Abstract
Objective
To investigate peripheral blood (PB) cell transcript profiles of systemic sclerosis (SSc) and its subtypes in direct comparison with systemic lupus erythematosus (SLE).
Methods
We investigated PB cell samples from 74 SSc patients, 21 healthy controls, and 17 SLE patients using Illumina Human Ref-8 BeadChips and quantitative polymerase chain reaction confirmation. None of the study participants were receiving immunosuppressive agents other than low-dose steroids and hydroxychloroquine. In addition to conventional statistical and modular analysis, a composite score for the interferon (IFN)–inducible genes was calculated. Within the group of patients with SSc, the correlation of the IFN score with the serologic and clinical subtypes was investigated, as were single-nucleotide polymorphisms in a selected number of IFN pathway genes.
Results
Many of the most prominently overexpressed genes in SSc and SLE were IFN-inducible genes. Forty-three of 47 overexpressed IFN-inducible genes in SSc (91%) were similarly altered in SLE. The IFN score was highest in the SLE patients, followed by the SSc patients, and then the controls. The difference in IFN score among all 3 groups was statistically significant (P < 0.001 for all 3 comparisons). SSc and SLE PB cell samples showed striking parallels to our previously reported SSc skin transcripts in regard to the IFN-inducible gene expression pattern. In SSc, the presence of antitopoisomerase and anti–U1 RNP antibodies and lymphopenia correlated with the higher IFN scores (P = 0.005, P = 0.001, and P = 0.004, respectively); a missense mutation in IFNAR2 was significantly associated with the IFN score.
Conclusion
SLE and SSc fit within the same spectrum of IFN-mediated diseases. A subset of SSc patients shows a “lupus-like” high IFN-inducible gene expression pattern that correlates with the presence of antitopoisomerase and anti–U1 RNP antibodies.
Systemic sclerosis (SSc) is a multisystem autoimmune disease of connective tissue characterized by immune dysregulation, obliterative vasculopathy, and fibrosis of skin and internal organs (1–3). Using microarrays, 4 previous studies have investigated the transcript profiles of SSc peripheral blood (PB) cells or their subpopulations. We first reported that, compared with PB cells from controls, PB cells from patients with early SSc have a distinct transcript pattern that includes dysregulation of interferon (IFN)–inducible genes (4). This observation was replicated by other investigators in PB mononuclear cells (PBMCs) (5), monocytes, and CD4+ T cells (6) and PB cells (7) from SSc patients. None of those studies found a correlation between the IFN signature and the clinical or serologic subtypes of SSc. Due to the heterogeneity of SSc, those studies may not have been sufficiently powered to assess subtle clinical and serologic differences in the transcript profile. However, Bos et al (7) reported that the expression levels of 5 IFN-inducible gene transcripts measured by quantitative polymerase chain reaction (PCR) in an extended group of 43 SSc patients were higher in patients without anticentromere antibodies (ACAs).
Several studies have shown a striking pattern of up-regulated type I IFN–inducible genes in PBMCs from patients with systemic lupus erythematosus (SLE) (8–10). In particular, the presence of anti-Ro, anti–U1 RNP, anti-Sm, and anti–double-stranded DNA (anti-dsDNA) is significantly associated with a high IFN score in SLE (11).
SSc shares several similarities with SLE, such as autoantibodies directed against nuclear antigens, and in some patients, overlapping clinical features. At the gene level, there is emerging evidence that SSc and SLE share common genetic associations, such as IRF5 (12–14) and PTPN22 (15–17). The risk alleles of the IRF5 and PTPN22 genes have been linked to higher serum levels of IFNα in SLE patients (18,19). Despite the evidence of similarities between SSc and SLE, a direct comparison of the transcript profiles of patients with the 2 diseases has not been undertaken.
We now report the results of such a comparative study of a large number of SSc patients with SLE patients and controls. We analyzed our results using conventional methods and a newly described modular analysis framework (10). We examined whether SSc patients with certain genetic, clinical, or serologic features are more likely to have a transcript profile that is similar to that of SLE. The results place a subset of SSc patients alongside SLE in the continuum of IFN-mediated autoimmune diseases.
PATIENTS AND METHODS
Study participants
The study participants were recruited prospectively from January 2005 to February 2008. All SSc patients met the 1980 American College of Rheumatology (ACR; formerly, the American Rheumatism Association) preliminary criteria for the classification of SSc (20). None of the SSc patients fulfilled the ACR classification criteria for SLE (21). We used the more stringent criterion of first Raynaud’s or non-Raynaud’s symptom as the disease onset date, because we were interested in exploring early immune dysregulations that lead to the different phenotypes of SSc. Patients with early SSc had a disease duration ≤5 years, while late SSc was defined as a disease duration ≥7 years. The healthy control subjects had no history of autoimmune diseases and were matched by age, sex, and ethnicity to patients with early SSc. All enrolled SLE patients met the ACR classification criteria for SLE (21) and had signs of active disease in at least 2 categories of the revised Systemic Lupus Activity Measure (SLAM-R) (22). SSc or SLE patients receiving immunosuppressive agents other than low-dose steroids (prednisone ≤5 mg/day) and hydroxychloroquine were excluded from the study.
The majority of SSc patients were recruited from the prospective outcome Genetics versus Environment in Scleroderma Outcome Study (GENISOS). The remaining patients were enrolled at the rheumatology outpatient clinics of the University of Texas Health Science Center at Houston (UTHSC-H) and the national and local meetings of the Scleroderma Foundation. The SLE patients were enrolled from the rheumatology outpatient clinics of the UTHSC-H and affiliated hospitals. Medical records for all participants were obtained to verify their diagnosis and to characterize the disease. We collected the following information about SSc patients: medication regimen, extent of skin involvement (limited versus diffuse cutaneous skin involvement [23]), Modified Rodnan Skin Thickness Score (MRSS) (24), date of onset of Raynaud’s phenomenon and first non-Raynaud’s symptoms, white blood cell (WBC) count and differential cell count obtained within 1 week of blood sample drawing, presence of pulmonary hypertension, pulmonary fibrosis/alveolitis on imaging, and pulmonary function test results. All study subjects provided written informed consent, and the study was approved by the institutional review boards of all participating centers.
Sample processing and microarray experiments
Autoantibodies, including antinuclear antibodies (ANAs), ACA pattern, antitopoisomerase, anti–RNA polymerase III (anti– RNAP III), anti–U1 RNP, and anti-Ro/La, were detected with commercial kits in all SSc serum samples at the laboratories of the UTHSC-H Division of Rheumatology.
Blood samples for transcript studies were drawn directly into PAXgene tubes (PreAnalytiX, Franklin Lakes, NJ). Total RNA was isolated according to the manufacturer’s protocol using the PAXgene RNA kit (PreAnalytiX). The RNA quality and yield were assessed using a 2100 Bioanalyzer (Agilent, Palo Alto, CA) and an ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). We did not conduct a globin reduction on our samples, because this procedure did not increase percent present calls in an experiment with 9 healthy control samples (further information is available online at http://www.uth.tmc.edu/scleroderma).
Two hundred nanograms of total RNA was amplified and purified using the Illumina TotalPrep RNA Amplification Kit (Applied Biosystems/Ambion, Austin, TX) in accordance with the manufacturer’s instructions. The amplified complementary RNA was hybridized on Illumina Human Ref-8 BeadChips, and the data were extracted with the Illumina Beadstudio software suite (Illumina, San Diego, CA).
Microarray data analysis
The initial analysis was performed with Beadstudio software. Raw data were also exported into and analyzed with BRB-ArrayTools, developed by Richard Simon and Amy Peng Lam (National Cancer Institute, Frederick, MD).
Any transcript with a P value for signal detection that was not significantly different from the negative controls (P = 0.01) was removed from the analysis. In addition, transcripts with missing or filtered out expression values in >50% of the experiments were excluded. Data were normalized using the median over the entire array. A transcript was defined as differentially expressed when the significance level for the comparison was P ≤ 0.01 and the false discovery rate was ≤0.10 using a random-variance t-test (25). Differentially expressed transcripts were also modeled in Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) (further information is available online at http://www.uth.tmc.edu/scleroderma). We also employed a modular data mining strategy described by Chaussabel et al (10) (further information is available online at http://www.uth.tmc.edu/scleroderma).
We calculated an IFN score according to the method outlined by Baechler et al based on a set of 286 IFN-inducible genes identified by treatment of healthy PBMCs with IFNα/β and IFNγ (8). The IFN score in our study was calculated based on the 43 transcripts from the IFN-inducible gene list that were differentially expressed in SSc and SLE patients when compared with healthy controls (further information is available online at http://www.uth.tmc.edu/scleroderma).
Real-time quantitative PCR
Quantitative PCR assays for the STAT1, IFI6, IFIT3, and TLR5 genes were designed to confirm the microarray results. Each sample was assayed in triplicate plus a control without reverse transcriptase to assess DNA contamination levels (further information is available online at http://www.uth.tmc.edu/scleroderma). An average gene expression value for IFN-inducible genes was calculated using a normalization method described by Niewold et al (18). The final IFN quantitative PCR score was calculated based on the average of relative values in STAT1, IFI6, and IFIT3.
Genotyping
The genomic DNA from the investigated samples had already been genotyped for selected single-nucleotide polymorphisms (SNPs) in a group of IFN-inducible genes as part of a larger candidate gene study. In an exploratory analysis, we investigated the correlation of these SNPs with the respective IFN scores (further information is available online at http://www.uth.tmc.edu/scleroderma).
Statistical analysis
The categorical dependent variables were analyzed by chi-square test or by Fisher’s exact test if the values were less than 5. Continuous variables were analyzed by t-test if the raw or log-transformed data had a normal distribution. If the assumption of equal variance was not met, we used the Aspin-Welch unequal variance t-test. The Mann-Whitney nonparametric test was used if the model assumptions for the t-test were not met. Furthermore, linear regression analysis was used to adjust for the effect of multiple independent variables. Two-sided P values less than 0.05 were considered significant. The analyses were performed using the NCSS 2007 statistical program (NCSS, Kaysville, UT).
RESULTS
Characteristics of the study participants
Sixty patients with early SSc, 14 patients with late SSc, 21 healthy controls, and 17 SLE patients with active disease were examined in this cross-sectional study. The demographic features of the subjects are shown in Table 1. As expected from the typical ages at disease onset, the patients with SLE were significantly younger than other study subjects. No other features differed significantly among the study groups.
Table 1.
Demographic features of the study participants*
| Patients with early SSc (n = 60) |
Patients with late SSc (n = 14) |
Healthy controls (n = 21) |
SLE patients (n = 17) |
Total (n = 112) |
|
|---|---|---|---|---|---|
| Age at enrollment, mean ± SD years | 48.94 ± 13.63 | 50.12 ± 13.37 | 53.53 ± 16.72 | 38.5 ± 12.64† | 48.37 ± 14.62 |
| No. of women/no. of men | 50/10 | 9/5 | 17/4 | 16/1 | 92/20 |
| Race | |||||
| Caucasian | 33 (55) | 7 (50) | 13 (61.9) | 6 (35.3) | 59 (52.7) |
| African American | 13 (21.7) | 4 (28.6) | 4 (19) | 2 (11.8) | 23 (20.5) |
| Hispanic | 12 (20) | 2 (14.3) | 4 (19) | 8 (47.1) | 26 (23.2) |
| Other | 2 (3.3) | 1 (7.1) | 0 | 1 (5.9) | 4 (3.6) |
| SSc-related autoantibodies | |||||
| ACAs | 9 (15) | – | – | – | – |
| Antitopoisomerase | 10 (16.7) | 14 (100) | – | – | – |
| Anti–RNAP III | 15 (25) | – | – | – | – |
| Anti–U1 RNP | 4 (6.7) | – | – | – | – |
Except where indicated otherwise, values are the number (%) of subjects. ACAs = anticentromere antibodies; anti–RNAP III = anti–RNA polymerase III.
The patients with systemic lupus erythematosus (SLE) were significantly younger than the patients with early systemic sclerosis (SSc), the patients with late SSc, and the healthy controls (P = 0.006, P = 0.0191, and P = 0.004, respectively).
In the patients with early SSc, ACAs, antitopoisomerase, anti–RNAP III, and anti–U1 RNP antibodies were present in 9 patients (15%), 10 patients (16.7%), 15 patients (25%), and 4 patients (6.7%), respectively. All patients with late SSc were positive for antitopoisomerase antibodies (Table 1). Furthermore, 2 patients with early SSc had 2 SSc-related antibodies, while 17 patients in this group were ANA positive but did not have any of the above autoantibodies; only 2 SSc patients were ANA negative. The mean ± SD disease duration in patients with early SSc was 2.94 ± 1.53 years, while that in patients with late SSc was 9.86 ± 1.58 years. Among all the SSc patients, 38 (51.4%) had diffuse cutaneous involvement, and 33 (44.6%) had limited disease. The extent of skin involvement could not be determined in 3 SSc patients. The majority of SLE patients had active disease, as demonstrated by a mean ± SD SLAM-R score of 11.19 ± 6.73.
Both SLE and SSc PB cells demonstrate dysregulation of IFN-regulated transcripts
A total of 8,172 transcripts were detected across all PB cell samples that passed our filtering criteria. Clustering analyses according to date of sample collection and hybridization did not indicate that the observed gene expression patterns resulted from a technical artifact.
In a 3-way comparison, there were 907 significantly differentially expressed transcripts among SSc patients, SLE patients, and controls. Notably, of the top 100 differentially regulated transcripts among the 3 groups, 35 (35%) were known to be regulated by type I and type II IFNs.
A comparison of SSc patients with controls demonstrated 297 differentially expressed transcripts, whereas 987 transcripts were differentially expressed between SLE patients and controls. The complete lists of differentially expressed genes are available online at http://www.uth.tmc.edu/scleroderma/Supplemental_data.html.
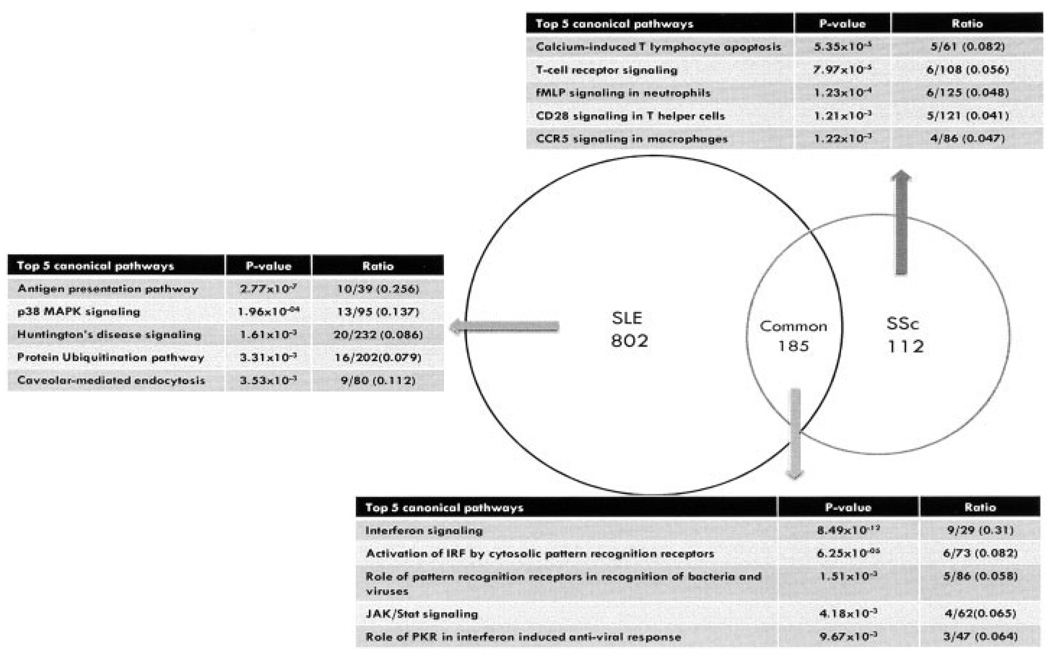
Intersection of the differentially expressed transcripts in SSc and SLE demonstrated 185 transcripts that the 2 diseases had in common (Figure 1). In other words, 62% (185/297) of the transcripts differentially expressed in SSc PB cells were also differentially expressed in SLE PB cells. Modeling of these data in the Ingenuity Pathway Analysis Canonical Pathway Knowledge Base revealed significant enrichment for transcripts involved in IFN signaling, regulation of IFN regulatory factors, recognition of pathogen-associated molecular patterns, and JAK/STAT signaling (Figure 1). A hierarchical clustering of the genes that were differentially expressed in SSc and SLE is available online at http://www.uth.tmc.edu/scleroderma.
Figure 1.
Differentially expressed pathways in systemic lupus erythematosus (SLE) patients and systemic sclerosis (SSc) patients compared with controls (further information is available online at http://www.uth.tmc.edu/scleroderma). IRF = interferon regulatory factor; PKR = RNA-dependent protein kinase.
We also modeled the differentially expressed transcripts that were unique to SLE and SSc, in the context of the Ingenuity Pathway Analysis Canonical Pathway Knowledge Base. The 802 unique SLE transcripts were enriched for transcripts involved in antigen presentation. This is consistent with the classic notion of SLE being an “antigen-driven” disease. In contrast, the 112 unique SSc transcripts demonstrated significantly decreased levels of genes involved in T lymphocyte apoptosis and T cell receptor and CD28 signaling compared with controls. However, transcripts belonging to the fMLP pathway for activation of granulocytes were up-regulated in SSc. Constitutive granulocyte activation leading to enhanced production of reactive oxygen species has been reported previously in SSc (Figure 1) (26–28).
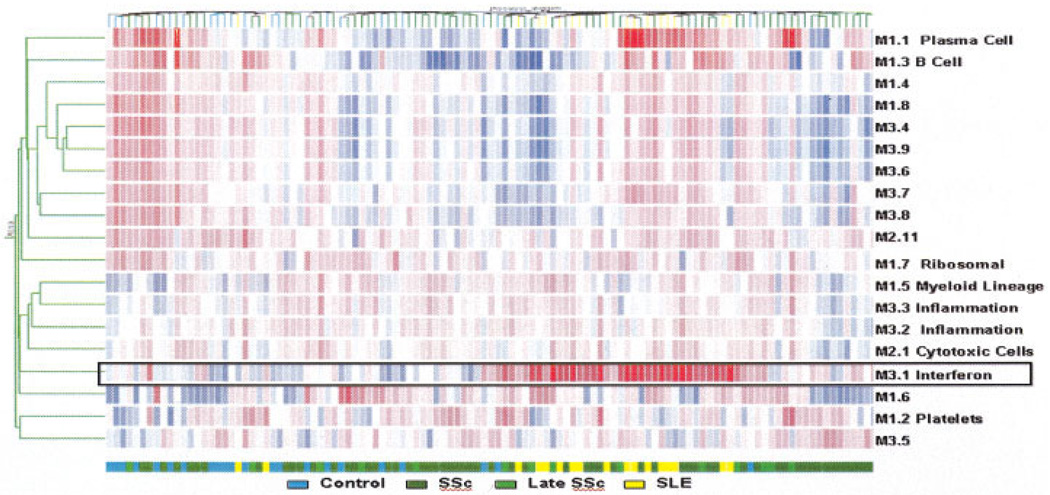
A modular data mining strategy described by Chaussabel et al showed similar results (10). For this analysis, we derived composite transcriptional vectors by averaging expression values for the genes forming each module. As a result, the global transcriptional activity of each patient is summed up in 28 vectors. Unsupervised hierarchical clustering arranged modules (rows) and samples (columns) based on patterns of modular activity (Figure 2). This analysis confirmed the presence of an IFN signature (M3.1) in the blood transcript profiles in a vast majority of SLE patients. Furthermore, it demonstrated that a subset of patients with SSc possessed a “lupus-like” signature, which in some patients included both IFN and plasma cell signatures (M3.1 and M1.1, respectively).
Figure 2.
Mapping modular blood transcriptional activity. The heatmap represents levels of activity for a set of predetermined transcriptional modules. Average gene expression levels were obtained for each module. For each patient, 28 vectors were thus obtained, and the data set was subjected to an unsupervised analysis: modules for which at least 1 sample showed a deviation from the median of >1.5-fold were selected and arranged based on their activity pattern across patients. Conversely, patients were arranged using the same hierarchical clustering algorithm based on their activity patterns across modules. Red indicates a relative increase in transcriptional activity; blue indicates a relative decrease. The distribution of the different study groups is indicated by a color code. Functional interpretations available for some of these modules are also provided. See Figure 1 for definitions.
We also reanalyzed microarray data from a previous skin biopsy study, performed by our group, of patients with early SSc (29). The skin biopsy sections used for this analysis had histologic findings that were typical for SSc, but they did not show increased staining for CD3 or CD20 markers compared with controls (29). From these data, we extracted a set of significantly differentially expressed transcripts using the same threshold criteria, and we modeled the expression data on the Ingenuity Pathway Analysis Canonical Pathway Knowledge Base. SSc skin showed striking parallels to SSc and SLE PB cells with regard to up-regulation of multiple transcripts involved in the IFN signaling pathway (further information is available online at http://www.uth.tmc.edu/scleroderma). One notable difference was that STAT2 was differentially up-regulated only in SSc. These data suggest that both SLE and SSc may belong to the same spectrum of IFN-mediated diseases. Furthermore, the data from SSc PB cells and skin strongly suggest that there is activation of IFN signaling pathways at the immune effector arm (circulating blood cells) as well as at the level of the target organ (skin) in SSc.
The IFN signature in SSc and SLE patients is a quantitative trait
A comparison between the SSc and SLE patient groups demonstrated that expression of all the IFN-inducible genes was significantly higher in SLE relative to SSc (http://www.uth.tmc.edu/scleroderma/Supplemental_data.html), suggesting a gradient of the IFN signature that is strongest in SLE patients, intermediate in SSc patients, and lowest in healthy controls. To better quantitate this, we cross-referenced our expression data with a set of 286 transcripts reported by Baechler et al (8) that were observed to be induced by treatment of normal PBMCs with IFNα/β and IFNγ. A total of 83 IFN-inducible genes were differentially expressed in SLE patients compared with controls (80 of 83 up-regulated), whereas 47 transcripts showed altered expression in SSc patients compared with controls (44 of 47 up-regulated). Forty-three genes (91%) were differentially expressed in both comparisons. We used these 43 transcripts to calculate an IFN score (see Patients and Methods; further information is available online at http://www.uth.tmc.edu/scleroderma). Similarly, 37 of the 40 genes (92.5%) that were uniquely differentially expressed among SLE patients were overexpressed in SSc patients, but these genes did not reach our selection criteria for differentially expressed genes in the group of SSc patients.
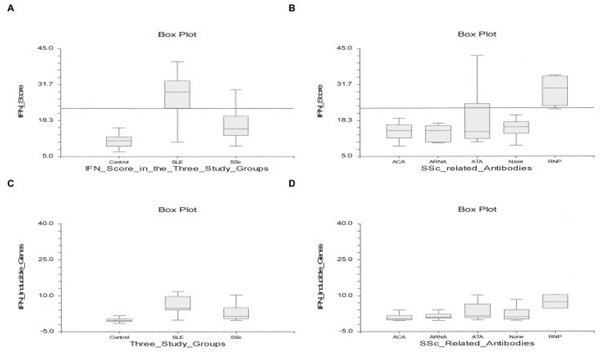
The IFN score was highest in SLE patients, followed by SSc patients and then controls (mean ± SD 28.1 ± 7.26, 17.6 ± 7.39, and 11.57 ± 10.91, respectively). Figure 3A shows the box plots of the IFN scores in the 3 study groups. The difference in IFN score among all 3 groups was statistically significant (P < 0.001 for all 3 comparisons). Furthermore, the IFN score correlated with the SLAM-R score in the SLE patients (P = 0.01). The IFN score was also dichotomized according to a threshold value corresponding to the 95th percentile in controls (horizontal line in Figures 3A and B); the IFN score values above this threshold were considered positive.
Figure 3.
A and B, Interferon (IFN) score based on 43 IFN-inducible genes in the 3 study groups (A) and in the serologic subtypes of SSc (B). The IFN score was also dichotomized according to a threshold value corresponding to the 95th percentile in controls (horizontal line). C and D, Average score of 3 IFN-inducible genes based on quantitative polymerase chain reaction in the 3 study groups (C) and in the serologic subtypes of SSc (D). Data are shown as box plots. Each box represents the 25th to 75th percentiles. The depth of the box is the interquartile range (IQR). The line inside the box represents the median. The upper adjacent value is the largest observation that is less than or equal to the 75th percentile plus 1.5 times the IQR. The lower adjacent value is the smallest observation that is greater than or equal to the 25th percentile minus 1.5 times the IQR. ACA = anticentromere antibody; ARNA = anti–RNA polymerase III; ATA = antitopoisomerase; RNP = anti–U1 RNP (see Figure 1 for other definitions).
Calculation of the IFN score between the 3 groups using quantitative PCR of selected IFN-regulated transcripts (STAT1, IFI6, and IFIT3) confirmed this observation. Graphs show the expression values of the 3 IFN-regulated genes in addition to TLR5 (negative control) in the investigated study groups (further information is available online at http://www.uth.tmc.edu/scleroderma). Similar to the microarray results, the expression levels of all 3 IFN-inducible genes were highest in SLE, followed by SSc. The expression level of TLR5 (negative control) did not differ among the 3 study groups. The expression levels of the 3 IFN-inducible transcripts were averaged. As shown in Figure 3C, the SLE patients had significantly greater expression of averaged IFN-inducible gene scores than controls (P < 0.001) and SSc patients (P = 0.003). The averaged IFN-inducible gene score was also significantly higher in SSc patients than in controls (P = 0.009).
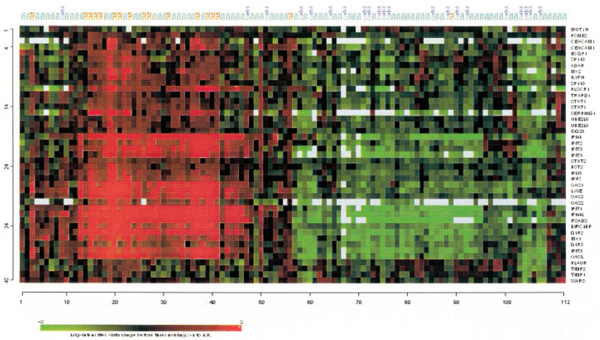
We performed unsupervised hierarchical clustering of the samples using the 43 IFN-regulated transcripts in individual samples across the 3 groups (Figure 4). This analysis revealed that the relative expression level of IFN-regulated transcripts was increased in 16 of 18 SLE patients (89%) in contrast to only 3 of 21 controls (14%). However, 35 of 74 SSc patients (47.3%) had increased expression of IFN-inducible genes, and these patients tended to segregate with SLE patients. These data confirm that the IFN signature is strongest and most prevalent in SLE, but the picture is more heterogeneous in SSc. We therefore undertook a subgroup analysis of the SSc patients to ascertain possible clinical, serologic, and genetic variables that underlie this observed heterogeneity.
Figure 4.
Unsupervised hierarchical clustering of 43 interferon-inducible transcripts. Samples are labeled according to study groups (SSc patients = green; SLE patients = red; controls = blue). See Figure 1 for definitions.
The IFN signature defines a particular serologic subset of SSc patients
Comparisons between patients with early antitopoisomerase-positive SSc and those with late antitopoisomerase-positive SSc, as well as comparisons between patients with late antitopoisomerase-positive SSc and the remaining SSc patients, using all genes that passed our filtering criteria, did not show any differentially expressed genes. Similarly, comparison of SSc patients with limited skin involvement and those with diffuse skin involvement (23) did not reveal any differentially regulated genes.
The IFN score showed an inverse correlation with the lymphocyte count (P < 0.001), while it was not associated with the WBC (P = 0.438), neutrophil (P = 0.624), monocyte (P = 0.806), eosinophil (P = 0.318), or basophil (P = 0.672) counts. The IFN score did not correlate with disease duration, the MRSS (24), predicted forced vital capacity, or diffusing capacity for carbon monoxide. Furthermore, IFN score positivity was not associated with disease type (limited versus diffuse) (P = 0.25), pulmonary fibrosis (P = 0.594), or pulmonary hypertension (P = 0.16). The IFN score correlated with the number of SLE criteria present (P = 0.003) (21) among the SSc patients. After ANA positivity, the most common SLE criteria were lymphopenia at 2 time points (41.8%), arthritis (9.1%), oral ulcers (1.8%), and pleural effusion (1.8%). The association of the IFN score with SLE criteria was driven mainly by lymphopenia (P = 0.004).
We also found that SSc patients with anti–U1 RNP antibodies were more likely to be IFN signature positive (P = 0.001). Similarly, SSc patients with antitopoisomerase antibodies had a higher likelihood of a positive IFN score than other SSc patients, when anti–U1 RNP–positive patients were excluded (P = 0.005). Two SSc patients had anti-Ro antibodies, and both belonged to the IFN signature–positive group (P = 0.044). The presence of ACAs or anti–RNAP III antibodies was not associated with IFN positivity (P = 0.108 and P = 0.247, respectively). Figure 3B shows the IFN score levels in the serologic subtypes of SSc. The results of quantitative PCR mirror the microarray results (Figure 3D). The IFN-inducible genes in SSc patients with anti–U1 RNP antibodies showed higher average expression values than those in the remainder of SSc patients (P = 0.007). The antitopoisomerase-positive patients had higher IFN-inducible gene expression than SSc patients with ACAs (P = 0.026) and showed higher expression levels of IFN-inducible genes than did the remainder of SSc patients (P = 0.015). The anti-Ro positivity in the patients with SSc was also associated with increased IFN-inducible gene expression levels (P = 0.035).
Class comparisons of all genes passing our filtration criteria between antitopoisomerase- and ACA-positive subsets revealed that 407 transcripts were differentially expressed. Modeling of these differentially expressed genes in the Ingenuity Pathway Analysis Canonical Pathway Knowledge Base showed significant up-regulation of transcripts involved in CD28 and ephrin signaling in antitopoisomerase-positive patients. Comparisons between the other serologic subtypes of SSc, including antitopoisomerase-positive SSc versus anti–RNAP III–positive SSc and ACA-positive SSc versus anti–RNAP III–positive SSc, did not reveal any significantly differentially expressed transcripts.
IFN score correlates with genetic variants of the IFN pathway
Among the patients with SSc, the presence of the GG or GT genotype of IFNAR2 rs7279064 was significantly associated with a higher IFN score (P = 0.008), which remained significant after correction for ethnicity (P = 0.032). The IFN score did not vary significantly by the investigated SNP in the control group. The other investigated SNPs in the IFNAR1, IFNAR2, IRF5, IRF7, STAT1, and STAT4 genes did not correlate with the IFN score (detailed results are available online at http://www.uth.tmc.edu/scleroderma).
DISCUSSION
This study represents the first direct comparison of SSc and SLE on the same platform. The results confirmed previously published data indicating possible similarities in increased expression of IFN-inducible genes in PB cells from SSc patients and SLE patients (4–6,8,9). Importantly, we demonstrated that there appears to be a gradient effect, with the magnitude of IFN-inducible gene activation strongest and most prevalent among SLE patients, followed by SSc patients. Qualitatively, the overall pattern of IFN-inducible gene activation was strikingly similar between SSc and SLE.
Our data showed increased expression of IFN-inducible genes activated by type I and type II IFNs in PB cells. Based on the IFN score, it is not clear which type of IFN signaling predominates (8). Further studies at the protein level would be required to validate this observation. Nonetheless, it is interesting to note that a randomized, placebo-controlled trial of subcutaneous IFNα in early SSc showed that IFNα treatment resulted in significant worsening of lung function and a trend toward skin deterioration (30). Type I IFNs, in particular, have been implicated in the pathogenesis of SLE (31,32). The plasmacytoid dendritic cells (PDCs) are thought to be one of the main sources of type I IFN (33). IFNs lead to production of autoantibodies by their effects on B cells, and in the setting of antigen excess this leads to formation of immune complexes containing DNA or RNA. In vitro studies have shown that the nucleic acid–containing immune complexes can lead to self-perpetuating, excessive IFN production (26–28,34–37). Though it has yet to be demonstrated in vivo, this process is thought to contribute to development of SLE in a susceptible host. Anecdotal reports of development of SLE and SSc in patients who were undergoing IFN treatment for other conditions lend some support to this notion (38–40).
In our study, 89% of SLE patients demonstrated up-regulation of IFN-inducible genes compared with 77–93% in other studies (8,9). Likely explanations for this high percentage are that the SLE patients were not receiving immunosuppressive agents and had high disease activity (average SLAM-R score 11.19) at enrollment. This is in agreement with observations that the presence of the IFN signature in SLE correlates with disease activity, renal involvement, and anti-dsDNA and complement levels (9,11).
We did not observe differences in the PB cell transcript profiles or IFN score in relation to SSc disease duration. These observations are consistent with a previous report that the IFNα-inducing activity of SSc sera does not correlate with disease duration (41). We also could not demonstrate any correlation of transcript profiles with various clinical manifestations of SSc or its disease activity. The latter is hampered by the lack of a standardized disease activity measure in SSc. The assessed clinical manifestations represent more accurately the irreversible disease damage rather than disease activity. This implies that a transient change in gene expression pattern correlating with the development of various clinical manifestations could have been missed in our cross-sectional design. Longitudinal studies are needed to investigate the predictive role of the IFN signature for development of various clinical complications of SSc.
Our data demonstrated that the presence of antitopoisomerase and anti–U1 RNP antibodies are associated with a higher IFN score. Furthermore, we were able to subgroup the SSc patients at the transcript level based on the presence of an IFN-inducible gene pattern. These data suggest that there is a gradient in the IFN signature among the serologic subtypes of SSc that could have implications for future studies of pathogenesis and therapeutic targets. Indeed, Kim et al demonstrated that >90% of antitopoisomerase-positive SSc sera had IFNα-inducing activity (41). This activity was contained in the IgG fraction. Furthermore, the anti–topoisomerase I levels correlated with IFNα induction in that study. Moreover, another study showed that the serologically heterogeneous group of ACA-negative SSc patients had higher levels of IFN-inducible gene expression (7).
We also observed a similar IFN signature in skin biopsy samples from SSc patients, but we did not find increased IFNα or IFNβ messenger RNA (mRNA) levels in PB cells from SSc or SLE patients, suggesting that cells may be responding to local IFNα at the target tissue level. In another study, increased levels of IFNα mRNA and PDCs were reported in the skin tissue of patients with diffuse SSc. An IFN-inducible gene pattern in the monocytes and T cells of a separate group of patients was also seen, but the corresponding sera did not have detectable IFNα that correlated with the IFN signature (6). A study by Milano et al (42) examining skin biopsy transcript profiles of SSc patients showed that a subgroup consisting of patients with limited and diffuse disease type showed an “inflammatory profile.” Although a correlation with different serologic subtypes of SSc was not conducted, it is notable that the samples classified as inflammatory had overexpression of multiple IFN-inducible genes as well.
We also found that the IFNAR2 rs7279064 SNP was significantly associated with a higher IFN score in the SSc patients, indicating that this polymorphism plays a role in the variance of IFN activity seen in this disease. Our genotype correlation study was hampered by our relatively small sample sizes and the difficulties in correcting for the effect of ethnicity on gene expression. Further studies are needed to investigate the effect of various SNPs on the expression of IFN-inducible genes.
In summary, our data show that SSc and SLE may belong in the same spectrum of IFN-mediated diseases. A subset of SSc patients has a “lupus-like” high IFN-inducible gene expression pattern that correlates with the presence of antitopoisomerase and anti–U1 RNP antibodies. The classification of SSc based on the presence of the IFN signature can provide opportunities for better understanding of its pathogenesis and for development of targeted therapeutic interventions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Emily C. Baechler for her helpful advice regarding RNA extraction protocol and calculation of the IFN score. We are also grateful to Dr. David Loose, Dr. Gregory Shipley, Mrs. Nancy Shipley, and Mrs. Jun Ying for their assistance in the design and performance of the laboratory studies.
Supported by the NIH (grants R01-AR-055258, UL1-RR-024148, and W81XWH-07-0111, NIH Centers for Research Translation grant P50-AR-054144, University Clinic Research Center grants M01-RR-00073 to the University of Texas Medical Branch at Galveston and M01-RR-01346 to the University of Texas Health Science Center at San Antonio, and the NIH Clinical and Translational Sciences Award, 1U54-RR-23417-01). Dr. Assassi was recipient of the American College of Rheumatology Clinical Investigator Fellowship Award.
Footnotes
Dr. Pascual has received consulting fees, speaking fees, and/or honoraria from MedImmune, Biogen, and Novartis (less than $10,000 each).
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Assassi had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Assassi, Mayes, Arnett, Gourh, Agarwal, Fischbach, Charles, Reveille, Tan.
Acquisition of data. Assassi, Mayes, Arnett, Gourh, McNearney, Fischbach, Shah, Charles, Reveille, Tan.
Analysis and interpretation of data. Assassi, Mayes, Gourh, Agarwal, Chaussabel, Oommen, Charles, Pascual, Tan.
REFERENCES
- 1.Prescott RJ, Freemont AJ, Jones CJ, Hoyland J, Fielding P. Sequential dermal microvascular and perivascular changes in the development of scleroderma. J Pathol. 1992;166:255–263. doi: 10.1002/path.1711660307. [DOI] [PubMed] [Google Scholar]
- 2.Silver RM, Medsger TA, Jr, Bolster MB. Systemic sclerosis and scleroderma variants: clinical aspects. In: Koopman WJ, Moreland LW, editors. Arthritis and Allied Conditions. 15th ed. Philadelphia: Lippincott Williams & Wilkins; 2005. pp. 1633–1680. [Google Scholar]
- 3.Steen VD. Autoantibodies in systemic sclerosis. Semin Arthritis Rheum. 2005;35:35–42. doi: 10.1016/j.semarthrit.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 5.York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and Toll-like receptor agonists. Arthritis Rheum. 2007;56:1010–1020. doi: 10.1002/art.22382. [DOI] [PubMed] [Google Scholar]
- 6.Duan H, Fleming J, Pritchard DK, Amon LM, Xue J, Arnett HA, et al. Combined analysis of monocyte and lymphocyte messenger RNA expression with serum protein profiles in patients with scleroderma. Arthritis Rheum. 2008;58:1465–1474. doi: 10.1002/art.23451. [DOI] [PubMed] [Google Scholar]
- 7.Bos CL, van Baarsen LG, Timmer TC, Overbeek MJ, Basoski NM, Rustenburg F, et al. Molecular subtypes of systemic sclerosis in association with anti-centromere antibodies and digital ulcers. Genes Immun. 2009;10:210–218. doi: 10.1038/gene.2008.98. [DOI] [PubMed] [Google Scholar]
- 8.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-α pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 12.Dieude P, Guedj M, Wipff J, Avouac J, Fajardy I, Diot E, et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 2009;60:225–233. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 13.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 14.Sigurdsson S, Nordmark G, Goring HH, Lindroos K, Wiman AC, Sturfelt G, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gourh P, Tan FK, Assassi S, Ahn CW, McNearney TA, Fischbach M, et al. Association of the PTPN22 R620W polymorphism with anti–topoisomerase I– and anticentromere antibody–positive systemic sclerosis. Arthritis Rheum. 2006;54:3945–3953. doi: 10.1002/art.22196. [DOI] [PubMed] [Google Scholar]
- 16.Dieude P, Guedj M, Wipff J, Avouac J, Hachulla E, Diot E, et al. The PTPN22 620W allele confers susceptibility to systemic sclerosis: findings of a large case–control study of European Caucasians and a meta-analysis. Arthritis Rheum. 2008;58:2183–2188. doi: 10.1002/art.23601. [DOI] [PubMed] [Google Scholar]
- 17.Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-α activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-α activity and low tumor necrosis factor α levels in patients with lupus. Arthritis Rheum. 2008;58:2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subcommittee for Scleroderma Criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–590. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 21.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 22.Bae SC, Koh HK, Chang DK, Kim MH, Park JK, Kim SY. Reliability and validity of Systemic Lupus Activity Measure-Revised (SLAM-R) for measuring clinical disease activity in systemic lupus erythematosus. Lupus. 2001;10:405–409. doi: 10.1191/096120301678646146. [DOI] [PubMed] [Google Scholar]
- 23.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, et al. Scleroderma (systemic sclerosis): classification, subsets, and pathogenesis. J Rheumatol. 1988;15:202–205. [PubMed] [Google Scholar]
- 24.Clements P, Lachenbruch P, Seibold J, White B, Weiner S, Martin R, et al. Inter and intraobserver variability of total skin thickness score (modified Rodnan TSS) in systemic sclerosis. J Rheumatol. 1995;22:1281–1285. [PubMed] [Google Scholar]
- 25.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 26.Maslen CL, Hall ND, Woolf AD, Maddison PJ. Enhanced oxidative metabolism of neutrophils from patients with systemic sclerosis. Br J Rheumatol. 1987;26:113–117. doi: 10.1093/rheumatology/26.2.113. [DOI] [PubMed] [Google Scholar]
- 27.Luczynska M, Szkudlarek U, Dziankowska-Bartkowiak B, Waszczykowska E, Kasielski M, Jozefowicz-Okonkwo G, et al. Elevated whole blood chemiluminescence in patients with systemic sclerosis. Clin Exp Rheumatol. 2005;23:173–179. [PubMed] [Google Scholar]
- 28.Kovacs IB, Meyrick Thomas RH, Mackay AR, Rustin MH, Kirby JD. Increased chemiluminescence of polymorphonuclear leucocytes from patients with progressive systemic sclerosis. Clin Sci (Lond) 1986;70:257–261. doi: 10.1042/cs0700257. [DOI] [PubMed] [Google Scholar]
- 29.Gardner H, Shearstone JR, Bandaru R, Crowell T, Lynes M, Trojanowska M, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum. 2006;54:1961–1973. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 30.Black CM, Silman AJ, Herrick AI, Denton CP, Wilson H, Newman J, et al. Interferon-α does not improve outcome at one year in patients with diffuse cutaneous scleroderma: results of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 1999;42:299–305. doi: 10.1002/1529-0131(199902)42:2<299::AID-ANR12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16:801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Ronnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 34.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, et al. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronnblom L, Alm GV. An etiopathogenic role for the type I IFN system in SLE. Trends Immunol. 2001;22:427–431. doi: 10.1016/s1471-4906(01)01955-x. [DOI] [PubMed] [Google Scholar]
- 37.Vallin H, Blomberg S, Alm GV, Cederblad B, Ronnblom L. Patients with systemic lupus erythematosus (SLE) have a circulating inducer of interferon-α (IFN-α) production acting on leucocytes resembling immature dendritic cells. Clin Exp Immunol. 1999;115:196–202. doi: 10.1046/j.1365-2249.1999.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beretta L, Caronni M, Vanoli M, Scorza R. Systemic sclerosis after interferon-alfa therapy for myeloproliferative disorders. Br J Dermatol. 2002;147:385–386. doi: 10.1046/j.1365-2133.2002.48901.x. [DOI] [PubMed] [Google Scholar]
- 39.Ioannou Y, Isenberg DA. Current evidence for the induction of autoimmune rheumatic manifestations by cytokine therapy [review] Arthritis Rheum. 2000;43:1431–1442. doi: 10.1002/1529-0131(200007)43:7<1431::AID-ANR3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 40.Solans R, Bosch JA, Esteban I, Vilardell M. Systemic sclerosis developing in association with the use of interferon α therapy for chronic viral hepatitis. Clin Exp Rheumatol. 2004;22:625–628. [PubMed] [Google Scholar]
- 41.Kim D, Peck A, Santer D, Patole P, Schwartz SM, Molitor JA, et al. Induction of interferon-α by scleroderma sera containing autoantibodies to topoisomerase I: association of higher interferon-α activity with lung fibrosis. Arthritis Rheum. 2008;58:2163–2173. doi: 10.1002/art.23486. [DOI] [PubMed] [Google Scholar]
- 42.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008;3:e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.