Gender differences in the incidence of cardiovascular disease may be ascribed at least in part to the protective effects of estrogen through both long-term and rapid ("nongenomic") actions.1 Nitric oxide (NO), generated by endogenous cardiac NO synthases (NOS; NOS1 or neuronal NOS [nNOS], NOS2 or inducible NOS, NOS3 or endothelial NOS), plays a major role in both normal cardiac physiology and cardioprotection (particularly myocardial ischemia/reperfusion injury; see the Figure),2 and the article by Lin et al3 in this issue of Circulation contributes to growing evidence that the cardioprotective functions of estrogen are conveyed in significant part by NO. A rapidly expanding body of studies indicates that NO acts in most cellular contexts largely through the covalent modification of protein Cys thiols (to generate an S-nitroso [SNO]-protein, designated S-nitrosylation),4 and recent analyses using a new generation of analytical approaches (see the Table) both confirm original measurements of SNO-proteins that have been long-standing sources of controversy in the field and point to important roles for S-nitrosylation in NO-derived cardioprotection (see the Figure).3,5,6
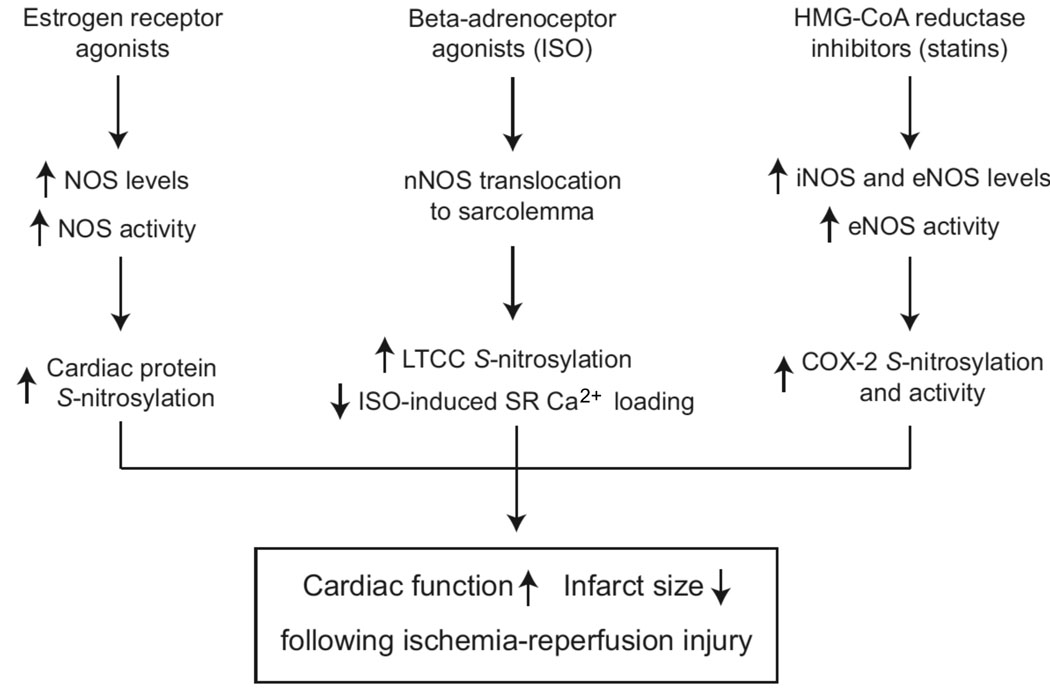
Figure.
NO-based mechanisms for preconditioning in ischemia/reperfusion injury. Accumulating evidence suggests a role for NO/SNO in estrogen and adrenergic receptor–mediated and statin-induced preconditioning in ischemia/reperfusion injury. Protein S-nitrosylation, resulting from increased NOS expression and activity and altered subcellular localization, appears to be a principal mediator of these effects.
Table.
Methodologies for the Detection and Absolute or Relative Quantification of SNO-Proteins and Other S-Nitrosothiols
| SNO Manipulation | Detection Method | Applicability |
|---|---|---|
| UV photolysis (with and without Hg2+)14 | Ozone-based chemiluminescence | Plasma, blood, cells, tissue homogenates |
| UV photolysis32 | Radicl trapping by nitrone (method in development) | |
| Cu/Cys (2C, 3C) or Cu/ascorbate reduction24,25 | Ozone-based chemiluminescence; NO electrode | Plasma, blood (cells, tissue homogenates?) |
| Decomposition by organoselenium26 | SNO sensor | Plasma (online) |
| Decomposition by gold nanoparticle27 | SNO sensor | Plasma |
| Decomposition by organomercury16 | HPLC/MS | Plasma |
| Hg2+ displacement28 | Colorimetric (Saville); fluorescent (DAN/DAF-2) | Cells |
| None29 | SNO-specific antibody | Cells, tissue homogenates |
| Denitrosylation by ascorbate | Affinity tagging or fluorescent labeling of nascent thiol21 (eg, BST); differential gel electrophoresis with multiple fluorescent tags3; labeling with isotope-coded affinity tags30 (eg, SNO-CAP); isobaric labeling of SNO-peptides after resin-assisted capture (SNO-RAC)31 |
Cells, tissue homogenates |
UV indicates ultraviolet; HPLC, high-performance liquid chromatography; and MS, mass spectrometry.
In the myocardium, endothelial NOS is associated primarily with sarcolemmal caveolae and perhaps β-arrestin, and endothelial NOS–derived NO influences β-adrenergic receptor stimulation of myocardial contractility,7 at least in part through S-nitrosylation and inhibition of the L-type Ca2+ channel5 and G-protein receptor kinase 2.8 In contrast, nNOS is localized primarily to the sarcoplasmic reticulum, and nNOS-derived NO S-nitrosylates and activates the sarcoplasmic reticulum–resident ryanodine receptor/Ca2+-channel (RyR2), resulting in cytosolic Ca2+ release and enhanced catecholamine-stimulated contractility.7 Mice lacking nNOS, but not those lacking endothelial NOS, exhibit hypo-S-nitrosylation of RyR2 and diastolic Ca2+ leakage with arrhythmia characteristic of sudden cardiac death syndrome.9 Hearts and myocytes from female versus male mice show less isoproterenol-induced sarcoplasmic reticulum Ca2+ loading, which is associated with translocation of nNOS to the sarcolemma and S-nitrosylation of the L-type Ca2+ channel in ischemia.5 The redistribution of nNOS and upregulation of NOS isoforms appear to contribute significantly to the protection of female hearts from ischemia/reperfusion injury,5 and upregulation of NOS can be ascribed to 17β-estradiol–dependent gene expression.10 However, sarcolemmal redistribution of nNOS also is observed in the hearts of humans with idiopathic dilated cardiomyopathy and of rodents with experimental myocardial infarction.7,11. Thus, S-nitrosylation of myocardial proteins may exert cardioprotective effects that are coupled to estrogen receptors and, when aberrant, may contribute to the characteristic dysfunction of the failing heart.
This scenario is reminiscent of the well-established cardioprotection conferred by circulating SNO-proteins, particularly S-nitrosoalbumin.12,13. However, these early studies were encumbered by controversy over methods of SNO-protein analysis, in which the importance of S-nitrosylation reactions in NO biology was challenged, but that controversy is now coming to resolution. Results obtained with a number of new methodologies to measure SNO-proteins in blood, plasma, and tissues (see the Table), including the approach taken by Lin et al,3 are providing support for the principal results obtained by a methodology known as Hg-coupled photolysis-chemiluminescence,14 which was used to detect the first endogenous SNO-proteins and SNO-peptides in extracellular fluids, cells, and cardiac tissues, including SNO-albumin, SNO-glutathione, SNO-hemoglobin, and SNO-RyR2, and which remains a gold standard. In addition, the recent appreciation of the differential reactivity of various SNOs4,15 and of the importance of preparative steps that stabilize rapidly degrading SNO-protein pools16,17 has revealed the basis of methodological flaws in some widely used assays (particularly triiodide chemiluminescence, which cannot be advocated for measurements of NO-derived species in any complex biological system).15,18 Limitations intrinsic to methods of SNO assay suggest that they are often better suited for either blood, plasma, or tissues, except photolysis-chemiluminescence, which has been used in all settings (see the Table).
These new methods also have provided novel insights into the basis of gender differences in long-QT syndrome, an inherited disease characterized by a prolonged QT interval that can result in fatal arrhythmia. S-nitrosylation can influence QT interval by altering the function of sodium channels that mediate late sodium currents (INa)19 and of the slowly activating delayed-rectifier K+ channel (IKs).20 In addition, gender differences in QT duration and in susceptibility to ventricular arrhythmia have been linked to differences in IKs currents.20 Taken together, these findings suggest that gender differences in cardiac function and pathophysiology can reflect differences in the localization and activity of NOS, which result in altered S-nitrosylation of critical cardiac proteins, and that these differences are likely to reflect in significant part the effects of estrogen receptor stimulation.
In the study by Lin et al,3 ovariectomized mice were infused for 2 weeks with vehicle, 17β-estradiol, or a selective agonist of the β subtype of estrogen receptor, 2,2,-bis(4-hydroxphenyl)-proprionitrile (DPN). Isolated hearts were then subjected to ischemia and reperfusion, and cardiac function and infarct severity were assessed. Treatment with either 17β-estradiol or DPN resulted in substantial improvements in functional recovery and decreased infarction. DPN had no cardioprotective effects in knockout mice lacking the β subtype of estrogen receptor (ER-β). Furthermore, cardioprotection by DPN was abolished by treatment with low doses of an NOS inhibitor. Thus, activation of ER-β confers significant cardioprotection in an ischemia/reperfusion model, and that protection is NO dependent.
To examine the possibility that differences in protein S-nitrosylation might underlie the protective effects of estrogen receptor stimulation, Lin et al3 surveyed total heart homogenates for S-nitrosylated proteins using a modification of the biotin-switch technique (BST),21,22 which is discussed further below. Mass spectrometric analysis identified 11 proteins for which S-nitrosylation was enhanced by treatment with 17β-estradiol or DPN and 3 proteins for which S-nitrosylation was suppressed. All identified proteins were affected by both 17β-estradiol and DPN, although the DPN effects were consistently greater. The enhancement of S-nitrosylation by DPN was eliminated in mice lacking ER-β and by pharmacological NOS inhibition.
The demonstration by Lin et al3 that stimulation of estrogen receptors conferred cardioprotection that was abrogated by NOS inhibition indicates a critical role for NO, at least in the case of the ER-β. Although the mechanism of enhanced protein S-nitrosylation was not determined (enhanced NOS expression and/or activation) and the relationship between NOS-dependent cardioprotection and S-nitrosylation is correlative in their study, the proteomic approach represented in this work presages a new era in the study of NO-based cellular mechanisms, which until now has been essentially phenomenological in the cardiovascular system. Indeed, the potentiated S-nitrosylation reported by Lin et al would be at least consistent with a causal role, inasmuch as S-nitrosylation of multiple proteins, including cyclooxygenase, hypoxia-inducible factor α, complex I, and caspase, has been shown to be cardioprotective. It is of note that the majority of substrates for which enhanced S-nitrosylation was reported by Lin et al consist of metabolic or mitochondrial enzymes that are present at high cellular abundance, which may suggest that energy conservation contributes in some way to cardioprotection.
Notably, the accumulation of evidence demonstrating the ubiquitous action of S-nitrosylation has been exponential because, although absolute quantification of SNO levels in cells, tissues, and purified proteins has long been possible (the Table), methodology only recently has emerged that may be applied facilely to identify individual S-nitrosylated substrates and the sites and degree of S-nitrosylation within those proteins.4 This history is hardly unique in form, given the importance of isotopic labeling for the analysis of post-translational protein modification by phosphorylation. The principal advance in methodology was provided by the introduction of the BST.21 In this approach, free Cys thiols within S-nitrosylated proteins are blocked chemically, the NO group is selectively removed from S-nitroso-cysteine with ascorbate, and the newly available Cys thiols are labeled with biotin to allow subsequent display or affinity purification. Multiple variants of the BST have been used (see the Table), but all are based on blocking free thiols and selectively removing the NO group from NO-modified thiols with ascorbate, and they differ only by the moiety, including fluorescent or other tags, used to subsequently label or bind nascent thiols.
To assess changes in S-nitrosylation of cardiac proteins in response to 17β-estradiol or DPN, Lin et al3 used differential gel electrophoresis for relative SNO-protein quantification (Huang et al23 carried out a similar analysis in endothelial cells to identify a large set of proteins in which S-nitrosylation was regulated by shear flow). In this approach, relative SNO-protein levels were assessed by labeling samples with 1 of 2 or 3 fluorescent dyes, each with a unique excitation/emission wavelength; then, samples were mixed and separated on a single 2-dimensional gel. This technique allowed the authors to compare differences in S-nitrosylation between DPN-treated hearts, DPN- and NOS inhibitor–treated hearts, and DPN-treated hearts in which extracts were first pretreated with ascorbate (as a negative control). Other recently published variations on the BST have used isotopically coded biotin tags or, alternatively, isobaric labeling coupled with solid-phase capture of SNO-proteins (see the Table), either of which can be used to perform relative quantification of individual SNO sites within proteins. Methods that allow quantification of >2 samples have the greatest advantages over more qualitative BST-based strategies because they allow inclusion of additional controls (eg, prephotolysis to homolytically cleave the SNO bond).22 In addition, methods using isotopic coding or isobaric labeling may be helpful in normalizing for differences in protein abundance, a problem that arises simply as a result of experimental variation in sample processing but also when treatments induce changes in both protein expression and S-nitrosylation, as Lin et al3 encountered. For all current BST-based methods, sensitivity of SNO-protein detection remains an issue, although improvements have recently been made (see SNO-RAC31). Photolysis-chemiluminescence is not limited by sensitivity and can readily provide SNO stoichiometry, but it is not adaptable to proteomic analysis. Several new techniques, including amperometric SNO sensors based on organoselenium and nanogold technology, have been adapted for online measurements in plasma (see the Table). These techniques have the added advantage of assaying in real time and indicate that SNO-proteins are highly abundant in the bloodstream (micromolar). It seems likely that in the near future additional methodological advances, conceivably involving the direct labeling of S-nitrosothiols in situ, will again provide a quantum improvement in our ability to characterize dynamic protein S-nitrosylation in the context of cellular signal transduction and disease.
Acknowledgements
Source of Funding
Dr Stamler’s work is supported by National Institutes of Health grant 5P01-HL075443.
Footnotes
Disclosures
Dr Stamler owns equity in LifeHealth, a company developing assays for the detection of NO-based molecules. The remaining authors report no conflicts.
References
- 1.Konhilas JP, Leinwand LA. The effects of biological sex and diet on the development of heart failure. Circulation. 2007;116:2747–2759. doi: 10.1161/CIRCULATIONAHA.106.672006. [DOI] [PubMed] [Google Scholar]
- 2.Jones SP, Bolli R. The ubiquitous role of nitric oxide in cardioprotection. J Mol Cell Cardiol. 2006;40:16–23. doi: 10.1016/j.yjmcc.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-β activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–254. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel α1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 6.Atar S, Ye Y, Lin Y, Freeberg SY, Nishi SP, Rosanio S, Huang MH, Uretsky BF, Perez-Polo JR, Birnbaum Y. Atorvastatin-induced cardioprotection is mediated by increasing inducible nitric oxide synthase and consequent S-nitrosylation of cyclooxygenase-2. Am J Physiol Heart Circ Physiol. 2006;290:H1960–H1968. doi: 10.1152/ajpheart.01137.2005. [DOI] [PubMed] [Google Scholar]
- 7.Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J Clin Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of β-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez DR, Beigi F, Treuer AV, Hare JM. Deficient ryanodine receptor S-nitrosylation increases sarcoplasmic reticulum calcium leak and arrhythmogenesis in cardiomyocytes. Proc Natl Acad Sci U S A. 2007;104:20612–20617. doi: 10.1073/pnas.0706796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambliss KL, Shaul PW. Estrogen modulation of endothelial nitric oxide synthase. Endocr Rev. 2002;23:665–686. doi: 10.1210/er.2001-0045. [DOI] [PubMed] [Google Scholar]
- 11.Bendall JK, Damy T, Ratajczak P, Loyer X, Monceau V, Marty I, Milliez P, Robidel E, Marotte F, Samuel JL, Heymes C. Role of myocardial neuronal nitric oxide synthase-derived nitric oxide in α-adrenergic hyporesponsiveness after myocardial infarction-induced heart failure in rat. Circulation. 2004;110:2368–2375. doi: 10.1161/01.CIR.0000145160.04084.AC. [DOI] [PubMed] [Google Scholar]
- 12.Ng ES, Jourd'heuil D, McCord JM, Hernandez D, Yasui M, Knight D, Kubes P. Enhanced S-nitroso-albumin formation from inhaled NO during ischemia/reperfusion. Circ Res. 2004;94:559–565. doi: 10.1161/01.RES.0000117771.63140.D6. [DOI] [PubMed] [Google Scholar]
- 13.Hallstrom S, Gasser H, Neumayer C, Fugl A, Nanobashvili J, Jakubowski A, Huk I, Schlag G, Malinski T. S-nitroso human serum albumin treatment reduces ischemia/reperfusion injury in skeletal muscle via nitric oxide release. Circulation. 2002;105:3032–3038. doi: 10.1161/01.cir.0000018745.11739.9b. [DOI] [PubMed] [Google Scholar]
- 14.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hausladen A, Rafikov R, Angelo M, Singel DJ, Nudler E, Stamler JS. Assessment of nitric oxide signals by triiodide chemiluminescence. Proc Natl Acad Sci U S A. 2007;104:2157–2162. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bramanti E, Cavallaro R, Onor M, Zamboni R, D'Ulivo A. Determination of thiolic compounds as mercury complexes by cold vapor atomic absorption spectrometry and its application to wines. Talanta. 2008;74:936–943. doi: 10.1016/j.talanta.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Zhang F, Wang Y, Krishnamoorthy M, Roy-Chaudhury P, Bleske BE, Meyerhoff ME. Photoinstability of S-nitrosothiols during sampling of whole blood: a likely source of error and variability in S-nitrosothiol measurements. Clin Chem. 2008;54:916–918. doi: 10.1373/clinchem.2007.102103. [DOI] [PubMed] [Google Scholar]
- 18.Gow A, Doctor A, Mannick J, Gaston B. S-nitrosothiol measurements in biological systems. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:140–151. doi: 10.1016/j.jchromb.2007.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci U S A. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai CX, Kurokawa J, Tamagawa M, Nakaya H, Furukawa T. Nontranscriptional regulation of cardiac repolarization currents by testosterone. Circulation. 2005;112:1701–1710. doi: 10.1161/CIRCULATIONAHA.104.523217. [DOI] [PubMed] [Google Scholar]
- 21.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 22.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang B, Chen SC, Wang DL. Shear flow increases S-nitrosylation of proteins in endothelial cells. Cardiovasc Res. 2009;83:536–546. doi: 10.1093/cvr/cvp154. [DOI] [PubMed] [Google Scholar]
- 24.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc Natl Acad Sci U S A. 2005;201:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandley RE, Tyurin VA, Huang W, Arroyo A, Daftary A, Harger G, Jiang J, Pitt B, Taylor RN, Hubel CA, Kagan VE. S-nitrosoalbumin-mediated relaxation is enhanced by ascorbate and copper: effects in pregnancy and preeclampsia plasma. Hypertension. 2005;45:21–27. doi: 10.1161/01.HYP.0000150158.42620.3e. [DOI] [PubMed] [Google Scholar]
- 26.Cha W, Anderson MR, Zhang F, Myerhoff ME. Amperometric S-nitrosothiol sensor with enhanced sensitivity based on organoselenium catalysts. Biosens Bioelectron. 2009;15:2441–2446. doi: 10.1016/j.bios.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia HY, Liu Y, Zhang XJ, Han L, Du LB, Tian Q, Xu YC. Potential oxidative stress of gold nanoparticles by induced-NO releasing in serum. J Am Chem Soc. 2009;131:40–41. doi: 10.1021/ja808033w. [DOI] [PubMed] [Google Scholar]
- 28.Marzinzig M, Nussler AK, Stadler J, Marzinzig E, Barthlen W, Nussler NC, Beger HG, Morris SM, Jr, Brückner UB. Improved methods to measure end products of nitric oxide in biological fluids: nitrite, nitrate, and S-nitrosothiols. Nitric Oxide. 1997;1:177–189. doi: 10.1006/niox.1997.0116. [DOI] [PubMed] [Google Scholar]
- 29.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 30.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senupta R, Billiar TR, Stoyanovsky DA. Studies toward the analysis of S-nitrosoproteins. Org Biomol Chem. 2009;7:232–234. doi: 10.1039/b817981f. [DOI] [PubMed] [Google Scholar]