Abstract
Protein kinases have emerged as one of the most frequently targeted families of proteins in drug discovery. While the development of small-molecule inhibitors that have the potency and selectivity necessary to be effective cancer drugs is still a formidable challenge, there have been several notable successes in this area over the last decade. However, in the course of the clinical use of these inhibitors, it has become apparent that drug resistance is a recurring problem. Because kinase inhibitors act by targeting a specific kinase or set of kinases, there is a strong selective pressure for the development of mutations that hinder drug binding but preserve the catalytic activity of these enzymes. To date, resistance mutations to clinically-approved kinase inhibitors have been identified in a number of kinases. This review will highlight recent work that has been performed to understand how mutations in the kinase catalytic domain confer drug resistance. In addition, recent experimental efforts to predict potential sites of clinical drug resistance will be discussed.
Keywords: DFG motif, Gatekeeper residue, Hydrogen bond, IC50, Imatinib, Kd, P-loop, Protein kinase
Reversible protein phosphorylation cascades represent a central theme in cellular signal transduction. Protein kinases are the single family of enzymes that catalyze the transfer of the γ-phosphate group from adenosine 5’-triphosphate (ATP) to a target protein, and thus are key regulators of these phosphorylation pathways (1). Due to the central role that these enzymes play in cellular behavior, it is not surprising that misregulated protein kinase activity contributes to a number of diseases including cancer, inflammation and diabetes (2). Currently, there are dozens of small-molecule protein kinase inhibitors undergoing clinical evaluation, with eleven approved for clinical use (3, 4).
The catalytic domains of protein kinases are bi-lobal with a smaller N-terminal lobe comprised mainly of β-strands and a larger α-helical C-terminal lobe (Figure 1, top left panel) (5, 6). These lobes are joined by a segment known as the hinge region, which outlines a narrow hydrophobic cleft where ATP binds. The adenine ring of ATP makes key hydrogen-bonding contacts with the amide backbone of the hinge region. The α- and β-phosphate groups of ATP are aligned for catalysis via an interaction with a divalent magnesium ion and a conserved catalytic lysine (7, 8). Protein substrates bind in an extended conformation along a shallow groove on the C-lobe, which allows the residue that will be phosphorylated to accept the γ-phosphate of ATP. Adjacent to the ATP-binding cleft is a 20–30 residue long activation loop that increases the catalytic activity of most kinases when phosphorylated (9). The activation loop contains the highly conserved Asp-Phe-Gly (DFG) motif, the conformation of which is directly coupled to the activation state of the kinase. The aspartate residue in the DFG motif of active kinases faces into the ATP-binding cleft, while the phenylalanine residue is buried in a hydrophobic pocket adjacent to this site (DFG-in). While the active conformation of most kinases are very similar due to the necessity of utilizing the same co-factor, ATP, as a substrate, their inactive conformations are more heterogeneous in nature (9).
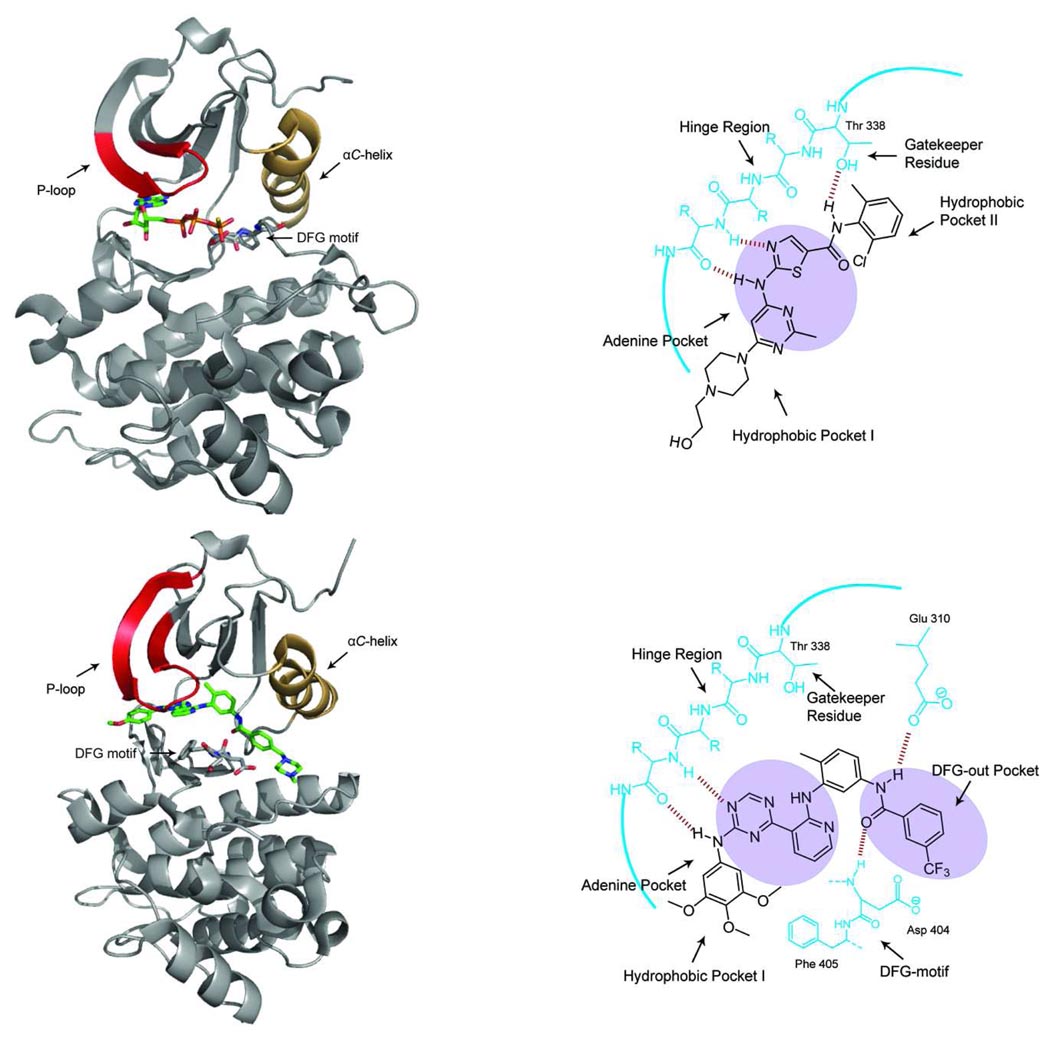
Figure 1.
(top left panel) Crystal structure of the tyrosine kinase SRC (the P-loop is shown in red and the αC-helix is shown in orange) bound to AMP-PNP (PDB ID 3DQW). The active conformation of the kinase places the aspartate residue of the DFG motif facing towards the ATP-binding pocket. (top right panel) A schematic representation of dasatinib, a type I kinase inhibitor, bound to the tyrosine kinase SRC. Type I inhibitors occupy the Adenine Pocket (shaded) in the ATP-binding cleft and make 1–3 hydrogen bonds with the amide backbone of the hinge region. Dasatinib makes an additional hydrogen bond with the side-chain of the threonine gatekeeper residue in SRC. Both hydrophobic pockets (Hydrophobic Pocket 1 and Hydrophobic Pocket 2) that are often exploited by small-molecule kinase inhibitors are labeled. (bottom left panel) Crystal structure of the tyrosine kinase SRC bound to a type II inhibitor (PDB ID 3G6G). SRC is in the DFG-out conformation with the DFG motif flipped relative to the active conformation. This structural rearrangement causes the Phe of the DFG motif to occupy the ATP-binding cleft. (bottom right panel) A schematic representation of a type II inhibitor bound to the tyrosine kinase SRC. This inhibitor forms two hydrogen bonds with the hinge region of the kinase. The characteristic set of hydrogen bonds that type II inhibitors form with the conserved glutamate residue in the αC-helix and the amide backbone of the aspartate of the DFG motif are shown. The DFG-out pocket that is generated by the movement of the phenylalanine residue of the DFG motif is shaded.
All clinically-approved small-molecule inhibitors of protein kinases, except for compounds that target mTOR, and most compounds in late stage clinical trials target some portion of the ATP-binding cleft (3, 10–12). Most of these inhibitors recognize the active conformation of their kinase target(s) and make a characteristic set of interactions with the ATP-binding cleft (type I inhibitors) (13). Type I inhibitors tend to make similar hydrophobic contacts as the adenine ring of ATP and form one to three hydrogen bonds with the backbone amides of the hinge region (Figure 1, top right panel). Affinity and selectivity is often achieved through specific interactions with hydrophobic pockets adjacent to the site of ATP binding (Figure 1, top right panel, Hydrophobic Pockets I and II). In contrast, type II inhibitors recognize a specific inactive conformation of protein kinases (DFG-out) (Figure 1, bottom left and right panels) (13–15). Currently, the number of kinases that are able to adopt the DFG-out conformation is not known, but for kinases that have been structurally characterized in this conformation, the distinctive orientation of the DFG motif is highly conserved. For kinases in the DFG-out conformation, the DFG motif is in a flipped orientation relative to the active form; with the phenylalanine residue rotated almost 180° and the aspartate side chain facing out of the active site. This rearrangement reveals an additional hydrophobic pocket (DFG-out pocket) that is exploited by type II inhibitors (Figure 1, bottom right panel). In addition to hydrophobic contacts with the DFG-out pocket, type II inhibitors usually make a characteristic set of hydrogen bonds with a conserved glutamate in the αC-helix and the backbone amide of the aspartate in the DFG-motif. Like type I inhibitors, type II inhibitors usually form hydrogen-bonding interactions with the amide backbone of the hinge region and hydrophobic contacts with the adenine site.
As kinases have become increasingly more prevalent as drug targets in human disease, significant success has been achieved in targeting kinases involved in cancer. In many cases this clinical success has been shown to exist within a limited timeframe, due to the development of drug resistance. As most kinase inhibitors exert their effects by targeting a specific kinase or set of kinases, there is strong selective pressure for the development of mutations that prevent drug binding. However, there is a limited spectrum of mutations that are available to a kinase for developing resistance due to the necessity of maintaining the catalytic activity of these enzymes. This review will highlight recent work that has been performed to determine the biochemical mechanisms that protein kinases have developed to gain resistance to small-molecule inhibitors. These studies provide information on the inherent structural plasticity of the catalytic domain of protein kinases and give insight into how active site mutations can affect ligand binding. While several routes are available for cells to gain resistance to targeted kinase inhibitors, this review will focus on the role of kinase domain mutations that hinder drug binding but preserve catalytic activity. For a more comprehensive overview of kinase drug resistance, the reader is referred to a recent review by Mansour and co-workers (16).
Resistance to Inhibitors of BCR-ABL
Chronic myelogenous leukemia (CML), which accounts for 15–20% of adult leukemia in Western populations, is a blood and bone marrow disease that is caused by unregulated proliferation of myeloid cells. In a majority of cases, CML coincides with a reciprocal translocation of chromosomes 9 and 22, which is referred to as the Philadelphia chromosome (17). This chromosomal abnormality results in the generation of a fusion gene, named BCR-ABL1, from the joining of the breakpoint cluster region (BCR) gene and the ABL tyrosine kinase gene. The protein product of the BCR-ABL1 gene, BCR-ABL, is a 210 kDa protein that contains the constitutively active tyrosine kinase domain of ABL fused to 902 or 927 amino acids of BCR. A large part of the pathogenesis of BCR-ABL1-positive leukemia is driven by the increased catalytic activity of the tyrosine kinase ABL, which phosphorylates a number of downstream substrates and results in cell transformation and proliferation. The small-molecule kinase inhibitor imatinib (Gleevec) has revolutionized the treatment of CML (Figure 2, top left panel) (18, 19). Imatinib is a 2-phenylaminopyrimidine derivative inhibitor that targets the ATP-binding site of ABL. While imatinib was originally designed to target the active conformation of the ATP-binding pocket of ABL kinase, it was later discovered that this inhibitor targets the DFG-out inactive form (Figure 1, bottom right panel and Figure 2, bottom left panel) (20–22). Despite the challenge in identifying kinase inhibitors with high selectivity, a number of in vitro and proteomic screens have demonstrated that imatinib only has sub-micromolar potency against several other kinases (PDGFR, ABL1, ABL2, c-KIT and DDR) besides BCR-ABL (23–26). This high degree of selectivity for inhibiting the kinase catalytic activity that is responsible for driving the pathogenesis of CML is believed to be at least partially responsible for the clinical success of this drug. More than 80% of patients that undergo imatinib treatment in the early stages of CML (chronic phase) show a complete cytogenetic response (no detectable levels of the Philadelphia chromosome) (27). This response has been found to be robust, with less than 3% of these patients progressing to more advanced stages of CML (accelerated or blast phase) after five years. However, imatinib therapy is not the equivalent of a cure for CML because residual leukemia cells persist in all patients and the recurrence of active leukemia is common amongst patients that cease treatment.
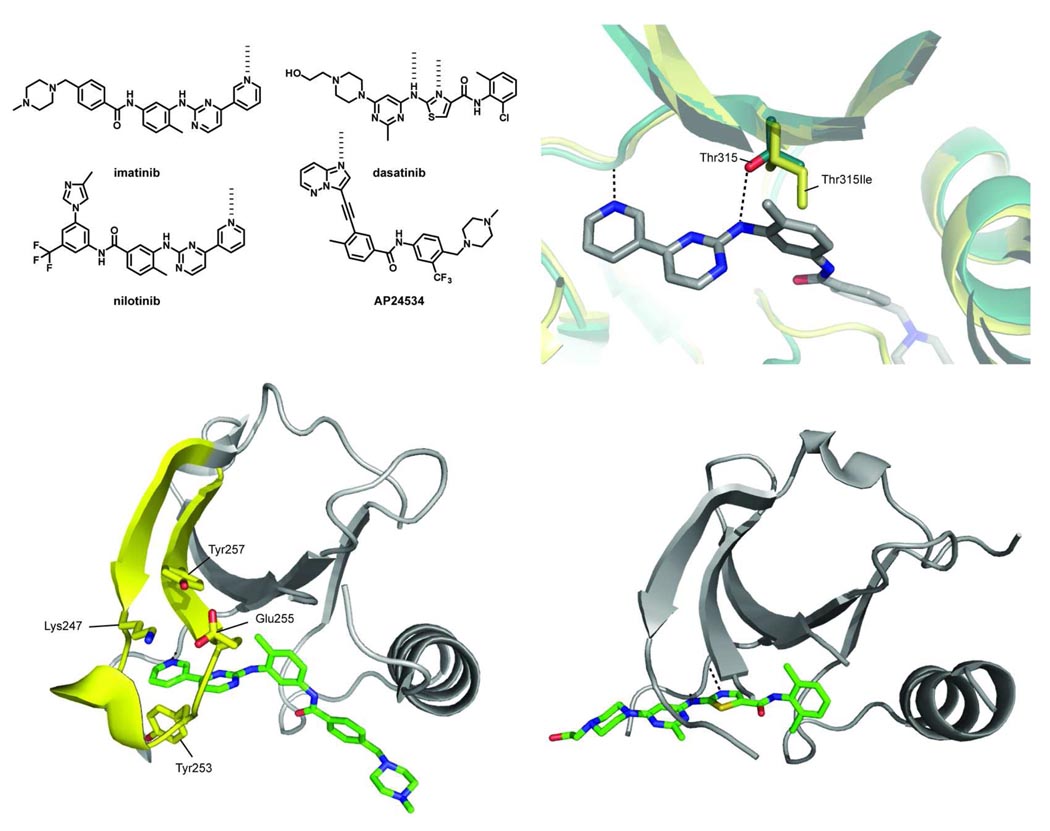
Figure 2.
(top left panel) Chemical structures of selected ABL kinase inhibitors. (bottom left) The crystal structure of imatinib bound to the tyrosine kinase ABL (PDB ID 1OPJ). In ABL, the P-loop (shown in yellow) adopts a unique kinked conformation, effectively shielding one face of the drug from solvent. P-loop residues Tyr253 and Glu255 are two key sites of resistance mutations. Mutation of Tyr253 to a Phe or His perturbs the face-to-edge interaction the side chain makes with the pyrimidine of imatinib and disrupts its hydrogen bond with Asn322 in the C-lobe. Glu255 makes two hydrogen bonds with Lys247 and Tyr257, stabilizing the kinked conformation of the P-loop. Mutating this residue to a Val or Lys disrupts these hydrogen-bonding interactions and most likely destabilizes the kinked P-loop conformation. (top right panel) An overlay of imatinib-bound ABL (green, PDB ID 2HYY) and the ABL Thr315Ile resistance mutant (yellow, PDB ID 2Z60). The key hydrogen-bonding interaction between the inhibitor and the alcohol side-chain of the gatekeeper threonine side chain is shown (dashed black line). Upon mutation to an isoleucine, this hydrogen bond is lost and the bulkier side chain creates a steric clash with the drug. (bottom right) A close-up view of dasatinib bound to the ATP-binding cleft of SRC (PDB ID 3G5D). The P-loop of SRC is disordered in this structure.
Despite the effectiveness of imatinib as a targeted therapeutic for the treatment of CML, the emergence of clinical resistance is an ongoing challenge. While relapse is infrequent for patients undergoing imatinib therapy during chronic phase, it is very common for those that are diagnosed and treated during advanced stages of the disease (28). Currently, it is estimated that about thirty percent of patients undergoing imatinib therapy will switch to an alternative treatment within five years due to side effects and the development of drug resistance (27, 29–31). For patients undergoing treatment with imatinib, relapse occurs through re-activation of the BCR-ABL pathway in the presence of the drug. The most frequent route (60–90% of all cases) for the development of resistance to imatinib is through mutations in the kinase domain of ABL (31–36). To date, over 50 different point mutations in the ABL kinase domain have been detected in imatinib-resistant CML patients. Despite the large number of mutations that have been identified, imatinib resistance frequently occurs through several common mechanisms.
While resistance mutations have been identified throughout the catalytic and regulatory domains of ABL, a large percentage localize to a region called the phosphate-binding loop (P-loop) or glycine-rich loop. The P-loop is a flexible, glycine-rich loop that makes contact with theα- and β-phosphates of ATP (Figure 1, top left panel) (37–41). X-ray crystal structures of the imatinib-ABL complex have demonstrated that the P-loop adopts a unique kinked conformation, which shields the pyridine and pyrimidine rings of the drug from solvent (20, 21, 42, 43) (Figure 2, bottom left panel). The ordered nature of the P-loop when ABL is bound to imatinib has been confirmed in solution by NMR spectroscopy (22). The two most commonly observed sites of mutation in the P-loop are Tyr253 and Glu255, which account for over 30% of all clinically-observed imatinib-resistance mutations (44). Commonly, Tyr253 is mutated to a His or Phe residue and Glu255 to a Lys or Val. In vitro activity assays with purified ABL kinase have demonstrated that the Tyr253His and Tyr253Phe mutations result in a >18- and 15-fold loss in drug sensitivity, respectively (45). Analysis of the imatinib-ABL complex has shown that there are likely two reasons that these mutations result in the observed loss in potency of imatinib. First, conversion of Tyr253 to a phenylalanine or histidine residue most likely leads to a less favorable face-to-edge aromatic interaction between this side-chain and the pyrimidine ring of the drug. In addition, these mutations remove the ability of this side-chain to hydrogen bond with Asn322 in the C-lobe which most likely results in disruption of the distorted conformation of the P-loop. Glu255 mutations result in a similar loss in potency, with the Glu255Val and Glu255Lys mutants of ABL showing 13-and >18-fold less sensitivity to imatinib, respectively (45). Unlike Tyr253, the side-chain of Glu255 does not make direct contact with the drug. Rather, the carboxylate from this residue forms a hydrogen-bonding network with Lys247 and Tyr257 that stabilizes the anti-parallel β-strand of the P-loop. Mutating Glu to a Lys or Val residue disrupts these interactions and most likely destabilizes the conformation of the P-loop. It has been hypothesized that mutations in the P-loop contribute to imatinib resistance by destabilizing the inactive DFG-out conformation of ABL. While this may be true in a cellular context, several recent studies show that this is unlikely for BCR-ABL in the absence of other interacting proteins. First, although there is conflicting data on the relative catalytic activities of P-loop mutants versus wild-type BCR-ABL, the kinetic constants (kcat and Km) for purified kinase constructs in activity assays are very similar. In addition, a series of inhibitors that bind the DFG-out conformation of ABL without interacting with the P-loop are minimally affected by mutations in Tyr253 and Glu255 (46). Furthermore, a recent study using hydrogen/deuterium exchange mass spectrometry (HX MS) shows that there are no detectable differences in the solution conformational dynamics of wild-type, Tyr253His and Glu255Val ABL (47).
Another common mutation that accounts for about 15% of all cases of imatinib-resistant CML is the Thr315Ile gatekeeper mutant (48). The gatekeeper residue controls access to a hydrophobic pocket that is adjacent to the adenine site, which is exploited by a number of kinase inhibitors. This residue is often a direct determinant of inhibitor selectivity and has been exploited for the generation of mutant kinases that are uniquely sensitive to a series of modified kinase inhibitors (49–52). In addition to BCR-ABL, mutations at the gatekeeper position of the tyrosine kinases c-KIT, PDGFRA and EGFR have been linked to the development of drug resistance (53, 54). X-ray structural analysis of the ABL-imatinib complex shows that the m-diaminophenyl group of imatinib sits in close proximity to the side-chain of Thr315. In addition, the nitrogen linking the pyrimidine ring and the m-diaminophenyl ring forms a critical hydrogen bond with the secondary alcohol of this residue. Conversion of the threonine residue to a bulkier isoleucine creates a steric clash with the drug and does not allow a hydrogen bond to be formed, which results in imatinib demonstrating a dramatic loss in affinity (>500-fold) for this mutant (Figure 2, top right panel). Several studies suggest that the Thr315Ile mutation also affects the conformational dynamics of the ABL kinase domain. For example, this mutant has been demonstrated to have higher basal catalytic activity and increased enzymatic activation in cells (55). Furthermore, HX MS analysis of the Thr315Ile ABL mutant shows that two regions of the kinase (a region that is in close proximity to imatinib binding and the more distant RT-loop of the regulatory SH3 domain) have increased conformational dynamics compared to the wild type enzyme (47). Thus, the highly resistant nature of the Thr315Ile mutant may be due to a combination of direct disruption of active site-drug interactions and subtle changes in the conformational dynamics of the catalytic domain.
The drugs dasatinib and nilotinib have been approved as second generation therapies for the treatment of imatinib-resistant CML (Figure 2, top left panel) (56–60). Both drugs are considerably more potent inhibitors of the catalytic activity of wild-type ABL than imatinib. Structural analyses of the nilotinib-ABL complex by X-ray crystallography and NMR spectroscopy have demonstrated that this drug binds to the DFG-out conformation of the catalytic domain in an analogous manner to imatinib (22, 56). The increased potency of nilotinib is due to a more optimal interaction between the 3,5-imidazole/trifluoromethyl substituent of this compound and the DFG-out pocket of ABL. The fact that nilotinib exploits many of the same contacts as imatinib is reflected in its similar kinase selectivity profile. Furthermore, while nilotinib is effective at inhibiting the Tyr253 and Glu255 P-loop mutants of ABL, these mutations cause this drug to have a similar fold loss in overall potency as imatinib (45, 56). In contrast to nilotinib, dasatinib was developed as a dual SRC and ABL inhibitor that targets the active conformation of the ATP-binding site. While it has been speculated that dasatinib should be capable of binding both the active and inactive conformations of the ATP-binding sites of these kinases, a recent NMR study of its interaction with ABL has demonstrated that this kinase is exclusively in the active form when bound to the drug (22). As many of the contacts that dasatinib makes with the active forms of SRC and ABL are conserved in a number of tyrosine kinases, this drug potently inhibits a number of members from this sub-family. Because dasatinib does not rely on interactions with the P-loop of ABL, this compound inhibits the Tyr253 and Glu255 mutants with a similar potency as the wild type enzyme (Figure 2, bottom right panel) (45).
Currently, there are no clinically-approved inhibitors that effectively target the Thr315Ile gatekeeper mutant of BCR-ABL. Nilotinib’s increased interaction with the DFG-out pocket is not able to overcome the energetic penalty of the steric clash from the isoleucine side-chain and loss of hydrogen-bonding interaction. Despite dasatinib targeting the active form of ABL, this drug occupies the hydrophobic pocket adjacent to the gatekeeper residue (Figure 1: bottom right panel). Conversion of the gatekeeper position to a bulkier residue obstructs access to this pocket and results in dasatinib being >500-fold less potent against this mutant. Several ATP-competitive type I inhibitors of ABL Thr315Ile have been described (61). VX-680 and PHA-739358 were originally developed as type I Aurora kinase inhibitors but were later found to potently block the catalytic activity of Thr315Ile BCR-ABL (62, 63). SGX393 is a highly selective type I inhibitor of BCR-ABL that is effective against the gatekeeper mutant (64). However, P-loop mutants of BCR-ABL show resistance to this compound. In addition to these type I inhibitors, several potent type II inhibitors of ABL Thr315Ile have been developed (46, 55). The most extensively characterized of these inhibitors is AP24534, which is a sub-nanomolar inhibitor of BCR-ABL (Figure 2, top left panel) (65). AP24534 contains an imidazo[1,2b]pyridazine core that is linked to a 3-trifluormethylphenyl group with an alkyne linker. The alkyne linker of this inhibitor provides a bridge between the imidazo[1,2b]pyridazine core, which makes a hydrogen bond with the hinge region, and the 3-trifluoromethylphenyl group, which makes extensive contacts with the DFG-out pocket, without clashing with the side-chain of the isolecuine gatekeeper residue. This lack of a steric clash is demonstrated by the only 6-fold loss in potency of AP24534 against the Thr315Ile mutant compared to wild-type BCR-ABL in an in vitro activity assay. Furthermore, AP24534 is a potent inhibitor of previously described P-loop mutants and no additional BCR-ABL variants that confer resistance to this compound were identified in an accelerated mutagenesis assay (66). Selectivity profiling of AP24534 with activity assays demonstrated that this compound potently inhibits a number of kinases despite targeting the DFG-out conformation of ABL. However, this decreased selectivity does not appear to be detrimental in a cellular context because this compound maintains a >1000-fold selectivity for Ph-positive cells in proliferation assays. It is interesting to note that all of the type II inhibitors that have been found to effectively target ABL Thr315Ile, to date, are less selective than imatinib or nilotinib. The success of dasatinib as a second generation therapy for the treatment of imatinib-resistant CML shows that a compound with a limited selectivity profile can still serve as an effective drug.
Resistance to Inhibitors of EGFR
The epidermal growth factor receptor (EGFR) is a cell-surface receptor tyrosine kinase (RTK) in the larger ErbB family of receptors (67). Upon binding of the epidermal growth factor, EGFR transitions from an inactive monomeric form to an active homo- or heterodimer to initiate intracellular signaling that results in cell growth, migration, differentiation and death. Mutations that occur in the EGFR kinase domain that cause the kinase to be over-expressed or hyperactive have been implicated in the development of cancer, particularly non-small cell lung carcinomas (NSCLC) (68, 69). To this end, a number of reversible ATP-competitive small-molecule kinase inhibitors have been developed to target EGFR. These inhibitors include the clinically-approved 4-anilinoquinazolines gefitinib (Iressa) (70), erlotinib (Tarceva) (71) and lapatinib (Tykerb) (72) and the clinical candidate AEE788 (73) (Figure 3, top left panel). In addition to inhibitors that interact with the ATP-binding site of EGFR in a reversible manner, several analogs that covalently modify the active site have been developed (74). An example of an inhibitor of this class is the 4-anilino-3-quinolinecarbonitrile inhibitor neratinib, which covalently modifies Cys797 in the ATP-binding site of EGFR (75).
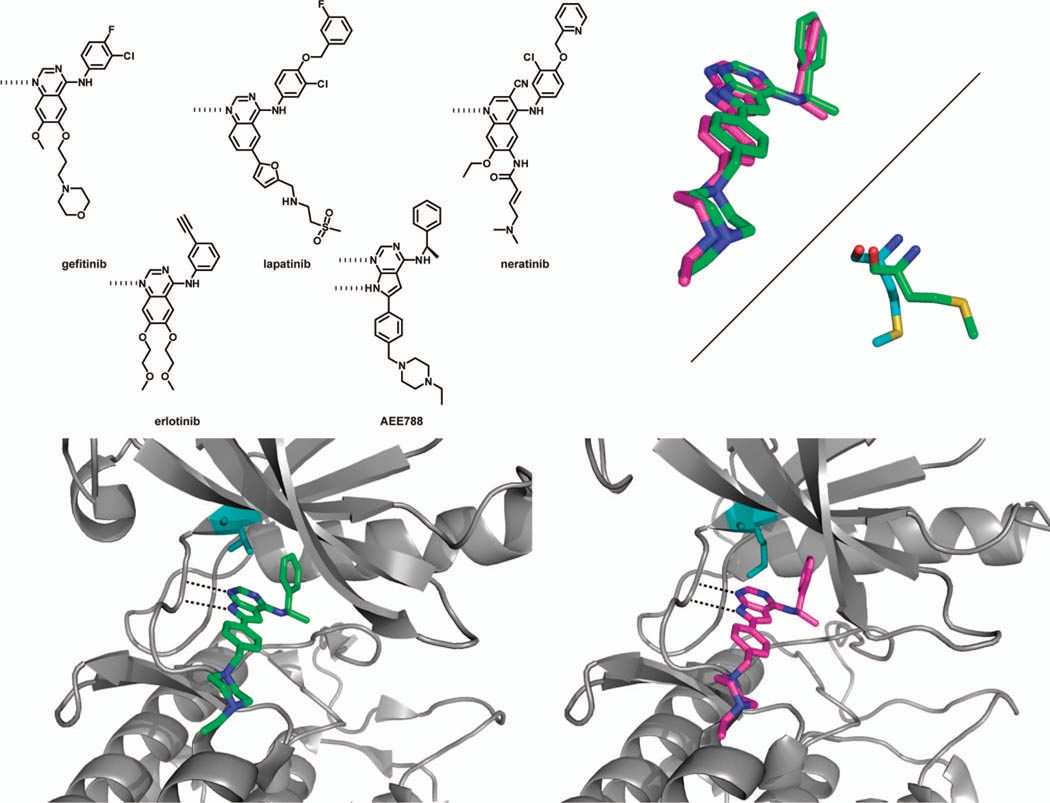
Figure 3.
(top left panel) Chemical structures of selected EGFR kinase inhibitors. (bottom left panel) Crystal structure of wild-type EGFR kinase bound to AEE788 (PDB ID 2J6M). The Thr gatekeeper residue is shown in teal. (bottom right panel) The Thr790Met EGFR mutant in complex with AEE788 (PDB ID 2JIU). The larger methionine gatekeeper residue shown in teal does not block access to the hydrophobic pocket occupied by the phenethylamine moiety of the inhibitor. (top right panel) (top) An overlay of the binding modes of AEE788 in wild-type EGFR (green) and the Thr790Met mutant (pink). In the gatekeeper mutant, the inhibitor maintains its hydrogen bonds with the hinge region, while the phenethylamine substituent rotates slightly to occupy Hydrophobic Pocket II. (bottom) Overlay of the methionine gatekeeper residue from apo-Thr790Met EGFR (green, PDB ID 2JIT) and Thr790Met bound to AEE788 (blue). In the inhibitor-bound conformation, this residue adopts a slightly different orientation to accommodate the phenethylamine moiety of the inhibitor.
Gefitinib, erlotinib and lapatinib are structurally related quinazoline-based compounds that display different anilines from the 4-position. These inhibitors interact with the ATP-binding pocket of EGFR in a similar manner, with the quinazoline core positioning itself along the hinge region. This orientation allows the nitrogen from the quinazoline core to form a hydrogen bond with the hinge region and the substituents at the 6- and 7-position to extend into the solvent. The small threonine gatekeeper residue of EGFR (Thr790) allows the aniline at the 4-position to form extensive interactions with the hydrophobic pocket adjacent to the adenine site, which contributes to the high selectivity exhibited by these compounds. Lapatinib, which contains a more extended 4-anilino substituent than erlotinib and gefitinib, binds to a unique inactive conformation of EGFR (76). Kinome-wide selectivity screens have demonstrated that these inhibitors are highly selective for EGFR and its ErbB family members, with lapatinib showing the highest selectivity (24, 26). The reversible inhibitor AEE788 binds to EGFR kinase in the active conformation, with the αC-helix pointing in towards the ATP binding pocket. Much like the quinazoline inhibitors, the pyrrolopyrimidine core of AEE788 makes hydrogen-bonding interactions with the backbone amides of the kinase hinge region. Mimicking the aniline groups of gefitinib and erlotinib, the phenethylamine substituent extends into a hydrophobic pocket guarded by the gatekeeper residue, while the ethylpiperazine moiety is directed out of the ATP-binding pocket, towards the solvent. AEE788 has been shown to be an extremely potent inhibitor of ErbB family kinases and VEGFR, with low nanomolar potency against wild-type EGFR kinase.
Several oncogenic mutations in EGFR have been identified that give rise to NSCLCs. These include exon 19 deletions and a point mutation in exon 21 that mutates Leu858 in the activation loop to an Arg, the latter accounting for approximately 40% of all mutations (77, 78). A third point mutation that occurs less frequently is the conversion of Gly719 in the P-loop to a Ser. Both Leu858Arg and Gly719Ser are gain-of-function mutations, and the success of gefitinib and erlotinib partially arises from their increased potency against these mutant kinases over the wild-type enzyme (almost 10- to 100-fold more potent in cells expressing EFGR mutants over those expressing wild-type kinase) (79, 80).
Several studies have been conducted to characterize the structure and activity of the Leu858Arg and Gly791Ser mutants of EGFR (78). Crystal structures of the Leu858Arg and Gly791Ser mutants bound to the non-hydrolyzable ATP analog AMP-PNP show that these kinases exist in an active conformation, similar to that of the wild-type kinase. To understand the mechanism of activation of the Leu858Arg mutant, crystal structures of wild-type EGFR bound to lapatinib were studied. Lapatinib binds to an inactive form of the kinase domain, with the activation loop segment forming a helical turn that displaces the αC-helix from the regulatory site. Leu858 is one of several hydrophobic residues on the activation loop that helps to stabilize this inactive conformation. Upon substitution of leucine to arginine, the charged residue is no longer favorably accommodated in the hydrophobic pocket, effectively destabilizing the inactive form of the kinase. Similar reasoning is applied to the Gly179Ser mutant; the serine residue destabilizes the inactive conformation of the P-loop. These structural changes results in the Leu858Arg and Gly791Ser mutants of EGFR having a ~50- and ~10-fold increase in activity over wild-type in the presence of excess ATP and peptide substrate, respectively. Further kinetic analysis demonstrated that these mutations result in a 10-to-20-fold increase in the kcat for ATP. However, this is compensated by a 5-to-10-fold higher Km for ATP. Because cellular concentrations of ATP (1–5 mM) are much higher than EGFR’s Km for ATP, the increase in kcat is the most relevant parameter in a cellular context.
Although patients with NSCLC that bear the Leu858Arg mutation respond well to gefitinib and erlotinib treatment, relapse due to drug resistance is common. Molecular analysis of tumor material obtained from patients with acquired resistance to gefitinib/erlotinib treatment has found that a single amino-acid substitution in the catalytic domain of EGFR coincides with a majority of cases of drug resistance; conversion of the Thr790 gatekeeper residue to methionine (68). Significantly, the Thr790Met mutant occurs in the context of the Leu858Arg sensitizing mutation. Therefore, it appears that the gatekeeper mutation eliminates the drug sensitivity that Leu858Arg confers. This resistance mutation has been identified in almost 50% of cases of acquired resistance, making it a significant target of research towards more effective therapies (81). In a more recent study involving tumor cells obtained from both treatment-naïve and treatment-experienced patients, low levels of the Thr790Met mutation were observed in 40% of the treatment-naïve patients (these patients remained susceptible to gefitinib and erlotinib therapy) (82). Although the resistance allele was detected in only a small number of cells, it remains possible that tyrosine kinase inhibitor therapy might select for those tumor cells harboring the pre-existing Thr790Met mutation.
It was originally believed that transformation of the threonine at the gatekeeper position to a bulkier methionine caused resistance to erlotinib and gefitinib through steric interference; analogous to how the ABL Thr315Ile mutation confers resistance to imatinib (83). However, this steric argument for EGFR resistance became tenuous upon the discovery that irreversible EGFR inhibitors can overcome the resistance caused by this mutation in cellular assays (68, 84). In order to further investigate this seemingly unique mechanism of resistance, Yun and co-workers employed a direct binding assay to determine the affinities of gefitinib and AEE788 for wild-type, Leu858Arg, Thr790Met and Leu858Arg/Thr790Met EGFR kinase (Table 1) (85). As expected, gefitinib has a low nanomolar affinity for the Leu858Arg mutant (Kd = 2.4 nM), which is a 15-fold increase in potency over the wild-type enzyme (Kd = 35 nM). The Thr790Met gatekeeper single mutant of EGFR is also quite sensitive to gefitinib, with a Kd = 4.6 nM. Surprisingly, the Thr790Met/Leu858Arg double mutant was found to have only a moderately lower binding affinity for gefitinib (Kd = 11 nM), which is only a 4-fold difference compared to the Leu858Arg single mutant. Clearly, conversion of the threonine gatekeeper residue to a methionine does not create a large steric clash that prevents inhibitor binding. Furthermore, the modest difference in binding affinity to the double mutant cannot fully explain the drug resistance that is observed in cellular assays and clinically.
Table 1.
Thermodynamic and kinetic parameters for wild-type EGFR and mutants.
In order to further study how EGFR can become resistant to small-molecule inhibition, crystal structures of the Thr790Met mutant, in the apo-form and bound to the inhibitors AEE788 (Figure 3, bottom panels) and neratinib, were obtained. As described earlier, AEE788 has similar binding interactions with the pocket adjacent to the gatekeeper residue as gefitinib. Like gefitinib, the binding affinity of AEE788 for Thr790Met (Kd = 28 nM) and Thr790Met/Leu858Arg (Kd = 19 nM) is very similar to wild-type EGFR (Kd = 5.3 nM). Consistent with conversion of the Thr gatekeeper to Met having only a minimal effect on binding affinity, the superimposed crystal structures of AEE788 bound to wild-type and Thr790Met EGFR show that there is little difference in the binding mode of the inhibitor (Figure 3, top right panel). The pyrrolopyrimidine scaffold of AEE788 is in an identical orientation when bound to wild-type and Thr790Met EGFR. Furthermore, there is no apparent steric clash between the bulkier methionine residue and phenethylamine substituent as it enters the hydrophobic pocket adjacent to the adenine site; the gatekeeper residue adopts a slightly different orientation that allows the phenethylamine access to the pocket (Figure 3, top right panel). Presumably, the gatekeeper residue of Thr790Met EGFR undergoes a similar conformational change when bound to gefitinib or erlotinib.
To gain a better understanding of how the Thr790Met mutation leads to drug resistance, kinetic characterization of wild-type, Leu858Arg, Thr790Met and Leu858Arg/Thr790Met EGFR was performed (Table 1) (85). Interestingly, the Leu858Arg mutant has a 30-fold higher Km for ATP than wild-type EGFR (148 µM versus 5.2 µM). However, the Thr790Met gatekeeper mutation restores the Km of Leu858Arg to 8.4 µM. Thus, it is the lower Km for ATP that causes the drug resistance conferred by the double mutant of EGFR. Notably, the gatekeeper mutation alone does not alter the Km of the kinase for ATP; the structural bases for how these mutations affect EGFR’s Km for ATP are not understood. Thus, the Leu858Arg mutation contributes to EGFR’s sensitivity to erlotinib, gefitinib and AEE788 by altering its Km for ATP, which allows these inhibitors to effectively outcompete the high intra-cellular concentrations of ATP (1–5 mM). Conversion of the gatekeeper residue of the Leu858Arg mutant from a threonine to a methionine restores this enzyme’s low micromolar Km for ATP and reduces the effectiveness of these inhibitors in cells. The Thr790Met gatekeeper represents a generic resistance mutation that will affect any ATP-competitive inhibitor, independent of which interactions they make with the ATP-binding cleft.
Pre-clinical cellular studies have shown that irreversible inhibitors such as neratinib (Figure 3, top left panel) and EKB-569 are able to effectively inhibit the Thr790Met mutant of EGFR kinase. These inhibitors are able to achieve greater ATP-binding site occupancy in this kinase by forming a covalent bond with an active site cysteine. For example, neratinib proved to be considerably more effective than gefitinib in suppressing EGFR auto-phosphorylation and phosphorylation of downstream effectors AKT and MAPK in a NCI-H1975 bronchoalveolar cancer cell line harboring the Leu858Arg/Thr790Met mutant (84). However, in clinical settings involving patients with the Thr790Met resistance mutation, irreversible inhibitors have demonstrated only limited success and dose-limiting toxicity has been observed (86, 87). A series of irreversible inhibitors that were specifically developed to target the Thr790Met mutant of EGFR were recently reported (88). These inhibitors, which are based on an anilinopyrimidine-based core rather than a 4-anilinoquinazoline scaffold, are significantly more potent against gefitinib-resistant cell lines than previously described irreversible inhibitors. Furthermore, these covalent inhibitors are selective for the Thr790Met EGFR mutant over the wild-type kinase. A crystal structure of an analog from this series bound to the Thr790Met EGFR kinase catalytic domain provides an explanation for the increased potency of the anilinopyrimidine-based inhibitors against the gatekeeper mutant. While gefitinib and other 4-anilinoquinazoline-based inhibitors are able to avoid a steric clash with the methionine gatekeeper residue, a substituent from the anilinopyrimidine-based core forms a favorable interaction with this residue. This interaction most likely contributes to the increased potencies observed for these inhibitors and helps explain their selectivity for the gatekeeper mutant over wild-type EGFR. Importantly, the most selective compound in this series was found to cause significant tumor regression in Thr790Met-containing murine models with a minimal amount of observed toxicity. While extensive testing is needed to determine if any of these inhibitors will be of clinical utility, the development of mutant-selective kinase inhibitors appears to be a promising strategy for overcoming clinical drug-resistance.
Identification of Potential Sites of Drug-Resistance Mutations: Aurora Kinases, MEK1 and the PI3Ks
The Aurora kinases are a family of serine/threonine kinases that are key regulators of eukaryotic cell mitosis. There are three Aurora kinases in humans that have been characterized to date: Aurora A, Aurora B and Aurora C. Aurora A is localized to centrosomes and spindle poles during various phases of mitosis and is closely associated with centrosome maturation (89). Aurora B localizes to microtubules and is responsible for histone H3 phosphorylation as well as spindle assembly checkpoint (SAC) and cytokinesis (90). Aurora C is thought to be a chromosome passenger, but little more is known about this third class of mitotic serine/threonine kinases (91). Overexpression of these enzymes is apparent in several human cancers, thus, these kinases have become popular targets for anti-cancer therapies (92, 93).
A number of ATP-competitive inhibitors of the Aurora kinases have been discovered that block such cellular actions as chromosome alignment, SAC and cell division. Some of these inhibitors include the small molecules ZM447439, VX-680 and Hesperadin (Figure 4, top left panel). ZM447439 is a quinazoline-based inhibitor, which is 20-fold more potent against Aurora B than Aurora A (94). Mammalian cells that are treated with ZM447439 enter mitosis but have a perturbed spindle assembly and chromosome alignment, inhibiting cytokinesis (92, 95). VX-680, a pyrimidinyl-based compound is a potent inhibitor of both Aurora A and B in cells. VX-680 is highly effective in blocking cell cycle progression and inducing apoptosis in a variety of developing tumors (62). In addition, VX-680 has been shown to have anti-tumor activity in rodent xenograft models. Hesperadin acts much like ZM447439, inhibiting chromosome alignment and segregation in the cell (96).
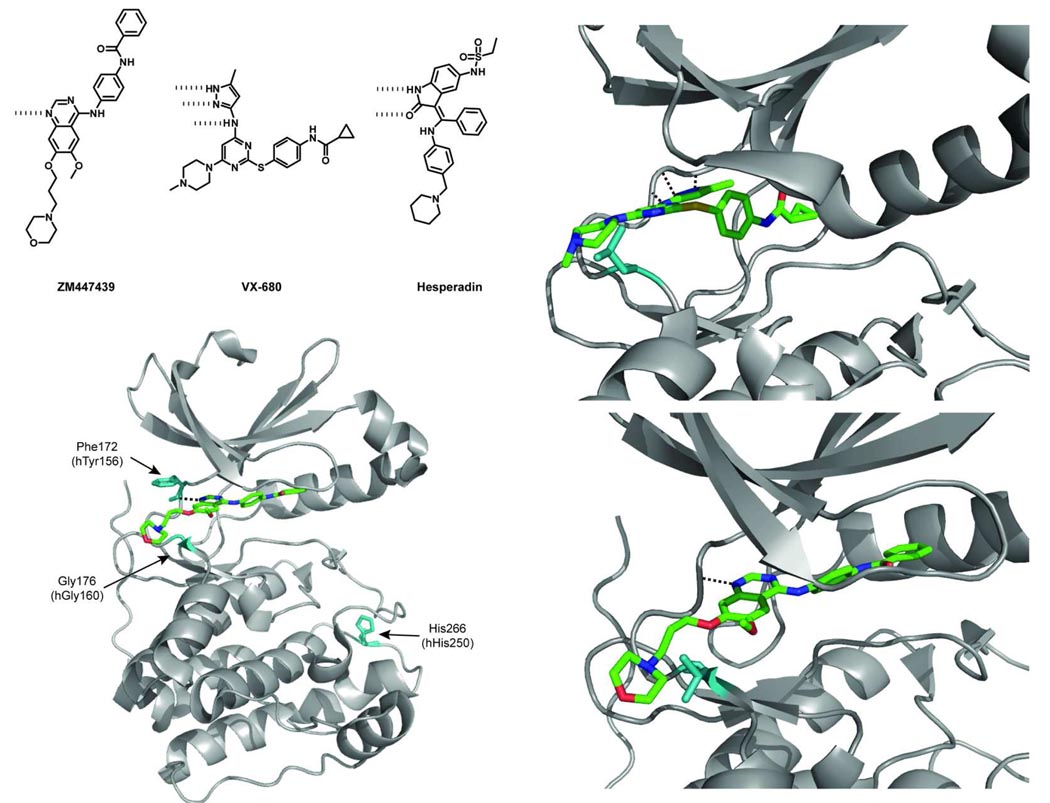
Figure 4.
(top left panel) Chemical structures of selected Aurora kinase inhibitors. (bottom left) A crystal structure of Xenopus laevis (Xl) Aurora B in complex with ZM447439 (PDB ID 2VRX). Sites of resistance mutations are shown in teal and labeled (the numbering on top is for Aurora B from Xenopus, while the lower numbering is for human Aurora B) (bottom right) A representation of the Gly176Val mutation in Xenopus laevis (Xl) Aurora B showing a potential steric clash between the bulkier valine residue and the morpholino moiety of ZM447439. (top right) A representation of the Gly216Leu mutation in Aurora A showing a potential steric clash between the leucine residue and the methylpiperazine of VX-680 (PDB ID 3E5A).
While no Aurora kinase inhibitors have yet been approved for clinical use, the lessons learned from the emergence of drug resistance to BCR-ABL and EGFR inhibitors stress the importance of anticipating which specific mutations, and their consequent effects, may arise. To this end, Girdler and co-workers developed a novel genetic screen to identify cell lines that are resistant to the Aurora kinase inhibitor ZM447439 (97). A key component of this screen is the use of HCT-116 cells, which are a hypermutagenic cell line due to a defect in DNA mismatch repair. In addition, these cells express low levels of drug transporters, which reduces the likelihood of resistance occurring through this mechanism. HCT-116 cells were treated with a 1 µM cytotoxic concentration of ZM447439 over a three week span. Seven cell lines were generated from those cells that maintained strong colony growth in the presence of ZM447439, with several of these cell lines maintaining high cell numbers in the presence of increasing concentrations of the drug. In comparison to parental control cells, two of the resistant cell lines maintained both cell division and histone H3 phosphorylation, indicating that Aurora B kinase was indeed active, and a mutation in this kinase might be the source of drug resistance.
Sequencing of Aurora cDNAs from the seven drug-resistant clones showed that all cell lines contained Aurora B genes with point mutations, giving rise to a total of five different amino-acid substitutions in the catalytic domain (Tyr156His, Gly160Glu, Gly160Val, His250Tyr and Leu308Pro). Three of the seven cell lines contained two different Aurora B single mutants; His250Tyr with Gly160Val (2x) and His250Tyr with Gly160Glu (1x). All of the Aurora mutants, with the exception of Leu308Pro, were ectopically expressed as Myc-tagged fusions in DLD-1 cells and shown to localize correctly and maintain normal kinase function. In the presence of ZM447439, phosphorylation of the Aurora B substrate histone H3(Ser10) was rescued in cells expressing drug-resistant mutants of this kinase. Expression of similar levels of wild-type Aurora B did not show a comparable effect. The most drug-resistant mutant proved to be the Gly160Val of Aurora B (>75% of the cells positive for phospho-histone H3 with 4 µM ZM447439), followed by Tyr156His and His250Tyr (80% and 45% of the cells positive with 2 µM ZM447439, respectively). In vitro activity assays using histone H3 as a substrate showed that the Tyr156His mutant is 10-fold less sensitive to the drug than wild-type kinase, while the Gly160Val and Gly160Glu mutants are completely resistant to 500 µM ZM447439. Most strikingly, these mutations were found to confer resistance to other known Aurora kinase inhibitors of unrelated structure to ZM447439. In the same in vitro activity assay, VX-680 was >20-fold less potent against the Tyr156His and Gly160Glu mutants of Aurora B than the wild-type kinase. Resistance mutations also diminished the ability of Hesperadin to block the catalytic activity of Aurora B. Consistent with the types of drug-resistance mutations that have been identified in BCR-ABL and EGFR, the Tyr156His, Gly160Val, Gly160Glu and His250Tyr mutants of Aurora B do not have compromised catalytic activity. In fact, an in vitro assay in the presence of 200 µM ATP demonstrated that these mutants of Aurora B have higher catalytic activities than the wild-type enzyme. Further analysis of the kinetic parameters of these Aurora B mutants was not performed.
Structural studies were performed to characterize the specific mechanism of resistance. A crystal structure of the Xenopus laevis (Xl) Aurora B:INCENP complex bound to ZM447439 shows that the inhibitor sits in the ATP-binding pocket with the quinazoline core lying against the hinge region of the kinase, the benzamide directed towards the αC-helix and the morpholino substituent directed out of the pocket into solvent (Figure 4, bottom left panel) (98). Mapping of the human Aurora mutations onto the Xenopus model places the Tyr156 residue at the hinge region of the kinase in close proximity to the aromatic quinazoline core of ZM447439 (Figure 4, bottom left panel). The authors hypothesize that mutation of the tyrosine to a histidine may weaken the van der Waals contacts that this hinge region amino acid makes with the small-molecule inhibitor. The Gly160 residue maps to the hinge loop as well. In a similar fashion to the Thr315Ile gatekeeper mutation that renders ABL insensitive to imatinib, substitution of glycine for a larger residue most likely introduces a steric clash with the bound inhibitor. From a model of human Gly160Val bound to ZM447439, it is apparent that the morpholinyl-propoxy moiety extends over the hinge loop and would be expected to collide with the valine or glutamate residue (Figure 4, bottom right panel). A similar steric clash would be expected to occur with the piperazine ring of VX-680 (Figure 4, top right panel). Based on the structure of Aurora bound to AMP-PNP and the kinetic data for these mutants, these substitutions do not affect the binding of ATP. Despite the diverse chemical structures of ZM477439, VX-680 and Hesperadin, these inhibitors exploit similar contacts with the ATP-binding pocket of Aurora, which leads to their uniform sensitivity to mutations in these region (99).
In a related study, Scutt and co-workers identified mutations in Aurora A that confer resistance to the inhibitor VX-680 (100). Upon structural analysis of the binding mode of VX-680 in Aurora A kinase, the analogous glycine residue that confers resistance to Aurora B was identified (Gly216). Mutation of this residue to a bulkier amino acid conferred resistance to VX-680 and ZM447439, with Gly216Leu showing the greatest loss in sensitivity compared to wild-type Aurora A (250-fold) (Figure 4, top right panel). However, these substitutions in Aurora A greatly reduced the overall activity of this enzyme, which is in contrast to their effect on the catalytic activity of Aurora B. Notably, the Gly216Leu, Gly216Val and Gly216Glu mutants of Aurora A were found to have 6%, <1% and 12% of the activity of the wild-type enzyme, respectively. Despite the overlapping inhibitor sensitivities and structural similarities between Aurora A and B, resistance mutations do not affect these enzymes uniformly.
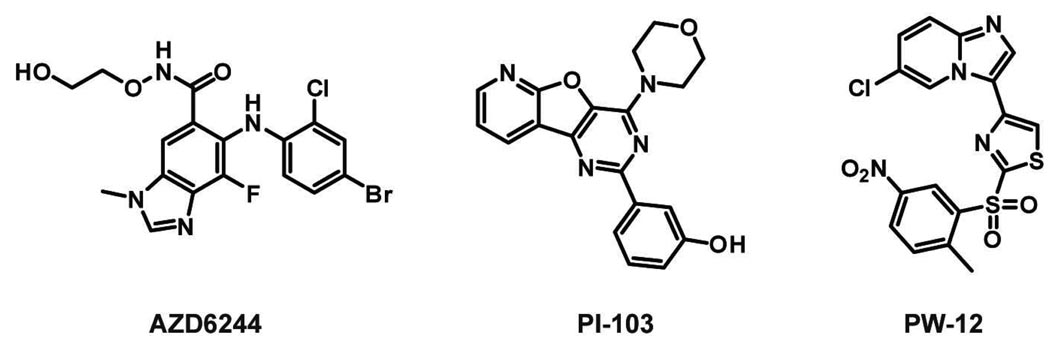
Like the Aurora family, several studies have been conducted with other disease-relevant protein kinases to anticipate potential mechanisms of resistance to their respective small molecule inhibitors. Upregulation of the mitogen-activated protein kinase (MAPK) pathway has been implicated in a number of human cancers. For example, a gain of function mutation in the MAPK kinase kinase B-RAF (Val600Glu) is found in many melanomas (50–70% of cases (101). Thus, small-molecule inhibitors that target proteins in the MAPK pathway, such as B-RAF and its downstream kinase substrate MEK1, are promising drug candidates. Potent and selective inhibitors of the catalytic activity of MEK1 have been developed, with a series of non-ATP-competitive inhibitors showing potential in clinical trials (102). Garraway and co-workers conducted a study to identify mutations that may arise to confer resistance to the non-ATP-competitive inhibitors AZD6244 or CI-1040 (Figure 5) (103). To do this, a random mutagenesis screen in melanoma cells harboring Val600Glu B-RAF was performed in the presence of cytotoxic concentrations of these drugs. Sequencing of resistant clones identified a set of MEK1 mutant alleles; a majority of which contained point mutations surrounding the site of inhibitor binding (primary). It is likely that these mutants confer resistance through direct interference with inhibitor binding or by altering the conformation of the αC-helix. In addition, several mutations were identified in regions of the catalytic domain that are not close to the site of site of drug binding (secondary); a subset of which may cause resistance by upregulating the intrinsic catalytic activity of MEK1. Several drug-resistant MEK1 mutants expressed in A375 melanoma cells showed increased AZD6244 GI50 values (50- to 1000-fold) relative to wild-type A375 cells. Analysis of cells expressing these resistant MEK1 mutants showed that phosphorylation of the downstream MAPK ERK was rescued in the presence of inhibitor. These results were compared to clinical resistance mutants by sequencing tumors from melanoma patients who had relapsed upon treatment with AZD6244. These efforts led to the identification of a Pro124Leu MEK1 mutant, which is analogous to two secondary mutations (Pro124Ser and Pro124Gln) that were discovered in the random mutagenesis screen. The Pro124Leu MEK1 mutant provided a modest increase (5-fold) in AZD6244 GI50 when expressed in parental A375 melanoma cells.
Figure 5.
Chemical structures of MEK1 inhibitor AZD6244 and PI3K p110α inhibitors PI-103 and PW-12.
A drug resistance study has also been performed with the phosphatidylinositol 3-kinase (PI3K) p110α, which is a lipid kinase that generates phosphatidylinositol-3,4,5-trisphosphate (PIP3) from phosphatidylinositol 4,5-bisphosphate (PIP2). p110α is the most frequently mutated gene in human cancer, with the activating mutation His1047Arg in the kinase domain being the most common. For this reason, a number of ATP-competitive small-molecule inhibitors of p110α have been developed and are undergoing clinical trials for the treatment of cancer (104). To facilitate the identification of p110α resistance mutations in vitro, Shokat and co-workers developed a PI3K inhibitor screen in the yeast S. cerevisiae. Over-expression of membrane-localized p110α inhibits the growth of S. cerevisiae, most likely because these yeast lack the ability to degrade any PIP3 that is generated (105). However, small-molecule inhibitors of PI3K can rescue growth. Through the use of replica plating and robotic pinning this screen allows the rapid assessment of a large number of mutants under various conditions. A library of high-copy plasmids (pURA3-2µ-GAL1) containing mutants of p110α-CAAX, which were generated by site-directed saturation mutagenesis, was transformed into the drug-permeable yeast strain YRP1. The library of p110α-CAAX variants was then screened on glucose and galactose media to determine which mutants retain catalytic activity. Active mutants that were growth-inhibited on galactose in the presence of high p110α inhibitor concentrations, for example PI-103 (Figure 5), were selected and sequenced. In contrast to protein kinases, the gatekeeper residue of p110α was found to be intolerant to mutation and, therefore, not a likely site of resistance. However, another residue that lines the ATP-binding pocket, Ile800, was found to confer resistance without compromising kinase activity. The identified resistance mutations did not affect all of the p110α inhibitors uniformly; one drug-resistant mutant, Ile800Leu, sensitized p110α to dual PI3K/mTOR inhibitor BEZ-235 and multi-targeted kinase inhibitor PW-12 (Figure 5). The functional relevance of these resistance mutations was validated with in vitro activity assays and in the non-tumorigenic mammary epithelial cell line MCF10A.
Conclusions
The emergence of drug resistance to targeted cancer therapies is an ongoing clinical problem. While resistance to small-molecule kinase inhibitors can be caused by the amplification of the oncogenic kinase gene being targeted or the re-wiring of signaling cascades, the emergence of mutations in the catalytic domain that hinder drug binding is a common mechanism. However, the range of mutations that are available to a kinase to confer drug resistance are limited due to the necessity of these enzymes maintaining their cellular functions. Several general themes emerge by comparing drug-resistance mutations in BCR-ABL, EGFR, MEK1, p110α and the Aurora kinases. First, point mutations that generate resistance to small-molecule kinase inhibitors do not greatly reduce the catalytic activities of these enzymes. In some cases, these kinase variants have greater catalytic activity than the wild-type enzyme. Second, interactions that contribute to inhibitor selectivity are often the main sites of resistance mutations. For example, a large part of imatinib’s selectivity for ABL over other closely related kinases is due to its unique interaction with the P-loop of this kinase but this segment is the most frequent site of resistance mutations. Finally, catalytic domain mutations can lead to drug resistance in unexpected ways. While mutating the gatekeeper position from a smaller residue to a larger one is a common route of drug resistance in BCR-ABL and EGFR, the mechanistic reasons for reduced inhibitor binding in cells are very different. The generality of the lessons learned from the kinases highlighted in this review will be tested as more kinase inhibitors enter clinical use and additional resistance mutations are identified. The ability to perform cellular screens that are able to predict which mutations will likely arise should greatly expedite this process.
Once new mechanisms of drug resistance have been identified and characterized, it will be important to develop effective strategies for targeting kinases that harbor these mutations. The rapid development of second generation inhibitors that target many drug-resistant BCR-ABL mutants provides precedent for future success. While there are still no clinically-approved inhibitors that effectively target the Thr315Ile gatekeeper mutant, several type I and type II inhibitors that are able to bypass the increased steric bulk of this substitution have been identified. In addition, several inhibitors that target sites outside of the ATP-binding pocket have been described (106, 107). Finally, the recently reported strategy of developing mutant-selective kinase inhibitors may prove to be an extremely effective tool for combating drug resistance (88).
Acknowledgements
D. Maly gratefully acknowledges financial support from the NIH (R01GM086858) and the University of Washington.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 4.Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SS, Bubis J, Toner-Webb J, Saraswat LD, First EA, Buechler JA, Knighton DR, Sowadski J. CAMP-dependent protein kinase: prototype for a family of enzymes. FASEB J. 1988;2:2677–2685. doi: 10.1096/fasebj.2.11.3294077. [DOI] [PubMed] [Google Scholar]
- 6.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 7.Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 8.Madhusudan, Trafny EA, Xuong NH, Adams JA, Ten Eyck LF, Taylor SS, Sowadski JM. cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 10.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Liao JJ. Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J. Med. Chem. 2007;50:409–424. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- 12.Vieth M, Sutherland JJ, Robertson DH, Campbell RM. Kinomics: characterizing the therapeutically validated kinase space. Drug Discov. Today. 2005;10:839–846. doi: 10.1016/S1359-6446(05)03477-X. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 14.Mol CD, Fabbro D, Hosfield DJ. Structural insights into the conformational selectivity of STI-571 and related kinase inhibitors. Curr Opin Drug Discov. Devel. 2004;7:639–648. [PubMed] [Google Scholar]
- 15.Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, Sim T, Xie Y, Wang X, Xia G, Spraggon G, Warmuth M, Liu Y, Gray NS. A general strategy for creating "inactive-conformation" abl inhibitors. Chem. Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Bikker JA, Brooijmans N, Wissner A, Mansour TS. Kinase domain mutations in cancer: implications for small molecule drug design strategies. J. Med. Chem. 2009;52:1493–1509. doi: 10.1021/jm8010542. [DOI] [PubMed] [Google Scholar]
- 17.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu. Rev. Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 18.Sawyers CL, Hochhaus A, Feldman E, Goldman JM, Miller CB, Ottmann OG, Schiffer CA, Talpaz M, Guilhot F, Deininger MW, Fischer T, O'Brien SG, Stone RM, Gambacorti-Passerini CB, Russell NH, Reiffers JJ, Shea TC, Chapuis B, Coutre S, Tura S, Morra E, Larson RA, Saven A, Peschel C, Gratwohl A, Mandelli F, Ben-Am M, Gathmann I, Capdeville R, Paquette RL, Druker BJ. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 19.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 20.Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 21.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 22.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Grzesiek S, Jahnke W. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J. Biol. Chem. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 23.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 24.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 25.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nature Biotechnology. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 26.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 27.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MWN, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell BL, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA, Investigators I. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. New Eng. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 28.Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 29.Marin D, Milojkovic D, Olavarria E, Khorashad JS, de Lavallade H, Reid AG, Foroni L, Rezvani K, Bua M, Dazzi F, Pavlu J, Klammer M, Kaeda JS, Goldman JM, Apperley JF. European LeukemiaNet criteria for failure or suboptimal response reliably identify patients with CML in early chronic phase treated with imatinib whose eventual outcome is poor. Blood. 2008;112:4437–4444. doi: 10.1182/blood-2008-06-162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, Gathmann I, Bolton AE, van Hoomissen IC, Goldman JM, Radich JP, Taylor K, Durrant S, Schwarer A, Joske D, Seymour J, Grigg A, Ma D, Arthur C, Bradstock K, Joshua D, Lechner K, Verhoef G, Louwagie A, Martiat P, Straetmans N, Bosly A, Shepherd J, Shustik C, Lipton J, Kovacs DM, Turner AR, Nielsen JL, Birgens H, Bjerrum OW, Guilhot F, Reiffers J, Rousselot P, Facon T, Harousseau JL, Tulliez M, Guerci A, Blaise D, Maloisel F, Michallet M, Fischer T, Hossfeld D, Mertelsmann R, Andreesen R, Nerl C, Freund M, Gattermann N, Hoeffken K, Ehninger G, Deininger M, Ottmann O, Peschel C, Fruehauf S, Neubauer A, Le Coutre P, Aulitzky W, Baccarani M, Saglio G, Fanin R, Rosti G, Mandelli F, Morra E, Carella A, Lazzarino M, Petrini M, Ferrini PR, Nobile F, Liso V, Ferrara F, Rizzoli V, Fioritoni G, Martinelli G, Cornelissen J, Ossenkoppele G, Browett P, Gedde-Dahl T, Tangen JM, Dahl I, Cervantes F, Odriozola J, Boluda JCH, Steegmann JL, Canizo C, Sureda A, Diaz J, Granena A, Fernandez MN, Simonsson B, Stenke L, Paul C, Bjoreman M, Malm C, Wadenvik H, Nilsson PG, Turesson I, Gratwohl A, Hess U, Solenthaler M, O'Brien SG, Russell N, Mufti G, Cavenagh J, Clark RE, Green AR, Holyoake TL, Lucas GS, Smith G, Milligan DW, Rule SJ, Burnett AK, Druker BJ, Larson RA, Moroose R, Wetzler M, Bearden J, Brown R, Lobell M, Cataland S, Rabinowitz I, Meisenberg B, Gabrilove J, Thompson K, Graziano S, Emanuel P, Gross H, Cobb P, Bhatia R, Dakhil S, Irwin D, Issell B, Pavletic S, Kuebler P, Layhe E, Butera P, Glass J, Moore J, Grant B, Niell H, Herzig R, Burris H, Kantarjian H, Peterson B, Powell B, Kalaycio M, Stirewalt D, Samlowski W, Berman E, Limentani S, Seay T, Shea T, Akard L, Smith G, Becker P, DeVine S, Hart R, Veith R, Wade J, Brunvand M, Silver R, Kalman I, Strickland D, Shurafa M, Bashey A, Shadduck R, Cooper S, Safah H, Rubenstein M, Collins R, Keller A, Stone R, Tallman M, Stevens D, Pecora A, Agha M, Holmes H, Grp IS. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. New Eng. J. Med. 2003;349:1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- 31.von Bubnoff N, Schneller F, Peschel C, Duyster J. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: a prospective study. Lancet. 2002;359:487–491. doi: 10.1016/S0140-6736(02)07679-1. [DOI] [PubMed] [Google Scholar]
- 32.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 33.Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K, Herrmann R, Lynch KP, Hughes TP. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 34.Al-Ali HK, Heinrich MC, Lange T, Krahl R, Mueller M, Muller C, Niederwieser D, Druker BJ, Deininger MW. High incidence of BCR-ABL kinase domain mutations and absence of mutations of the PDGFR and KIT activation loops in CML patients with secondary resistance to imatinib. Hematol. J. 2004;5:55–60. doi: 10.1038/sj.thj.6200319. [DOI] [PubMed] [Google Scholar]
- 35.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 36.Hochhaus A, Kreil S, Corbin AS, La Rosee P, Muller MC, Lahaye T, Hanfstein B, Schoch C, Cross NC, Berger U, Gschaidmeier H, Druker BJ, Hehlmann R. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 37.Grant BD, Hemmer W, Tsigelny I, Adams JA, Taylor SS. Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry. 1998;37:7708–7715. doi: 10.1021/bi972987w. [DOI] [PubMed] [Google Scholar]
- 38.Aimes RT, Hemmer W, Taylor SS. Serine-53 at the tip of the glycine-rich loop of cAMP-dependent protein kinase: role in catalysis, P-site specificity, and interaction with inhibitors. Biochemistry. 2000;39:8325–8332. doi: 10.1021/bi992800w. [DOI] [PubMed] [Google Scholar]
- 39.Hirai TJ, Tsigelny I, Adams JA. Catalytic assessment of the glycine-rich loop of the v-Fps oncoprotein using site-directed mutagenesis. Biochemistry. 2000;39:13276–13284. doi: 10.1021/bi001216g. [DOI] [PubMed] [Google Scholar]
- 40.Wong L, Jennings PA, Adams JA. Communication pathways between the nucleotide pocket and distal regulatory sites in protein kinases. Acc. Chem. Res. 2004;37:304–311. doi: 10.1021/ar020128g. [DOI] [PubMed] [Google Scholar]
- 41.Barouch-Bentov R, Che J, Lee CC, Yang Y, Herman A, Jia Y, Velentza A, Watson J, Sternberg L, Kim S, Ziaee N, Miller A, Jackson C, Fujimoto M, Young M, Batalov S, Liu Y, Warmuth M, Wiltshire T, Cooke MP, Sauer K. A conserved salt bridge in the G loop of multiple protein kinases is important for catalysis and for in vivo Lyn function. Mol. Cell. 2009;33:43–52. doi: 10.1016/j.molcel.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, Rheinberger P, Centeleghe M, Fabbro D, Manley PW. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr. D Biol. Crystallogr. 2007;63:80–93. doi: 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 44.Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, Iacobucci I, Amabile M, Abruzzese E, Orlandi E, Radaelli F, Ciccone F, Tiribelli M, di Lorenzo R, Caracciolo C, Izzo B, Pane F, Saglio G, Baccarani M, Martinelli G. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin. Cancer Res. 2006;12:7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 45.O'Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, Cowan-Jacob SW, Lee FY, Heinrich MC, Deininger MW, Druker BJ. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 46.Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl. Acad. Sc. U.S.A. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg. Med. Chem. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 50.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr. Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 51.Eyers PA, Craxton M, Morrice N, Cohen P, Goedert M. Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem. Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 52.Blencke S, Zech B, Engkvist O, Greff Z, Orfi L, Horvath Z, Keri G, Ullrich A, Daub H. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem. Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 53.Tamborini E, Bonadiman L, Greco A, Albertini V, Negri T, Gronchi A, Bertulli R, Colecchia M, Casali PG, Pierotti MA, Pilotti S. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 54.Cools J, Stover EH, Boulton CL, Gotlib J, Legare RD, Amaral SM, Curley DP, Duclos N, Rowan R, Kutok JL, Lee BH, Williams IR, Coutre SE, Stone RM, DeAngelo DJ, Marynen P, Manley PW, Meyer T, Fabbro D, Neuberg D, Weisberg E, Griffin JD, Gilliland DG. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRalpha-induced myeloproliferative disease. Cancer Cell. 2003;3:459–469. doi: 10.1016/s1535-6108(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 55.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, Kung AL, Mestan J, Daley GQ, Callahan L, Catley L, Cavazza C, Azam M, Neuberg D, Wright RD, Gilliland DG, Griffin JD. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 57.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, Fairchild C, Hunt JT, Inigo I, Johnston K, Kamath A, Kan D, Klei H, Marathe P, Pang S, Peterson R, Pitt S, Schieven GL, Schmidt RJ, Tokarski J, Wen ML, Wityak J, Borzilleri RM. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 58.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 59.Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, Tanaka C, Manley P, Rae P, Mietlowski W, Bochinski K, Hochhaus A, Griffin JD, Hoelzer D, Albitar M, Dugan M, Cortes J, Alland L, Ottmann OG. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 60.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, Blackwood-Chirchir MA, Iyer V, Chen TT, Huang F, Decillis AP, Sawyers CL. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 61.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, 3rd, Edeen PT, Floyd M, Ford JM, Grotzfeld RM, Herrgard S, Insko DE, Mehta SA, Patel HK, Pao W, Sawyers CL, Varmus H, Zarrinkar PP, Lockhart DJ. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JM, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 63.Modugno M, Casale E, Soncini C, Rosettani P, Colombo R, Lupi R, Rusconi L, Fancelli D, Carpinelli P, Cameron AD, Isacchi A, Moll J. Crystal structure of the T315I Abl mutant in complex with the aurora kinases inhibitor PHA-739358. Cancer Res. 2007;67:7987–7990. doi: 10.1158/0008-5472.CAN-07-1825. [DOI] [PubMed] [Google Scholar]
- 64.O'Hare T, Eide CA, Tyner JW, Corbin AS, Wong MJ, Buchanan S, Holme K, Jessen KA, Tang C, Lewis HA, Romero RD, Burley SK, Deininger MW. SGX393 inhibits the CML mutant Bcr-AblT315I and preempts in vitro resistance when combined with nilotinib or dasatinib. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5507–5512. doi: 10.1073/pnas.0800587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, Metcalf CA, III, Tyner JW, Loriaux MM, Corbin AS, Wardwell S, Ning Y, Keats JA, Wang Y, Sundaramoorthi R, Thomas M, Zhou D, Snodgrass J, Commodore L, Sawyer TK, Dalgarno DC, Deininger MW, Druker BJ, Clackson T. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradeen HA, Eide CA, O'Hare T, Johnson KJ, Willis SG, Lee FY, Druker BJ, Deininger MW. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108:2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell. Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 69.Johnson BE, Janne PA. Epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Cancer Res. 2005;65:7525–7529. doi: 10.1158/0008-5472.CAN-05-1257. [DOI] [PubMed] [Google Scholar]
- 70.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 71.Pollack VA, Savage DM, Baker DA, Tsaparikos KE, Sloan DE, Moyer JD, Barbacci EG, Pustilnik LR, Smolarek TA, Davis JA, Vaidya MP, Arnold LD, Doty JL, Iwata KK, Morin MJ. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J. Pharmacol. Exp. Ther. 1999;291:739–748. [PubMed] [Google Scholar]
- 72.Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, Gilmer TM. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol. Cancer Ther. 2001;1:85–94. [PubMed] [Google Scholar]
- 73.Traxler P, Allegrini PR, Brandt R, Brueggen J, Cozens R, Fabbro D, Grosios K, Lane HA, McSheehy P, Mestan J, Meyer T, Tang C, Wartmann M, Wood J, Caravatti G. AEE788: a dual family epidermal growth factor receptor/ErbB2 and vascular endothelial growth factor receptor tyrosine kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2004;64:4931–4941. doi: 10.1158/0008-5472.CAN-03-3681. [DOI] [PubMed] [Google Scholar]
- 74.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, Liang MC, Perera SA, Zaghlul S, Borgman CL, Kubo S, Takahashi M, Sun Y, Chirieac LR, Padera RF, Lindeman NI, Janne PA, Thomas RK, Meyerson ML, Eck MJ, Engelman JA, Shapiro GI, Wong KK. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 75.Tsou HR, Overbeek-Klumpers EG, Hallett WA, Reich MF, Floyd MB, Johnson BD, Michalak RS, Nilakantan R, Discafani C, Golas J, Rabindran SK, Shen R, Shi X, Wang YF, Upeslacis J, Wissner A. Optimization of 6,7-disubstituted-4-(arylamino)quinoline-3-carbonitriles as orally active, irreversible inhibitors of human epidermal growth factor receptor-2 kinase activity. J Med Chem. 2005;48:1107–1131. doi: 10.1021/jm040159c. [DOI] [PubMed] [Google Scholar]
- 76.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, Shewchuk L. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 77.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer -- search and destroy. Eur. J. Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 78.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 81.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M "gatekeeper" mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol. Cancer. Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 84.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, Harris PL, Driscoll DR, Fidias P, Lynch TJ, Rabindran SK, McGinnis JP, Wissner A, Sharma SV, Isselbacher KJ, Settleman J, Haber DA. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wong KK. HKI-272 in non small cell lung cancer. Clin. Cancer Res. 2007;13:s4593–s4596. doi: 10.1158/1078-0432.CCR-07-0369. [DOI] [PubMed] [Google Scholar]
- 87.Eskens FA, Mom CH, Planting AS, Gietema JA, Amelsberg A, Huisman H, van Doorn L, Burger H, Stopfer P, Verweij J, de Vries EG. A phase I dose escalation study of BIBW 2992, an irreversible dual inhibitor of epidermal growth factor receptor 1 (EGFR) and 2 (HER2) tyrosine kinase in a 2-week on, 2-week off schedule in patients with advanced solid tumours. Br. J. Cancer. 2008;98:80–85. doi: 10.1038/sj.bjc.6604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Jänne PA. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J. Cell. Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 90.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell. Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 91.Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev. Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 92.Girdler F, Gascoigne KE, Eyers PA, Hartmuth S, Crafter C, Foote KM, Keen NJ, Taylor SS. Validating Aurora B as an anti-cancer drug target. J. Cell. Sci. 2006;119:3664–3675. doi: 10.1242/jcs.03145. [DOI] [PubMed] [Google Scholar]
- 93.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 94.Gadea BB, Ruderman JV. Aurora kinase inhibitor ZM447439 blocks chromosome-induced spindle assembly, the completion of chromosome condensation, and the establishment of the spindle integrity checkpoint in Xenopus egg extracts. Mol. Biol. Cell. 2005;16:1305–1318. doi: 10.1091/mbc.E04-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell. Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell. Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Girdler F, Sessa F, Patercoli S, Villa F, Musacchio A, Taylor S. Molecular basis of drug resistance in aurora kinases. Chem. Biol. 2008;15:552–562. doi: 10.1016/j.chembiol.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 98.Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 99.Cheetham GM, Charlton PA, Golec JM, Pollard JR. Structural basis for potent inhibition of the Aurora kinases and a T315I multi-drug resistant mutant form of Abl kinase by VX-680. Cancer Lett. 2007;251:323–329. doi: 10.1016/j.canlet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 100.Scutt PJ, Chu ML, Sloane DA, Cherry M, Bignell CR, Williams DH, Eyers PA. Discovery and exploitation of inhibitor-resistant aurora and polo kinase mutants for the analysis of mitotic networks. J. Biol. Chem. 2009;284:15880–15893. doi: 10.1074/jbc.M109.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 102.Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, Hamid O, Varterasian M, Asbury P, Kaldjian EP, Gulyas S, Mitchell DY, Herrera R, Sebolt-Leopold JS, Meyer MB. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J. Clin. Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 103.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, MacConaill LE, Zhang J, Gray NS, Sellers WR, Dummer R, Garraway LA. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim. Biophys. Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 105.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell. 2008;14:180–192. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adrián FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat. Chem. Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 107.Gumireddy K, Baker SJ, Cosenza SC, John P, Kang AD, Robell KA, Reddy MV, Reddy EP. A non-ATP-competitive inhibitor of BCR-ABL overrides imatinib resistance. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1992–1997. doi: 10.1073/pnas.0408283102. [DOI] [PMC free article] [PubMed] [Google Scholar]