Summary
IL-10R2 is a shared cell surface receptor required for the activation of five class 2 cytokines (IL-10, IL-22, IL-26, IL-28, and IL-29) that play critical roles in host defense. To define the molecular mechanisms that regulate its promiscuous binding, we have determined the crystal structure of the IL-10R2 ecto-domain at 2.14Ǻ resolution. IL-10R2 residues required for binding were identified by alanine scanning and used to derive computational models of IL-10/IL-10R1/IL-10R2 and IL-22/IL-22R1/IL-10R2 ternary complexes. The models reveal a conserved binding epitope that is surrounded by two clefts that accommodate the structural and chemical diversity of the cytokines. These results provide a structural framework for interpreting IL-10R2 single nucleotide polymorphisms associated with human disease.
Introduction
IL-10R2 is a ubiquitously expressed and essential receptor chain for at least five IL-10 family cytokines that share ~10–20% sequence identity and distinct 3D structures (Donnelly et al., 2004; Walter, 2004). IL-10R2, first called CRF2–4, was originally discovered as an essential component of the IL-10 hetero-dimeric signaling complex formed between the IL-10R1 chain and IL-10R2 (Kotenko et al., 1997). Solution and cell binding studies have shown IL-10R1 exhibits high affinity (nM) for IL-10, whereas IL-10R2 chain affinity is very low (~mM) (Ding et al., 2000; Logsdon et al., 2002; Yoon et al., 2005; Yoon et al., 2006). IL-10 mediated assembly of the IL-10R1/IL-10R2 complex activates intracellular kinases, JAK1 and TYK2, which phosphorylate the intracellular domains of the receptor as well as STAT3 (Moore et al., 2001). Subsequent studies have revealed IL-10R2 also forms IL-22R1/IL-10R2 (Kotenko et al., 2001; Xie et al., 2000), IL-20R1/IL-10R2 (Sheikh et al., 2004) and IL-28R1/IL-10R2 (Kotenko et al., 2003; Sheppard et al., 2003) heterodimers that are activated by IL-22, IL-26, and IL-28/ IL-29, respectively (Fig S1). Thus, IL-10R2 functions as a common signaling chain in the class 2 cytokine family as observed for gp130, IL-2γc, and GM-CSF βc chains in the class 1 cytokine family (Wang et al., 2009).
Cytokines that signal via IL-10R2 induce pleotropic activities that protect the host from over-exuberant immune responses and activate innate immunity programs in epithelial cells. IL-10 inhibits macrophage and dendritic cell function, prevents pro-inflammatory cytokine synthesis (e.g. TNF-α, IFN-γ, IL-12, IL-l), and blocks antigen presentation (Moore et al., 2001). As a result of its potent immunosuppressive functions, numerous pathogens, including HIV, evade host immune responses by increasing the production of cellular IL-10 (Blackburn and Wherry, 2007; Redpath et al., 2001). In addition, cytomegalovirus (CMV) and Epstein Barr virus (EBV) express their own virally encoded IL-10s (cmvIL-10 and ebvIL-10) that disrupt immune function (Slobedman et al., 2009). IL-22, produced by TH17 cells, induces epithelial cells to produce anti-microbial peptides (Liang et al., 2006) but also exhibits protective functions in the gut, lung, and liver (Radaeva et al., 2004; Zenewicz et al., 2007). IL-26 is also produced by TH17 cells, up-regulates pro-inflammatory genes in intestinal epithelial cells, and its expression is increased in active Chron’s disease (Dambacher et al., 2009). Finally, IL-28 and IL-29 form the recently discovered type III interferon family (Kotenko et al., 2003; Sheppard et al., 2003). IL-28 and IL-29 exhibit potent antiviral activities in keratinocytes and dendritic cells and may play an important role in resolving hepatitis C infection (Ge et al., 2009).
The molecular basis, whereby IL-10R2 is able to form promiscuous receptor-ligand interactions is unknown. In addition, single nucleotide polymorphisms (SNPs) in the IL-10R2 extracellular domain are associated with hepatitis virus subtype B (HBV) persistence (Frodsham et al., 2006), early-onset colitis (Glocker et al., 2009), graft-versus-host disease related to transplant rejection (Lin et al., 2005; Sivula et al., 2009), and systemic sclerosis (Hikami et al., 2008); an autoimmune disease characterized by fibrosis of skin and other organs. Thus, structure-function studies on IL-10R2 may be useful in understanding SNPs that alter immune function and result in human disease.
To define the molecular basis for promiscuous binding to diverse cytokine complexes, the crystal structure of the soluble extracellular IL-10R2 chain (sIL-10R2) has been determined at 2.14Ǻ resolution. A series of sIL-10R2 mutants were evaluated for binding to three different binary complexes (BCs, IL-22/sIL-22R1, cmvIL-10/sIL-10R1, hIL-10/sIL-10R1) using tri-molecular surface plasmon resonance (SPR) assays. The binding data provided input for computational docking studies to generate IL-10 and IL-22 ternary complex models. These studies reveal IL-10R2 uses a central common binding epitope, centered on Y82IL-10R2, which is surrounded by two clefts that allow the IL-10R2 chain to scan the unique structures and chemistries of helices A and D on the cytokines. The studies reveal IL-10R2 shares a conserved recognition motif with gp130 and IL-2γc suggesting a common origin for the promiscuous class 1 and class 2 cytokine receptors. Despite these common features, sIL-10R2 forms structurally and energetically distinct contacts with IL-10/sIL-10R1 and IL-22/sIL-22R1 BCs, which could assist in the prediction of IL-10R2 SNP function.
Results
Structure Determination
The sIL-10R2 chain crystallized in spacegroup P6 with 2 molecules (chain A and B) in the asymmetric unit (Fig S2). Crystals of sIL-10R2 were first obtained from protein expressed in insect cells where N-linked glycosylation sites N49sIL-10R2, N68sIL-10R2, N102sIL-10R2 and N161sIL-10R2, were mutated to glutamine (sIL-10R2-NQ). sIL-10R2-NQ binds to the IL-22/sIL-22R1 complex with twofold weaker affinity (Kd = 14µM) than glycosylated sIL-10R2 (Kd = 7µM) expressed in insect cells. For phasing, sIL-10R2, containing a C106sIL-10R2 to Ser mutation, was expressed in E. coli as a seleno-methionine labeled protein and refolded from insoluble inclusion bodies. Larger crystals, used for data collection and refinement (crystal 2, Table 1), were obtained with a sIL-10R2 double mutant (C106SsIL-10R2/S126CsIL-10R2). The structure of was solved by SAD phasing and refined at 2.14Å resolution to Rcryst and Rfree values of 21.7% and 23.7%, respectively (Table 1). Electron density was observed for residues 20–220 in chain A and 20–215 in chain B. sIL-10R2 residue numbering is based on the sequence found in the Uniprot database code Q08334.
Table 1.
sIL-10R2 Data Collection, Phasing, and Refinement Statistics
| Crystal 1 | Crystal 2 | |
|---|---|---|
| Space group | P6 | P6 |
| Cell a,b | 124.27 | 124.84 |
| Cell c | 83.70 | 84.22 |
| Wavelength (Å) | 0.97934 (peak) | 1.00000 |
| Resolution | 50–2.5 | 50–2.14 |
| Highest resolution | 2.59–2.50 | 2.22–2.14 |
| Completeness (%) | 100 (100) | 97.5 (84.6) |
| Redundancy | 10.8 (10.7) | 11.0 (5.1) |
| I/σ | 72 (26) | 50 (5.9) |
| Rsym (%) | 7.1 (15.7) | 7.1 (28.9) |
| Phasing statistics | ||
| # Se (SOLVE) | 12 | |
| FOM (SOLVE) | 0.48 | |
| FOM (CNS DM) | 0.91 | |
| Refinement statistics | ||
| Resolution (Å) | 50–2.14 | |
| Highest resolution | 2.14–2.16 | |
| Rwork (%) | 21.0 (27.7) | |
| No. of reflections (work) | 38,362 | |
| Rfree (%) | 23.9 (36.5) | |
| No. of reflections (free) | 2,016 | |
| Residues in model | A: 20–220 | |
| B: 20–215 | ||
| No. of protein atoms | 3,284 | |
| No. of water atoms | 212 | |
| No. of sulfate | 10 | |
| No. of glycerol | 1 | |
| Rmsd bond distance (Å) | 0.0085 | |
| Rmsd bond angles (°) | 1.43 | |
| Average B factor (Å2) | 37.3 | |
| Ramachandran plot | ||
| Most favored (%) | 88.1 | |
| Additionally allowed (%) | 9.6 | |
| Generally allowed (%) | 1.4 | |
| Disallowed (%) | 0.8 |
Structure of sIL-10R2 and comparison with high affinity R1 chains
The overall structure of sIL-10R2 is similar to the previously determined structures of sIL-10R1 and sIL-22R1 (Bleicher et al., 2008; Jones et al., 2008; Josephson et al., 2001). sIL-10R2 consists of two fibronectin type III domains (FBNIII) each comprising 7 β-strands (Fig 1A). The N-terminal domain, D1, and C-terminal domain, D2, adopt an inter-domain angle of approximately 95° (elbow angle, Fig S2) which is similar to the high affinity sIL-10R1 and sIL-22R1 chains (Figs 1B, S2). As seen in Figure 1B, the overall domain orientations of sIL-10R2 are closer to sIL-22R1 than sIL-10R1.
Figure 1. Structure of the sIL-10R2 chain.
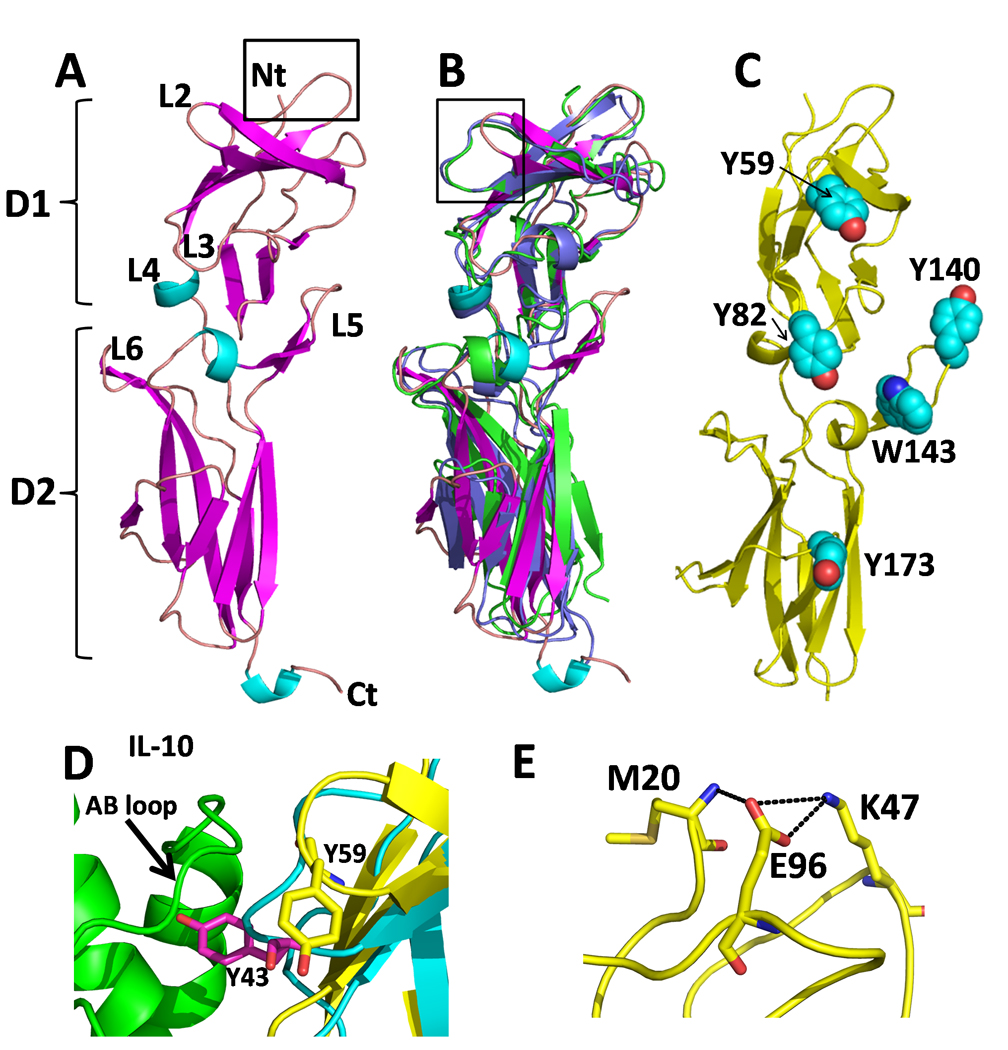
(A) Ribbon diagram of the sIL-10R2 chain colored by secondary structure with binding loops labeled. Box shows the location of Figure F. (B) Superposition of sIL-10R2 (colored as in A) with sIL-10R1 (green) and sIL-22R1 (purple). Box shows the location of Figure D. (C) Location of aromatic residues on sIL-10R2 on sIL-10R2. (D) Comparison of the high affinity site 1 interaction between sIL-10R1 Y43sIL-10R1 and the AB loop of IL-10 (green) with the sIL-10R2 L2 loop and Y59sIL-10R2 (yellow). (E) Interaction network for K47sIL-10R2. Replacement of K47sIL-10R2 with a glutamic acid is associated with persistent HBV infection (Frodsham et al., 2006).
The sIL-10R2 β-strands are connected by a series of loops (L2-L6), which are predicted to mediate ligand binding (Fig 1A, (Jones et al., 2008; Pletnev et al., 2005)). sIL-10R2 L2 and L5 loops exhibit large structural differences compared to L2 and L5 in the sIL-10R1 and sIL-22R1 chains (Figs 1B and 2). L2sIL-10R2 adopts a β-hairpin structure that is 2 residues shorter than observed for the L2 loops in sIL-10R1 and IL-22R1. As a result, L2 residue Y59sIL-10R2 cannot form the canonical high affinity “R1 like” interaction, which consists of tyrosine residues on the L2 loops of the R1 chains (Y43sIL-10R1 and Y60sIL-22R1) that insert into clefts formed by the AB loops of the cytokines in the IL-10/sIL-10R1 and IL-22/sIL-22R1 complexes (Fig 1D, (Bleicher et al., 2008; Jones et al., 2008; Josephson et al., 2001)). Thus, the conformation of sIL-10R2 L2 is consistent with the low affinity of the sIL-10R2 chain for its various binding partners.
Figure 2. Unique clefts identified in sIL-10R2.
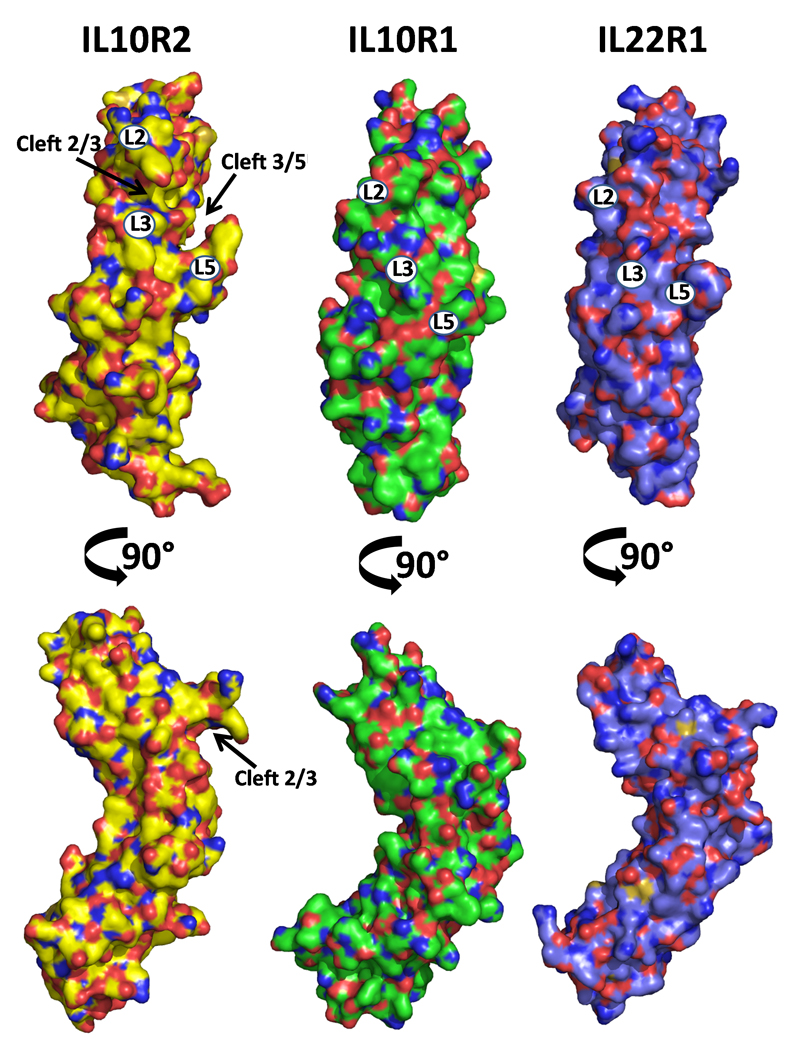
The structures of sIL-10R2, sIL-10R1, and sIL-22R1 are shown as molecular surfaces. Images in the top row are orientated as found in Figure 1A. The surfaces are colored by atom type with oxygens red, nitrogens blue, and sulfur orange. Carbons are colored yellow, green, and purple in sIL-10R2, sIL-10R1, and sIL-22R1, respectively. Numbers on the surfaces correspond to loop positions.
The sIL-10R2 L5 loop forms a “thumb-like” structure that extends away from the rest of the molecule (Figs 1A and 2). Despite conformational differences at the tip of L5 in chains A and B due to crystal contacts, both L5 loops exhibit the same overall “thumb” structure (Fig S2). The novel conformations of the sIL-10R2 L2 hairpin and L5 thumb give rise to two distinct clefts (Fig 2). The first is formed between the L2 and L3 loops (L2/3 cleft) and the second is between L3 and L5 (L3/L5 cleft). In contrast to sIL-10R2, no clefts are found in the high affinity IL-10R1 chain, and only very small L2/L5 and L3/L5 clefts are observed in sIL-22R1 (Fig 2). Interestingly, IL-22R1 is also a shared receptor, which forms IL-22R1/IL-10R2 and IL-22R2/IL-20R2 heterodimers (Dumoutier et al., 2001). These structural features, combined with the presence of four surface exposed aromatic residues (Y59, Y82, Y140, and W143) on the L2, L3, and L5 loops (Fig 1C), suggest the clefts may play an important role in the promiscuous binding properties of sIL-10R2.
Identification of sIL-10R2 residues required for ternary complex formation
To identify sIL-10R2 residues required for promiscuous interactions with structurally and chemically diverse cytokines, a SPR assay was designed to evaluate the relative binding strength of sIL-10R2 alanine mutants to IL-10/sIL-10R1, cmvIL-10/sIL-10R1, and IL-22/IL-22R1 BCs (Figs 3, S3-S5). Of the 22 point mutants tested, only Y82AsIL-10R2 displayed drastically reduced (~10–20% of sIL-10R2WT) binding to all 3 BCs (Fig 3). Y59AsIL-10R2, located on the L2 loop, is the only other mutant that exhibited significantly reduced binding (~50% of WT) to all three complexes, but to a lesser extent than Y82AsIL-10R2. Other mutants disrupted sIL-10R2 recognition of cmvIL-10/sIL-10R1 and hIL-10/sIL-10R1 complexes or IL-22/sIL-22R1 complexes, but not both. For example, Y56AsIL-10R2 and Y140AsIL-10R2 exhibit diminished binding (~20% of WT) to hIL-10/cmvIL-10 BCs, but had no impact on the recognition of IL-22/sIL-22R1. In contrast, W143AIL-10R2, and to a lesser extent E141IL-10R2, selectively disrupted IL-22/IL-22R1 recognition, but not hIL-10/cmvIL-10/sIL-10R1 binding. Together, these studies suggest L3 residue Y82sIL-10R2 provides a centrally located common binding epitope, while different residues on L2 and L5 make distinct energetic contributions to binding in different binary complexes.
Figure 3. Binding analysis of sIL-10R2 alanine mutants by SPR.
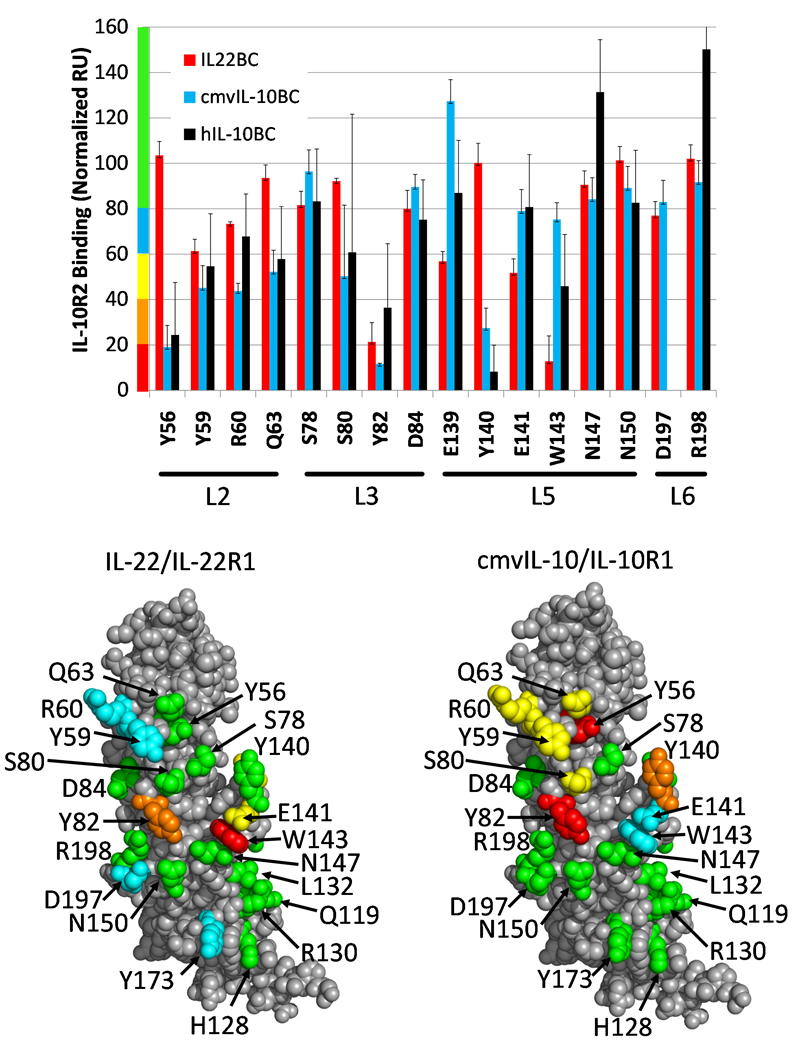
(A) Relative binding of sIL-10R2 alanine mutants to IL-22/IL-22R1 (red), cmvIL-10/IL-10R1 (cyan), and hIL-10/IL-10R1 (black) binary complexes. sIL-10R2 mutant binding is presented in normalized response units (RU) relative to wild-type sIL-10R2 binding at concentrations of 150µM. Results are expressed as the mean of multiple measurements ±standard deviation. Residues tested for binding to IL-22/sIL-22R1 (B) and cmvIL-10/IL-10R1 (C) are mapped onto sIL-10R2 surfaces and colored according to the y axis in Figure A (See also Figures S3-S5).
Six of the 22 sIL-10R2 mutants (Q119A, H128A, R130A, L132A, K135A, and Y173A), located on β-strands A, B and E of D2, targeted putative receptor-receptor contacts (Jones et al., 2008; Pletnev et al., 2005) between the D2 of the R1 chains and D2sIL-10R2. Y173AsIL-10R2 exhibited reduced binding (65% of WT) to IL-22/IL-22R1 but did not reduce binding to cmvIL-10/sIL-10R1 or hIL-10/sIL-10R1 complexes. The other five mutants had no impact on IL-10R2 binding to any binary complex (data not shown). Thus, from a limited analysis of D2 residues, we have not identified an essential sIL-10R2 residue (such as Y82sIL-10R2 described above) involved in R1/R2 D2-D2 contacts. It is unclear if this is due to the limited number of residues analyzed or specific characteristics of the D2 binding interfaces themselves.
Generation of IL-10 and IL-22 ternary complexes by computational docking
To further understand the mechanisms which allow promiscuous IL-10R2 recognition, the mutagenesis data (Fig 3, Table S1, (Logsdon et al., 2004; Wolk et al., 2004; Wu et al., 2008; Yoon et al., 2006)) was used to guide computer-based docking of sIL-10R2 onto IL-22/sIL-22R1, cmvIL-10/sIL-10R1, and hIL-10/sIL-10R1 crystal structures (Jones et al., 2002; Jones et al., 2008; Josephson et al., 2001). Top ranked sIL-10R2 docking solutions, consistent with the mutagenesis data, were identified for IL-22/sIL-22R1 and cmvIL-10/sIL-10R1 complexes. However, sIL-10R2 docking to the hIL-10/sIL-10R1 complex was unsuccessful (Table S2, Fig S6).
IL-22/sIL-22R1/sIL-10R2 and cmvIL-10/sIL-10R1/sIL-10R2 ternary complexes (TC) are assembled from three major interaction surfaces labeled site 1, site 2, and site 3 (Figs 4 and 5, Tables S3, S4). Site 1 forms the high affinity cytokine/R1 contact surface, which is identical to what has been previously described for the IL-22/sIL-22R1 and cmvIL-10/sIL-10R1 binary complexes (Bleicher et al., 2008; Jones et al., 2002; Jones et al., 2008). The sIL-10R2 binding site 2 is centered on cytokine helices A and D, but also consists of receptor-receptor contacts in the cmvIL-10 TC (Figs 4, 5). Site 3 is formed from contacts between R1 and sIL-10R2 D2 domains near the C-termini of the receptors and in close proximity to the cell membrane (Figs 4, 5). While the overall structures of the IL-22 and cmvIL-10 TCs are similar, contacts in the site 2 and site 3 interfaces are considerably different as reflected in the sIL-10R2 binding data (Fig 3).
Figure 4. IL-22 Ternary complex.
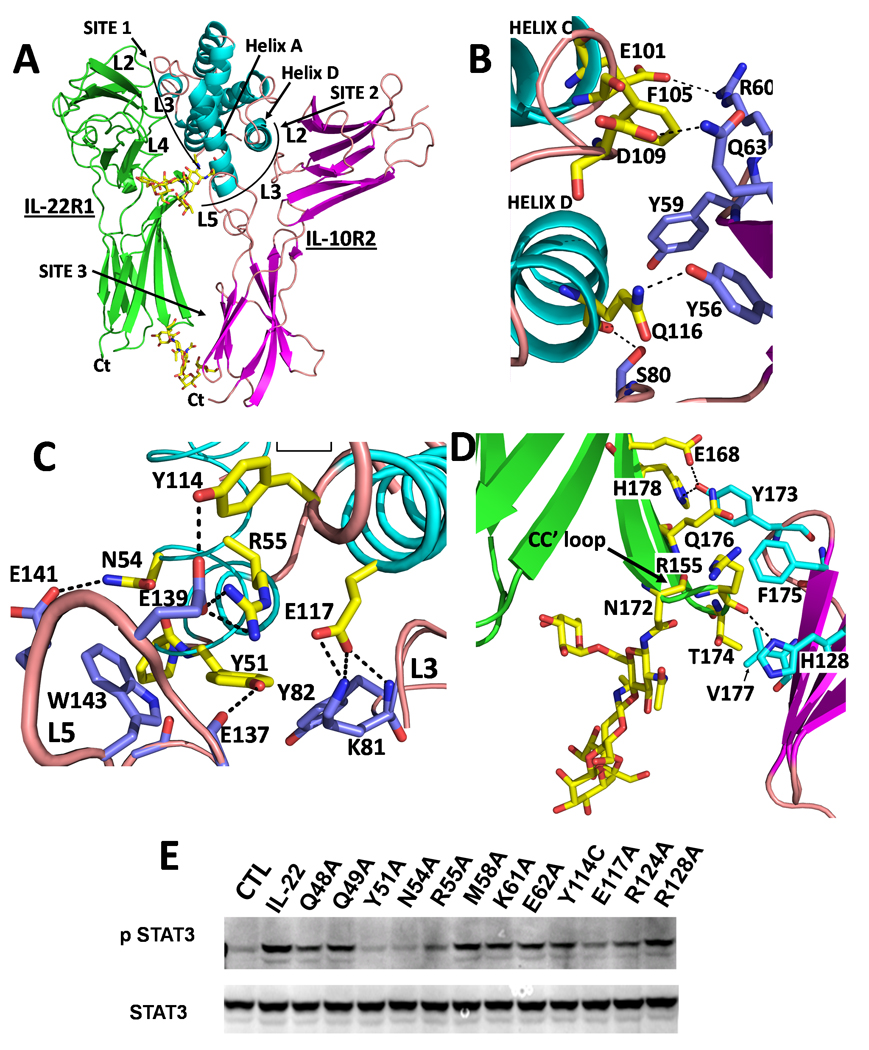
(A) Overall structure of the IL-22 ternary complex. (B) Site 2 interactions between helix D and the L2/L3 cleft. (C) Site 2 interactions between helix A and L3/L5 cleft. (D) Site 3 contacts (E) Anti-Phospho-STAT3 western blot from lysates prepared from HepG2 cells stimulated with IL-22 or IL-22 mutants (See Also Figure S6 and Tables S1-S4).
Figure 5. cmvIL-10 Ternary complex.
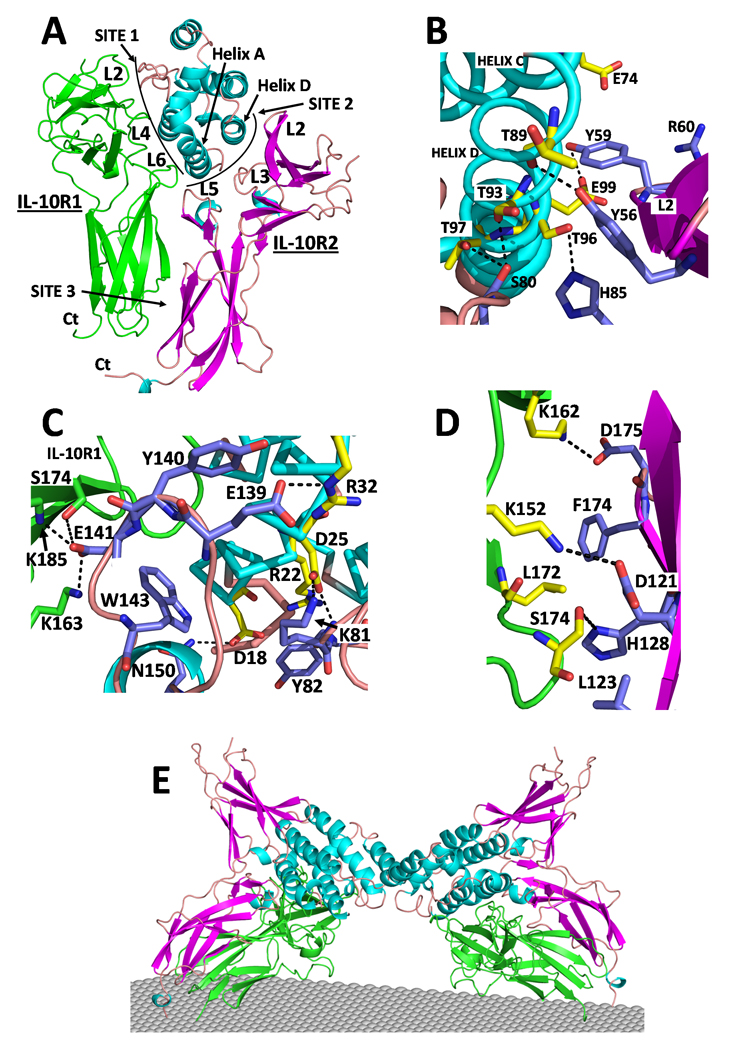
(A) Overall structure of the cmvIL-10 ternary complex. (B) Site 2 interactions between helix D and the L2/L3 cleft. (C) Site 2 interactions between helix A and the L3/L5 cleft. (D) Site 3 contacts. (E) Dimeric cmvIL-10 signaling complex. (See also Figures S6 and Tables S1-S4).
IL-22/sIL-22R1/sIL-10R2 complex
The IL-22/sIL-10R1/sIL-10R2 site 2 interface (910 Ǻ2) is formed exclusively between IL-22 and sIL-10R2. The major feature of site 2 is the protruding IL-22 “knob” surface formed by Y51IL-22, which is sandwiched between Y82sIL-10R2 and W143sIL-10R2 in the sIL-10R2 L3/L5 cleft (Fig 4C). L5sIL-10R2 does not contact sIL-22R1 in the ternary complex but is positioned adjacent to glycosylated helix A residue N54IL-22. This creates an opening between sIL-22R1 and sIL-10R2 for the N54IL-22-linked carbohydrate without disrupting complex formation (Fig 4A). N54IL-22 and R55IL-22 form interactions with L5sIL-10R2 residues E141sIL-10R2 and E139sIL-10R2, respectively (Fig 4C, Table S3). Interactions made by Y82sIL-10R2, W143sIL-10R2, Y51IL-22, and N54IL22 in the IL-22 TC provide an explanation for why these residues are critical for sIL-10R2 recognition of IL-22/sIL-22R1 (Fig 3). To further characterize the validity of model, IL-22 mutants described by Logsdon et al., were evaluated for their ability to activate STAT3 in human HepG2 liver cells, which express human IL-22R1 and IL-10R2 chains (Fig 4E). These studies reveal Y51IL-22, N54IL-22, R55IL-22, and to a lesser degree E117IL-22, are most important for activating IL-22 STAT3 activity in these cells. The biological activity data further supports the contacts observed in the IL-22 TC model (Fig 4), which includes mainchain and sidechain interactions between E117IL-22 and K81sIL-10R2 in the L3/L5 cleft (Fig 4C).
The site 3 interface (315Ǻ2) occurs between β-strand C’ and the CC” loop on sIL-22R1 (residues 173–178), and β-strands B and E on sIL-10R2 (Fig 4D). The carbohydrate, attached to N172sIL-22R1, is also important in site 3 because it stabilizes the CC’ loop for sIL-10R2 binding. When the carbohydrate attached to N172sIL-22R1 is removed, crystallographic studies reveal CC’ loop residues 172–175, that contact sIL-10R2 in our model, become disordered (Bleicher et al., 2008). Although carbohydrate was not included in the docking experiments, it fits nicely between sIL-22R1 and sIL-10R2 D2 domains, suggesting it might also participate in sIL-22R1/sIL-10R2 site 3 interactions (Fig 4D). Y173sIL-10R2, the only site 3 residue to significantly disrupt sIL-10R2 binding to IL-22/sIL-22R1, buries 89Ǻ2 of surface area and participates in two hydrogen bonds with E168sIL-22R1 and H179sIL-22R1. Other sIL-10R2 site 3 residues tested by mutagenesis (Q119A, H128A, R130A, L132A, K135A) form less extensive site 3 contacts and do not disrupt sIL-10R2 binding.
cmvIL-10/sIL-10R1/sIL-10R2 complex
The cmvIL-10 TC site 2 and site 3 interfaces bury 1,172Ǻ2 and 368Ǻ2 of accessible surface area, respectively (Fig 5). In contrast to the IL-22 TC, sIL-10R2 (L5sIL-10R2) is positioned adjacent to the cmvIL-10/sIL-10R1 interface. As a result, receptor-receptor contacts in the cmvIL-10 TC are more than double (705 Ǻ2) the size of those in the IL-22 TC (315 Ǻ2). The increase is largely due to L5sIL-10R2 residues Y140sIL-10R2 and E141sIL-10R2, which bury 121Ǻ2 and 78Ǻ2 of buried surface area, respectively, into the cmvIL-10 helix A / sIL-10R1 interface (Fig 5C). This structural feature provides an explanation for the importance of Y140sIL-10R2 in cmvIL-10/sIL-10R1 and hIL-10/sIL-10R1 recognition, while it does not contribute to IL-22/sIL-22R1 binding (Fig 3).
There is no protruding knob, corresponding to Y51IL-22, in cmvIL-10. Thus, few contacts are formed between the sIL-10R2 L3/L5 cleft and cmvIL-10. However, more extensive interactions are made between the sIL-10R2 L2/L3 cleft and cmvIL-10 helix D (Fig 5B). The L2/L3 cleft interactions are centered on Y56sIL-10R2, which forms a hydrogen bond with the mainchain oxygen of T89cmvIL-10. Consistent with the structural model, the mutagenesis data demonstrates Y56sIL-10R2 is critical for binding to the cmvIL-10/sIL-10R1 complex, but not to IL-22/sIL-22R1 (Fig 3, Fig S3-S4). Despite differences in the interactions of Y56sIL-10R2 and Y140sIL-10R2, some contacts in the IL-22 and cmvIL-10 TCs are conserved. First, Y82sIL-10R2, located in the center of site 2, packs against R22cmvIL-10, between helices A and D, in essentially the same position as observed in the IL-22TC (Fig 5C). In addition, energetically important binding residues Y59sIL-10R2 and R60sIL-10R2, located at the tip of L2sIL-10R2, form similar contacts in both TCs. Y59sIL-10R2 packs against helix D and R60sIL-10R2 is positioned to form salt bridges with the conserved helix C residues E74cmvIL-10 and E101IL-22 (Figs 4B, 5B).
The amount of surface area buried in the cmvIL-10 TC site 3 (368 Ǻ2) is similar to the IL-22 TC (315Ǻ2). However, sIL-10R1 site 3 residues are donated from three β-strands (C, C’, and E) rather than one (C’) in the IL-22 TC. This is caused by the different D1/D2 inter-domain angle of sIL-10R1 compared to sIL-22R1 (Fig 1B, Fig S2). In contrast to the tightly clustered site 3 residues in the IL-22 TC, site 3 contacts in the cmvIL-10 TC are dispersed over the entire surface with no obvious energetically important interaction to stabilize the complex.
The crystal structure of the cmvIL-10/sIL-10R1 complex revealed cmvIL-10 forms a cysteine-linked dimer that binds two sIL-10R1 chains (Jones et al., 2002). As a result, the cell surface receptor complex presumably consists of two IL-10R1 and two IL-10R2 chains. A model of this complex was generated by superimposing 2 docking models (Fig 5A) onto the dimeric cmvIL-10/sIL-10R1 complex (Fig 5E). In the resulting complex, the twofold-related IL-10R1 and IL-10R2 chains are separated at their C-termini by 107Ǻ and 122Ǻ, respectively, and do not form contacts with one another (e.g. they are independent). The space between the two R1/R2 heterodimers is presumably occupied by the intracellular kinases JAK1 and TYK2, which associate with IL-10R1 and IL-10R2, respectively (Finbloom and Winestock, 1995). For each R1/R2 heterodimer, the spacing between the C-termini of the D2 domains is ~23Ǻ (Table S2). Furthermore, the C-terminus of the IL-10R2 (last residue in D2, T194sIL-10R2) is positioned ~9Ǻ higher, relative to the putative position of the membrane, than the equivalent residue in sIL-10R1 (L205sIL-10R1). Similar D2 asymmetries have been observed in the growth hormone receptor complex, and other ternary complexes (de Vos et al., 1992; Wang et al., 2009). The asymmetry is proposed to be induced by cytokine binding, which provides a structural mechanism by which JAK kinase activation may occur (Brown et al., 2005).
hIL-10/sIL-10R1/sIL-10R2 ternary complex
In contrast to docking studies performed with IL-22/sIL-22R1 and cmvIL-10/sIL-10R1, a high quality docking solution was not obtained using the hIL-10/sIL-10R1 complex. Additional computational studies were performed to try to determine why the docking failed. For example, was it due to subtle conformational differences in sIL-10R1s in the hIL-10/sIL-10R1 and cmvIL-10/sIL-10R1 BCs? To ask this question, sIL-10R1 from the cmvIL-10/sIL-10R1 structure was used to form a hybrid hIL-10/sIL-10R1cmv complex. However, a better docking solution was not obtained using this hybrid complex. In addition, docking studies were performed using an entire hIL-10 dimer and one sIL-10R1 chain to ensure the entire sIL-10R2 binding epitope was included in the experiment. However, these experiments were also unsuccessful.
Despite the inability to obtain a de novo docking solution, structural similarity between cmvIL-10/sIL-10R1 and hIL-10/sIL-10R1 complexes (.e.g. ½ of the dimeric complexes), and similar binding properties of sIL-10R2 mutants for both complexes (Fig 3), suggest sIL-10R2 contacts hIL-10/sIL-10R1 in essentially the same manner as observed in the cmvIL-10 TC. Based on these observations, a “best” hIL-10 TC model was generated by superposition of hIL-10/sIL-10R1 onto the cmvIL-10 TC. Contacts made by sIL-10R2 in this hIL-10 TC model support current and prior hIL-10 mutagenesis studies. In particular, the most important residue identified for the hIL-10/sIL-10R2 interaction is M22hIL-10 (Yoon et al., 2006), which packs against similarly important Y82sIL-10R2 in the hIL-10TC. Additional hIL-10 and sIL-10R2 residues important for sIL-10R2 binding that form putative hydrogen bonds in the hIL-10TC include N21hIL-10/Trp143sIL-10R2, R32hIL-10/S78sIL-10R2, and Ser93hIL-10/Y56sIL-10R2.
Discussion
This study was initiated to understand the molecular mechanisms that allow the sIL-10R2 common chain to promiscuously bind to at least five different class 2 cytokines, which induce diverse cellular activities. The questions we sought to address were: 1) what are the structural features of sIL-10R2 that facilitate promiscuous binding? 2) Does sIL-10R2 form energetically and structurally identical, or distinct, interactions when engaging different binding partners? In addition to determining the molecular basis for promiscuous low affinity protein-protein interactions (14–250µM), for which there is a paucity of literature, these studies also have implications for interpreting IL-10R2 SNPs which cause human disease. In particular, SNPs in sIL-10R2 result in numerous pathologies including early-onset colitis (Glocker et al., 2009) and persistent HBV infection (Frodsham et al., 2006). Several more IL-10R2 SNPs have been identified in patients with inflammatory bowel disease (personal communication, Christoph Klein, Hannover, de), which further argues for the need to understand the structure and mechanism of IL-10R2 binding.
High affinity interactions between hIL-10/sIL-10R1, cmvIL-10/sIL-10R1, and IL-22/sIL-22R1 binary complexes depend on a conserved L2 loop tyrosine, which inserts itself into a cleft between helices A and B on the respective cytokines (Fig 1D). This paradigm is reversed for sIL-10R2, which forms L2/L3 and L3/L5 clefts (Fig 2) that selectively recognize protruding surfaces on helix DcmvIL-10, hIL-10 and helix AIL-22, respectively (Figs 4, 5). Thus, sIL-10R2 site 2 recognition is largely defined by the distinct interhelix angles, between helices A and D, found in the different cytokines (Presnell and Cohen, 1989; Walter, 2004), Figs 4A, 5A). Additional contacts are made by tyrosine, arginine, and glutamate residues located at the tips of L2sIL-10R2 and L5sIL-10R2.
While the sIL-10R2 clefts provide the ability to recognize distinct cytokine features, Y82sIL-10R2 appears to be the common sIL-10R2 epitope required for interaction with each binary receptor complex, much like the conserved L2 tyrosine of sIL-10R1 and sIL-22R1 is highly conserved and required for high affinity IL-22 and IL-10 interactions (Fig 1D). Interestingly, prior structure-function studies on the shared receptors, gp130 and γc, has shown aromatic residues (F169gp130 and Y103γc) form conserved contacts in the center of the IL-6/gp130, IL-2/γc, and IL-4/γc interfaces (Wang et al., 2009). Superposition of the D1 domain of sIL-10R2, with gp130, and γc reveals L3 Y82sIL-10R2 is structurally conserved with F169gp130 and Y103γc (Fig 6A). Furthermore, superposition of IL-6 from the IL-6/IL-6R/gp130 structure and IL-22 from the IL-22/sIL-22R1/sIL-10R2 model, reveals Y82sIL-10R2 and F169gp130 are separated by 4Ǻ (Cα atoms) in their respective complexes (Fig 6B). This result provides additional independent data to confirm the IL-22 TC model (Fig 4), and suggests the structurally conserved L3 aromatic residues (Tyr or Phe) form the critical common binding epitope used by the shared cytokine receptors, gp130, IL-2γc, and IL-10R2, to recognize diverse cytokine surfaces. Mutation of F169gp130 and Y103γc functionally disrupts receptor-ligand interactions and subsequent cell signaling (Kurth et al., 1999; Middleton et al., 1996). Together, these data suggest Y82sIL-10R2 is critical for sIL-10R2 function and hints at a common origin of promiscuous shared cytokine receptors.
Figure 6. Common binding epitope identified between the promiscuous shared cytokine receptors.
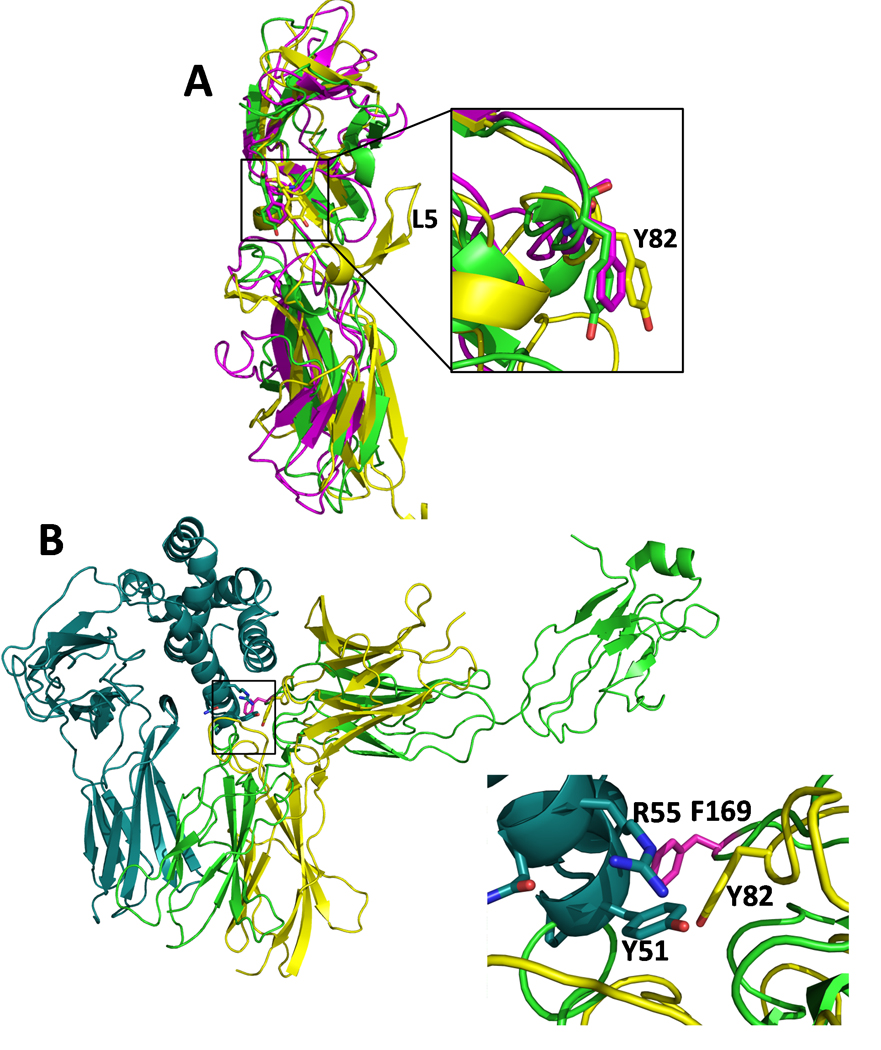
(A) Superposition of sIL-10R2 (yellow), gp130 (magenta), and γc chain (green). The inset shows the structural similarity of L3 loop residues Y82sIL-10R2, F169gp130 (F191gp130 in uniprot database P40189), and Y103γc. EPOR and the growth hormone receptor (GHR) also have aromatic L3 residues (F93EPOR and W104GHR) which have diverged from the structural alignment shown in Figure 6A (See Figure S7). (B) The position of Y82sIL-10R2 and F169gp130 are conserved in their respective ternary complexes. Ribbon diagram of the IL-22TC model (Fig 4) with IL-22/sIL-22R1 colored cyan and sIL-10R2 colored yellow. IL-6 from the IL-6/IL-6R/gp130 complex (pdbid 1P9M, (Boulanger et al., 2003)) was superimposed onto IL-22 from the IL-22TC model (Fig 4) and the position of gp130 (green) is shown. The inset shows IL-22 residues Y51IL-22, and R55IL-22 along with Y82sIL-10R2 (yellow) and F169gp130 (magenta).
In addition, Olosz and Malek (Olosz and Malek, 2000) pointed out Y103γc is structurally conserved with the erythropoietin receptor (EPOR) residue, F93. However, the position of F93EPOR has diverged significantly from the tight cluster of common chain aromatic residues shown in Figure 6A (Fig S7). Nonetheless, it is notable that F93EPOR is found in the interface of the EPO/EPOR complex (Syed et al., 1998) and in the interface of the EPOR dimer structure, which presumably mimics the unbound EPOR cell surface complex (Livnah et al., 1999). In contrast to IL-10R2, gp130, and γc, EPOR, as well as the growth hormone receptor (GHR), form homodimeric receptor complexes (e.g. 1:2 EPO/EPOR or GH/GHR, (de Vos et al., 1992; Syed et al., 1998)). GHR also contains a functionally critical aromatic residue in its L3 chain (W104GHR, (Clackson and Wells, 1995)), but its position has diverged even further away from Y82sIL-10R2 than F93EPOR (Fig S7). This suggests L3 aromatic residues are generally critical for recognition of diverse class 1 and class 2 cytokines. However, EPOR and GHR, which must form high and low affinity binding interfaces, presumably require greater structural diversity in the positions of their aromatic residues (F93EPOR and W104GHR) than the specialized common chain receptors (IL-10R2, gp130, γc), which only form promiscuous low affinity contacts.
Since IL-10R2 activates multiple cytokine complexes, the cellular phenotype of an IL-10R2 SNP can be caused by disrupting the function of one or more cytokines. Thus, we sought to determine if sIL-10R2 mutants will have the same binding phenotype is each complex. Our studies found select sIL-10R2 residues (Y56sIL-10R2, Y140sIL-10R2, and W143sIL-10R2) have distinct energetic functions in the IL-22 and cmvIL-10/hIL-10 TCs (Figs 3, S5). This result suggests specific sIL-10R2 SNPs could selectively disrupt IL-10 or IL-22 signaling and suggests caution should be applied in attributing biological outcomes to specific cytokines. Additional studies will be required to characterize how IL-26 and IL-28/IL-29 are recognized by IL-10R2.
The studies reported here have focused on sIL-10R2 residues directly involved in cytokine and R1 chain recognition. However, other mechanisms that disrupt IL-10R2 function are possible. For example, the K47EsIL-10R2 SNP, which is associated with failure to clear HBV, has been shown to increase IL-10R2 expression levels on cells (Frodsham et al., 2006). Consistent with this interpretation, we found E47sIL-10R2 and K47sIL-10R2 had no impact on sIL-10R2 binding to IL-22/sIL-22R1 (data not shown). From a structural perspective, K47sIL-10R2 is considered the preferred residue since it forms a salt bridge with E96sIL-10R2, which would be disrupted when replaced with a glutamate residue (Fig 1E). How this mutation increases IL-10R2 surface expression remains unknown.
A key feature of IL-10R2 promiscuity is its very low affinity for the cellular cytokine complexes that it activates. This presents technical challenges that have, to date, prevented crystal structure analysis of the class 2 cytokine ternary complexes. To overcome this problem, we used computational docking studies to generate IL-22 and cmvIL-10 ternary complex models. Although not crystal structures, these models provide clear molecular explanations for how sIL-10R2 distinctly recognizes IL-22 and IL-10 binary complexes, which are consistent with experimental data. However, with the current docking parameters, we failed to generate an independent hIL-10 TC model. Interestingly, the success, or failure, of the docking experiments correlates with sIL-10R2 affinity for IL-22/sIL-22R1 (~14µM), cmvIL-10/sIL-10R1 (~80µM), and hIL-10/sIL-10R1 (~250µM) binary complexes (Logsdon et al., 2004; Yoon et al., 2005). This suggests successful sIL-10R2 docking requires a Kd of at least ~100µM using the current docking parameters and restraints.
In summary, our structural, biochemical, modeling, and activity data provide novel insights into the promiscuous binding of the IL-10R2 common chain. In particular, the studies demonstrate that despite being a shared weak-binding receptor, sIL-10R2 residues make distinct energetic and structural contributions to binding. This provides an molecular frame work to characterize additional IL-10R2 SNPs and might be used to design protein therapeutics that restore one or more cytokine activities in patients with defective IL-10R2 signaling complexes.
Experimental Procedures
Expression and purification of sIL-10R2
For crystallization, sIL-10R2, with mutations N49Q, N68Q, N102Q, and N161Q (sIL-10R2NQ) and sIL-10R2NQ C106S/S126C, were expressed in insect cells as C-terminal his6 fusion proteins. Mutagenesis was performed using QuikChange site-directed mutagenesis kit (Stratagene). sIL-10R2 was purified by nickel affinity chromatography and the his6 tag was removed using Factor Xa protease (Novagen). The cleaved protein was further purified by anion exchange chromatography (Poros HQ/H 10/100) and gel filtration chromatography using Superdex 200 HR 10/30 columns.
For selenomethionine incorporation, pET-21a plasmid sIL-10R2NQ C106S was transformed into E. coli strain, B834 (DE3) (Novagen). Cells were grown using M9 SeMet high yield growth medium (Medicilon) at 37°C to OD600 = 0.6, and protein expression was induced with 1 mM IPTG for 8 hours. sIL-10R2NQ C106S inclusion bodies were purified and solubilized as previously described (Yoon and Walter, 2007). Solubilized sIL-10R2NQ C106S was refolded by rapid 10-fold dilution in refolding buffer containing 50 mM NaCl, 100 mM Tris-HCl (pH 8.0), 2.5 mM EDTA, 0.2 mM oxidized glutathione, 2 mM reduced glutathione, and 600 mM arginine and purified as described above.
Crystallization and data collection
sIL-10R2NQ C106S/S126C and SeMet-sIL-10R2NQ C106S proteins were concentrated to 10 mg/ml in 20 mM Tris pH 8.0, 150 mM NaCl and crystallized by hanging drop vapor diffusion methods with 0.95 M Li2SO4, 0.1 M Hepes, pH 7.5. Crystals were placed into cryoprotectant consisting of 20% glycerol, 1.4 M Li2SO4, and 0.1 M Hepes pH 7.5, and flash-frozen under a N2 gas stream at 100° K. X-ray diffraction data were collected at the Advance Photon Source (SER-CAT ID22 beamline). All data were processed with HKL2000 (Otwinowski and Minor, 1997).
Structure determination and refinement
SAD diffraction data was collected on a single crystal of SeMet-sIL-10R2NQ C106S (Crystal 1, Table 1) and SeMet positions were identified using the program SOLVE (Terwilliger and Berendzen, 1999). The resulting phases were combined with structure factor amplitudes obtained from sIL-10R2NQ C106S/S126C crystal (Crystal 2, Table 1) and refined by solvent leveling/flipping using CNS version 1.1 (Abrahams and Leslie, 1996; Brunger et al., 1998). The sIL-10R2 model was built automatically using ArpWarp implemented in the CCP4i suite (Collaborative Computational Project 4, 1994; Lamzin and Wilson, 1993; Perrakis et al., 1997) and refinement was completed using CNS version 1.1 (Brunger et al., 1998).
SPR experiments
All sIL-10R2 proteins used for SPR studies (WT and mutants) contained the same NQ mutations as sIL-10R2-NQ used for structure determination. Each protein was expressed and purified as previously described (Logsdon et al., 2002). SPR data were obtained using a Biacore 2000 as previously described (Logsdon et al., 2004; Yoon et al., 2005; Yoon et al., 2006), except sIL-10R2, sIL-22R1, and sIL-10R1 were simultaneously injected over cytokines amine coupled to CM-5 sensor chips. IFN-τ4 (control surface), IL-22, cmvIL-10, and hIL-10 were coupled at final surface densities of 280–293RU, 220–274RU, 237–291RU, and 601–823RU, respectively. Maximal sIL-10R2 binding, in response units (RU), was determined by subtracting a baseline response obtained by injecting 1µM sIL-10R1 and 500nM sIL-22R1 over the chip surface (Fig S3). Additional injections contained 1µM sIL-10R1, 500nM sIL-22R1, and 150µM sIL-10R2 or sIL-10R2 mutant (Supplemental Experimental Procedures, Figs S3-S4). An additional 25µM sIL-10R2 concentration was also tested with equivalent results (Fig S5). Absolute RUs were normalized to wild-type sIL-10R2. All sIL-10R2 mutants were tested at least twice. The error in the assay was determined by comparing 10 different wild-type sIL-10R2 experiments and also by evaluating replicates of the sIL-10R2 mutants. Both analyses revealed equivalent overall errors of 7%, 10%, and 23% for IL-22, cmvIL-10, and hIL-10 chip surfaces, respectively.
HADDOCK docking experiments
Docking studies were performed using the program HADDOCK (Dominguez et al., 2003) running on the UAB Cheaha computing grid (http://docs.uabgrid.uab.edu/wiki/Cheaha). Experiments consisted of rigid body docking of 1000 possible solutions evaluated using 180° sampling. The top 200 solutions were subjected to semi-flexible simulated annealing using CNS for atoms within 5Ǻ of the interfaces (van Dijk et al., 2006). Finally, flexible explicit solvent refinement was performed to provide a final ranking of the solutions (van Dijk and Bonvin, 2006). Default energetic, refinement, and scoring parameters were used for all runs. Ambiguous interaction restraints (AIRs, Table S1) were chosen based on hIL-10 (Yoon et al., 2006), IL-22 (Logsdon et al., 2004; Wolk et al., 2004; Wu et al., 2008), and sIL-10R2 mutagenesis studies (Fig 3). Unambiguous restraints were applied to each Cα atom in the binary IL-22/sIL-22R1, hIL-10/sIL-10R1, and cmvIL-10/sIL-10R1 complexes resulting in 342 to 349 additional restraints. Solutions were clustered using an r.m.s.d. cut off of 7.5Ǻ and evaluated graphically with PYMOL (DeLano, 2002). Buried surface areas were calculated with NACCESS (Hubbard and Thorton, 1993). Superpositions were performed by STAMP (Russell and Barton, 1992) as implemented in VMD (Humphrey et al., 1996).
Stat3 Activity Assay
Human hepatoma HepG2 cells (ATCC, Manassas, VA) were cultured in serum-free EMEM medium overnight followed by stimulation with IL-22 or IL-22 mutants (10 ng/ml) for 30 minutes. Cells were then washed with cold phosphate buffered saline, lysed, and protein extracts (30 µg) were prepared and used for Western blot analyses using anti-phospho-STAT3 (Tyr705) antibodies (Cell Signaling Technology, Beverly, MA).
Highlights
The crystal structure of the IL-10R2 is distinct from IL-10R1 and IL-22R1 chains.
IL-10R2 residues required for binding multiple cytokines have been identified.
Docking studies provide structures of the promiscuous recognition paradigm.
Class 1 and Class 2 receptor common chains exhibit conserved binding epitopes.
Supplementary Material
Acknowledgments
Use of the Advance Photon Source (SER-CAT, ID-22) was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. This work was funded by grants AI043700 and AI043700-08S1 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The coordinates for the structure of IL-10R2 have been deposited in the protein databank with the accession code 3LQM.
Supplemental Data
Supplemental data includes seven figures and four tables.
References
- Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 2007;15:143–146. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985–2992. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. 2005;12:814–821. doi: 10.1038/nsmb977. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project 4, C. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dambacher J, Beigel F, Zitzmann K, De Toni EN, Goke B, Diepolder HM, Auernhammer CJ, Brand S. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–1217. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- Ding Y, Qin L, Kotenko SV, Pestka S, Bromberg JS. A single amino acid determines the immunostimulatory activity of interleukin 10. J Exp Med. 2000;191:213–224. doi: 10.1084/jem.191.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004 doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- Dumoutier L, Leemans C, Lejeune D, Kotenko SV, Renauld JC. Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J Immunol. 2001;167:3545–3549. doi: 10.4049/jimmunol.167.7.3545. [DOI] [PubMed] [Google Scholar]
- Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–1090. [PubMed] [Google Scholar]
- Frodsham AJ, Zhang L, Dumpis U, Taib NA, Best S, Durham A, Hennig BJ, Hellier S, Knapp S, Wright M, et al. Class II cytokine receptor gene cluster is a major locus for hepatitis B persistence. Proc Natl Acad Sci U S A. 2006;103:9148–9153. doi: 10.1073/pnas.0602800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikami K, Ehara Y, Hasegawa M, Fujimoto M, Matsushita M, Oka T, Takehara K, Sato S, Tokunaga K, Tsuchiya N. Association of IL-10 receptor 2 (IL10RB) SNP with systemic sclerosis. Biochem Biophys Res Commun. 2008;373:403–407. doi: 10.1016/j.bbrc.2008.06.054. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thorton JM. Depart. Biochem., Molec. Biol. London: University College; 1993. NACCESS Computer Program. [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci U S A. 2002;99:9404–9409. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–1344. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta ) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J Biol Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. Embo J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Horsten U, Pflanz S, Dahmen H, Kuster A, Grotzinger J, Heinrich PC, Muller-Newen G. Activation of the signal transducer glycoprotein 130 by both IL-6 and IL-11 requires two distinct binding epitopes. J Immunol. 1999;162:1480–1487. [PubMed] [Google Scholar]
- Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Storer B, Martin PJ, Tseng LH, Grogan B, Chen PJ, Zhao LP, Hansen JA. Genetic variation in the IL-10 pathway modulates severity of acute graft-versus-host disease following hematopoietic cell transplantation: synergism between IL-10 genotype of patient and IL-10 receptor beta genotype of donor. Blood. 2005;106:3995–4001. doi: 10.1182/blood-2004-11-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- Logsdon NJ, Jones BC, Allman JC, Izotova L, Schwartz B, Pestka S, Walter MR. The IL-10R2 binding hot spot on IL-22 is located on the N-terminal helix and is dependent on N-linked glycosylation. J Mol Biol. 2004;342:503–514. doi: 10.1016/j.jmb.2004.07.069. [DOI] [PubMed] [Google Scholar]
- Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res. 2002;22:1099–1112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- Middleton SA, Johnson DL, Jin R, McMahon FJ, Collins A, Tullai J, Gruninger RH, Jolliffe LK, Mulcahy LS. Identification of a critical ligand binding determinant of the human erythropoietin receptor. Evidence for common ligand binding motifs in the cytokine receptor family. J Biol Chem. 1996;271:14045–14054. doi: 10.1074/jbc.271.24.14045. [DOI] [PubMed] [Google Scholar]
- Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Olosz F, Malek TR. Three loops of the common gamma chain ectodomain required for the binding of interleukin-2 and interleukin-7. J Biol Chem. 2000;275:30100–30105. doi: 10.1074/jbc.M004976200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, editors. Processing x-ray diffraction data collected in oscillation mode. Pasadena, CA: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- Pletnev S, Magracheva E, Wlodawer A, Zdanov A. A model of the ternary complex of interleukin-10 with its soluble receptors. BMC Struct Biol. 2005;5:10. doi: 10.1186/1472-6807-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell SR, Cohen FE. Topological distribution of four-alpha-helix bundles. Proc Natl Acad Sci U S A. 1989;86:6592–6596. doi: 10.1073/pnas.86.17.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001;9:86–92. doi: 10.1016/s0966-842x(00)01919-3. [DOI] [PubMed] [Google Scholar]
- Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Baurin VV, Lewis-Antes A, Shah NK, Smirnov SV, Anantha S, Dickensheets H, Dumoutier L, Renauld JC, Zdanov A, et al. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J Immunol. 2004;172:2006–2010. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Sivula J, Turpeinen H, Volin L, Partanen J. Association of IL-10 and IL-10Rbeta gene polymorphisms with graft-versus-host disease after haematopoietic stem cell transplantation from an HLA-identical sibling donor. BMC Immunol. 2009;10:24. doi: 10.1186/1471-2172-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83:9618–9629. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, et al. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk AD, Bonvin AM. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22:2340–2347. doi: 10.1093/bioinformatics/btl395. [DOI] [PubMed] [Google Scholar]
- van Dijk M, van Dijk AD, Hsu V, Boelens R, Bonvin AM. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34:3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MR. Structural analysis of IL-10 and Type I interferon family members and their complexes with receptor. Adv Protein Chem. 2004;68:171–223. doi: 10.1016/S0065-3233(04)68006-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K, Witte E, Reineke U, Witte K, Friedrich M, Sterry W, Asadullah K, Volk HD, Sabat R. Is there an interaction between interleukin-10 and interleukin-22? Genes Immun. 2004 doi: 10.1038/sj.gene.6364144. [DOI] [PubMed] [Google Scholar]
- Wu PW, Li J, Kodangattil SR, Luxenberg DP, Bennett F, Martino M, Collins M, Dunussi-Joannopoulos K, Gill DS, Wolfman NM, Fouser LA. IL-22R, IL-10R2, and IL-22BP binding sites are topologically juxtaposed on adjacent and overlapping surfaces of IL-22. J Mol Biol. 2008;382:1168–1183. doi: 10.1016/j.jmb.2008.07.046. [DOI] [PubMed] [Google Scholar]
- Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Jones BC, Logsdon NJ, Walter MR. Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure. 2005;13:551–564. doi: 10.1016/j.str.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Logsdon NJ, Sheikh F, Donnelly RP, Walter MR. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J Biol Chem. 2006;281:35088–35096. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Walter MR. Identification and characterization of a +1 frameshift observed during the expression of Epstein-Barr virus IL-10 in Escherichia coli. Protein Expr Purif. 2007;53:132–137. doi: 10.1016/j.pep.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


