Abstract
There have been relatively few publications using linear regression models to predict a continuous response based on microarray expression profiles. Standard linear regression methods are problematic when the number of predictor variables exceeds the number of cases. We have evaluated three linear regression algorithms that can be used for the prediction of a continuous response based on high dimensional gene expression data. The three algorithms are the least angle regression (LAR), the least absolute shrinkage and selection operator (LASSO), and the averaged linear regression method (ALM). All methods are tested using simulations based on a real gene expression dataset and analyses of two sets of real gene expression data and using an unbiased complete cross validation approach. Our results show that the LASSO algorithm often provides a model with somewhat lower prediction error than the LAR method, but both of them perform more efficiently than the ALM predictor. We have developed a plug-in for BRB-ArrayTools that implements the LAR and the LASSO algorithms with complete cross-validation.
Keywords: regression model, gene expression, continuous outcome
Background
DNA microarray technology has been proven to be a powerful tool for exploring gene expression patterns in biological systems in the past decade. Many medical applications of microarrays involve class prediction, that is, prediction of a categorical class or phenotype based on the expression profile of the patient. The classes often represent diagnostic categories or binary treatment response. For example, Wang et al1 developed a gene-expression based predictor of whether a patient with advanced melanoma would respond to IL2-based treatment.
Challenges are experienced where the development and validation of predictive models for settings where the number of candidate predictors ( p) is much larger than the number of cases (n). Many algorithms have been studied for developing and evaluating gene-expression-based predictors of a categorical class variable. Classification methods widely used include the compound covariate predictor,2 diagonal linear discriminant analysis,3 nearest neighbor4 and shrunken centroid methods,5 support vector machines,6 and random forests,7 all of which are available in the BRB-ArrayTools software, provided without charge for non commercial purposes by the National Cancer Institute.8 Sophisticated methods of complete cross-validation or bootstrap re-sampling efficiently utilize the data and avoid biased estimates of predictive accuracy.9 Methods for predicting survival risk based on censored survival times and microarray data have been described by several authors and recently compared by Bovelstad et al.10 Methods of complete cross-validation are much less developed for such settings and most published studies involving survival prediction transform the outcome data into discrete categories (see the review by Dupuy & Simon).11
There have been relatively few publications using linear regression models to predict a continuous response based on microarray expression profiles. Standard linear regression methods are problematic when the number of predictor variables exceeds the number of cases because X’X is singular, where X is the design matrix. Software available to biomedical investigators has not included the more sophisticated methods needed for developing and properly validating continuous response models in the p > n setting. One example of such a study is that of Bibikova et al12 who identified a group of 16 genes significantly associated with Gleason scores for prostatic carcinomas. They avoided the p > n problem by first identifying 16 genes which individually appeared predictive of the Gleason score, and then fitting single variable linear regression models for each of the 16 genes. The final predicted Gleason grade for each sample was the average of 16 independently derived predicted values from each model. Although this method has the merit of simplicity, the method of validation they used was problematic and consequently their model requires further validation with an independent data set.
To properly estimate the accuracy of a prediction model, the test set cannot be used for selecting the genes to be included in the model or for estimating the parameters of the model. This key principle of separating the data used for model development from the data used for model validation must be carefully observed in using either a split-sample or cross validation approach of estimating prediction accuracy. The simulation study13 shows the importance of cross validating all steps of model building in estimating the error rate, especially the feature selection step that is often overlooked. Enormous bias in estimation of prediction error can result if the full dataset is used for gene selection and sample splitting or cross-validation applied to fitting a model based on those selected genes. Unfortunately, the survey by Dupuy and Simon11 indicated that improper use of incomplete cross-validation is prevalent in the published literature with class prediction methods and this problem also occurred in the study of Bibikova et al.12
We have evaluated three linear regression algorithms that can be used for prediction of a continuous response based on high dimensional gene expression data. The first two algorithms are Least Angle Regression (LAR)14 and LASSO.15 LASSO is a penalized regression method. It identifies regression coefficients for all genes to minimize a weighted average of mean squared prediction error for cases in the training set plus the sum of absolute values of all regression coefficients. The weighting factor is optimized by cross-validation. LAR can be viewed as an accelerated version of forward stagewise regression.16,17 The algorithm developed by Efron et al14 is highly efficient and can also be used to find the LASSO solution. Both methods develop relatively parsimonious models and do not require the prior step of gene selection. There have been many applications of LASSO in different fields such as on protein mass spectrometry data18 and SNP data.19 To our knowledge, the use of these models with gene expression profiles to predict continuous outcome have not been reported. The third algorithm we evaluated is the averaged linear regression (ALM) method used in Bibikova et al.12 We used an unbiased complete cross validation approach in order to get a correct error estimate for the model. All methods were tested using simulations in which the gene expression levels were based on a real dataset and analysis of two sets of real gene expression data.
Methods
Data sets
We simulated continuous response yi by applying the following formula to the publicly available gene expression dataset of Beer et al,20 which is curated in the BRB-ArrayTools Data Archive.21
| (1) |
where ɛij~N(0, σ2). Various values of noise variance were used for the simulations. The value of σ is calculated from the real data, which is 0.63 on the log 2 scale; xij are the expression levels for sample i (from 1 to 96) in the first ten genes with no missing values. Independent Gaussian noise is added. The data set contains 86 lung cancer samples and 10 normal samples using the Affymetrix HuGeneFL chip with 7129 probe sets. We filtered out probe sets with missing data, so the final data set contained 3501 probe sets.
The first real data set we analyzed with the three algorithms relates gene expression to cytotoxic activity of the anti-cancer agent paclitaxel in lung cancer cell lines.22 The data set contains 29 lung cancer cell lines with 22,282 transcripts (HG-U133 A, Affymetrix, Santa Clara, CA). The drug activity data is measured as growth inhibitory activity GI50. The log 2 based GI50 is used as continuous response. One cell line (H69) is excluded in this study because its GI50 is beyond the detection limit.
The second real data set used to evaluate the algorithms relates gene expression, measured with a DASL array, to the Gleason score of human prostate cancers.12 The data set contains 70 prostate tumor patients with Gleason scores. There are 512 genes on each chip. Since the pre-processing steps in that paper were not described in sufficient detail for our application, we use the summary of Intensity data generated by the Illumina’s BeadArray package as gene measurement input.
Algorithms
LAR and LASSO
For a linear regression model in microarray gene expression data
| (2) |
where Y is the outcome vector with length n, X is an n by p matrix of expression levels of p genes and n samples, β is the regression coefficient vector, and ɛ is the normal noise vector. LASSO is designed to minimize
where ||Y − X β || denotes the sum of squares of residuals, the summation is over the genes j = 1,…,p, and θ is a positive scalar.
LASSO is a regression with an L1 penalty. LAR can be viewed as a version of forward stagewise regression that uses mathematical formulas to accelerate the computation.17 It first selects the predictor most correlated with the response. It brings that predictor into the model only to the extent that it remains most correlated with the response. At each stage, the variable most correlated with the residuals of the current model is included.17
The LARS algorithm builds a sequence of models in a stepwise manner which are indexed in terms of a parameter representing the fraction (f) of algorithmic steps relative to the model containing n genes, where n is the number of samples. The LARS algorithm can also be used to generate a sequence of models containing increasing numbers of variables; each model representing a linear model which is a Lasso solution. That is, the LARS algorithm can be used to generate a sequence of models, each of which minimizes the sum of squared residuals plus a weight times the sum of absolute values of the regression coefficients. The models of the sequence correspond to decreasing values of the weight penalty.
The weighting factors for LAR and LASSO are optimized by cross-validation. The LARS algorithm can be used to generate a sequence of models indexed by a tuning parameter f. For each value of f, a cross-validated estimate of the squared prediction error is obtained for each round of the cross-validation. After the entire sequence of models is built, the final model can be selected based on the f value for which the estimated total squared prediction error is minimized.
We use the ‘LARS’ function with method ‘LASSO’ and ‘LAR’ in the library of the R statistical package. We have implemented it into BRB-ArrayTools8 as a plug-in.
Averaged linear regression predictor (ALM)
The algorithm was proposed in Bibikova et al.12 We implemented their approach but incorporated a complete cross validation step so that model error can be correctly estimated.
With K-fold validation, the samples are randomly partitioned into K (approximately) equal size groups S1,S2,…,SK. One of the K subsets is omitted, say subset k. We fitted a simple linear model for each gene using a training set consisting of samples in the union of the other K-1 subsets, denoted S̄k . For each gene j = 1, …, M (M is the total number of genes), we fitted the univariate linear regression model yij = αkj + βkjxij +ɛij for all samples, i ∈ S̄k . where ɛij are independent Gaussian errors. This provides estimates of the regression parameters α○, β̂, and a significance level for testing the hypothesis βkj = 0 .
Only the variables that have significance levels less than a threshold are selected. The threshold can either be pre-specified or optimized by cross validation within the training set S̄k . The predicted continuous outcome for a sample i* in the omitted subset Sk is the average.
| (3) |
where the summation is over the genes j whose significance levels in the training set S̄k is less than the threshold and mk is the number of such genes. The prediction errors are recorded for the samples in this subset Sk . This is done K times, omitting each of the K subsets one at a time and the errors for the samples in each subset are obtained and totaled into an overall error.
Cross validation
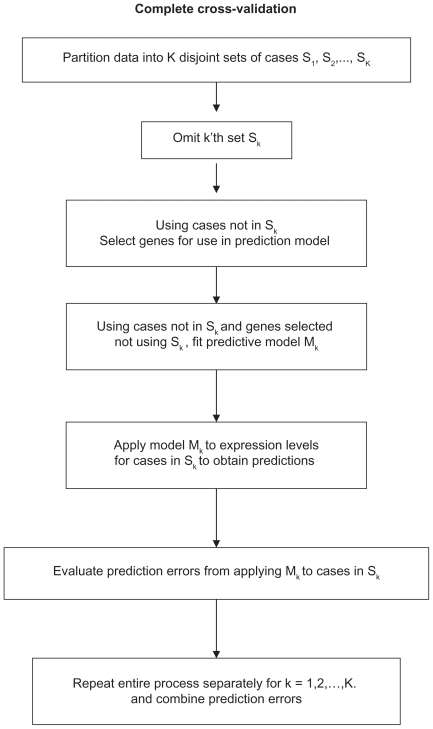
When comparing different regression algorithms, we use 10-fold cross validation. Each time, 10% of the samples are omitted, the model is built using the remaining 90% of the samples. The prediction errors are recorded for the samples withheld. This is done 10 times, omitting each of the 10 subsets one at a time and the errors for the samples in each subset are obtained and totaled into an overall error. The flow chart of the cross validation approach is shown in Figure 1.
Figure 1.
Flow chart of complete cross validation.
Results
Simulated data with no noise
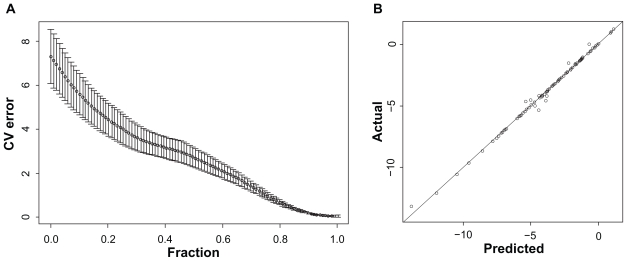
We first evaluated the three linear regression algorithms using simulated data. The simulated response was first generated as described in the methods section with no noise added. Figure 2A shows the relationship between the 10 fold cross-validated estimate of prediction error and model size for the LASSO models. The confidence bars are output by the R function ‘cv.lars’. The global minimum occurs for a model with 10 variables and a squared prediction error of approximately 0.04. Figure 2B shows the relationship of predicted and observed response for the LASSO model containing 10 variables. These 10 variables are exactly the 10 genes used to generate the data and their coefficients are almost the same as used in generating the data (Table 1). The R2 between the predicted and observed response is 0.99.
Figure 2.
Simulated data with no noise using LASSO. Part A shows the relationship between the cross-validated estimate of prediction error to model size for the corresponding models. The confidence bars are output by the R function ‘cv.lars’. The x-axis stands for fraction, which refers to the ratio of the L1 norm of the coefficient vector relative to the norm at the full LS solution for the model with the maximum steps used. Part B shows the relationship of predicted and observed response for the optimal model.
Table 1.
List of coefficients of the variables (genes) in each model using no noise and noise added simulated data. When generating the simulated continuous response data, we use −1 for the coefficients of the first five variables and −1 for the next five variables. For ALM, the selected and unselected genes in the final model are marked as “Yes” and “No”.
| True coefficient | No noise |
1 SD noise |
2 SD noise |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LAR | LASSO | ALM | LAR | LASSO | ALM | LAR | LASSO | ALM | ||
| Gene 1 | 1 | 1 | 0.977 | No | 0.836 | 0.654 | No | 0.673 | 0.449 | No |
| Gene 2 | 1 | 1 | 0.991 | Yes | 0.833 | 0.886 | Yes | 0.870 | 0.948 | Yes |
| Gene 3 | 1 | 1 | 0.989 | Yes | 0.565 | 0.657 | No | 0.394 | 0.439 | No |
| Gene 4 | 1 | 1 | 0.993 | Yes | 0.926 | 0.857 | Yes | 0.641 | 0.572 | Yes |
| Gene 5 | 1 | 1 | 0.987 | Yes | 0.468 | 0.512 | Yes | 0.357 | 0.438 | Yes |
| Gene 6 | −1 | −1 | −0.979 | No | −0.243 | −0.453 | No | 0 | 0 | No |
| Gene 7 | −1 | −1 | −0.976 | No | 0 | 0 | No | 0 | 0 | No |
| Gene 8 | −1 | −1 | −0.989 | No | −1.139 | −0.931 | No | −0.715 | −1.060 | No |
| Gene 9 | −1 | −1 | −0.975 | No | 0 | −0.183 | No | 0 | 0 | No |
| Gene 10 | −1 | −1 | −0.987 | No | −1.035 | −1.016 | No | −1.009 | −1.100 | No |
For LAR models, the cross-validated prediction error has a minimum of 0 (Fig. 3A). The R2 between the predicted and observed response is 1 (Fig. 3B). The optimized LAR model includes 10 variables which also are the 10 genes used to generate the continuous response (Table 1). The coefficients of the ten variables are the same as used in generating the data.
Figure 3.
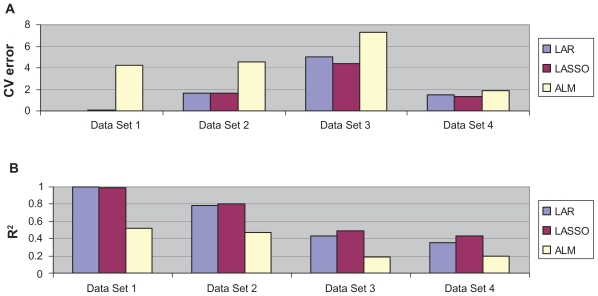
Comparison of LAR, LASSO, and ALM on simulated and real data sets. Data Set 1: Simulated data with no noise; Data Set 2: Simulated data with 1 SD noise; Data Set 3: Simulated data with 2 SD noise; Data Set 4: real data.22 Part (A) shows the cross validated global minimum estimated squared prediction errors. Part (B) shows the association between observed and predicted responses (R2).
Results for the ALM model are shown in Figures 3A and 3B. The minimum cross-validated prediction error is 4.23 and occurred for a model with only 5 variables, containing only four genes used to generate the response (Table 1). The R2 between the predicted and observed is only 0.52.
For the simulated data with no noise, the LASSO model and the LAR model both appear to be highly effective. The ALM algorithm is much less effective.
Simulated data with noise
We also compared the performances of the three algorithms when one or two standard deviations (SD) of noise are added to the simulated responses (i.e. σ = 1 or σ = 2). For data with 1 SD noise added, Figure 3A shows the cross-validated estimates of prediction error for LAR models is approximately 1.66. That model contains 8 of the 10 genes used to generate the data. The coefficients for these 8 genes in the model are listed in Table 1. The R2 between the predicted and observed response is 0.78. For data with 2 SD noise added, the global minimum occurs with a squared prediction error of approximately more than tripled at 5.02 using the LAR model in Figure 3A. This model contains only 7 of the 10 genes used to generate the data (Table 1). The R2 between the predicted and observed response decreases to 0.43 (Fig. 3B).
The LASSO models show similar trends but with slightly better results on noise added simulated data. The cross-validated prediction accuracy decreases continuously to a minimum of 1.64 for 1 SD noise added data (Fig. 3A). The optimized LASSO model includes 9 out of the 10 genes used to generate the continuous response for 1 SD noise added data. The coefficients for these 9 genes in the model are also listed in Table 1. The R2 between the predicted and observed response is 0.80 (Fig. 3B). When larger noise (i.e. 2 SD) is added, the global minimum occurs with a squared prediction error of approximately 4.37 using the LASSO model as seen in Figure 3A. This model still contains 7 of the 10 genes used to generate the data (Table 1). The R2 between the predicted and observed response decreases to 0.49 (Fig. 3B).
For the ALM model, the minimum cross-validated prediction error is 4.55 and occurs for a model with 4 variables for 1 SD noise added data (Fig. 3). For 2 SD noise added data, the minimum cross-validated prediction error is 7.26 and occurs for a model with 6 variables. Under both conditions (different noise levels), the optimal models both contain only three gene used to generate the response (Table 1). The R2 between the predicted and observed are 0.47 and 0.19, respectively (Fig. 3B).
When the different levels of noise are added, all models are gradually less effective than for the simulated data without noise. Among them, LASSO and LAR perform similarly robust to 1 SD noise. LASSO performs slightly better than LAR with 2 SD noise. ALM is again the least effective algorithm among the three.
Predicting cytotoxicity of paclitaxel against lung cancer cell lines
We applied the three methods to predict the growth inhibitory activity (GI50) of paclitaxel in cancer cell lines.22 The comparison of the cross-validation estimate of prediction errors is shown in Figure 3 for the three models. The minimum cross-validation error values are 1.46 for LAR, 1.30 for LASSO and 1.89 for ALM. The R2 values are 0.35, 0.43 and 0.20 for LAR, LASSO and ALM models respectively. Thirteen variables are in the LAR model with the minimum cross-validated error. The optimal LASSO model contains 27 variables, including 11 of the 13 variables in the optimal LAR model. The optimal ALM model contains 55 variables.
Predicting Gleason score of human prostate cancer tumors12
The global minimum estimated squared prediction errors of both LAR and LASSO models occur at fraction zero, which means that the null model yields the minimum prediction error. Consequently, when properly cross-validated, there is no evidence that the Gleason score can be predicted from the gene expression profile. We get similar result using the ALM algorithm, with the null model having the minimum estimated squared prediction error.
Simulation study with models including nonlinear terms
The original LARS method is for linear regression, but it can be generalized to fit additive models with pre-defined nonlinear terms. For example, with p variables, the nonlinear terms could be squared values and pairwise cross-product terms of the p variables. When p is very large, however, as with microarray data, this is problematic. The LARS algorithm can be used in a two stage mode in which n linear terms are selected from all p candidate variables in the first step. A second step would fit an optimally tuned LARS model containing the variables selected in the first step plus nonlinear terms based on these variables to the continuous outcome.
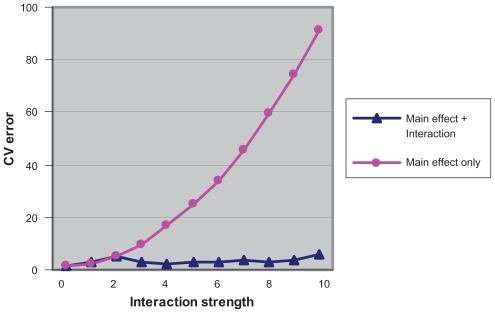
We applied the above two step approach to the lung cancer cytotoxicity data set,22 embedding the entire two stage algorithm in a leave-one-out cross validation to estimate the prediction error. We simulated a series of responses by adding different levels of strength of a two-way interaction to the original true response, which is the growth inhibitory activity (GI50) of paclitaxel in cancer cell lines. The simulation result is shown in Figure 4. When the added interaction is zero or small, the cross validated error for the model including two way interactions and quadratic terms is slightly larger than the one for the original LARS model including only linear terms. In cases where nonlinear terms have strong effects, the two stage application of LARS to include quadratic terms provides improved predictions. In other cases, however, including large numbers of nonlinear candidate variables can cause overfitting. This results in an increased value of the cross-validated prediction error.
Figure 4.
Comparison of the performances of the models including main effects only and the model including nonlinear terms on the lung cancer cyto-toxicity data set.22 The x axis is the levels of strength of the two way interaction added to the true response. The y axis is the cross validated estimate of prediction error. The line with squares stands for the models with main effects only, while the line with triangles stands for the models including main effects and interactions.
Discussion
We evaluated three linear regression algorithms using both simulated data and real data. We find that LAR and LASSO perform effectively and similarly in all data sets, consistent with the findings of Efron and Tibshirani.14 In the simulated data without noise, both LAR and LASSO select exactly the original ten genes that were used to generate the data. In the simulated data with noise, the LASSO model tends to select more variables and achieve a somewhat lower prediction error. The LASSO model selected more true variables (9 out of 10 with 1 SD noise added data and 7 out of 10 with 2 SD noise added data, (Table 1)) used to generate the data, but with more noise variables included. Failure to include the informative variables is often more serious than including more noise variables in prediction, and this was reflected in the lower prediction error for LASSO (Fig. 2). In the simulated data with 1 SD and 2 SD noise, LAR and LASSO still perform reasonably well and effectively while ALM performs poorly and ineffectively. We noted, however, in some cases, that LAR failed to converge in our simulation study using LOOCV.
When applied to the data set concerning cytotoxicity of lung cancer cell lines, the three methods performed reasonably well. The LASSO algorithm again provided a model with somewhat lower prediction error than LAR, but both performed much better than ALM.
In our comparisons we selected the final model from each sequence of models with the minimum cross-validated squared prediction error. Alternatively, we could select the model containing the fewest variables for which the cross-validated squared prediction error is no greater than a certain percentage (e.g. 10%) above the minimum. The estimate of cross-validated prediction error can be noisy and hence using a tolerance percentage above the minimum may in some cases provide a more parsimonious model (containing fewer genes) without loss of true prediction accuracy. This option is provided for the implementation in BRB-ArrayTools. For the value of f selected, the model fitted to the full dataset is reported; i.e. which genes are included in the model and what their regression coefficients are. The cross-validated predictions for models with that f value are graphed versus the observed values. These cross validated predictions are based on models with the selected f value, but the actual model differs for each loop of the cross validation. It should be noted, that the cross-validated predictions used in our implementation of LAR and LASSO in BRB-ArrayTools are based on “complete cross validation” in the sense that the genes are re-selected using LARS for each loop of the cross-validation.
Because gene expression profiles contain thousands of genes as potential variables, it is essential to carefully separate the data used for any aspect of model building from the data used for evaluating prediction accuracy. This means that when cross-validation is used, variable selection must be repeated from scratch for each loop of the cross validation. The large number of variables does not guarantee to the ability to build a good model. In a previous publication,12 a set of 16 genes from a data set of prostate cancer were selected to predict the Gleason score. When we build the model without cross validation, the LAR/LASSO model fits the Gleason scores almost perfectly. We believe that the gene selection procedure in the paper by Bibikova12 may be biased. We therefore designed an unbiased approach to evaluate their averaged linear regression predictor method. Based on our findings, no genes are informative for predicting the Gleason score for the prostate cancer data set, given the fact that all three linear regression methods selected the null model based on minimizing a properly cross- validated prediction error. The ALM algorithm is computationally efficient and reminiscent of weighted voting for classification. It may be of value for other datasets.
Conclusions
We describe an evaluation and comparison of methods for developing parsimonious models for predicting a quantitative response in high dimensional settings. It is based on both simulated and real gene expression data. We described how signal to noise can affect model performance and demonstrated the importance of complete cross-validation in evaluating the performance of a quantitative response prediction model. To our knowledge, there are no other publications that address these issues in a form accessible to bioinformatics professionals involved in the analysis of high dimensional data. Because of the complexity of using linear regression approaches with high dimensional data and obtaining proper estimates of prediction error, particularly for biomedical scientists, we have developed a plug-in for BRB-ArrayTools that implements LAR and LASSO algorithms with complete cross-validation.
Availability and Requirements
All calculations in this manuscript were done using R version 2.9 and BRB-ArrayTools. The plug-in of the least angle regression and lasso algorithms is freely available in the BRB-ArrayTools 3.8.1 stable release for non-commercial users. The link for BRB-ArrayTools downloading website is: http://linus.nci.nih.gov/BRB-ArrayTools.html
Acknowledgements
We thank Dr. Jian-Bing Fan at Illumina Inc. for providing us with the gene expression data set of prostate cancer. We thank Dr. Ming-Chung Li for providing support in implementing the BRB-ArrayTools plug-in.
List of Abbreviations
- LAR
least angle regression
- LASSO
least absolute shrinkage and selection operator
- ALM
averaged linear regression
- SD
standard deviation
- GI50
50% growth inhibitory
- LS
Least Squares
Footnotes
Authors’ Contributions
Yingdong Zhao and Richard Simon conceived the study and participated in the design, analyses and interpretation of data. Both authors have been involved in drafting and revising the manuscript. Both authors read and approved the final manuscript.
Disclosures
This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Wang E, Miller LD, Ohnmacht GA, et al. Evolving molecular portraits of metastatic melanoma. Cancer Res. 2002;62:3581–6. [PMC free article] [PubMed] [Google Scholar]
- 2.Radmacher MD, McShane LM, Simon RM. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9:505–11. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 3.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. Journal of the American Statistical Association. 2002;97:77–87. [Google Scholar]
- 4.Fix E, Hodges JL. Technical Report 4. USAF School of Aviation Medicine; Randolph Field, Texas: 1951. Discriminatory analysis, nonparametric discrimination: Consistency properties. [Google Scholar]
- 5.Tibshirani R, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–14. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Yu CY, Singer B. Cell and tumor classification using gene expression data: construction of forest. Proc Natl Acad Sci U S A. 2003;100:4168–72. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon R, Lam AP, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-ArrayTools. Cancer Informatics. 2007;2:1–7. [PMC free article] [PubMed] [Google Scholar]
- 9.Molinaro A, Simon R, Pfeiffer R. Prediction error estimation: A comparison of resampling methods. Bioinformatics. 2005;21:3301–7. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 10.Bovelstad HM, Nygard S, Storvold HL, et al. Predicting survival from microarray data–a comparative study. Bioinformatics. 2007;23:2080–7. doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- 11.Dupuy A, Simon RM. Critical review of published Microarray studies for cancer outcome and guidelines on statistical analysis and reporting. Journal of the National Cancer Institute. 2007;99:147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 12.Bibikova B, et al. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–72. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Simon R, Radmacher M, Dobbin K, McShane LM. Pitfalls in the Use of DNA Microarray Data for Diagnostic and Prognostic Classification. Journal of the National Cancer Institute. 2003;95:14–8. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 14.Efron B, Hastie T, Johnstone I, Tibshirani R. Least Angle Regression. Annals of Statistics. 2004;32:407–99. [Google Scholar]
- 15.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58:267–88. [Google Scholar]
- 16.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning; Data mining, Inference and Prediction. Springer Verlag; New York: 2001. [Google Scholar]
- 17.Hesterberg T, Choi NH, Meier L, Fraley C. Least angle and ℓ1 penalized regression: A review. Statist Surv. 2008;2:61–93. [Google Scholar]
- 18.Tibshirani B, Hastie T, et al. Sample classification from protein mass spectrometry, by ‘peak probability contrasts’. Bioinformatics. 2004;20:3034–44. doi: 10.1093/bioinformatics/bth357. [DOI] [PubMed] [Google Scholar]
- 19.Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 25:714–21. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beer, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Simon R. BRB-ArrayTools Data Archive for Human Cancer Gene Expression: A Unique and Efficient Data Sharing Resource. Cancer Informatics. 2008;6:9–15. doi: 10.4137/cin.s448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gemma A, Li C, Sugiyama Y, et al. Anticancer drug clustering in lung cancer based on gene expression profiles and sensitivity database. BMC Cancer. 2006;6:174–81. doi: 10.1186/1471-2407-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]