Abstract
Obesity is a complex phenotype affected by genetic and environmental influences such as sociocultural factors and individual behaviors. Previously, we performed two separate genome-wide investigations for adiposity-related traits (BMI, percentage body fat (%BF), abdominal circumference (ABDCIR), and serum leptin and serum adiponectin levels) in families from American Samoa and in families from Samoa. The two polities have a common evolutionary history but have lately been influenced by variations in economic development, leading to differences in income and wealth and in dietary and physical activity patterns. We now present a genome-wide linkage scan of the combined samples from the two polities. We adjust for environmental covariates, including polity of residence, education, cigarette smoking, and farm work, and use variance component methods to calculate univariate and bivariate multipoint lod scores. We identified a region on 9p22 with genome-wide significant linkage for the bivariate phenotypes ABDCIR–%BF (1-d.f. lod 3.30) and BMI–%BF (1-d.f. lod 3.31) and two regions with genome-wide suggestive linkage on 8p12 and 16q23 for adiponectin (lod 2.74) and the bivariate phenotype leptin–ABDCIR (1-d.f. lod 3.17), respectively. These three regions have previously been reported to be linked to adiposity-related phenotypes in independent studies. However, the differences in results between this study and our previous polity-specific studies suggest that environmental effects are of different importance in the samples. These results strongly encourage further genetic studies of adiposity-related phenotypes where extended sets of carefully measured environmental factors are taken into account.
INTRODUCTION
Obesity has become a global health problem, contributing to the rise in non-communicable diseases, and is associated with nutrition transition changes in physical activity and diet (1–3). Overweight and obesity are remarkably frequent in adults residing on the Samoan islands. In 2002, 89% of men and 92% of women in American Samoa had a BMI >26 kg/m2. In 2003, in Samoa, 68% of men and 84% of women had a BMI >26 kg/m2 (3,4). Body composition studies of Polynesians suggests that a BMI >26 kg/m2 defines overweight and a BMI >32 kg/m2 defines obesity (5).
American Samoans and Samoans have a common evolutionary history of ~3,000 years but have, during the past century, been differently influenced by political economic development, resulting in variation in environmental factors such as income and wealth and nutritional behaviors (4,6,7). Their relative isolation, large family sizes, and recent exposure to modernization make these two polities advantageous populations for genetic studies of complex phenotypes (6–9). To study adiposity-related traits, including BMI, percentage body fat (%BF), abdominal circumference (ABDCIR), fasting serum leptin levels (LEPTIN), and fasting serum adiponectin levels (ADIPONEC), we previously conducted separate genome-wide scans for each of the two polities (10,11).
Here we present a genome-wide investigation to detect quantitative trait loci (QTLs) for the five adiposity-related phenotypes in a combined study sample from American Samoa and Samoa. In an attempt to handle environmental factors that affect the phenotypes investigated, we adjust for environmental covariates, including polity of residence, education, cigarette smoking, and farm work, and use variance component methods and LOKI (12)/SOLAR (http://www.sfbr.org/solar) (13,14) software to calculate univariate and bivariate multipoint lod scores.
METHODS AND PROCEDURES
Study sample
The study population derives from two polities, American Samoa and Samoa. The 2000 census population in American Samoa was 57,291, of whom 88.2% were ethnic Samoans (15). The 2001 census population in Samoa was 177,714, with 92.6% ethnic Samoans (16). American Samoa has higher education levels, a higher proportion of adults in wage and salary occupations, and higher economic and material lifestyle indicators than Samoa (4,6,7,17). For example, the per capita income for American Samoa rose from $596 in 1969 to $3,039 in 1989, to $5,800 in 2005 (18), whereas the per capita income in Samoa in 2007 was $2,020 (19).
This study includes 71 pedigrees containing 1,164 genotyped individuals (534 men and 630 women) aged ≥ 18 years. Twenty families contain genotyped members from both American Samoa and Samoa. All individuals included in the study self-reported as being of Samoan ethnicity (see Supplementary Methods and Procedures online).
Phenotypes
Standard procedures were applied to measure the phenotypes BMI, %BF, ABDCIR, LEPTIN, and ADIPONEC (see Supplementary Methods and Procedures online). Interviews were used to collect information on educational level and physical activity from farm work. Farm work was a dichotomous variable based on self-reported weekly subsistence work at a farm, and in a few cases fishing (4). In addition, the participants were asked whether they currently were cigarette smokers.
Genotypes
In total, 387 microsatellite markers from the ABI PRISM linkage mapping set v2.5 MD10 (Applied Biosystems, Foster City, CA) were genotyped. For the samples from American Samoa, some markers were genotyped with an ABI PRISM 3100 genetic analyzer and some markers with an ABI PRISM 3130XL (Applied Biosystems), and the Samoan samples were genotyped using the ABI PRISM 3130XL for all markers (see Supplementary Methods and Procedures online).
As our study sample contains pedigrees with individuals genotyped using different instruments, to be able to perform linkage analysis it is important to ensure that the same allele label in the two sets defines identical alleles. This issue has recently been addressed by others (20,21), but to our knowledge no software is yet available that can handle extended pedigrees. We therefore merged the two data sets according to the minimal differences in marker allele frequencies (S), as shown in the following equation:
where the summation is overall available alleles for a specific marker and where xi1 is the frequency of allele i in data set 1 and xi2 is its frequency in data set 2.
For each marker, we evaluated S when a single shift (any integer from −10 to 10) was applied to all alleles in one of the two data sets. If the smallest S was greater than a certain cutoff value (≥0.08), we evaluated S again using two shifts, which allows the smaller alleles to be shifted differently than the larger alleles. We searched for the minimized S by altering the two shifts as well as altering the breakpoint between the two shifts. The breakpoint could be placed between any two adjacent alleles except in the terminal intervals on either end. Shifts changing the allele order were eliminated from evaluation. In addition, all allele alignments were plotted and visually evaluated.
Error checking and data handling
Adiposity phenotypes were checked for data-entry mistakes and for outliers. Box–Cox power transformations (22) were applied to the adiposity phenotypes that were not normally distributed.
The polity-specific study samples had been extensively checked for genotype errors and errors of pedigree structure before the separate genome scans (10,11). For the combined study sample, we performed additional checks for inconsistencies in pedigree structure using RELPAIR v2.0.1 (http://csg.sph.umich.edu/index.php). No errors in the pedigree structures were found, presumably because our earlier data cleaning resolved all such relationship errors (see Supplementary Methods and Procedures online).
Multipoint linkage analysis
Marker allele frequencies were estimated from our pedigree data using LOKI (12), while simultaneously estimating the identity-by-descent sharing matrices required by SOLAR (13,14). In this way, proper modeling of the variability in the estimates of the allele frequencies occurs. To minimize the effect of the variation in lod score from run to run caused by variations in LOKI’s stochastic estimates of the identity-by-descent matrix, we report the average univariate maximum lod scores from 10 runs. We have applied the same statistical strategy and genetic map as we previously used for our polity-specific investigations (10,11). The genetic map, based on Kosambi cM, was derived from the Rutgers Combined Linkage-Physical Map (23).
Univariate multipoint linkage analysis
We used the multipoint variance component linkage analysis as implemented in SOLAR to search for QTLs for adiposity-related phenotypes on the autosomes. This maximum-likelihood approach takes full advantage of phenotypic and genotypic information from all pedigree members, and the expected genetic covariances between relatives are specified as a function of the observed identity by descent at a given position in the genome as estimated from the marker data. Before the variance component linkage analysis, we used the “polygenic” command in SOLAR to include covariates and fit the variance component model. In this study three sets of covariates were used for linkage analyses: the basic set, the environmental set, and the extended set (Table 1) (see Supplementary Methods and Procedures online).
Table 1.
Description of covariate sets used in univariate linkage analysis
| Covariate set | Age | Sex | Polity of residence |
Education | Farm work | Cigarette smoking |
BMI |
|---|---|---|---|---|---|---|---|
| Basic | + | + | + | ||||
| Environmental | + | + | + | + | + | + | |
| Extended | + | + | + | + | + | + | + |
X-linked analysis
The variance component linkage analysis as implemented in the current release of SOLAR does not correctly carry out multipoint X-linked analysis. Instead, we broke our pedigrees into nuclear families using Mega2 (http://watson.hgen.pitt.edu) (24) and applied models for mapping X-linked QTLs that are implemented in Mendel v6.0.1 (http://www.genetics.ucla.edu/software) (25).
Bivariate analysis
For all chromosomes that, in the univariate analyses, obtained QTLs with lod score ≥1.175 for two or more phenotypes within the same region we performed bivariate multipoint linkage analysis using SOLAR. The bivariate analysis tests for simultaneous linkage of two phenotypes to a single genetic region. We here report the bivariate lod score transformed to 1 degree of freedom, which is comparable to a univariate lod score. The bivariate linkage variance component analysis as implemented in SOLAR is a multivariate analysis that appropriately handles the correlation between the traits, and thus no additional correction for multiple non-independent tests is necessary (26). Covariate screening in bivariate models is not supported in SOLAR. To improve the power to detect susceptibility loci, we included the union of significant covariates from the environmental set for the two traits, using the “polygenic -all” command in SOLAR, that were included in the univariate analyses.
We carried out two separate likelihood-ratio tests at the location of the maximum multipoint bivariate lod score, one to test whether the QTL signal is due to pleiotropy (i.e., a major gene affects both phenotypes) and the other to test whether it is due to co-incident linkage (i.e., a set of clustered genes, each influencing a particular trait). We used a χ2-test to evaluate the results. A P-value of 0.05 was used to reject either pleiotropy or co-incident linkage (27) (see Supplementary Methods and Procedures online).
RESULTS
As the combined study sample includes pedigrees with individuals who were genotyped using different instruments, we merged the data sets according to the minimal differences in allele frequencies. After initial alignment, we observed 10 out of 387 markers with large differences in allele frequencies between the sets. New allele calls were generated for these markers (see Supplementary Methods and Procedures online). After visual evaluation of the alignments from the recalled markers, these recalled markers were also judged acceptably aligned and were included in the analyses. None of the recalled markers was one of the six markers that, after application of our alignment algorithm, had the greatest S values (see Supplementary Figure S1 online).
Descriptions of socio-demographic characteristics and phenotypes (traits and covariates) for men and women in the combined study sample, and for American Samoans and Samoans separately, are presented in Supplementary Table S1 online. Mean BMI across sex and residence subgroups ranges from 28.9 to 36.6 kg/m2. LEPTIN is 2.7–3.8 times higher in women than in men in all subsamples. In Samoa, 84% of the men do farm work, but in American Samoa only 53% do. The mean values of BMI and %BF for men tend to be lower in Samoa than in American Samoa. However, the mean value for BMI for both sexes is considered overweight or obese in all three subsamples (see Supplementary Table S1 online).
We used three sets of covariates in the heritability and linkage analyses (Table 1). Heritability estimates for the five adiposity phenotypes—BMI, %BF, ABDCIR, LEPTIN, and ADIPONEC—range from 0.42 to 0.45 when adjusted for the basic covariates (see Supplementary Table S2 online). When additional covariates were included, the heritability estimates slightly decreased for all traits but ADIPONEC. The proportion of variance for each trait explained by the covariates is listed in Supplementary Table S2.
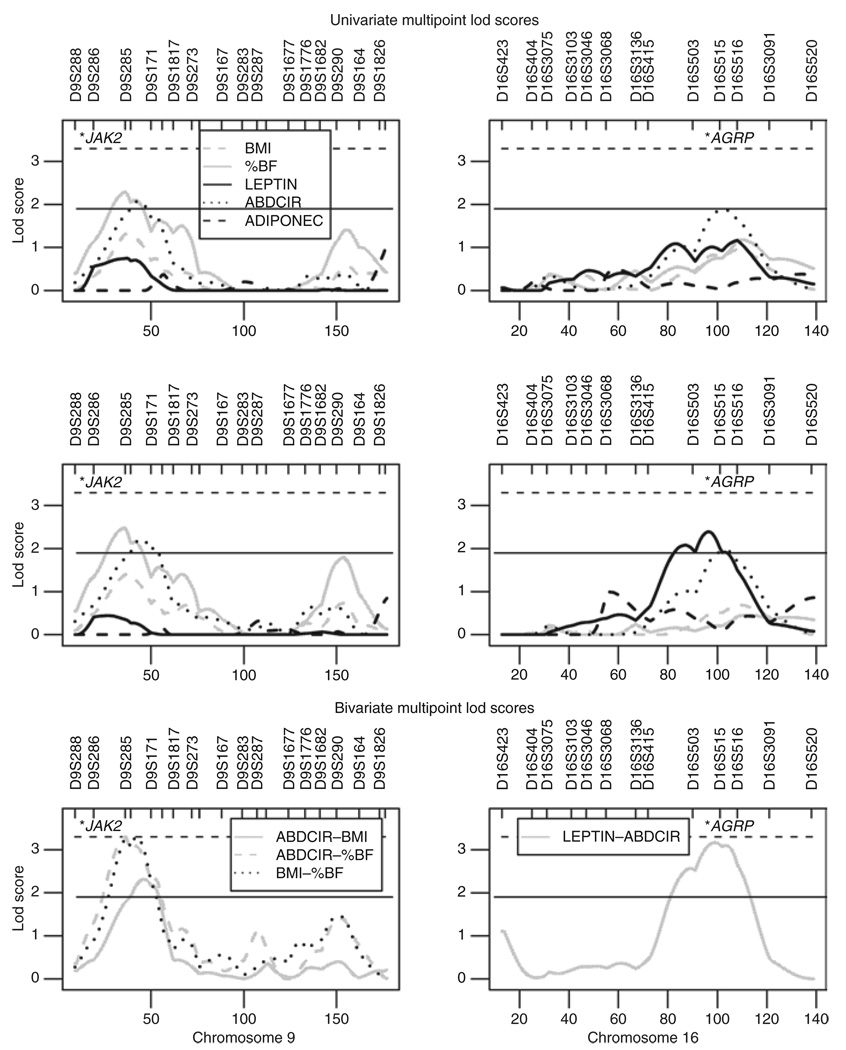
Univariate multipoint lod scores from one representative run for the three covariate sets are displayed in Supplementary Figure S2. Regions with univariate multipoint lod ≥1.5 for at least one model are listed in Table 2. When the environmental set was used, we detected two regions (9p22.3–p21.3 and 16q21–q23.1) with suggestive linkage (lod ≥01.9). On 9p we observed linkage for %BF (lod 2.48) and ABDCIR (lod 2.14). In this region, BMI had a lod score of 1.40. On 16q we detected linkage for LEPTIN (lod 2.33) and ABDCIR (lod 1.81). The top and middle rows of Figure 1 show univariate lod scores from one representative run for chromosomes 9 and 16.
Table 2.
Summary of chromosomal regions with an average univariate multipoint lod score ≥1.5 in at least one model
| Cytogenetic position |
Trait | Closest marker(s) |
Lod score (screened for basic set)a |
Lod score (screened for environmental set)a |
Lod score (screened for extended set)a |
|---|---|---|---|---|---|
| 1p21.2 | ABDCIR | D1S206 | 1.40 (1.28–1.49)—A,S,R |
1.66 (1.48–1.79)—A,S,R,F,E |
|
| 8p12 | ADIPONEC | D8S505 |
2.74 (2.68–2.90)—A,S,R,C,B |
||
| 8q12.2–q21.3 | ADIPONEC | D8S260–D8S270 | 1.50 (1.45–1.58)—A,S,R,C,B |
||
| 9p22.3–p21.3 | %BF | D9S285 |
2.30 (2.25–2.36)—A,S,R |
2.48 (2.43–2.57)—A,S,R,F,E |
|
| 9p22.3–p21.3 | BMI | D9S285–D9S157 | 1.31 (1.28–1.35)—A,S,R |
1.40 (1.38–1.47)—A,S,R,F,E |
|
| 9p22.3–p21.3 | ABDCIR | D9S157–D9S171 |
2.04 (1.88–2.14)—A,S,R |
2.14 (2.00–1.25)—A,S,R,F,E |
|
| 9q34.11–q34.2 | %BF | D9S290–D9S164 | 1.36 (1.29–1.41)—A,S,R |
1.75 (1.64–1.80)—A,S,R,F,E |
|
| tel-12p13.33 | ABDCIR | D12S352 | 1.40 (1.36–1.41)—A,S,R |
1.64 (1.61–1.65)—A,S,R,F,E |
|
| tel-12p13.33 | BMI | D12S352 | 1.22 (1.18–1.24)—A,S,R,F,E |
||
| 13q31.3 | LEPTIN | D13S265 | 1.67 (1.60–1.74)—A,S,R,F,E,C |
||
| 16q23.1 | LEPTIN | D16S515 |
2.33 (2.22–2.39)—A,S,R,F,E,C |
||
| 16q23.1 | ABDCIR | D16S515–D16S516 | 1.74 (1.46–1.86)—A,S,R |
1.81 (1.55–1.94)—A,S,R,F,E |
|
| 18p11.31 | LEPTIN | D18S452 | 1.25 (1.21–1.31)—A,S,R |
1.73 (1.62–1.87)—A,S,R,F,E,C |
QTLs that obtained suggestive linkage (lod score ≥1.9) are indicated in boldface. Lod scores that did not obtain potential linkage (<1.17) are not shown. Mean and range for 10 independent runs are shown.
Significant covariates (P value < 0.1) included in polygenic model are listed next to lod score.
A, age; B, body mass index; C, cigarette smoker; E, education; F, farm work; R, polity of residence; S, sex.
Figure 1.
Multipoint linkage results from chromosomes 9 (left column) and 16 (right column). The top row shows univariate linkage when adjusted for the basic covariate set. The middle row shows univariate linkage results when the environmental set was screened for. The bottom row shows bivariate multipoint lod scores. Genome-wide significance (lod 3.3) is indicated by the horizontal dashed line, and suggestive linkage is indicated by the horizontal solid line (lod 1.9). The locations of potential candidate genes, JAK2 and AGRP, are indicated with asterisks.
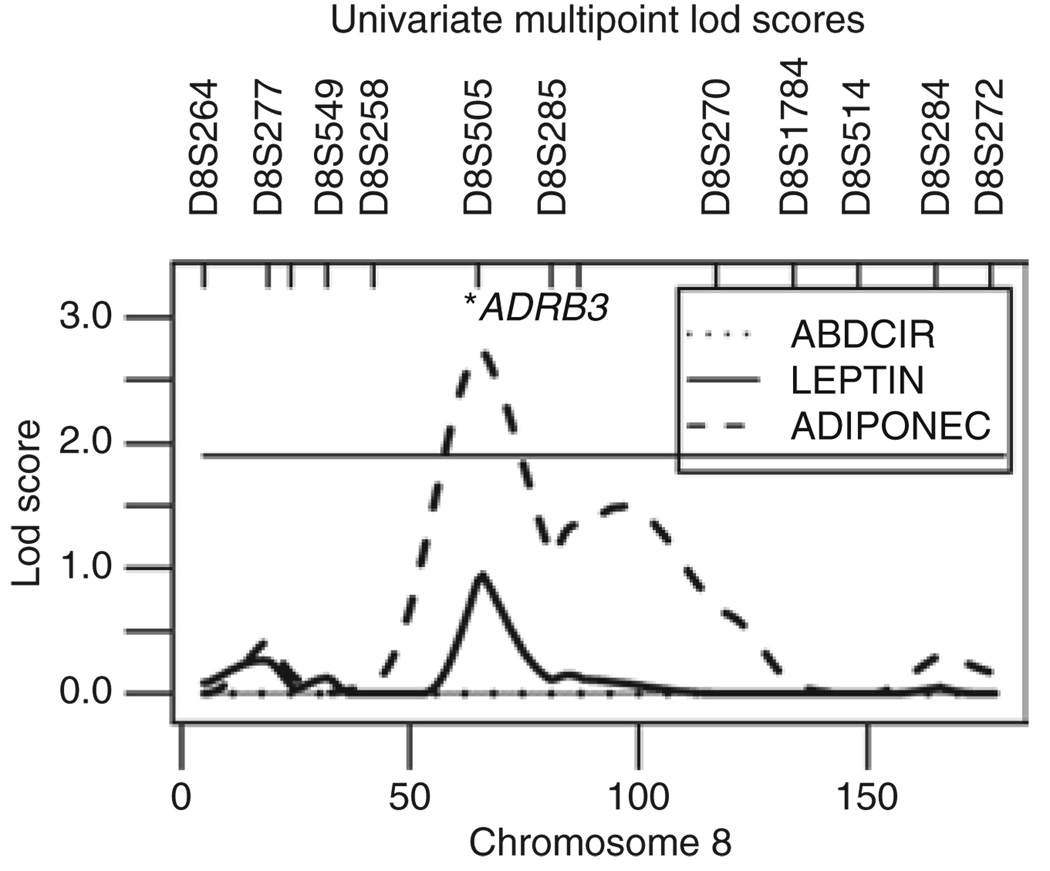
When we used the extended set of covariates, including BMI in addition to all other covariates, we observed linkage for ADIPONEC (lod 2.74) on 8p12 (Figure 2). This QTL was not detected with the other two covariate sets (Table 2).
Figure 2.
Multipoint linkage results from chromosome 8 when the extended covariate set was screened for. Suggestive linkage is indicated by the horizontal line (lod 1.9). The location of the potential candidate gene, ADRB3, is indicated with an asterisk.
Bivariate lod scores (transformed to 1 d.f.) are shown on the bottom row of Figure 1. On 9p we observed maximum bivariate lod scores that reached genome-wide significance (lod ≥3.3) (28) for two trait combinations (Table 3): for ABDCIR–%BF (lod 3.30) and BMI–%BF (lod 3.31). In this region ABDCIR–BMI showed a bivariate lod score of 2.31. On 16q LEPTIN–ABDCIR had a bivariate lod score of 3.17 (Table 3). Co-incident linkage was rejected for all four bivariate analyses (P-value <0.004), but pleiotropy was not (P-value >0.05) (Table 3).
Table 3.
Pleiotropy and co-incident linkage test for loci with bivariate multipoint lod ≥1.9
| Traits | Cytogenetic position |
Closest marker(s) |
Bivariate lod score (1 d.f.) |
Covariatesa | Pleiotropyb | Co-incident linkageb |
|---|---|---|---|---|---|---|
| ABDCIR and %BF | 9p22.3 | D9S285 | 3.30 | A,S,R,F,E | Possible 0.22 | Rejected 9.42 × 10−14 |
| BMI and %BF | 9p22.3–p21.3 | D9S285–D9S171 | 3.31 | A,S,R,F,E | Possible 0.14 | Rejected 0.0038 |
| BMI and ABDCIR | 9p22.2–p21.3 | D9S157–D9S171 | 2.31 | A,S,R,F,E | Possible 0.29 | Rejected 0.0016 |
| LEPTIN and ABDCIR | 16q23.1 | D16S515 | 3.17 | A,S,R,F,E,C | Possible 0.26 | Rejected 1.78 × 10−10 |
Covariates included in the polygenic model.
Interpretation and P value for tests of complete pleiotropy and co-incident linkage.
A, age; C, cigarette smoker; E, education; F, farm work; R, polity of residence; S, sex.
On the X chromosome no lod score >1.09 was detected.
DISCUSSION
In the combined study sample from American Samoa and Samoa we detected three QTLs for adiposity phenotypes in our genome-wide linkage analysis. One region, on 9p, showed significant linkage, and two regions, on 8p and 16q, showed suggestive linkage to adiposity-related phenotypes. Although the genotype information from the two study samples was in part derived using two different instruments, we combined the data on the basis of the minimal differences in allele frequencies. We are confident this strategy is suitable for the study samples, as our previous work demonstrated a common population history and similar marker genotype frequencies (9,29).
The highest univariate linkage peak (lod 2.74) was detected for ADIPONEC, on 8p12, when the trait was screened for the covariate set including BMI (Table 2). As low ADIPONEC characterizes the obese state (30–33), we included BMI as a covariate in an attempt to detect genes influencing ADIPONEC independent of overall adiposity. When BMI was not adjusted for, we detected no lod score of interest in this region. The adrenergic beta-3-receptor (ADRB3) gene located in the 8p12 region (Figure 2) has been suggested as a potential candidate gene for adiposity-related traits (34). Across the centromere, on 8q12.1–q21.3, we detected a second minor peak for ADIPONEC (lod 1.5). In this region we previously detected a QTL (lod 1.87) for ADIPONEC in the adult study sample from Samoa (11). This region was recently reported as being linked to ADIPONEC in Hispanic children (35).
The second-highest univariate linkage peak (lod 2.48) was detected for %BF on 9p22.3–p21.3, using the environmental covariates set (Table 2). In this region ABDCIR had a lod score of 2.14 and BMI had a lod score of 1.40. When bivariate linkage analyses were carried out with combinations of the traits detected in this 9p region, the lod scores increased to reach the level of genome-wide significance for ABDCIR–%BF (lod 3.30) and for BMI–%BF (lod 3.31). The bivariate lod score for ABDCIR–BMI was 2.31. χ2 statistics suggest that pleiotropy rather than co-incident linkage occurs for %BF, ABDCIR, and BMI on 9p22.3–p21.3 (Table 3).
In adults from Samoa, we detected linkage to ABDCIR (lod 2.14) and %BF (lod 1.76) within the 9p region (11). Other studies have reported linkage to adiposity-related traits such as ADIPONEC (36) and low-density lipoprotein particle size (37), but to our knowledge these Samoan studies are the first reports of linkage to %BF, ABDCIR, and BMI on 9p22.3–p21.3. No candidate gene has been reported in this region. However, the Janus kinase 2 (JAK2) gene that is near 9p22 has been suggested as a potential candidate (Figure 1). The JAK2 gene product mediates downstream signaling when leptin binds to LEPR (38).
The third locus with suggestive linkage was 16q23.1. We detected univariate linkage for LEPTIN (lod 2.33) and ABDCIR (lod 1.81) when using the environmental covariate set (Table 2). When no environmental covariates were adjusted for, no lod score of interest (≥1.18) was detected in this region, which suggests that environmental covariates have an effect on the traits. Bivariate linkage analysis of LEPTIN and ABDCIR together increased the lod score to 3.17, and χ2 statistics suggest that pleiotropy rather than co-incident linkage occurs in this adiposity QTL at 16q23.1 (Table 3). In our study of American Samoan adults, suggestive linkage for ABDCIR was reported at 16q23.1 (lod 1.95), and slightly centromeric of this region, in 16q21, suggestive linkages for LEPTIN (lod 2.98) and %BF (lod 2.24) were reported (10). In a Nigerian study sample a QTL for resting energy expenditure has been reported at 16q23.3 (39). A promising candidate gene for these QTLs is the agouti related protein homolog (AGRP) (Figure 1). The agouti-related protein is a neuropeptide implicated in control of feeding and body weight, via leptin signaling. Serum levels of the agouti-related protein are correlated with obesity in men, and polymorphisms in AGRP have been associated with body weight (40).
The regions on 9p and 16q detected in the combined sample were previously detected in one of the polity-specific study samples (see Supplementary Table S3 and Supplementary Figure S3 online), but the region on 8q was not. In addition, we detected potential linkage for ABDCIR and %BF on 1p21 and 9q34. In neither of these regions did we detect suggestive linkage in the separate studies of the Samoan polities, nor have we found other studies in support of these findings. We therefore reason that these regions are of minor importance worldwide for the adiposity-related traits studied, or could be unique to this population. As multipoint linkage analysis may inflate the lod score toward the telomeres, the potential linkage peak found on tel-12p13 requires further investigation. On chromosomes 13q31 and 18p11 we detected potential linkage for LEPTIN. A suggestive QTL for LEPTIN and for ADIPONEC on 13q was previously detected in both the Samoan (11) and American Samoan samples (10), but no previous linkage to LEPTIN was detected on 18p. The highest univariate lod score detected for adiposity-related phenotypes in the three Samoan study samples was a QTL for LEPTIN (lod 3.83) located on 6q23.2 in the American Samoan sample (10). This locus was not detected in the Samoan study sample or in the combined sample. As the two Samoan polities are known to have a common evolutionary history, this suggests that significant environmental effects not adjusted for, independently or interactively with the genetic effect, may influence LEPTIN. Another possible explanation for this lack of overlap between the study samples is that the false negative rate per study may be relatively large, and thus the lack of overlap could be due to a false negative result in one of the two samples. Chromosomal regions with suggestive linkage detected in at least two Samoan samples are shown in Supplementary Table S3 online. A graph of the genome-wide multipoint lod score curves, from a single run, detected in the three study samples from the Samoan islands is presented in Supplementary Figure S3 online.
As an afterthought, we also investigated the effects of allowing for interactions between covariates within the basic and the environmental covariate set on the adiposity traits. These analyses increased heritability estimates and slightly altered the lod scores but had no significant effect on the general conclusions (see Supplementary Methods and Procedures and Tables S4 and S5 online).
In summary, we identified one region on chromosome 9p with genome-wide significant linkage and two regions, on 8p and 16q, with suggestive linkage for adiposity-related phenotypes in an isolated, fairly homogeneous population from the Samoan islands. All three loci have previously been reported as linked to adiposity-related phenotypes in non-Polynesian study samples. The QTLs found among Samoans are not unique and may be important for understanding global diversity in the genetics of adiposity. Within the regions on 8p (ADRB3) and 16q (AGRP), promising candidate genes are located. However, the discrepant results from this study compared with our previous genome-wide investigations of the polity-specific study samples (10,11) support the importance of environmental factors and possible gene–environment interactions for adiposity-related traits. Here we adjusted for polity of residence, education, farm work, and cigarette smoking in a first attempt to adjust for environmental influences. Our results strongly encourage further studies of the Samoan population with extended sets of carefully measured environmental factors, along with more genotypic data made possible by technological advances.
Supplementary Material
ACKNOWLEDGMENTS
Our work is supported by NIH grant R01-DK59642 (STM PI). K.Å. was supported by the Sweden-America Foundation and the Swedish Research Council. We thank the Department of Health, American Samoa Government, and the Ministry of Health, Government of Samoa, the local political officials, and the study participants for their contributions to this research.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- 2.Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999–2002. JAMA. 2004;291:2847–2850. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- 3.Popkin BM, Gordon-Larsen P. The nutrition transition: worldwide obesity dynamics and their determinants. Int J Obes Relat Metab Disord. 2004;28 Suppl 3:S2–S9. doi: 10.1038/sj.ijo.0802804. [DOI] [PubMed] [Google Scholar]
- 4.Keighley ED, McGarvey ST, Turituri P, Viali S. Farming and adiposity in Samoan adults. Am J Hum Biol. 2006;18:112–122. doi: 10.1002/ajhb.20469. [DOI] [PubMed] [Google Scholar]
- 5.Swinburn BA, Ley SJ, Carmichael HE, Plank LD. Body size and composition in Polynesians. Int J Obes Relat Metab Disord. 1999;23:1178–1183. doi: 10.1038/sj.ijo.0801053. [DOI] [PubMed] [Google Scholar]
- 6.Galanis DJ, McGarvey ST, Quested C, Sio B, Afele-Fa’amuli SA. Dietary intake of modernizing Samoans: implications for risk of cardiovascular disease. J Am Diet Assoc. 1999;99:184–190. doi: 10.1016/s0002-8223(99)00044-9. [DOI] [PubMed] [Google Scholar]
- 7.McGarvey ST. Obesity in Samoans and a perspective on its etiology in Polynesians. Am J Clin Nutr. 1991;53:1586S–1594S. doi: 10.1093/ajcn/53.6.1586S. [DOI] [PubMed] [Google Scholar]
- 8.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 9.Tsai HJ, Sun G, Smelser D, et al. Distribution of genome-wide linkage disequilibrium based on microsatellite loci in the Samoan population. Hum Genomics. 2004;1:327–334. doi: 10.1186/1479-7364-1-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai F, Keighley ED, Sun G, et al. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int J Obes (Lond) 2007;31:1832–1842. doi: 10.1038/sj.ijo.0803675. [DOI] [PubMed] [Google Scholar]
- 11.Dai F, Sun G, Aberg K, et al. A whole genome linkage scan identifies multiple chromosomal regions influencing adiposity-related traits among Samoans. Ann Hum Genet. 2008;72:780–792. doi: 10.1111/j.1469-1809.2008.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 15.Washington DC: US Department of Commerce; Census of Population and Housing American Samoa 2000. 2004
- 16.Apia, Samoa: Government Printing House; Census of Population and Housing 2001. 2003
- 17.McGarvey ST. Cardiovascular disease (CVD) risk factors in Samoa and American Samoa, 1990–95. Pac Health Dialog. 2001;8:157–162. [PubMed] [Google Scholar]
- 18.American Samoa Government Statistical Report for Labor and Income, Table 10.13. 2007 < http://www.asdoc.info/>.
- 19.World Bank Country Statistical Information: Samoa. 2005 < http://ddp-ext.worldbank.org/ext/CSIDB/getCountryStatInfo>.
- 20.Presson AP, Sobel E, Lange K, Papp JC. Merging microsatellite data. J Comput Biol. 2006;13:1131–1147. doi: 10.1089/cmb.2006.13.1131. [DOI] [PubMed] [Google Scholar]
- 21.Chang YP, Kim JD, Schwander K, et al. The impact of data quality on the identification of complex disease genes: experience from the Family Blood Pressure Program. Eur J Hum Genet. 2006;14:469–477. doi: 10.1038/sj.ejhg.5201582. [DOI] [PubMed] [Google Scholar]
- 22.Box GEP, Cox DR. An analysis of transformations (with discussion) J Royal Stat Soc B. 1964;26:211–252. [Google Scholar]
- 23.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 25.Lange K, Cantor R, Horvath S, et al. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genetics. 2001;69:A1886. [Google Scholar]
- 26.Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 29.Deka R, Mc Garvey ST, Ferrell RE, et al. Genetic characterization of American and Western Samoans. Hum Biol. 1994;66:805–822. [PubMed] [Google Scholar]
- 30.Yang WS, Lee WJ, Funahashi T, et al. Weight reduction increases plasma levels of an adipose-derived anti-inflammatory protein, adiponectin. J Clin Endocrinol Metab. 2001;86:3815–3819. doi: 10.1210/jcem.86.8.7741. [DOI] [PubMed] [Google Scholar]
- 31.Tagami T, Satoh N, Usui T, Yamada K, Shimatsu A, Kuzuya H. Adiponectin in anorexia nervosa and bulimia nervosa. J Clin Endocrinol Metab. 2004;89:1833–1837. doi: 10.1210/jc.2003-031260. [DOI] [PubMed] [Google Scholar]
- 32.Valle M, Martos R, Gascon F, Canete R, Zafra MA, Morales R. Low-grade systemic inflammation, hypoadiponectinemia and a high concentration of leptin are present in very young obese children, and correlate with metabolic syndrome. Diabetes Metab. 2005;31:55–62. doi: 10.1016/s1262-3636(07)70167-2. [DOI] [PubMed] [Google Scholar]
- 33.Weyer C, Funahashi T, Tanaka S, et al. Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab. 2001;86:1930–1935. doi: 10.1210/jcem.86.5.7463. [DOI] [PubMed] [Google Scholar]
- 34.Hao K, Peng S, Xing H, et al. β3-Adrenergic receptor polymorphism and obesity-related phenotypes in hypertensive patients. Obes Res. 2004;12:125–130. doi: 10.1038/oby.2004.17. [DOI] [PubMed] [Google Scholar]
- 35.Tejero ME, Cai G, Goring HH, et al. Linkage analysis of circulating levels of adiponectin in hispanic children. Int J Obes (Lond) 2007;31:535–542. doi: 10.1038/sj.ijo.0803436. [DOI] [PubMed] [Google Scholar]
- 36.Lindsay RS, Funahashi T, Krakoff J, et al. Genome-wide linkage analysis of serum adiponectin in the Pima Indian population. Diabetes. 2003;52:2419–2425. doi: 10.2337/diabetes.52.9.2419. [DOI] [PubMed] [Google Scholar]
- 37.Badzioch MD, Igo RP, Jr, Gagnon F, et al. Low-density lipoprotein particle size loci in familial combined hyperlipidemia: evidence for multiple loci from a genome scan. Arterioscler Thromb Vasc Biol. 2004;24:1942–1950. doi: 10.1161/01.ATV.0000143499.09575.93. [DOI] [PubMed] [Google Scholar]
- 38.Fruhbeck G. Intracellular signalling pathways activated by leptin. Biochem J. 2006;393:7–20. doi: 10.1042/BJ20051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Luke A, Cooper RS, et al. A genome scan among Nigerians linking resting energy expenditure to chromosome 16. Obes Res. 2004;12:577–581. doi: 10.1038/oby.2004.66. [DOI] [PubMed] [Google Scholar]
- 40.Marks DL, Boucher N, Lanouette CM, et al. Ala67Thr polymorphism in the Agouti-related peptide gene is associated with inherited leanness in humans. Am J Med Genet A. 2004;126:267–271. doi: 10.1002/ajmg.a.20600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.