies (Figure 2). Two-way ANOVA analysis with Bonferonni post-tests to compare LTS and HTS results indicates that, where comparison is possible (with the more common transcripts), there are no significant differences in the estimates obtained by the two methods. Thus, we validated both the method and the subsequent filters applied to the data. Two-way ANOVA analysis indicates that genotype has no effect on editing frequency variation (HTS P-value = 0.4369; LTS P-value = 0.0812). Bonferroni post-tests to perform pairwise comparisons of genotype effect confirmed that there is no significant between-genotype difference in editing frequencies measured by the two methods (Figure 2A). Furthermore, two-way ANOVA analysis (HTS P-value = 0.9976; LTS P-value = 0.4011) comparing common transcripts detected in both genotypes with both methods shows no significant effect of genotype on common transcript frequency differences, and Bonferroni post-tests confirm no significant effect of genotype on transcript frequencies (Figure 2B). Additionally, we would point out that HTS produces transcript frequency estimates that range over approximately five orders of magnitude, in contrast to LTS, where the estimates range over one order of magnitude. Not surprisingly, in measuring the frequencies of rare transcripts, HTS is clearly superior to LTS (Figure 2C). HTS allows statistical comparison between wildtype and knockout animals with all 23 rare transcripts in question. LTS, in contrast, does not detect any one of the 23 rare transcripts in both wild-type and knockout animals, making any statistical comparison for the rare transcripts impossible. Two-way ANOVA analysis of the HTS data shows no effect of genotype on transcript frequency (P = 0.9867), which is confirmed by Bonferroni post-tests.
Figure 2.
(A) Comparison of editing frequencies by site as measured by LTS and HTS. The LTS and HTS approaches produce comparable estimates of editing frequencies by site. Two-way ANOVA analysis (HTS P-value = 0.4369; LTS P-value = 0.0812) and Bonferroni post-tests indicate that there is no effect of genotype on site editing frequency. (B) Comparison of common transcript frequencies as measured by LTS and HTS. Estimates derived by LTS and HTS are comparable. Two-way ANOVA analysis (HTS P-value = 0.9976; LTS P-value = 0.4011) and Bonferroni post-tests comparing common transcripts detected in both genotypes with both methods show no significant effect of genotype on transcript frequency differences. (C) Comparison of rare transcript frequencies as measured by LTS and HTS. No rare transcript is detected in both genotypes by LTS, so statistical comparison between genotypes is not possible. HTS, on the other hand, permits more sensitive estimation of rare transcript frequencies. Two-way ANOVA analysis (P-value = 0.9867) and Bonferroni post-tests of the HTS-generated rare transcript data shows that genotype has no significant effect on transcript frequency. LTS: N = 78 wild-types, N = 88 knockouts; HTS: N = 3 littermate pairs, 8 647 465 sequences total. Transcript frequencies are presented as means, expressed as a percentage of the total population of transcripts, ±SEM.
Multiplexing HTS technology to simplify the measurement of 5-HT2C RNA editing under different experimental conditions
Since lowering endogenous brain serotonin via a genetic strategy did not significantly alter 5-HT2C RNA editing, we wondered if pharmacologic manipulations might alter 5-HT2C RNA editing. Simultaneously, we optimized our experimental protocol to generate more sequence reads with a lower error rate and to further reduce the cost to measure editing in each sample. To optimize the number of sequence reads generated, we altered the forward primer to incorporate a sequence which allows us to use the Illumina-optimized sequencing primer. To further reduce costs, we incorporated a sample identification tag in the forward primer, using a different forward primer/identification tag for each sample. As a result, we were able to multiplex 26 samples in one lane, reducing the cost of processing one sample by approximately a factor of 26. It should be noted that this experimental design necessitates a longer read length (76 bp). With these modifications, we generated 71 187 995 reads after filtering (Figure 1). We also calculated new transcript frequency thresholds for the multiplexed sequencing protocol. Notably, these new thresholds were considerably lower than in the original protocol (none >15%, and all but two of the rest <5%).
We proceeded to examine the effect of a variety of chronic treatment regimens. These included chronic agonist [LSD, 0.25 mg/kg (31); DOI, 1 mg/kg (32); MK-212, 5 mg/kg (33)], inverse agonist [SB206553, 5 mg/kg (34,35)], antimanic [lithium 200 mg/kg (36,37); valproate, 300 mg/kg (36,38,39)], antipsychotic [clozapine, 10 mg/kg (40); olanzapine 5 mg/kg (41–43)] and antidepressant [fluoxetine, 10 mg/kg (41,44–46); amitriptyline, 10 mg/kg (47)]. First, we treated C57BL/6 mice daily with saline, fluoxetine, amitriptyline or olanzapine for a 28-day period, the rationale being that fluoxetine is a selective serotonin reuptake inhibitor (SSRI), which should raise endogenous brain serotonin levels, and amitriptyline is a tricyclic antidepressant which also inhibits serotonin reuptake. Olanzapine is an atypical antipsychotic drug, which is also a non-selective 5-HT2C antagonist that would be predicted to have no effect on 5-HT2C editing based on our aforementioned Pet-1 results. We proceeded to measure the editing frequency at the A, B, C, D and E sites in three brain regions: striatum, hippocampus and cortex. Antidepressant effects are thought to involve changes in hippocampal function, and a previous study has examined the effect of fluoxetine on cortical 5-HT2C RNA editing (21). We examined striatal editing because 5-HT2C receptors are known to be highly expressed in this region (28,48,49). Interestingly, we found that fluoxetine-treated mice exhibited an increase in A and B site editing relative to their saline-treated littermates, with no change in C, D or E site editing in striatum and hippocampus, but not cortex (Figure 3A-C). Chronic amitriptyline, on the other hand, led to an increase in A and B site editing only in hippocampus. Olanzapine had no significant effect on 5-HT2C editing.
Figure 3.
(A–C) Column 1—comparison of editing frequencies by site after treatment with saline or drug daily for 28 days, in striatum, hippocampus and cortex. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that chronic fluoxetine and amitriptyline treatment leads to an increase in the proportion of transcripts edited at the A and B sites in striatum (fluoxetine) and hippocampus (fluoxetine and amitriptyline), with no effect on other sites. Column 2—comparison of transcript frequencies by transcript group after treatment with saline or fluoxetine daily for 28 days in striatum, hippocampus, and cortex. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that fluoxetine (striatum and hippocampus) and amitriptyline (hippocampus) treatment lead to a decrease in the proportion of transcripts unedited at both the A and B sites (AA***), an increase in the proportion of transcripts edited at both the A and B sites (GG***), and no change in transcripts edited at either the A or B site (AG*** and GA***). N = 4 for each treatment regimen, with the estimates for each sample obtained by analyzing an average of 431 442 sequences. Editing frequencies and transcript frequencies are presented as means, expressed as a percentage of the total population of transcripts, ±SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To further characterize our results, we examined whether our overall editing frequencies calculated at each site were consistent with findings at the transcript level. We reasoned that an increase in A and B site editing should lead to a decrease in transcripts that are unmodified at both sites (AA***), with a concomitant increase in deaminated transcripts (AG***, GA***, and/or GG***). As seen in Figure 3A–C, while AA*** frequency decreases and GG*** increases as predicted, AG*** and GA*** remain unchanged. Thus, fluoxetine and amitriptyline increase the editing frequencies at the A and B sites by increasing the simultaneous editing of those two sites, most likely by regulating ADAR1 activity. Furthermore, the increase in A and B site editing resulted in significant changes at the transcript level in either two or three out of 32 transcripts after fluoxetine treatment (Striatum: AAAAA-INI, Saline—7.85% > Fluoxetine—5.70%, P < 0.01; AAAAG-INV, Saline—8.83% > Fluoxetine—5.35%, P < 0.001; GGAAG-VNV, SALINE—32.50% < Fluoxetine—35.42%, P < 0.001; Hippocampus: AAAAG-INV, Saline—17.84% > Fluoxetine—12.22%, P < 0.001; GGAAG-VNV, Saline—27.17% < Fluoxetine—31.57%, P < 0.001) and four of 32 transcripts after amitriptyline treatment (Hippocampus: AAAAA-INI, Saline—10.59% > Amitriptyline—6.94%, P < 0.05; AAAAG-INV, Saline—17.84% > Amitriptyline—8.36%, P < 0.001; GGAAG-VNV, Saline—27.17% < Amitriptyline—35.21%, P < 0.001; GGAGG-VSV, Saline—13.59% < Amitriptyline—17.26%, P < 0.05). There were no significant changes in any of the other 28 transcripts after chronic fluoxetine or amitriptyline treatment, and no significant changes whatsoever after olanzapine treatment.
We next examined several other drugs which can directly (e.g. LSD, DOI, SB206553, MK-212, clozapine) or indirectly (e.g. lithium, valproate) modulate 5-HT2C signaling. The treatment regimens were 10-day saline, LSD and DOI; and 14-day saline, SB206553, MK-212, clozapine, lithium and valproate. SB206553, a relatively selective 5-HT2C inverse agonist, caused a small but significant increase in 5-HT2C editing at the A and B sites in striatum only (Figure 4A), decreasing the proportion of AA*** transcripts and increasing the proportion of GG*** transcripts as expected. Lithium, which does not act directly at 5-HT2C receptors, but which can alter 5-HT2C signaling (50), increased editing at the C and D sites, which are edited by ADAR2, in cortex only, resulting in the expected changes in the proportions of ***AA and ***GG transcripts (Figure 4B). LSD, DOI, MK-212, clozapine and valproate did not alter 5-HT2C editing in any of the brain regions examined (Table 5).
Figure 4.
(A) Comparison of editing frequencies by site after treatment with saline or drug daily for 14 days, in striatum. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that chronic SB206553 (14 D) treatment leads to an increase in the proportion of transcripts edited at the A and B sites in striatum, with no effect on other sites. Comparison of transcript frequencies by transcript group after treatment with saline or SB206553 daily for 14 days in striatum, hippocampus and cortex is consistent with the increase in A and B site editing. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that SB206553 treatment leads to a decrease in the proportion of transcripts unedited at both the A and B sites (AA***), an increase in the proportion of transcripts edited at both the A and B sites (GG***), and no change in transcripts edited at either the A or B site (AG*** and GA***). (B) Comparison of editing frequencies by site after treatment with saline or drug daily for 14 days in cortex. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that chronic lithium treatment leads to an increase in the proportion of transcripts edited at the C and D sites in striatum, with no effect on other sites. Comparison of transcript frequencies by transcript group after treatment with saline or lithium daily for 14 days in cortex is consistent with the increase in C and D site editing. Two-way ANOVA analysis (P < 0.0001) and Bonferroni post-tests for pair-wise comparisons indicates that lithium treatment leads to a decrease in the proportion of transcripts unedited at both the C and D sites (***AA), an increase in the proportion of transcripts edited at both the C and D sites (***GG), and no change in transcripts edited at either the C or D site (***AG and ***GA). N = 4 for each treatment regimen, with the estimates for each sample obtained by analyzing an average of 431 442 sequences. Editing frequencies and transcript frequencies are presented as means, expressed as a percentage of the total population of transcripts, ±SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 5.
Summary of findings from multiplexed experiment
| Drug | Treatment length | Striatum | Hippocampus | Cortex |
|---|---|---|---|---|
| Fluoxetine | 28 D | * | * | NS |
| Amitriptyline | 28 D | NS | * | NS |
| Olanzapine | 28 D | NS | NS | NS |
| SB206553 | 14 D | * | NS | NS |
| Lithium | 14 D | NS | NS | * |
| Clozapine | 14 D | NS | NS | NS |
| MK-212 | 14 D | NS | NS | NS |
| Valproate | 14 D | NS | NS | NS |
| LSD | 10 D | NS | NS | NS |
| DOI | 10 D | NS | NS | NS |
*The treatment regimen significantly altered editing.
NS, no significant effect of the treatment regimen on editing.
DISCUSSION
In this article, we developed and utilized an ultra HTS approach using the Illumina Genome Analyzer II platform to analyze 5-HT2C RNA editing and provide quantitative estimates of editing site frequencies and transcript/isoform frequencies. We also compared our ultra high-throughput approach to the most commonly used low-throughput approach and show that our high-throughput approach is superior in part because it detects all transcripts, facilitating between-genotype comparisons of even rare transcripts, which is either impossible or prohibitively expensive and laborious by the more common low-throughput approach. We then used our novel approach to assess the effect of endogenous serotonin on RNA editing of the 5-HT2C receptor by comparing RNA editing frequencies in the hippocampi of Pet-1 wild-type and knockout mice, which have a defective serotonin system due to dysfunctional serotonergic raphe neurons that produce almost no serotonin. Surprisingly, we found that abnormally low levels of brain serotonin had no significant effect on 5-HT2C receptor RNA editing. We also show that the editing frequencies at each of the five individual editing sites are unchanged and that there is little or no alteration in the frequency of any one of the 32 5-HT2C transcripts in the absence of a normally functioning central serotonergic system. An examination of the effect on 5-HT2C editing of 10 different drugs with varying mechanisms of action indicated that their ability to modulate editing could not be predicted in a straightforward manner based on their activity at 5-HT2C receptors. Notably, our results in untreated mice are consistent with previous studies measuring the proportions of transcripts edited at the five edited sites (21,24).
Prior studies have yielded inconsistent results regarding the effects of various pharmacological manipulations of the serotonergic system on 5-HT2C RNA editing. As an example, one study reported increases in editing frequencies at the A, B, C and D sites after fluoxetine treatment of BALB/c mice, and no RNA editing changes after fluoxetine treatment in C57BL/6 mice (though there were consistent trends towards a decrease at the A, B, C and D sites). It should be noted that these mice were subjected to a modified forced swim test (FST) on their last 2 days of treatment (51). Another study in rats reported decreases in A, B and E site editing frequencies after fluoxetine treatment (52).
A potential shortcoming of all of these previous studies is the small number of sequences that was sampled, typically 50 or so samples per animal, with three or four animals in each treatment group. Small sample sizes are inevitable due to the labor-intensive nature of methods that rely on sequencing transcripts derived from individual bacterial clones (one clone = one transcript). The small sample sizes make measuring editing at the E site difficult because of the very low frequency of editing at this site (<5%). Not surprisingly, therefore, the most common change reported between treatment groups is at the E site, which is the most difficult to measure accurately. Furthermore, previous studies generally focus only on common transcripts, because rare transcripts are impossible to identify and quantify given the small sample sizes. Other studies rely on primer-extension analysis, which does not allow for the comprehensive assessment of individual transcript frequencies, but merely editing frequencies at each site. Two very recent articles apply next-generation sequencing technology to measuring or discovering RNA editing (53,54). Both sequenced only a few hundred to a few thousand 5-HT2C transcripts—an approach comparable to what can be done with presently established methods. In addition, neither study examined all 32 conceivable transcripts and no comparison was made with a gold standard method to validate the results.
Our novel HTS method has a number of advantages with respect to the previously established methods for quantifying RNA editing. First, HTS is many orders of magnitude less expensive per sequence. Furthermore, the Genome Analyzer II, on which our HTS experiment was performed, is amenable to multiplexing 26 or more samples in each lane of a flow cell, which cuts the cost per experiment by an order of magnitude, making RNA editing analysis by HTS considerably less expensive on a per animal basis than RNA editing analysis by LTS. Second, measuring RNA editing by HTS is less labor-intensive than LTS. For HTS, PCR fragments are simply generated and loaded into the Cluster Station and then the Genome Analyzer II, which sequences individual fragments directly. To generate similar information by LTS, fragments must be first ligated into a vector, the vector transformed into bacteria, the bacteria plated, individual bacterial clones picked and grown, plasmids purified, and finally the inserted fragment sequenced by Sanger sequencing. Third, HTS is advantageous as compared to primer-extension analysis and other similar PCR-based strategies in that data is digital rather than analog, with at least some of the advantages associated with digital information (55). Fourth, HTS permits the analysis of rare transcripts, which is either impossible or prohibitively expensive and labor intensive by the LTS method. The analysis of rare transcripts is also impossible by analog methods such as primer-extension analysis. We have also compared our results, where possible, to gold standard LTS, thus validating our method and the sequence quality filters we used.
To our knowledge, this study represents the first comprehensive measurement of the frequencies of all 32 possible 5-HT2C transcripts. Although we detected many rare transcripts, it is not clear to what extent rare transcripts may or may not be important in vivo. Experiments with mice expressing only the fully edited VGV isoform, however, have indicated that mutant VGV mice exhibit increases in total 5-HT2C expression as compared to wild-type mice, along with dramatic reductions in fat mass despite hyperphagia (18). This suggests the possibility that alterations in rare transcript frequencies may have important physiological consequences.
Finally, we would like to note that our data are easily reconcilable with the known mechanisms by which the 5-HT2C receptor pre-mRNA is edited by ADAR1, which edits the A and B sites, and ADAR2, which edits the C and D sites (16). Our data suggest that fluoxetine/amitriptyline and SB206553, which have the opposite effects on signaling through the 5-HT2C receptor but nonetheless all increase editing frequency at the A and B sites in at least one brain region, modulate 5-HT2C RNA editing by influencing ADAR1 function at the receptor without affecting function of ADAR2. In contrast, lithium appears to affect only the ability of ADAR2 to edit 5-HT2C pre-mRNA. Furthermore, our data indicate that chronic treatment leads to reciprocal changes in transcripts that are either unedited or fully edited at the A and B or C and D sites, with no change in transcripts edited at only one or the other site. We also show region-specific changes with respect to the handful of drugs which do affect editing (fluoxetine, amitriptyline, SB206553 and lithium) Finally, although we report significant changes in editing after some drug regimens, most of these changes are relatively small in magnitude (typically <10%) and it is unclear if such small changes are physiologically significant.
In summary, we have developed and optimized a novel ultra high-throughput method for measuring RNA editing digitally by adapting newly developed genome-wide sequencing tools. Our method is inexpensive, technically feasible for most laboratories, and provides more comprehensive information regarding 5-HT2C receptor editing than either existing analog methods or digital low-throughput methods. We applied our newly developed ultra high-throughput method to assess whether or not modulating endogenous brain serotonin would alter 5-HT2C RNA editing. We demonstrated through our more powerful measurement method that lowering endogenous brain serotonin levels does not affect 5-HT2C RNA editing in vivo. In contrast, treating mice with chronic fluoxetine, amitriptyline, SB206553 and lithium increased editing at a subset of the sites in one more brain regions, whereas chronic LSD, DOI, MK-212, valproate, clozapine and olanzapine had no effect. Our data suggest that the ability of a drug to alter 5-HT2C RNA editing cannot be predicted from its activity at 5-HT2C receptors. Given its considerable advantages, massively parallel HTS is likely to rapidly become the method of choice for quantifying RNA editing as next-generation sequencing platforms become more widely available.
FUNDING
NIMH61887, U19MH82441; NIMH Psychoactive Drug Screening Program (to A.A., D.U., M.S.F., N.H.J., W.K.K., B.L.R.); NARSAD Distinguished Investigator (to B.L.R.); CWRU MSTP and National Institutes of Health (T32 GM007250 to A.A.); UNC Chapel Hill and the Lineberger Cancer Center. Funding for open access charge: National Institutes of Health (U19MH82441, RO1MH61887).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Evan Deneris for providing us with the Pet-1 knockout mice on which the studies were performed. We are also grateful to the UNC Genome Analysis Facility for performing the Sanger sequencing for the LTS experiments and the UNC High Throughput Sequencing Facility for their help in performing the HTS studies.
REFERENCES
- 1.Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 3.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 4.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 8.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kikuno R, Nagase T, Waki M, Ohara O. HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 2002;30:166–168. doi: 10.1093/nar/30.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 14.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 15.Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol. Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 17.Berg KA, Cropper JD, Niswender CM, Sanders-Bush E, Emeson RB, Clarke WP. RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br. J. Pharmacol. 2001;134:386–392. doi: 10.1038/sj.bjp.0704255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 20.Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol. Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 21.Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J. Neurosci. 2002;22:10529–10532. doi: 10.1523/JNEUROSCI.22-24-10529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardiner K, Du Y. A-to-I editing of the 5HT2C receptor and behaviour. Brief. Funct. Genom. Proteom. 2006;5:37–42. doi: 10.1093/bfgp/ell006. [DOI] [PubMed] [Google Scholar]
- 23.Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DE, Humby T, Wilkinson LS, Isles AR. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum. Mol. Genet. 2009;18:2140–2148. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Y, Davisson MT, Kafadar K, Gardiner K. A-to-I pre-mRNA editing of the serotonin 2C receptor: comparisons among inbred mouse strains. Gene. 2006;382:39–46. doi: 10.1016/j.gene.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Rueter SM, Burns CM, Coode SA, Mookherjee P, Emeson RB. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995;267:1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto K, Bundo M, Kato T. Estimating RNA editing efficiency of five editing sites in the serotonin 2C receptor by pyrosequencing. RNA. 2005;11:1596–1603. doi: 10.1261/rna.2114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 28.Molineaux SM, Jessell TM, Axel R, Julius D. 5-HT1c receptor is a prominent serotonin receptor subtype in the central nervous system. Proc. Natl Acad. Sci. USA. 1989;86:6793–6797. doi: 10.1073/pnas.86.17.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruparel H, Bi L, Li Z, Bai X, Kim DH, Turro NJ, Ju J. Design and synthesis of a 3′-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc. Natl Acad. Sci. USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckholtz NS, Freedman DX, Middaugh LD. Daily LSD administration selectively decreases serotonin2 receptor binding in rat brain. Eur. J. Pharmacol. 1985;109:421–425. doi: 10.1016/0014-2999(85)90407-8. [DOI] [PubMed] [Google Scholar]
- 32.Buckholtz NS, Zhou DF, Freedman DX. Serotonin2 agonist administration down-regulates rat brain serotonin2 receptors. Life Sci. 1988;42:2439–2445. doi: 10.1016/0024-3205(88)90342-6. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham KA, Callahan PM, Appel JB. Discriminative stimulus properties of the serotonin agonist MK 212. Psychopharmacol. 1986;90:193–197. doi: 10.1007/BF00181240. [DOI] [PubMed] [Google Scholar]
- 34.Griebel G, Perrault G, Sanger DJ. A comparative study of the effects of selective and non-selective 5-HT2 receptor subtype antagonists in rat and mouse models of anxiety. Neuropharmacol. 1997;36:793–802. doi: 10.1016/s0028-3908(97)00034-8. [DOI] [PubMed] [Google Scholar]
- 35.Heisler LK, Tecott LH. A paradoxical locomotor response in serotonin 5-HT(2C) receptor mutant mice. J. Neurosci. 2000;20:RC71. doi: 10.1523/JNEUROSCI.20-08-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR, Caron MG. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc. Natl Acad. Sci. USA. 2004;101:5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaulieu JM, Marion S, Rodriguiz RM, Medvedev IO, Sotnikova TD, Ghisi V, Wetsel WC, Lefkowitz RJ, Gainetdinov RR, Caron MG. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell. 2008;132:125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- 38.Chen B, Wang JF, Young LT. Chronic valproate treatment increases expression of endoplasmic reticulum stress proteins in the rat cerebral cortex and hippocampus. Biol. Psychiatr. 2000;48:658–664. doi: 10.1016/s0006-3223(00)00878-7. [DOI] [PubMed] [Google Scholar]
- 39.Hao Y, Creson T, Zhang L, Li P, Du F, Yuan P, Gould TD, Manji HK, Chen G. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. J. Neurosci. 2004;24:6590–6599. doi: 10.1523/JNEUROSCI.5747-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbas AI, Yadav PN, Yao WD, Arbuckle MI, Grant SG, Caron MG, Roth BL. PSD-95 is essential for hallucinogen and atypical antipsychotic drug actions at serotonin receptors. J. Neurosci. 2009;29:7124–7136. doi: 10.1523/JNEUROSCI.1090-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kodama M, Fujioka T, Duman RS. Chronic olanzapine or fluoxetine administration increases cell proliferation in hippocampus and prefrontal cortex of adult rat. Biol. Psychiatr. 2004;56:570–580. doi: 10.1016/j.biopsych.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Seeman P, Weinshenker D, Quirion R, Srivastava LK, Bhardwaj SK, Grandy DK, Premont RT, Sotnikova TD, Boksa P, El-Ghundi M, et al. Dopamine supersensitivity correlates with D2High states, implying many paths to psychosis. Proc. Natl Acad. Sci. USA. 2005;102:3513–3518. doi: 10.1073/pnas.0409766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stockton ME, Rasmussen K. Electrophysiological effects of olanzapine, a novel atypical antipsychotic, on A9 and A10 dopamine neurons. Neuropsychopharmacol. 1996;14:97–105. doi: 10.1016/0893-133X(94)00130-R. [DOI] [PubMed] [Google Scholar]
- 44.Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J. Neurosci. 2000;20:9104–9110. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 46.Svenningsson P, Tzavara ET, Witkin JM, Fienberg AA, Nomikos GG, Greengard P. Involvement of striatal and extrastriatal DARPP-32 in biochemical and behavioral effects of fluoxetine (Prozac) Proc. Natl Acad. Sci. USA. 2002;99:3182–3187. doi: 10.1073/pnas.052712799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heal DJ, Hurst EM, Prow MR, Buckett WR. An investigation of the role of 5-hydroxytryptamine in the attenuation of presynaptic alpha 2-adrenoceptor-mediated responses by antidepressant treatments. Psychopharmacol. 1990;101:100–106. doi: 10.1007/BF02253725. [DOI] [PubMed] [Google Scholar]
- 48.Clemett DA, Punhani T, Duxon MS, Blackburn TP, Fone KC. Immunohistochemical localisation of the 5-HT2C receptor protein in the rat CNS. Neuropharmacol. 2000;39:123–132. doi: 10.1016/s0028-3908(99)00086-6. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Gimenez JF, Tecott LH, Palacios JM, Mengod G, Vilaro MT. Serotonin 5- HT (2C) receptor knockout mice: autoradiographic analysis of multiple serotonin receptors. J. Neurosci. Res. 2002;67:69–85. doi: 10.1002/jnr.10072. [DOI] [PubMed] [Google Scholar]
- 50.Basselin M, Chang L, Seemann R, Bell JM, Rapoport SI. Chronic lithium administration potentiates brain arachidonic acid signaling at rest and during cholinergic activation in awake rats. J. Neurochem. 2003;85:1553–1562. doi: 10.1046/j.1471-4159.2003.01811.x. [DOI] [PubMed] [Google Scholar]
- 51.Englander MT, Dulawa SC, Bhansali P, Schmauss C. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 2005;25:648–651. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwamoto K, Nakatani N, Bundo M, Yoshikawa T, Kato T. Altered RNA editing of serotonin 2C receptor in a rat model of depression. Neurosci Res. 2005;53:69–76. doi: 10.1016/j.neures.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 54.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]