Abstract
To understand how adherent cells regulate traction forces on their surrounding extracellular matrix (ECM), quantitative techniques are needed to measure forces at the cell–ECM interface. Microcontact printing is used to create a substrate of 1 μm diameter circles of ECM ligand to experimentally study the reconstruction of traction stresses at constrained, point-like focal adhesions. Traction reconstruction with point forces (TRPF) and Fourier transform traction cytometry (FTTC) are used to calculate the traction forces and stress field, respectively, at isolated adhesions. We find that the stress field calculated with FTTC peaks near the center of individual adhesions but propagates several microns beyond the adhesion location. We find the optimal set of FTTC parameters that yield the highest stress magnitude, minimizing information lost from over-smoothing and sampling of the displacement or stress field. A positive correlation between the TRPF and FTTC measurements exists, but integrating the FTTC stress field over the adhesion area yields only a small fraction of the force calculated by TRPF. An effective area similar to that defined by the width of the stress distribution measured with FTTC is required to reconcile these measurements. These measurements set bounds on the spatial resolution and precision of FTTC measurements on micron-sized adhesions.
1. Introduction
Cells exert traction forces on the extracellular matrix (ECM) to which they are adhered through integrin-mediated focal adhesions [1–4]. Spatiotemporal regulation of traction forces is crucial in cell adhesion, migration, division and remodeling of the surrounding ECM [5–12]. For quantitative measurements of the direction and magnitude of cellular traction force, biophysical techniques are required [1, 13, 14]. Optimally these techniques would be able to probe length scales smaller than an individual focal adhesion (~0.5 μm) and as large as the size of an entire cell (~30 μm) and have the capability of resolving forces in the nano-Newton regime.
A variety of methods have been developed to probe cellular traction forces [15]. These methods have largely involved developing calibrated, compliant substrates to which adherent cells exert surface stresses. By imaging the resultant deformations and using the physical characteristics appropriate to each substrate, the cell-generated traction forces can be calculated. In some methods, these substrates consist of an array of discrete, isolated force sensors made from hard or soft micro-fabrication techniques [14, 16–19]. Since adhesions are attached to discrete locations, the spatial resolution of the force measurement is well defined by the geometry of the sensor array. However, this geometry also puts geometrical constraints on cellular adhesions and introduces topographical cues that may alter native cellular physiology [18].
Other methods use continuous gels, comprised typically of either polyacrylamide (PAA) or polydimethylsiloxane (PDMS) embedded with fiducial markers (e.g. fluorescent beads) to visualize the gel displacement field induced by cellular traction. Computational routines are then utilized to convert the displacements of the gel surface to estimate the magnitude and direction of cell-induced traction stresses. The boundary element method (BEM) [13] and Fourier transform traction cytometry (FTTC) [20] both use the displacement field and solve for the force per unit area on a discrete grid; these methods have been shown to yield similar results if the appropriate regularization scheme is used [21]. In current implementations, FTTC is computationally more efficient than BEM [21], and thus, can be easily implemented in different laboratories. However, it has been found that both FTTC and BEM methods potentially suffer from underestimating the actual traction force as the size of the adhesion approaches the displacement grid size [21]. A third method, traction reconstruction with point forces (TRPF), calculates forces at discrete, user-specified points of adhesion. Thus, TRPF requires knowledge of the location of focal adhesions but is not significantly impacted by the density of displacement vectors [1, 21].
Traction force microscopy methods are widely used to assess the changes in the average traction force exerted by individual cells under different pharmacological inhibitors or genetic perturbations [22–24]. Sub-cellular variations in traction stresses can also be qualitatively assessed by FTTC or BEM methods, with spatial resolutions of 5–10 μm easily resolved by these methods [2, 20, 25]. The high density of focal adhesions in certain regions of cells, however, can prohibit measurements of single focal adhesions. Measurements of the forces exerted by individual focal adhesions have been made with several groups using either TRPF or using methods with discretized force sensors [1, 14, 26]. Recent improvements to FTTC have improved the spatial resolution of this method to several microns [21] but experimental validation of consistency between FTCC and TRPF to resolve traction stresses at micron-sized focal adhesions is currently unknown. Knowledge of how these methods can be optimized to identify the location and magnitude of traction force vectors is essential for improving the resolution of traction force methodologies that can be easily implemented to resolve stresses at individual focal adhesions.
In this paper we explore the spatial resolution and consistency in the calculation of traction forces exerted at micron-sized adhesions using FTTC, and compare the results with those obtained using TRPF. To obtain a homogeneous distribution of small (1 μm2), spatially distinct adhesions, we used microcontact printing to control spatial organization of fibronectin on a polyacrylamide hydrogel. The stress field across individual adhesions was calculated by FTTC and we determined how the magnitude and the distribution of the traction stress were affected by the density of sampling of the substrate displacement and degree of smoothing used in the force reconstruction. We then compared these optimized FTTC measurements against forces calculated with TRPF to assess how the force extraction methods can be utilized for the best spatial resolution and the most accurate measurement of traction forces of small focal adhesions.
2. Methods
2.1. Cell culture
U2OS human osteosarcoma cells (ATCC) were cultured in McCoy’s 5A medium (Sigma) supplemented with 10% fetal bovine serum (HyClone), 2 mM L-glutamine (Gibco) and penicillin–streptomycin (Gibco). Cells were transfected with plasmid DNA constructs encoding for GFP-actin (gift of the Gary Borisy Lab, Northwestern University) and mApple-paxillin (gift of the Mike Davidson Lab, Florida State University) using FuGENE 6 (Roche) transfection reagent. After 24 h, cells were re-plated onto polyacrylamide gels coupled to glass coverslips for 16–20 h. Coverslips were then mounted in a Warner Perfusion Chamber (Warner Instruments) and imaged in McCoy’s culture medium supplemented with 10 mM HEPES and 30 μl ml−1 Oxyrase (Oxyrase, Inc.).
2.2. Polyacrylamide substrates for traction force microscopy
Polyacrylamide (PAA) substrates containing far red 40 nm fluorescent microbeads were prepared on coverslips using previously published methods [21]. The PAA gel consisted of 7.5% acrylamide cross-linked with 0.1% bis-acrylamide, resulting in an elastic gel with a shear elastic modulus of 2.8 kPa [27]. Previous work has shown that spatial resolution of traction stresses can be improved with the addition of two-colored beads [21]; here, to facilitate multi-spectral imaging of the F-actin and focal adhesions, we have chosen to use one color of bead. Thus, results reported here could be optimized further by choosing to add another bead color to enhance density of displacement vectors.
Microcontact printing of PAA gels was performed as described previously [28]. Briefly, the PAA gel surface was activated by incubating in pure hydrazine hydrate (Sigma) overnight for 16 h, followed by a 1 h incubation in 5% acetic acid (Fisher Scientific), and a 1 h wash in double-distilled water. A 10 μg ml−1 Fibronectin (Sigma) solution was prepared in sodium acetate buffer (pH 4.5). Oxidization of carbohydrate groups on the fibronectin was achieved by addition of sodium meta-periodate (Thermo Scientific) to the fibronectin solution for a final concentration of 40 μg ml−1 and incubation at room temperature for 30 min. To make a gel with a uniform coating, this oxidized fibronectin solution was pipetted directly onto the gel surface and incubated for 1 h. After the incubation the gel was thoroughly washed in phosphate-buffered saline (PBS) and sterilized under the UV-lamp of a tissue culture hood. For microcontact printing, the top surface of a PDMS stamp containing 1 μm circular features separated by 2 μm (gift of Kweku Addae-Mensah, Columbia University) was immersed in the fibronectin solution for 1 h, after which the stamp was dried with a stream of lab air. Excess water was removed from the PAA gel using a home-built coverslip spinner and the PDMS stamp was pressed gently onto the gel for 90 s. The PAA gel-coated coverslip was then washed thoroughly in PBS and sterilized.
2.3. Immunofluorescence
Cells were washed briefly in cytoskeletal buffer, fixed in 4% paraformaldehyde (Electron Microscopy Sciences) and permeabilized in 0.5% Triton X-100 (Fisher). The fixed and permeabilized cells were then incubated for 1 h in a 1.5% BSA in PBS solution containing 1:400 Alexa Fluor 488 labeled phalloidin, 1:400 polyclonal (rabbit) anti-fibronectin antibody (Sigma), and 1:400 monoclonal (mouse) anti-vinculin antibody (Sigma). After washing in PBS, cells were incubated in a secondary solution containing Cy5 labeled anti-mouse antibody (Jackson Laboratories) and Alexa 560 labeled anti-rabbit antibody to enable visualization of the primary antibodies. After a final wash, the coverslips were mounted on slides using ProLong Gold anti-fade reagent (Invitrogen).
2.4. Live cell imaging
Cells adhered to PAA substrates were imaged in a perfusion chamber at 37 °C on a multi-spectral spinning disk confocal microscope consisting of a Ti–E microscope body (Nikon), 60× 1.2 NA Plan Apo WI objective (Nikon), CSU-X confocal scanner (Yokogawa) and an HQ2 cooled CCD camera (Roper Scientific) controlled with Metamorph acquisition software (MDS Analytical Technologies). After imaging, 0.05% trypsin (Gibco) was perfused into the cell chamber to detach cells from the PAA substrate and an image of the cell-free bead positions were obtained for analysis.
2.5. Displacement analysis
Images of fluorescent beads embedded in the polyacrylamide gel were aligned to compensate for experimental drift and the bead displacement field was calculated between pairs of images comparing the bead images obtained after the cell had been removed to images obtained with an attached cell. Displacement fields were calculated using particle imaging velocimetry (PIV) software in Matlab (available at http://www.oceanwave.jp/softwares/mpiv/), using the minimum quadratic differences (MQD) algorithm which calculates the shift necessary to produce the minimum cross-correlation coefficient between a small region of the experiment image and the reference image. The software then uses recursive super-resolution PIV, which iteratively computes the displacement in a smaller grid spacing using information from the previous computations to filter noisy and spurious displacement vectors. Displacement vectors were filtered and interpolated using the kriging interpolation method. In this work we use four displacement grid sizes, ranging from 0.43 to 3.72 μm.
2.6. Force reconstruction
With the displacement data, FTTC and TRPF were then used to estimate traction stress and force at focal adhesions [21]. The FTTC method uses a discrete computational mesh to compute the stress as a function of position on the image. This method solves the problem of reconstructing the stress from given displacement data by assuming the substrate is a infinite elastic half-space and converting the problem into Fourier space, where it is more easily solved. The solution is then transformed back to real space. This method makes use of a zeroth order Tikhonov regularization scheme which minimizes χ2 of |G⃡ F⃗ − u⃗|2 under the condition that the computed force not grow too large:
In the above equation, F⃗ and u⃗ are vectors representing the force solution and displacement data, respectively; G⃡ is a matrix computed from the Green’s function and relates u⃗ to F⃗. The parameter λ2, the regularization parameter, governs the relative importance placed on agreeing with the input displacement data (the first term) and regularizing the solution (the second term). Regularization effectively constrains the amplitude of the solution, thus reducing the contribution from higher frequencies (presumed to be mostly noise) to the traction stress field [29]. The regularization parameter was varied from 10−8 to 10−1.
The TRPF method relies on user-supplied points where high traction force is expected, identified by a fluorescently tagged focal adhesion protein [29]. The traction force at each point is computed assuming the displacement field arose only from forces at the given points. The same regularization scheme as in FTTC is also used here to suppress noise-dominated high frequency contributions to the result. A criterion for choosing this regularization parameter is the L-curve, which plots |F⃗| as a function of |G⃡F⃗ − u⃗| to show the most reasonable trade-off between smooth data and faithful reconstruction of the forces [30].
Areas of focal adhesions were measured using intensity-based thresholding of GFP-paxillin images. Profiles of FTTC data were found by interpolating the grid of traction stress vectors onto a line scan across individual adhesions using a Gaussian weight function.
3. Results
3.1. Micro-patterned surface induces homogeneous distribution of small, well-separated focal adhesions
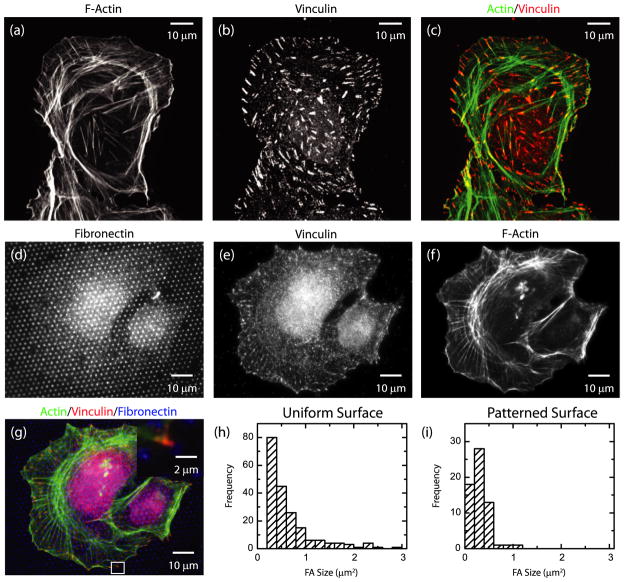
When U2OS osteosarcoma cells were plated on substrates uniformly coated with fibronectin, F-actin bundles that terminate in focal adhesions, visualized by immunofluorescence of vinculin, were observed throughout the cell body (figures 1(a)–(c)). The distance between focal adhesions was quite variable, with some regions near the cell periphery containing numerous focal adhesions separated by less than 1 μm. Moreover, the distribution of focal adhesion size was also broad, ranging from less than 0.5 to greater than 3 μm2 (figure 1(h)). Both the variation in focal adhesion size and the proximity of neighboring focal adhesions make it difficult to isolate forces from single focal adhesions.
Figure 1.
(a)–(c) Immunofluorescence image of a U2OS cell plated on a substrate uniformly coated with fibronectin. (a) F-actin; (b) vinculin; (c) color combine with F-actin in green and vinculin in red. (d)–(g) Immunofluorescence image of a U2OS cell plated on a substrate micro-patterned with fibronectin. (d) Fibronectin; (e) vinculin; (f) F-actin; (g) color combine with actin in green, vinculin in red, fibronectin in blue. Inset: magnified image of region indicated by white box. (h) Histogram of focal adhesion areas on a uniform substrate. (i) Histogram of focal adhesion areas on a patterned substrate as in (d).
To control the spacing and size of focal adhesions, cells were plated on polyacrylamide gels on which a patterned array of 1 μm diameter filled circles of fibronectin separated by 2 μm had been formed using microcontact printing (figure 1(d)). U2OS cells spread on these substrates and F-actin bundles terminated at vinculin-rich focal adhesions that co-localized with fibronectin circles (figures 1(e)–(g)). On these micro-patterned surfaces, focal adhesion size was tightly distributed around an area approximately 0.5 μm2 (figure 1(i)). On the patterned substrates, focal adhesions were separated by several microns such that contributions of traction stress from individual focal adhesions could be identified. Thus, utilization of micro-patterned substrates facilitated our ability to obtain a large population of well-separated focal adhesions with a uniform size distribution.
3.2. Traction force microscopy of individual focal adhesions
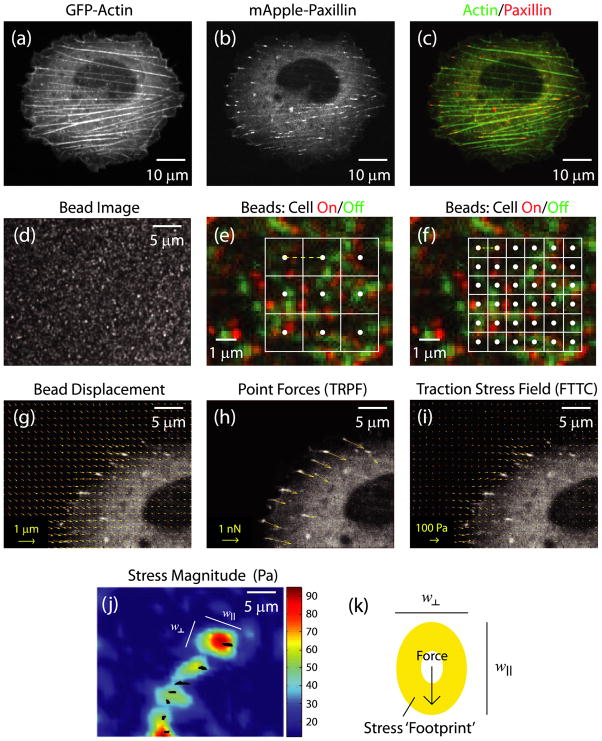
For traction force microscopy experiments, U2OS cells transfected with cDNA plasmids for GFP-actin and mApple-paxillin were plated on the polyacrylamide gels containing far red beads and coated with a micro-pattern of fibronectin. After 18 h, actin, paxillin and beads were imaged using a spinning disk confocal microscope (figures 2(a)–(d)). After image acquisition, cells were detached from the gel surface to obtain a reference bead image. The typical displacements in regions of high traction were on the order of 8–12 pixels. By color combining the reference bead image (green) to a bead image with the cell-on (red) the typical gel deformations can be visualized (figures 2(e) and (f)). To calculate displacement, a small region is selected in the reference image and the displacement which maximizes a cross-correlation to the ‘cell-on’ image is determined; typical sizes of square regions used in cross-correlation analysis are shown by white boxes in figures 2(e) and (f). By performing this across the entire image, a displacement field of the beads between the ‘cell-on’ and reference image is obtained (figure 2(g)). The density of the beads at the top surface of the gel was sufficient to allow the gel displacement field to be computed with a minimal spacing of displacement vectors as small as 0.43 μm (grid size indicated by yellow dashed line, figures 2(e) and (f)). The substrate displacement grid size was increased incrementally up to 3.42 μm by changing parameters in the PIV software.
Figure 2.
Live cell image of a U2OS cell expressing (a) GFP-actin and (b) mApple-paxillin. (c) Color combine with actin in green, paxillin in red. (d) Image of 40 nm far red fluorescent beads used as fiducial markers to track substrate displacements. (e) and (f) Magnified view of a small region of a bead image with the beads from the ‘cell-off’ image in green and beads from the ‘cell-on’ image in red. Overlaid are grids with a spacing of 0.86 μm (e) and 1.71 μm (f). The white dots at the center of each box indicate the origin of the displacement vector and the yellow dashed line represents the grid size. (g)–(j) Magnified image of upper left part of the cell shown in (a)–(c) overlaid with (g) bead displacement vectors, (h) point force vectors using the TRPF method and (i) stress vector field from FTTC method. (j) Heat map of stress magnitude using data from (i). The black marks indicate the location of focal adhesions. The two lengths (w|| and w⊥) are defined as the principle axes of the ellipsoidal area subtended by the region around focal adhesions where the traction stress is larger than the background. w|| indicates the length of the axes in the direction parallel to the traction stress vector, while w⊥ indicates the length of the axes in the direction perpendicular to the application of stress. (k) Cartoon of a focal adhesion (white circle) applying a force to the substrate, and the resultant stress footprint from the FTTC method.
The forces at the focal adhesions were calculated using TRPF assuming that the displacement field results from forces only at locations of individual focal adhesions. Here, a regularization parameter was chosen to maximize the force magnitude but minimize the noise in force direction (data not shown). The forces exerted at the adhesions were directed towards the cell interior and ranged in magnitude from 1 to 5 nN (figure 2(h)), comparable in magnitude with forces measured with other discretized force measurements [1, 14].
The traction stress vector field was calculated from the displacement field by using FTTC [21]. The resulting stress vector field for a gel displacement grid size of 1.28 μm and regularization parameter of 7 × 10−5 is shown in figure 2(i), with the corresponding magnitudes of the stress vectors shown as a heat map in figure 2(j). Similar to the TRPF method, the direction of the stress vectors points towards the cell center. The stress magnitudes range from less than 20 Pa approximately 5 μm away from focal adhesion sites to 40–90 Pa at adhesion sites (figure 2(j)). Noticeably, the stress vector field does not drop down to background immediately outside of the paxillin-rich focal adhesions, identified by the blackened regions in figure 2(j). Instead, a larger stress ‘footprint’ appears, with a diameter larger than the physical size of the focal adhesion (figures 2(j) and (k)). The two lengths (w|| and w⊥) are defined as the principle axes of the ellipsoidal area subtended by the region around focal adhesions where the traction stress is larger than the background. The length, w||, indicates the length of the axes in the direction parallel to the traction stress vector, while w⊥ indicates the length of the axes in the direction perpendicular to the application of stress.
If the focal adhesion is the only location of force transmission between the cell and the substrate, this result is surprising. While the displacement field of a point force is de-localized, ideally the traction force reconstruction routine would localize the origin of the point force. This feature provides motivation to understand and characterize the behavior of the FTTC force reconstruction technique.
3.3. Displacement grid size and smoothing parameter strongly affect peak traction stress
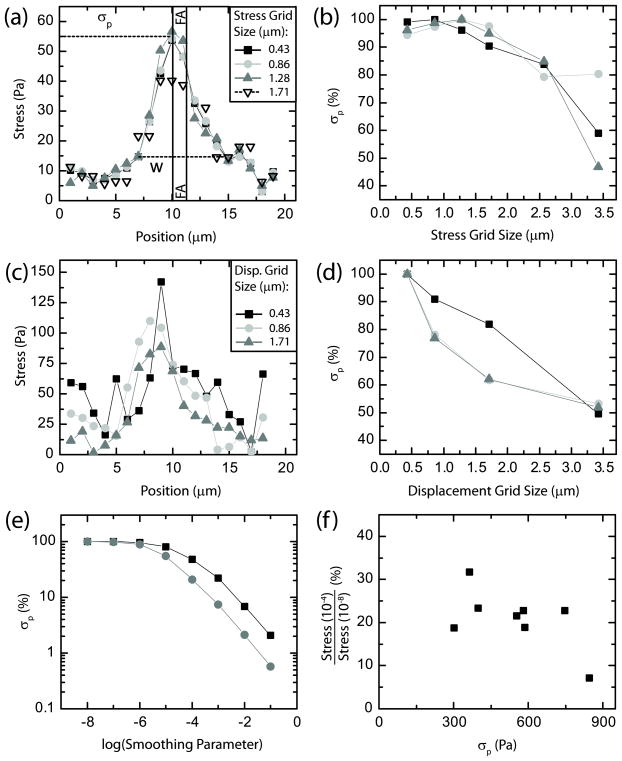
To assess the traction stress profile across individual adhesions, stresses were interpolated onto a line spanning the adhesion in the direction of traction stress. The stress profile across individual adhesions was Gaussian like, with the peak stress, σp, co-localized very closely to the physical location of the adhesion (black vertical lines), as shown in figure 3(a). The stress decays to background levels (~20 Pa) approximately 3 μm away from the peak. This distance sets a reasonable length scale for the ability to resolve neighboring adhesions. Previous work with very similar FTTC routines, but displacement data approximately fourfold denser, demonstrated a theoretical spatial resolution of 1 μm [21]. Interestingly, the full width, w, of the stress distribution was significantly larger, 7 μm, than the adhesion size of 1 μm (figure 3(a)). Thus, the peak stress determined by FTTC has a precision to the center of the adhesion sites on the order of 1 μm and has the ability to resolve focal adhesions spaced <4 μm apart. The traction stress distribution, however, is broadened over a larger area than the physical size of focal adhesions.
Figure 3.
(a) Traction stress as a function of position across a single focal adhesion for four different stress grid sizes. Lines on the plot indicate the peak stress (σp) and the distribution width (w). The vertical lines indicate the boundaries of the focal adhesion as determined by the fluorescence image of GFP-paxillin. Displacement grid size: 1.71 μm; smoothing parameter: 7 × 10−5. (b) Peak stress plotted as a function of stress grid size for three different focal adhesions. The measured peak stress decreases as the grid spacing increases. Displacement grid size: 1.71 μm; smoothing parameter: 7 × 10−5. (c) Traction stress as a function of position across a focal adhesion varying the displacement grid size. Stress grid size: 1.28 μm; smoothing parameter: 7 × 10−5. (d) Peak stress plotted as a function of displacement grid size for three different focal adhesions. Stress grid size: 1.28 μm; smoothing parameter: 7 × 10−5. (e) Peak stress plotted as a function of regularization (smoothing) parameter for two groups of focal adhesions, normalized and then averaged. Displacement grid size of 1.71 μm (black squares) and 0.86 μm (dark gray circles) are shown. Displacement grid size: 1.71 μm; stress grid size: 1.28 μm. (f) The ratio of the peak stresses measured with a smoothing parameter of 10−4 to those measured with a smoothing parameter of 10−8 are plotted against the peak stress. Displacement grid size is 0.86 μm.
To determine how parameters used in reconstruction the traction stress field impact σp and w, we systemically changed the stress grid size, the displacement grid size and the regularization parameter. The choices of varying either the displacement or stress grid size were designed to span from near our resolution limit (0.4 μm) to significantly larger than the size of individual focal adhesions. The base width, w, of the stress profile is not impacted as the stress grid size is increased from 0.4 to 1.71 μm (figure 3(a)). The peak stress, σp, measured is only weakly sensitive to stress grid sizes up to 1.7 μm, but rapidly decreases as the stress grid size becomes much larger than the adhesion size (figure 3(b)). Thus, as long as the stress grid size is sufficient such that a grid point is located in close proximity to a focal adhesion, the magnitude of the traction stress calculated is not particularly sensitive to the choice of stress vector spacing.
We also changed the spacing of displacement vectors, thereby increasing the density of displacement vectors as the displacement grid size was decreased. As the displacement grid size was decreased from 1.7 to 0.4 μm, the base width of the stress profile remained relatively constant, but the noise associated with the smallest grid spacing visually increased (figure 3(c)). The peak stress decreased by approximately 50% as the displacement grid size was increased from 0.4 to 3.5 μm (figure 3(d)). The dependence observed is consistent with previous simulations, which predicted that, in order for the calculated stress to be independent of the chosen displacement grid size, sufficient sampling of approximately three displacement vectors per adhesion site was needed [21]. Thus, calculated stresses for micron-sized focal adhesions are highly dependent on the density of displacement vectors.
Previously, we showed that the optimal regularization scheme identified for FTTC calculations is one that minimizes gradients in stress for higher degrees of regularization and choice of smoothing parameter [21]. When the magnitude of the smoothing parameter is less than 10−5, the peak traction is insensitive to further decreases in this parameter (figure 3(e)). However, there is approximately a 50-fold decrease in the peak traction stress calculated as the smoothing parameter is increased from 10−5 to 10−1 (figure 3(e)). The rate of decrease of the peak traction stress is weakly sensitive to the displacement grid size such that data calculated with a higher density of displacement vectors at falls off more rapidly than that obtained at a lower density of displacement vectors (figure 3(e)). This effect is likely to result due to the higher stresses that are calculated with smaller grid spacing.
To explore if a given regularization parameter would differentially impact traction stresses of different magnitudes, the ratio of the traction stresses calculated using two different regularization parameters was computed over a number of adhesions of different magnitudes. No strong correlation was observed for traction stresses ranging from 300 to 600 Pa (figure 3(f)), indicating stresses of different magnitudes within a single cell are impacted similarly for a given choice of displacement grid size and regularization.
As expected, as the regularization parameter decreases, the noise in the stress line scans becomes quite large and eventually dominates the signal (data not shown). The width of the stress profile, however, does not decrease significantly. Thus, while over-smoothing (with a large regularization parameter) can completely mitigate the magnitude of the measured traction stress, some amount of smoothing is necessary to reduce random noise in the measurement. The ideal set of these parameters will produce the most faithful stress reconstruction while limiting noise, or random deviations in the line’s stress profile.
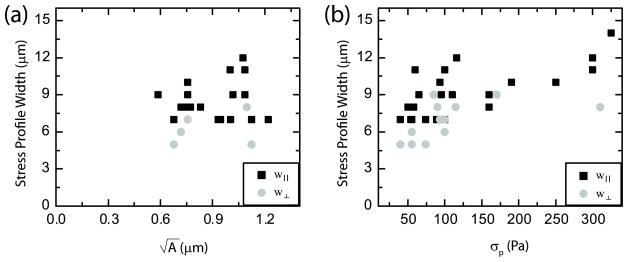
3.4. Stress footprint width proportional to traction stress magnitude
The behavior of the width of the stress profile is also significant, as it directly affects the ability to resolve neighboring focal adhesions. The width of the stress profile was not significantly impacted by modifications to the displacement or stress grid size or the regularization parameter. The width of the stress profile in the direction parallel to the traction stress (w|| in figure 2(k)) is approximately five- to seven-fold larger than the physical size of the adhesion and does not vary significantly with the range of adhesion sizes observed in these experiments (black squares, figure 4(a)). By contrast, w|| increases weakly with peak stress (black squares, figure 4(b)), indicating that spatial resolution decreases slightly as the magnitude of the traction stress increases. Similar behavior is observed with the width of the stress profile perpendicular to the direction of traction (gray circles, figures 4(a) and (b)). As noted previously, the width of the stress profiles is much larger than the length of the focal adhesion, indicating that the reconstructed stress field is broadened as a result of the FTTC calculation.
Figure 4.
The parallel and perpendicular width of the stress distributions, as defined in figures 2(j) and (k), as a function of (a) the square root of focal adhesion area and (b) the peak stress.
Interestingly, the width in the direction parallel to the application of stress is slightly larger than the width normal to the application of stress (figure 4(b)), indicating that the shape of the stress pattern is not perfectly circular but is more ellipsoidal. This broad distribution must be taken into consideration when attempting to resolve stresses from neighboring adhesion sites.
3.5. FTTC underestimates forces, as measured by TRPF, at micron-sized adhesions
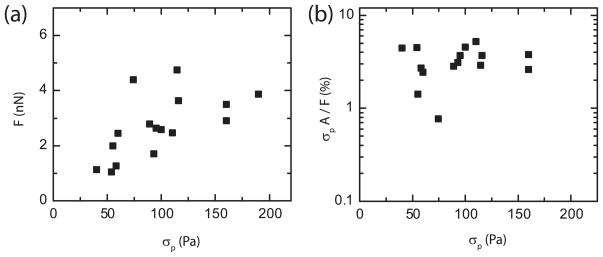
The TRPF method has previously been shown in simulations to precisely measure the correct force magnitude as long as the focal adhesion location is precisely determined [21]. To make a quantitative comparison between TRPF and FTTC, the FTTC measurement was optimized with a displacement grid size of 0.9 μm and a regularization parameter of 10−5. At each focal adhesion, the peak stresses obtained by FTTC, σp, were well correlated to the forces, F, calculated with the TRPF measurement (figure 5(a)). Deviations from this direct behavior were observed at large stresses, where the force measured by TRPF approached a plateau.
Figure 5.
(a) Plot of force, F, as measured with the TRPF method (y-axis) against the peak stress (σp) as measured by the FTTC method. (b) Plot of the ratio between σp multiplied by focal adhesion area, A, and F.
To compare the force magnitude predicted by FTTC results to those obtained with TRPF, we multiplied the peak stress, obtained using optimized FTTC parameters, by the physical area of the focal adhesion. Surprisingly, we measured that the force estimated from the FTTC measurement accounted for only 3% of that measured by TRPF (figure 5(b)).
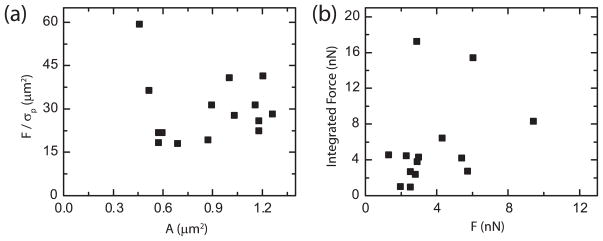
A method to examine this discrepancy is to identify an effective area required to make the FTTC results consistent with the TRPF measurement by dividing the force, as measured by TRPF, by the peak optimized stress, as measured by FTTC. This area, typically on the order of 30 μm2, is approximately 30-fold larger than the physical size of the adhesions (figure 6(a)). The width of the traction stress distribution across single focal adhesions is nearly five-fold larger than the actual length of localized GFP-paxillin intensity, providing a 25-fold scaling factor of area. The correspondence in the area scaling between the focal adhesion and the stress footprint as well as between the TRPF force and FTTC stress measurement implies a possible approach to resolving the discrepancy: integrating the stress profile across the whole area of the stress footprint. Doing so yields good agreement between the two methods, within a factor of two; however, the relationship between the force and the stress integrated over the footprint of an individual focal adhesion level is quite scattered (figure 6(b)) and further experiments and analysis are required to confirm the consistency between these measurements.
Figure 6.
(a) Plot of the force (F), determined by TRPF, divided by the peak stress (σp), determined by FTTC, as a function of focal adhesion area (A). (b) Plot of the integrated force found by integrating the stress distribution determined by the FTTC method against the force as calculated by the TRPF method.
4. Discussion
In this work, we have used microcontact printing in order to spatially control the adhesion of the cell to the substrate. The array of small fibronectin dots enables quantitative analysis of single, isolated, point-like focal adhesions. These micro-patterned substrates provided excellent platforms to fine-tune traction force reconstruction methods and learn how future results for cells on uniform or other patterned substrates might be interpreted. Both computation methods utilized, Fourier transform traction cytometry and traction reconstruction with point forces, are relatively easy to implement and computer time efficient, making them attractive to laboratories working on forces in cell adhesion.
We have addressed how FTTC can be optimized to resolve the stresses exerted at small, micron-sized focal adhesions. The peak traction stress localized to within approximately 1 μm of the centroid of individual adhesions and we estimate the ability to resolve neighboring adhesions is approximately 3 μm. With very similar computational methods, further enhancing bead density has been shown to theoretically improve this resolution to 1 μm [21]. The traction stress magnitude decreases by about 25% as the grid size is increased from approximately half to twice the adhesion size (0.5–2.0 μm). The stress grid size impacts the stress measurement even less, decreasing by approximately 10% across a similar range. Furthermore, the stress grid measurement displays a more random behavior, as measurements can still be relatively high or low if a grid point happens to fall near a point of peak stress; however as the grid grows more sparse this coincidence grows more unlikely.
The regularization parameter displays the most dramatic change of any of the parameters. For low values the stress measurement is insensitive to the magnitude of the smoothing parameter, but at a certain value the peak stress measured begins to fall rapidly. The particular value of the regularization parameter where the stress measurement begins its descent can be altered by choices of strain grid size. Using the largest value of regularization parameter prior to this steep descent in peak stress produces the truest measure of the traction stress while minimizing random noise in the measurement.
Reconciling the FTTC measurement with the measured forces from the TRPF method requires additional analysis as the peak stresses applied across the area of the focal adhesions is not similar to the forces measured by TRPF. Integrating the stress across the area of the stress footprint, approximately 30 μm2, results in the correct scale of the forces, but is still sensitive to choices of regularization parameter and displacement grid size. Furthermore, the significance of the extended stress profile found in the FTTC method remains unclear. In this computational method the extended profile appears as a result of using a finite sampling frequency which cannot capture all the details at the adhesion. Gauging biological significance represents more of a challenge. In general, it is possible that forces are transmitted to the underlying substrate at regions beyond focal adhesion plaques, as imaged by expression of GFP labeled focal adhesion proteins. For example, it has been shown recently by a combination of laser cutting and theoretical analysis of actin stress fiber retraction dynamics that forces are also transmitted to the substrate along stress fibers where focal adhesions are not visualized [31]. Additionally, traction force microscopy on migrating cells shows a significant traction at the base of the lamellipodium, which is associated with diffuse puncta of diffraction-limited focal adhesions [32]. These challenges will be addressed in the future and reveal more interesting biology of how sub-cellular variations in cytoskeletal dynamics and mechanics impact how cells generate traction on their external environment.
Acknowledgments
This work was supported by a NIH Director’s Pioneer Award (DP10D00354) to M L Gardel and the National Science Foundation Materials Research Science and Engineering Center at The University of Chicago (grant No. NSF-DMR-0820054).
References
- 1.Balaban NQ, et al. Nat Cell Biol. 2001;3:466. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 2.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. J Cell Biol. 2001;153:881. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bershadsky AD, Balaban NQ, Geiger B. Annu Rev Cell Dev Biol. 2003;19:677. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 4.Berrier AL, Yamada KM. J Cell Physiol. 2007;213:565. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 5.Pelham RJ, Jr, Wang Y. Proc Natl Acad Sci USA. 1997;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. J Cell Biol. 1998;141:539. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger B, Bershadsky A. Cell. 2002;110:139. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 8.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 9.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 10.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Dev Cell. 2006;10:11. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz U. Soft Matter. 2007;3:263. doi: 10.1039/b606409d. [DOI] [PubMed] [Google Scholar]
- 12.Lemmon CA, Chen CS, Romer LH. Biophys J. 2009;96:729. doi: 10.1016/j.bpj.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dembo M, Oliver T, Ishihara A, Jacobson K. Biophys J. 1996;70:2008. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci USA. 2003;100:1484. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JH, Lin JS. Biomech Model Mechanobiol. 2007;6:361. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 16.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proc Natl Acad Sci USA. 2005;102:2390. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sniadecki NJ, Chen CS. Methods Cell Biol. 2007;83:313. doi: 10.1016/S0091-679X(07)83013-5. [DOI] [PubMed] [Google Scholar]
- 18.Yang MT, Sniadecki NJ, Chen CS. Adv Mater. 2007;19:3119. [Google Scholar]
- 19.Galbraith CG, Sheetz MP. Curr Opin Cell Biol. 1998;10:566. doi: 10.1016/s0955-0674(98)80030-6. [DOI] [PubMed] [Google Scholar]
- 20.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Am J Physiol Cell Physiol. 2002;282:C595. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 21.Sabass B, Gardel ML, Waterman CM, Schwarz US. Biophys J. 2008;94:207. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Y, et al. Biophys J. 2006;91:3907. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paszek MJ, et al. Cancer Cell. 2005;8:241. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Gavara N, Sunyer R, Roca-Cusachs P, Farre R, Rotger M, Navajas D. J Appl Physiol. 2006;101:512. doi: 10.1152/japplphysiol.00185.2006. [DOI] [PubMed] [Google Scholar]
- 25.Beningo KA, Hamao K, Dembo M, Wang YL, Hosoya H. Arch Biochem Biophys. 2006;456:224. doi: 10.1016/j.abb.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. J Cell Biol. 2006;172:259. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung T, et al. Cell Motil Cytoskeleton. 2005;60:24. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 28.Damljanovic V, Lagerholm BC, Jacobson K. Biotechniques. 2005;39:847. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA. Biophys J. 2002;83:1380. doi: 10.1016/S0006-3495(02)73909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen PC. Rank-Deficient and Discrete Ill-Posed Problems: Numerical Aspects of Linear Inversion. Philadelphia: Society for Industrial and Applied Mathematics; 1998. p. 247. [Google Scholar]
- 31.Colombelli J, et al. J Cell Sci. 2009;122:1665. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 32.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. J Cell Biol. 2008;183:999. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]