Abstract
Aedes triseriatus mosquitoes transovarially transmit (TOT) La Crosse virus (LACV) to their offspring with minimal damage to infected ovaries. Ae. triseriatus inhibitor of apoptosis 1 (AtIAP1) is a candidate gene conditioning the ability to vertically transmit LACV. AtIAP1 was amplified and sequenced in adult mosquitoes reared from field-collected eggs. Sequence analysis revealed that AtIAP1 has much higher levels of genetic diversity than genes found in other mosquitoes. Despite this large amount of diversity, strong purifying selection of polymorphisms located in the BIR domains and to a lesser extent in the 5’ untranslated region seem to indicate that these portions of AtIAP1 are the most important. These results indicate that the 5’UTR plays an important role in transcription/translation and that the BIR domains are important functional domains in the protein. Single nucleotide polymorphisms (SNPs) were compared between LACV positive and negative mosquitoes to test for associations between segregating sites and the ability to be transovarially infected with LACV. Initial results indicated that five SNPs were associated with TOT of LACV, however, these results failed to hold up with larger sample sizes.
Keywords: Aedes triseriatus, inhibitor of apoptosis, association mapping, heated oligonucleotide ligation assay (HOLA)
1. Introduction
The Eastern treehole mosquito, Aedes (Ochlerotatus) triseriatus (Say), is the primary vector of La Crosse virus (LACV), the leading cause of pediatric arboviral encephalitis in the Unites States (Watts et al. 1972). An important part of the LACV transmission cycle in the field involves the infection of ovaries in an infected mosquito and subsequent transovarial and transtadial transmission of the virus to her adult offspring, which are then infected and capable of transmission. Transovarial transmission (TOT) is also an important part of LACV overwintering in temperate climates (Watts et al. 1973; Watts et al. 1974; Watts et al. 1975; Beaty and Thompson 1975; McGaw et al. 1998). TOT refractory and permissive strains of Ae. triseriatus have been selected (Graham et al. 1999), and three quantitative trait loci were mapped and shown to contribute additively to a female’s ability to TOT LACV (Graham et al. 2003).
In order for LACV to be transmitted transovarially, the virus must infect but not disrupt ovarian tissues. The LACV s-segment encodes a small non-structural protein (NSs) similar to the Drosophila pro-apoptotic protein, Reaper (Colon-Ramos et al. 2003). In mammalian cells and tissues, NSs expression or LACV infection may promote apoptosis. In contrast, LACV induced apoptosis has not been detected in LACV infected mosquito tissues. A candidate protein that may suppress apoptosis in infected tissues is the Aedes triseriatus inhibitor of apoptosis protein 1 (AtIAP1) (Blitvich et al. 2002), which is an ortholog of the well-characterized Drosophila inhibitor of apoptosis 1 (DIAP1). DIAP1 ubiquitinates the apical caspase Dronc to stop activation of downstream caspases that would eventually lead to apoptosis (Palaga and Osborne 2002). For apoptosis to occur, Reaper, Hid, Grim, and Sickle proteins must bind at their IAP binding motifs (IBMs) to the Baculovirus inhibitor of apoptosis repeat (BIR) domains of DIAP1 (Bergmann et al. 2003). This binding blocks the ability of DIAP1 to inactivate Dronc and the apoptotic cascade begins (Wang et al. 1999; Chai et al. 2000; Liu et al. 2000; Wu et al. 2000). AtIAP1 may act in a similar fashion to DIAP1 to counter the potential apoptotic effect of LACV in mosquitoes.
Previous observations concerning AtIAP1 have also led us to consider it a candidate gene affecting LACV TOT. LACV is known to scavenge the 5’ methylated guanine cap plus the adjacent oligonucleotide from host mRNAs to prime transcription of viral mRNAs (Beaty et al. 2000). Dobie et al. (1997) found that LACV predominantly scavenged the cap from an mRNA similar to AtIAP1 in a persistently infected Ae. albopictus larval cell line and in Ae. triseriatus eggs emerging from diapause (Dobie et al. 1997; Borucki et al. 2002).
The biology of the LACV TOT system provides a unique opportunity to exploit association mapping to determine if specific AtIAP1 genotypes condition efficient TOT and overwintering. Ae. triseriatus eggs were collected from oviposition sites throughout southwestern Wisconsin, southeastern Minnesota, and northeastern Iowa. These were hatched, reared to adults, tested for LACV infection and then separated into TOT+ (infected) and TOT− (uninfected) groups. The AtIAP1 gene of individual mosquitoes from both groups was amplified by polymerase chain reaction (PCR) and sequenced. The purpose of this study was to determine whether specific polymorphisms in the AtIAP1 gene condition whether an Ae. triseriatus mosquito will become transovarially infected with LACV (in eggs being laid by an infected female). An association between specific polymorphisms and increased TOT potential would allow mosquito control agencies to focus more effort on controlling Ae. triseriatus populations that contain these polymorphisms in a large number of individuals. While testing this hypothesis, several additional genetic analyses were performed on the AtIAP1 sequence.
2. Materials and Methods
A. Mosquito Collection and DNA Extraction
Aedes triseriatus eggs were collected by the La Crosse County Health Department from LACV endemic areas in southwestern Wisconsin, southeastern Minnesota, and northeastern Iowa where La Crosse encephalitis cases were reported. The eggs were collected from June through August of 2004 in cans that were painted black, half filled with tap water, and lined with seed germination paper as an oviposition substrate. Five traps were used at each site and placed at or slightly above ground level. The egg liners were collected after 10 days and sent to Colorado State University where the eggs were hatched and reared to adults. Adults were sacrificed and assayed for LACV using an immunofluorescence assay (Beaty and Thompson 1975). DNA was extracted from the thorax of each mosquito, using the salt extraction method (Black and DuTeau 1997), dissolved in 200 µL Tris-EDTA buffer (10 mM Tris, 1 mM EDTA), pH 8.0 and stored at −70° C.
B. PCR and DNA Sequencing
The AtIAP1 gene was amplified from each sample using three overlapping primer sets (Table 1 and Fig. 1). IAP1F and IAP2R amplified the region from the beginning of the 5’ UTR to the middle of the first BIR domain. IAP3F and IAP4R were designed to overlap the region amplified by IAP1F and IAP2R. These primers amplify a region beginning at the first BIR domain to near the end of the second BIR domain. IAP5F and IAP6R amplified a region from the middle of the second BIR domain to the 3’ polyadenylation site of the gene and thus overlapped the domain amplified by IAP3F and IAP4R (Fig. 1). PCR was completed with the following thermocycling parameters 1 min at 95° C, a 1 min at 51° C, and 2 min at 72° C; this was repeated 35 times. Products were separated on a 1% agarose gel containing tris-acetate-EDTA buffer (40 mM tris-acetate, 1 mM EDTA, pH 8.3). DNA bands were excised and purified using the Qiaquick gel extraction kit (Qiagen, Valencia, CA). Gel-extracted PCR products were sequenced using both PCR primers at Colorado State University’s Macromolecular Resources (Fort Collins, CO). 22 samples had an insertion/deletion polymorphism (one to three bases in length and primarily located in the 5’UTR) in one allele. This is problematic for direct sequencing because PCR amplification of the genome creates two alleles that exist in different reading frames. These samples were cloned into the pCR2.1-Topo vector (Invitrogen, Carlsbad, CA) and the inserts of several clones were sequenced to ensure that data was obtained for each allele. Each PCR product (or plasmid) was sequenced with both the forward and reverse PCR primers. Sequence trace files were aligned using Seqman II version 5.01 (DNAstar Inc., Madison, WI). Following alignment the chromatogram ends were trimmed so that only high quality sequence information was used. In addition, chromatogram alignments were visually analyzed and when there was a discrepancy between forward and reverse sequences the results were corrected to reflect those indicated by the better quality sequence. Generally, when this occurred one of the chromatograms showed a well defined peak while the other sequence had some anomaly (such as high background or a random signal spike). The complete AtIAP1 gene sequence was ascertained by assembling the three overlapping pieces. Some sequences were missing up to 50 base pairs from either end of the gene. This missing data was treated as unknown and was ignored in the subsequent analyses. Near complete sequences were determined for 45 LACV+ and 46 LACV− mosquitoes. Genotypes were recorded using the coding scheme in PGenome (Gorrochotegui-Escalante et al. 2005).
Table 1.
Primer sequences used for PCR amplification of the AtIAP1 gene.
| Primer Name | Sequence (5' to 3') | Optimal Annealing Temp |
|---|---|---|
| IAP0F | ACCATAAATGCATCTTCCAC | 51 |
| IAP1F | GGACCAAGAGTAGACGAAGAG | 51 |
| IAP2R | GCCCGACATAGTAAAAGC | 51 |
| IAP3F | GGACGGTTTTGTTCATCA | 51 |
| IAP4R | TACCACATGGCATGCTGT | 51 |
| IAP5F | CCTCAAGGATTGGGAAGC | 51 |
| IAP6R | CCAAAAACGATCACCTTTATTTTA | 51 |
Figure 1. Aedes triseriatus inhibitor of apoptosis 1 gene.
Nucleotides in the 5’ UTR are labeled with negative numbers. The 3’ UTR begins at nucleotide 1,213. BIR domain amino acids appear in bold underline (nucleotides 124 – 318 and 601 – 798), amino acids in the serine-rich domain appear in bold (nucleotides 436 – 513 and 883 – 1011), and amino acids in the zinc-ring finger motif are in italics (nucleotides (1063 – 1170) (Blitvich et al. 2002). Polymorphic nucleotides are highlighted in gray and listed using the following code: R = A or G, K = G or T, M = C or A, Y = C or T, W = A or T, S = G or C, H = A, C, or T, B = C, G, or T, V = A, G, or C, N = A, G, C, or T, 1 = A or −, 2 = C or −, 3 = G or −, 4 = T or −, 5 = G, A, or −, 6 = A, T, or −, 7 = G, C, or −, 8 = C, T, or −, and 9 = A, G, C, or −. Boxed amino acids represent positions where nonsynonymous substitutions occur (amino acids are listed according to proportion with the greatest at the left to the least at the right). A “~” indicates a frameshift mutation and a “*” indicates a stop codon. Primer sequences are italicized and underlined. Potential QTNs conditioning transovarial infection with LACV, as determined by complete sequence analysis with PGtheta, are found at positions −361, −268, 542, 555, and 570.
C. Analysis of Sequence Variability
The computer program DnaSP 4.10 (Rozas et al. 2003) was used for several genetic analyses of AtIAP1 sequences from 91 individuals (45 LACV+ and 46 LACV−). These sequences were used to estimate nucleotide diversity (π) (Nei 1987), the standard deviation of π (Nei 1987) and the F* test for neutrality (Fu and Li 1993). In addition, polymorphisms in the coding region of AtIAP1 were used to calculate the ratio of non-synonymous to synonymous polymorphisms (kA/kS) and the transition to transversion ratio. Finally, this program compared the level of intraspecific synonymous and non-synonymous polymorphisms within the AtIAP1 gene to interspecific polymorphisms between AtIAP1 and the Ae. aegypti and Ae. albopictus IAP1 genes (AeIAP1 AaIAP1, respectively) (genbank accession nos. DQ993355 and AF488809 respectively) using the McDonald-Kreitman test. Neutral evolution theory predicts that the ratio of synonymous to non-synonymous polymorphisms within one species will be identical in mean to the ratio seen between similar species.
D. Linkage Disequilibrium Analysis
Linkage disequilibrium among all pairs of segregating sites was analyzed with the program PGLD (Gorrochotegui-Escalante et al. 2005) to calculate Ohta’s five D-statistics (Ohta 1982a; Ohta 1982b). The disequilibrium between two segregating sites (D2ST) was estimated and a χ2 analysis of this result was performed. After applying Bonferroni’s correction, a half matrix of the results of pairwise comparisons was plotted. In addition, D2ST was regressed on the number of nucleotides between segregating sites. A priori sites that are closer together are expected to be in greater disequilibrium than those sites that are farther apart. The significance of this regression using Mantel’s test was assessed (Mantel 1967).
E. Analysis of Genotype Frequencies
Wright’s FIS summarizes the relationship between observed and expected heterozygotes at each segregating site (Wright 1965).
where Ho(i) and He(i) are respectively the observed and expected frequencies of heterozygotes containing nucleotide i at a segregating site. Weir and Cockerham’s f is an estimator of FIS that is unbiased by small or unequal sample sizes (Weir and Cockerham 1984) and is calculated as:
where
is the frequency of nucleotide i at a segregating site and ny is the size of collection y.
F. Association Mapping Based on Allele and Genotype Frequencies
The sequenced AtIAP1 gene from 91 individual mosquitoes was analyzed using PGTheta (Gorrochotegui-Escalante et al. 2005). This program compares nucleotide frequencies at segregating sites among, in this case, TOT+ and TOT− mosquitoes (and can be downloaded at: http://www.evolcafe.com/popgen/download.html). At each segregating site, θ (Weir and Cockerham 1984) was estimated and its consistency was assessed with 10,000 permutations according to the procedure of Doerge and Churchill (1996) as follows: The original dataset was permuted by randomly assigning the genotype of one mosquito to another mosquito. After all genotypes were shuffled, θ was estimated between phenotypic groups and stored in memory. After 10,000 permutations, all θ were sorted. The 9,500th and 9,900th largest values respectively defined the 95% and 99% thresholds at each segregating site. Potential quantitative trait nucleotides (QTNs) were assigned when the original estimate of θ exceeded the 95% threshold of θ calculated by permutation. PGCon (Gorrochotegui-Escalante et al. 2005) is a program designed to perform contingency χ2 analysis of genotypes as segregating sites with probabilities adjusted using Bonferroni’s correction to determine if TOT rates are significantly different.
G. AtIAP1 Heated Oligonucleotide Ligation Assay
The heated oligonucleotide ligation assay (Lynd et al. 2005; Black et al. 2006) was used to determine the genotypes of 300 additional mosquitoes (150 TOT+ and 150 TOT−) at the 5 putative QTNs identified by PGTheta in the analysis of full sequences. These mosquitoes were collected from the same regions as the initial samples. PCR on each sample used primers IAP0F (located 199 bp upstream of the sequence in Fig. 1) and IAP4R to amplify a portion of the AtIAP1 gene that contained all five putative QTNs. HOLA reactions were conducted as described by Black et al. (2006) with the oligonucleotides in Table 2.
Table 2.
Reporter and detector oligonucleotides used in genotyping AtIAP1 segregating sites −361, −208, 542, 555 and 570.
| Oligonucleotide Name |
Sequence (5' to 3') | Optimal Ligation Temp (°C) |
|---|---|---|
| IAP-361dtcA | Biotin-TGATGATGGCRAYGATGCACT | 60 |
| IAP-361dtcC | Biotin-TGATGATGGCRAYGATGCACG | |
| IAP-361rpt | PO4-GAAAGATGCTGATGCTGT-Fluorescein | |
| IAP-208dtcA | Biotin-TCAATCTTGTTCTCCGGGGGT | 51 |
| IAP-208dtcC | Biotin-TCAATCTTGTTCTCCGGGGGG | |
| IAP-208rpt | PO4-TTTTCCGCRGTTCAGCTC-Fluorescein | |
| IAP542dtcC | Biotin-TTTCTGCTGWGGCAGCGCGGGCG | 58 |
| IAP542dtcT | Biotin-TTTCTGCTGWGGCAGCGCGGGCA | |
| IAP542rpt | PO4-CAGGAAGTTGTTGGGGGA-Fluorescein | |
| IAP555dtcA | Biotin-ATTCYGGCCGTTTCTGCTGT | 58 |
| IAP555dtcT | Biotin-ATTCYGGCCGTTTCTGCTGA | |
| IAP555rpt | PO4-GGCAGCGCGGGCRCAGGA-Fluorescein | |
| IAP570dtcA | Biotin-ATGGCATAGTTGGGATATTCT | 58 |
| IAP570dtcG | Biotin-ATGGCATAGTTGGGATATTCC | |
| IAP570rpt | PO4-GGCCGTTTCTGCTGWGGC-Fluorescein |
Genotypic results from HOLA analysis were combined with the genotypic results from the previous sequence analysis to give a total 195 TOT+ and 196 TOT− mosquitoes. Goodness of fit χ2 analyses were performed to determine if any of these five putative QTNs could be used as indicators for susceptibility to transovarial infection with LACV.
3. Results
A. Analysis of Sequence Variability
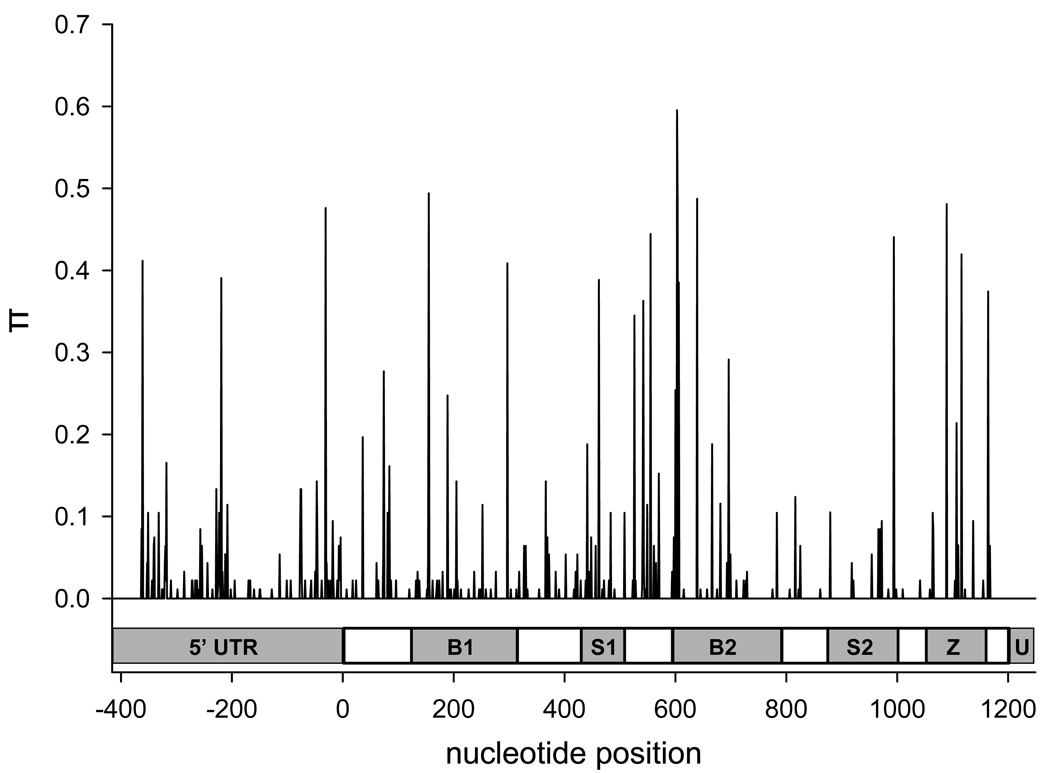
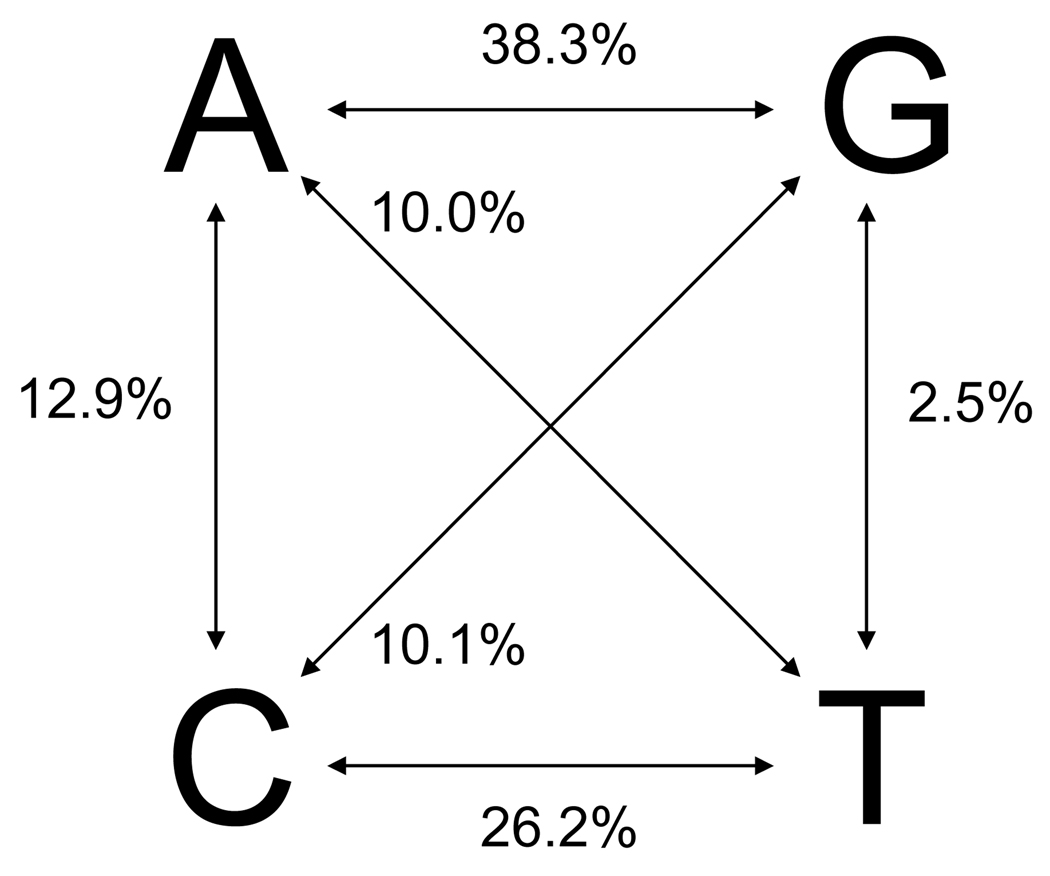
The amplified AtIAP1 sequence was 1,665 nucleotides in length with 416 bases in the 5’UTR, 1,212 bases in the coding region, and 37 bases in the 3’UTR (Fig. 1). In total, 113 segregating sites were found in the 5’UTR, 144 sites occurred in the coding region, and a single site was found in the 3’UTR for a total of 258 segregating sites (Supplemental Table 1). The overall nucleotide diversity (π) was 0.01133 and θ/site was estimated at 0.026 (Table 3). The average number of nucleotide differences (k) among pairs of mosquitoes was 17.432 (Table 3). π varied between 0.000 and 0.595 across the gene (Fig. 2 and Table 3) with π being ~13.5 times greater in synonymous vs. non-synonymous substitutions. DnaSP 4.10 does not estimate nucleotide diversity at aligned sites where any sequence is missing data, so the first 53 and the last 25 nucleotides were not included in this analysis. The coding region of AtIAP1 (nucleotides 1 – 1,212) has a non-synonymous to synonymous polymorphism (kA/kS) ratio of 0.235 and a transition to transversion ratio of 2.03 (Fig. 3).
Table 3.
Genetic Diversity in Individual Domains of the AtIAP1 gene.
| Domain | Nucleotide Position |
Nucleotide Diversity (π) (Range) |
θ/Site | Average Nucleotide Differences (k) |
|---|---|---|---|---|
| 5’ UTR | −416 to −1 | 0.014 (0.000 – 0.476) | 0.04313 | 4.626 |
| 1 to 123 | 0.008 (0.000 – 0.277) | 0.02181 | 0.990 | |
| 1st BIR Domain |
124 to 318 | 0.010 (0.000 – 0.494) | 0.03106 | 1.942 |
| 319 to 435 | 0.005 (0.000 – 0.143) | 0.02071 | 0.638 | |
| 1st Serine Rich Domain |
436 to 513 | 0.013 (0.000 – 0.388) | 0.02884 | 1.044 |
| 514 to 600 | 0.024 (0.000 – 0.444) | 0.03183 | 2.100 | |
| 2nd BIR Domain |
601 to 798 | 0.015 (0.000 – 0.600) | 0.02196 | 2.923 |
| 799 to 882 | 0.004 (0.000 – 0.124) | 0.01442 | 0.326 | |
| 2nd Serine Rich Domain |
883 to 1011 | 0.007 (0.000 – 0.441) | 0.01342 | 0.856 |
| 1012 to 1062 | 0.001 (0.000 – 0.022) | 0.00679 | 0.033 | |
| Zinc Finger Domain |
1063 to 1170 | 0.018 (0.000 – 0.481) | 0.02243 | 1.956 |
| 1171 to 1212 | 0.000 (0.000 – 0.000) | 0.00000 | 0.000 | |
| 3’ UTR | 1213 to 1261 | 0.000 (0.000 – 0.000) | 0.00000 | 0.000 |
| Whole Gene | −416 to 1261 | 0.011 (0.000 – 0.595) | 0.02622 | 17.432 |
Figure 2. Plot of nucleotide diversity (π) across AtIAP1.
This figure shows the nucleotide diversity seen across the entire AtIAP1 gene. The abbreviations in the genetic schematic represent individual domains as follows. 5’UTR = 5’ untranslated region, B1 = 1st BIR domain, S1 = 1st serine rich domain, B2 = 2nd BIR domain, S2 = 2nd serine rich domain, Z = Zing ring finger motif, and U = 3’ untranslated region. The coding region begins at nucleotide 0 and ends at nucleotide 1,212.
Figure 3. Percentage of transitions and transversions in the coding sequence of the AtIAP1 gene.
FIS varies between −1 and 1 with a positive value indicating an excess of homozygotes and a negative value indicating an excess of heterozygotes. Segregating sites in the AtIAP1 gene had a consistent excess of homozygotes. Of the 258 segregating sites only 25 had a negative Fis (data not shown). The majority of these sites were found within the coding sequence and more specifically near the 1st serine rich domain.
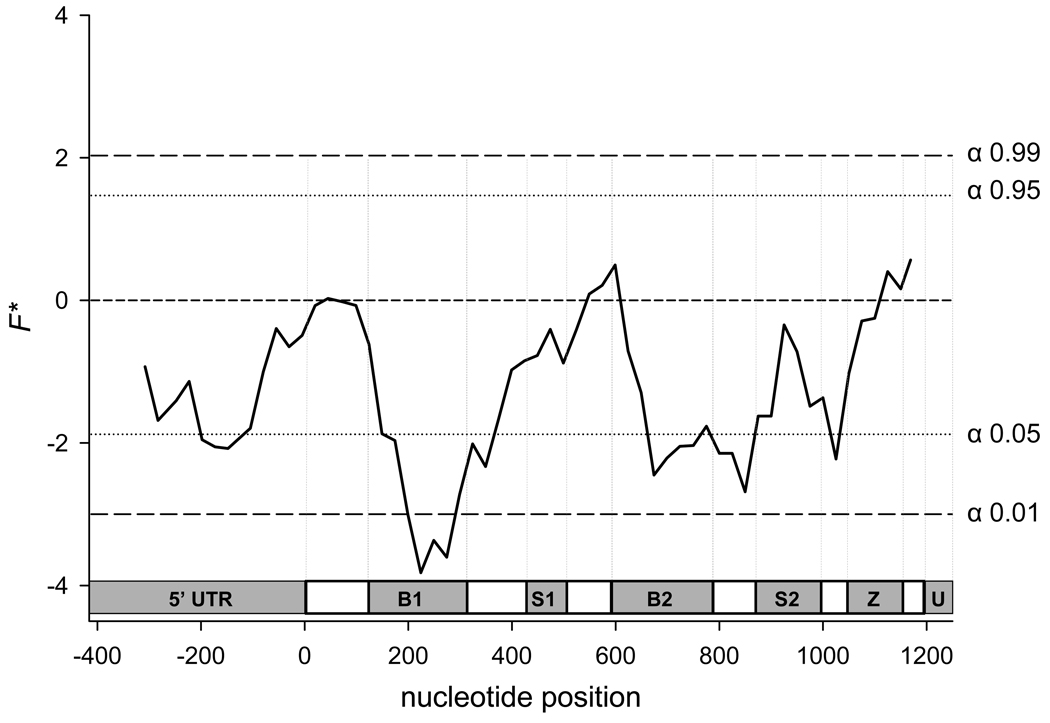
Fu and Li’s F* is a normalized comparison of all mutations (η) relative to the number of those appearing once (“singletons”- ηs). The underlying assumption of this test is that F* = 0 (η = ηs) under neutrality. F* > 0 (η > ηs) under balancing selection and F* < 0 (η < ηs) under purifying selection. Our analysis (Fig. 4) shows that the majority of the gene has a negative F* value, however, these values were only significant in a few small parts of the gene. In particular the negative F* values are significant in polymorphisms located in the BIR domains and to a lesser extent in the 5’UTR suggesting that polymorphisms in these regions may compromise fitness and are rapidly eliminated through purifying selection. These observations are consistent with the FIS analysis because an excess of homozygotes is also indicative of purifying selection.
Figure 4. Plot of Fu and Li’s F* across AtIAP1.
A significant negative value indicates that polymorphic sites are under purifying selection. A significant positive value indicates that polymorphic sites are under balancing selection. The abbreviations in the genetic schematic represent individual domains as follows. 5’UTR = 5’ untranslated region, B1 = 1st BIR domain, S1 = 1st serine rich domain, B2 = 2nd BIR domain, S2 = 2nd serine rich domain, Z = Zing ring finger motif, and U = 3’ untranslated region. The coding region begins at nucleotide 0 and ends at nucleotide 1,212.
The neutral mutation hypothesis was tested using IAP1 sequences from Ae. albopictus and Ae. aegypti. The McDonald-Kreitman (MK) test measures whether there are differences in the ratio of synonymous vs. non-synonymous polymorphisms within species compared to the ratio between species. The MK test (performed on the 1227 sites in the coding sequence) showed no significant difference (p = 0.38) in the ratio of synonymous to non-synonymous polymorphisms within Ae. triseriatus samples (206 synonymous to 56 non-synonymous) and between Ae. triseriatus, Ae. aegypti, and Ae. albopictus samples (100 synonymous to 34 non-synonymous). These results indicate that a general pattern of neutral evolution is responsible for the large amount of polymorphisms seen in the coding region of AtIAP1.
B. Linkage Disequilibrium Analysis
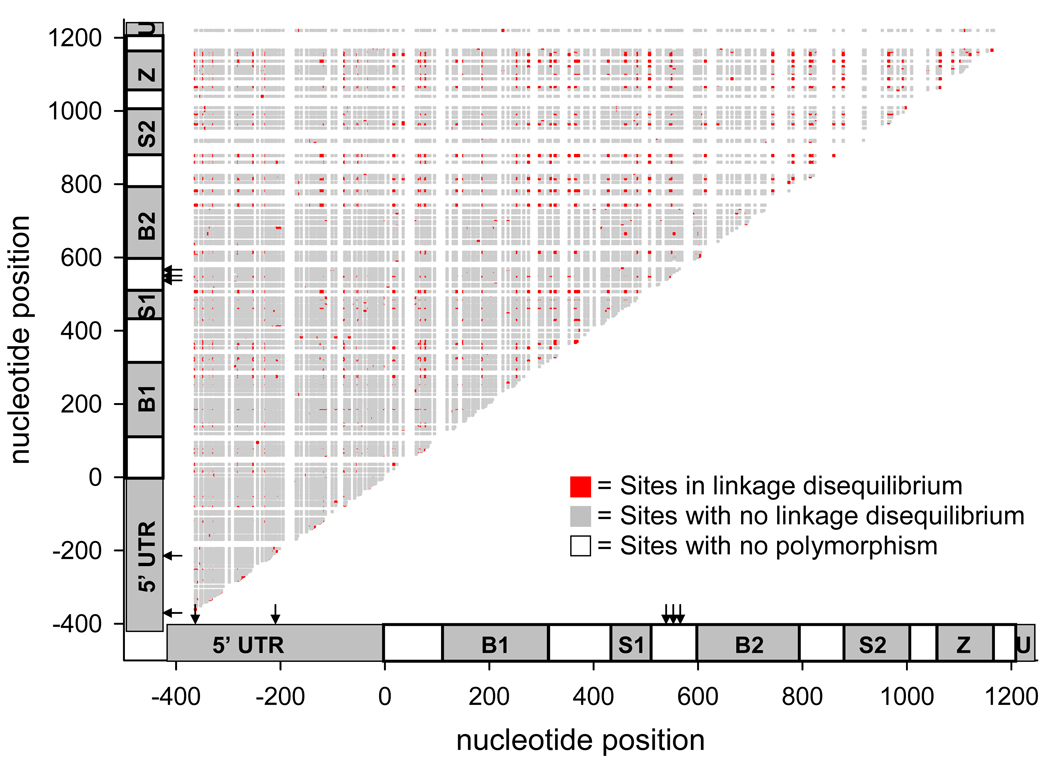
Polymorphisms occurring between the first BIR and serine-rich domains were more likely to be in disequilibrium amongst themselves than with other segregating sites (Fig. 5). There was little linkage disequilibrium among segregating sites in the 5’UTR. Regression analysis of D2ST on the number of nucleotides between segregating sites indicated that neither the y- intercept nor the slopes were significantly greater than 0 meaning the recombination frequency between polymorphisms occurring at nearby sites is similar to the recombination frequency between those polymorphisms that occur farther apart.
Figure 5. A half-matrix showing linkage disequilibrium coefficient D2ST between AtIAP1 segregating sites.
The abbreviations in the genetic schematic represent individual domains as follows. 5’UTR = 5’ untranslated region, B1 = 1st BIR domain, S1 = 1st serine rich domain, B2 = 2nd BIR domain, S2 = 2nd serine rich domain, Z = Zing ring finger motif, and U = 3’ untranslated region. The coding region begins at nucleotide 0 and ends at nucleotide 1,212. Potential QTNs conditioning transovarial infection with LACV, as determined by complete sequence analysis with PGtheta, are found at positions −361, −268, 542, 555, and 570 and are marked with arrows.
C. Association Mapping Based on Allele and Genotype Frequencies
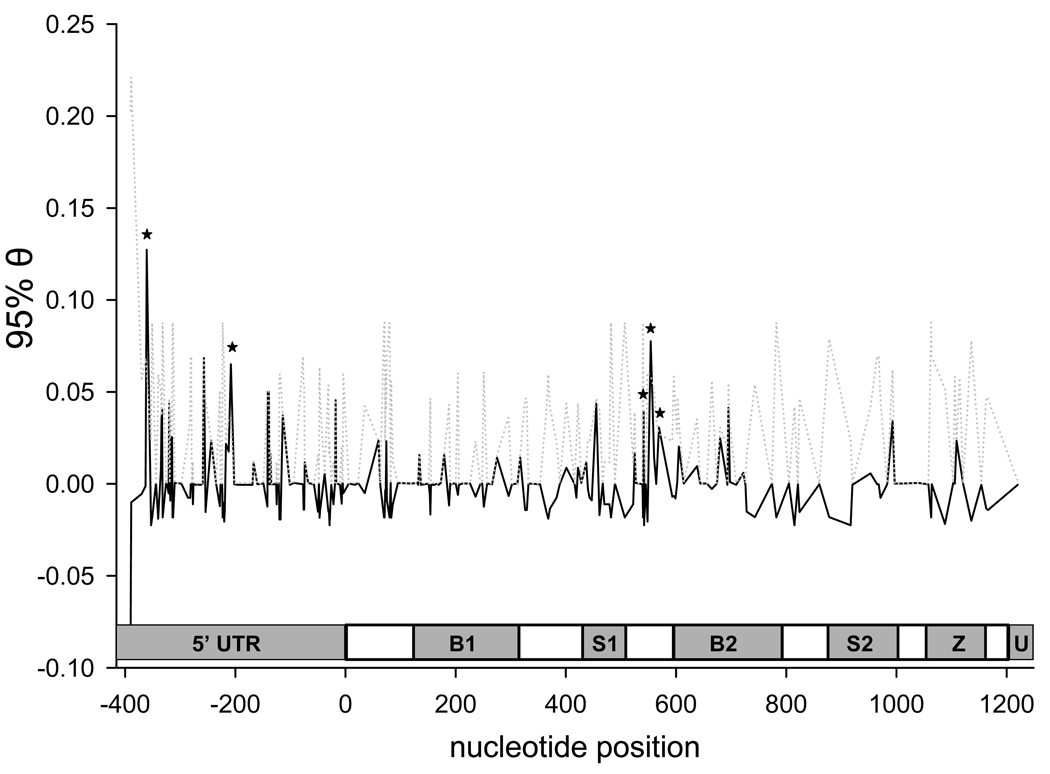
PGTheta identified five sites (at nucleotides −361, −208, 542, 555, and 570) in which θs estimated from the original dataset were greater than 95% of permuted θs (Fig. 6). Two of these putative quantitative trait nucleotides (QTNs) were found in the 5’UTR, the other three were found between the first serine-rich domain and the second BIR domain. QTNs 555 and 570 encode synonymous substitutions. QTN 542 encodes a transition in the second codon (GCG ⇔ GTG) causing an A ⇔ V amino acid substitution. Because the two other positions in this codon are also polymorphic, leucine and proline are also possible amino acid substitutions. QTNs 542 and 555 were in disequilibrium. The LACV infection rate was significantly different (p < 0.05) among genotypes at QTNs −361, 555, and 570, but not at sites −208 and 542 (data not shown).
Figure 6. Association mapping of AtIAP1 comparing polymorphic nucleotide frequencies between TOT+ and TOT− mosquitoes using PGtheta.
An asterisk indicates a position where the estimated θ (as shown by solid lines) exceeds 95% of the permuted θ values (as shown by dotted lines). The abbreviations in the genetic schematic represent individual domains as follows. 5’UTR = 5’ untranslated region, B1 = 1st BIR domain, S1 = 1st serine rich domain, B2 = 2nd BIR domain, S2 = 2nd serine rich domain, Z = Zing ring finger motif, and U = 3’ untranslated region. The coding region begins at nucleotide 0 and ends at nucleotide 1,212.
D. Association Mapping Based on Single Nucleotide Polymorphisms
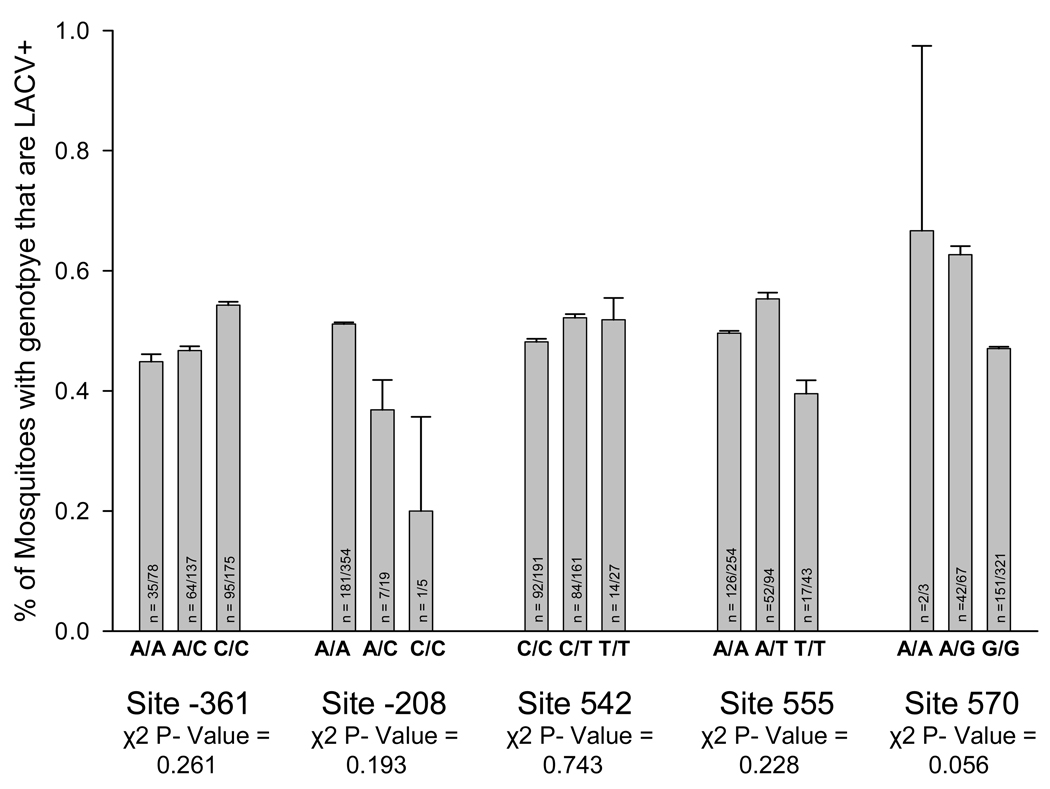
To further evaluate these five putative QTN’s, heated oligonucleotide ligation analysis (HOLA) was performed on an additional 150 LACV+ and 150 LACV− mosquitoes. These results were analyzed separately and then combined with the results from the fully sequenced mosquitoes. The transovarial infection rate was not statistically different among genotypes at any of the putative QTNs (Fig. 7).
Figure 7. Association mapping comparing genotype frequencies of LACV+ and LACV− samples at putative QTNs.
Potential QTNs conditioning transovarial infection with LACV were identified by complete sequence analysis with PGtheta of 45 LACV+ and 46 LACV− mosquitoes. 150 LACV+ and 150 LACV− samples, that were previously unsequenced, were genotyped at potential QTNs using the heated oligonucleotide ligation assay. Results from samples genotyped by sequencing and by HOLA were combined and graphed in terms of percent of each genotype that was LACV+. A chi-square goodness of fit analysis was used to determine if LACV infection rates differed between genotypes. The number on each bar represents the number of LACV positive individuals/number of individuals with the genotype. In all 391 samples were genotyped by either sequence analysis or by HOLA at each site, however occasionally results from the HOLA analysis were inconclusive which is why the number tested does not always add up to 391 at each site.
4. Discussion
The genetic analysis of AtIAP1 revealed 258 segregating sites. Of the 144 sites in the coding region, 37 were nonsynonymous. The overall nucleotide diversity (π) was 0.011 and θ/site was 0.026. Figure 2 indicates that variation is uniformly distributed across the gene. In comparison with the Ae. aegypti Early trypsin gene, π was nearly equivalent at 0.012 but θ/site was 0.010, only one third of that found in AtIAP1. In the Ae. aegypti abundant trypsin gene, π was only slightly lower at 0.009 and the θ/per site (0.009) was again one third of that found in AtIAP1. Thus the average diversity per site is one third as large in AtIAP1.
Previous studies indicate that these collections come from a large panmictic population with low levels of genetic drift. Neutral theory predicts that with a large effective population size, the large number of polymorphisms can be maintained only if positive directional selection is weak (Kimura 1985). Otherwise the large numbers of singletons and the greater θ/site seen in AtIAP1 are difficult to explain. This greater diversity could arise due to relaxation of selection, or through balancing or diversifying selection, but the high proportion of synonymous substitutions is not consistent with diversifying or balancing selection. Furthermore, the overall negative values of F* (Fig. 4) indicate that the numbers of singletons exceeded the overall numbers of shared polymorphisms which is evidence of purifying selection. Significant negative F* estimates occurred in polymorphisms located in the BIR domains. In Drosophila, BIR domains are targeted by pro-apoptotic caspases to neutralize DIAP1. Amino acid substitutions in this portion of the gene would compromise this function in AtIAP1 and probably the fitness of Ae. triseriatus.
Analysis of the AtIAP1 coding region showed that the ratio of non-synonymous to synonymous polymorphisms (Ka/Ks) is 0.235. This ratio is higher than the Ka/Ks ratio seen in the mosquitoes Anopheles funestus (0.181) (Wondji et al. 2007), An. gambiae (0.192) (Morlais et al. 2004), and Ae. aegypti (.204) (Morlais and Severson 2003). The ratio is much higher compared to that of D. melanogaster (0.115) (Moriyama and Powell 1996). A higher Ka/Ks ratio in AtIAP1 could result from either more non-synonymous polymorphisms or less synonymous polymorphisms than other mosquito genes. AtIAP1 has, on average, 1 SNP every 8.4 base pairs in the coding region while An. funestus, An. gambiae, and Homo sapiens have 1 SNP every 138, 125, and 1,000 base pairs respectively (Wang et al. 1998; Morlais et al. 2004; Wondji et al. 2007). This means that greater non-synonymous polymorphisms, rather than fewer synonymous polymorphisms, are probably responsible for the increased Ka/Ks ratio in AtIAP1.
An. funestus, An. gambiae, Ae. aegypti, and D. melanogaster all have lower genetic diversity per site (π) in their non-coding regions than in synonymous polymorphisms found in the coding region (Moriyama and Powell 1996; Morlais and Severson 2003; Morlais et al. 2004; Wondji et al. 2007). This trend is also seen in the AtIAP1 gene where π/site in the 5’UTR is 0.014 while π/site in synonymous sites is 0.035. This result indicates that the non-coding region is under greater purifying selection than the synonymous sites in the coding region, which makes sense if the 5’UTR plays an important role in transcription or translation of AtIAP1.
The BIR region of DIAP1 has been shown to be sufficient for preventing apoptosis and the BIR domain alone is even more efficient at inhibition of apoptosis then the full-length DIAP1 protein (Hay et al. 1995; Vucic et al. 1998). Perhaps the reason for the high level of diversity seen in the AtIAP1 gene is that the BIR domains are the only portion of the gene that have any selection pressure acting upon them. This could explain why McDonald-Kreitman tests show a general pattern of neutral selection acting on the coding sequence while Fu and Li’s F* analysis indicates that the AtIAP1 gene is under relatively little purifying selection, apart from the BIR domains.
There are several other possible reasons for the high level of nucleotide diversity observed in the AtIAP1 gene. One is that we are actually sequencing the same gene from several Ae. triseriatus subpopulations, however, previous studies reveal that there are no barriers to gene flow in the upper Midwest and that the mosquito exists as one panmictic population (Beck et al. 2005). Another possibility is that we have actually collected two different species of mosquitoes. Again, this seems unlikely because Ae. triseriatus mosquitoes are relatively easy to identify compared to other mosquitoes present in the study area with one exception. Ae. hendersoni can be found in the same geographic area and is closely related (in fact, the two mosquitoes can interbreed to form viable hybrids) (Munstermann et al. 1982). However, there are very few documented examples of interspecific hybrids from the field, which is most likely because these mosquitoes occupy different niches (Truman and Craig 1968; Grimstad et al. 1974). Ae. hendersoni mosquitoes feed and breed in the tree canopies while Ae. triseriatus mosquitoes remain relatively close to the ground (Copeland and Craig 1990). Because our samples were collected at ground level it is very unlikely that we have collected Ae. hendersoni along with Ae. triseriatus. A final possibility is that several IAP paralogs are present and that this sequence information represents data from multiple genes in the same family. This possibility is not unrealistic as 7 IAP genes have been identified in the principal vector of malaria, Anopheles gambiae (Christophides et al. 2002). However, these genes can be distinguished from one another and throughout the course of this study there was no information to indicate that multiple undistinguishable paralogs were sequenced. This possibility is difficult to test without the genome sequence of this mosquito. So while it is possible that we have sampled multiple subpopulations, species, or IAP paralogs these explanations seem less likely than the AtIAP1 gene having high levels of diversity.
Five significant quantitative trait nucleotides (QTNs) associated with TOT were detected in a sequence analysis of 91 mosquitoes. However, the subsequent prospective case control study failed to validate any of these 5 QTNs (Fig. 7). Interestingly, 19 of the 258 polymorphic sites observed during sequence analysis were not in Hardy-Weinberg equilibrium (this includes four of the potential QTNs with the fifth having a Hardy-Weinberg equilibrium probability of 0.0534) (Supplemental Table 1). While the sequence chromatograms show no obvious reason for doubting their validity, sequencing error is the most likely reason for this divergence from equilibrium. In addition to the positive FIS values this data indicates that sequence analysis likely misrepresented the genotype of several heterozygotes as homozygotes. This would indicate that the HOLA assay has a lower error rate than direct sequencing of PCR products and that the follow-up case control study likely represents the truth about the relationship of these SNPs and their effect on transovarial transmission. It is also a possibility that the initial sequencing study was performed with too few individuals causing some of these sites to appear as QTNs when in fact they were not. Irrespective of the reasons, this study did not find an association between AtIAP1 polymorphisms and TOT in field collected Ae. triseriatus.
Implicit assumptions in our study design reduced its power to detect valid QTNs. The design assumes that the Ae. triseriatus mothers of the offspring in this study were uniformly susceptible to infection with LACV and that all mothers were exposed to LACV. If true, then the only differences among reared TOT+ and TOT− adults would arise from genetic differences among mothers in the genes that condition TOT. The first assumption is probably valid; typically Ae. triseriatus are uniformly susceptible to oral or vertical infection with LACV (Woodring et al. 1998). The second assumption is obviously valid with TOT+ offspring but is probably false among TOT− offspring. The minimal field infection rate of Ae. triseriatus with LACV is 3.4 − 12.7/1000; making it difficult to verify whether offspring are uninfected because their mothers were never exposed to LACV or because the mothers were genetically incapable of TOT (Clark et al. 1983). This assumption could have been eliminated by returning mosquitoes to the laboratory, uniformly exposing them to LACV with either oral or intrathoracic inoculation, and then collecting eggs and analyzing the resulting offspring. However, this is problematic because Ae. triseriatus is a difficult species to colonize directly from the field.
Problems arising from non-uniform exposure of a study group do not preclude association mapping studies. Human genetic epidemiologists have to deal with non-uniform exposure in identifying genetic factors that condition genetic susceptibility to heritable or infectious diseases. Nevertheless, non-uniform exposure does lower the power of association mapping to detect valid QTNs. Only QTNs with large effects on phenotype are likely to be detected.
This study has shown that polymorphisms in the AtIAP1 gene likely do not have a significant effect on TOT of LACV. However, these results show that there is much greater diversity seen in this gene compared to genes studied in D. melanogaster, An. funestus, An. gambiae, and H. sapiens. These results also indicate that although the AtIAP1 gene is highly polymorphic it is generally evolving according to neutral theory. However, the 5’UTR and the two BIR domains are under stronger purifying selection than the remainder of the coding sequence. For this reason, it is likely that the 5’UTR and the BIR domains play major roles in transcription/translation efficiency and protein function, respectively.
Supplementary Material
REFERENCES
- Beaty BJ, Rayms-Keller A, Borucki MK, Blair CD. La Crosse encephalitis virus and mosquitoes: A remarkable relationship. ASM News. 2000;66:349–357. [Google Scholar]
- Beaty BJ, Thompson WH. Emergence of La Crosse virus from endemic foci: Fluorescent antibody studies of overwintered Aedes triseriatus. Am J Trop Med Hyg. 1975a;24:685–691. doi: 10.4269/ajtmh.1975.24.685. [DOI] [PubMed] [Google Scholar]
- Beck ET, Bosio CF, Geske DA, Blair CD, Beaty BJ, Black WC., IV An analysis of gene flow among Midwestern populations of the mosquito Ochlerotatus triseriatus. Am J Trop Med Hyg. 2005;73:534–540. [PubMed] [Google Scholar]
- Bergmann A, Yang AY-P, Srivastava M. Regulators of IAP function: Coming to grips with the grim reaper. Curr Opin Cell Biol. 2003;15:717–724. doi: 10.1016/j.ceb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Black WC, IV, DuTeau NM. RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton J, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors: A Methods Manual. New York: Chapman and Hall; 1997. pp. 361–373. [Google Scholar]
- Black WC, IV, Gorrochotegui-Escalante N, Duteau NM. Heated oligonucleotide ligation assay (HOLA): an affordable single nucleotide polymorphism assay. J Med Entomol. 2006;43:238–247. doi: 10.1603/0022-2585(2006)043[0238:holaha]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Blitvich BJ, Blair CD, Kempf BJ, Hughes MT, Black WC, IV, Mackie RS, Meredith CT, Beaty BJ, Rayms-Keller A. Developmental- and tissue-specific expression of an inhibitor of apoptosis protein 1 homologue from Aedes triseriatus mosquitoes. Insect Mol Biol. 2002;11:431–442. doi: 10.1046/j.1365-2583.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- Borucki MK, Kempf BJ, Blitvich BJ, Blair CD, Beaty BJ. La Crosse virus: Replication in vertebrate and invertebrate hosts. Microbes Infect. 2002;4:341–350. doi: 10.1016/s1286-4579(02)01547-2. [DOI] [PubMed] [Google Scholar]
- Chai JJ, Du CY, Wu JW, Kyin S, Wang XD, Shi YG. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Clark GG, Pretula HL, Rohrer WH, Harroff RN, Jakubowski T. Persistence of La Crosse virus (California encephalitis serogroup) in north-central Illinois. Am J Trop Med Hyg. 1983;32:175–184. doi: 10.4269/ajtmh.1983.32.175. [DOI] [PubMed] [Google Scholar]
- Colon-Ramos DA, Irusta PM, Gan EC, Olson MR, Song J, Morimoto RI, Elliott RM, Lombard M, Hollingsworth R, Hardwick JM, Smith GK, Kornbluth S. Inhibition of translation and induction of apoptosis by bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol Biol Cell. 2003;14:4162–4172. doi: 10.1091/mbc.E03-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RS, Craig GB., Jr Habitat segregation among treehole mosquitoes (Diptera: Culicidae) in the Great Lakes region of the United States. Ann Entomol Soc Am. 1990;83:1063–1073. [Google Scholar]
- Dobie DK, Blair CD, Chandler LJ, Rayms-Keller A, McGaw MM, Wasieloski LP, Beaty BJ. Analysis of La Crosse virus S mRNA 5' termini in infected mosquito cells and Aedes triseriatus mosquitoes. J Virol. 1997;71:4395–4399. doi: 10.1128/jvi.71.6.4395-4399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochotegui-Escalante N, Lozano-Fuentes S, Bennett KE, Molina-Cruz A, Beaty BJ, Black WC., IV Association mapping of segregating sites in the early trypsin gene and susceptibility to dengue-2 virus in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2005;35:771–788. doi: 10.1016/j.ibmb.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Graham DH, Holmes JL, Beaty BJ, Black WC., IV Quantitative trait loci conditioning transovarial transmission of La Crosse virus in the eastern treehole mosquito, Ochlerotatus triseriatus. Insect Mol Biol. 2003;12:307–318. doi: 10.1046/j.1365-2583.2003.00412.x. [DOI] [PubMed] [Google Scholar]
- Graham DH, Holmes JL, Higgs S, Beaty BJ, Black WC., IV Selection of refractory and permissive strains of Aedes triseriatus (Diptera: Culicidae) for transovarial transmission of La Crosse virus. J Med Entomol. 1999;36:671–678. doi: 10.1093/jmedent/36.6.671. [DOI] [PubMed] [Google Scholar]
- Grimstad PR, Garry CE, DeFoliart GR. Aedes hendersoni and Aedes triseriatus (Diptera: Culicidae) in Wisconsin: Characterization of larvae, larval hybrids, and comparison of adult and hybrid mesoscutal patterns. Ann Entomol Soc Am. 1974;67:795–804. [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Sun CH, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Lynd A, Ranson H, McCall PJ, Randle NP, Black WC, IV, Walker ED, Donnelly MJ. A simplified high-throughput method for pyrethroid knock-down resistance (kdr) detection in Anopheles gambiae. Malar J. 2005;4:16. doi: 10.1186/1475-2875-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- McGaw MM, Chandler LJ, Wasieloski LP, Blair CD, Beaty BJ. Effect of La Crosse virus infection on overwintering of Aedes triseriatus. Am J Trop Med Hyg. 1998;58:168–175. doi: 10.4269/ajtmh.1998.58.168. [DOI] [PubMed] [Google Scholar]
- Moriyama E, Powell J. Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- Morlais I, Poncon N, Simard F, Cohuet A, Fontenille D. Intaspecific nucleotide variation in Anopheles gambiae: New insights into the biology of malaria vectors. Am J Trop Med Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- Morlais I, Severson DW. Intraspecific DNA variation in nuclear genes of the mosquito Aedes aegypti. Insect Mol Biol. 2003;12:631–639. doi: 10.1046/j.1365-2583.2003.00449.x. [DOI] [PubMed] [Google Scholar]
- Munstermann LE, Taylor DB, Matthews TC. Population genetics and speciation in the Aedes triseriatus group. In: Steiner WW, editor. Recent developments in the genetics of insect disease vectors. Champaign, IL: Stipes; 1982. pp. 433–453. [Google Scholar]
- Ohta T. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc Natl Acad Sci USA. 1982a;79:1940–1944. doi: 10.1073/pnas.79.6.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Linkage disequilibrium with the island model. Genetics. 1982b;101:139–155. doi: 10.1093/genetics/101.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T, Osborne B. The 3D's of apoptosis: death, degradation and DIAPs. Nat Cell Biol. 2002;4:E149–E151. doi: 10.1038/ncb0602-e149. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Truman JW, Craig GB., Jr Hybridization between Aedes hendersoni and Aedes triseriatus. Ann Entomol Soc Am. 1968;61:1020–1025. doi: 10.1093/aesa/61.4.1020. [DOI] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Miller LK. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol Cell Biol. 1998;18:3300–3309. doi: 10.1128/mcb.18.6.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HAJ, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Watts DM, Pantuwatana S, Defoliart GR, Yuill TM, Thompson WH. Transovarial transmission of La Crosse virus (California Encephalitis Group) in the mosquito, Aedes Triseriatus. Science. 1973;182:1140–1141. doi: 10.1126/science.182.4117.1140. [DOI] [PubMed] [Google Scholar]
- Watts DM, Pantuwatana S, Yuill TM, Defoliart GR, Thompson WH, Hanson RP. Transovarial transmission of La Crosse virus in Aedes triseriatus. Annals New York Acad Sci. 1975;266:135–143. doi: 10.1111/j.1749-6632.1975.tb35094.x. [DOI] [PubMed] [Google Scholar]
- Watts DM, Thompson WH, Yuill TM, Defoliart GR, Hanson RP. Overwintering of La Crosse virus in Aedes triseriatus. Am J Trop Med Hyg. 1974;23:694–700. doi: 10.4269/ajtmh.1974.23.694. [DOI] [PubMed] [Google Scholar]
- Watts DM, Wright RE, Hanson RP, Defoliart GR, Morris CD. Transmission of La Crosse virus (California Encephalitis Group) by mosquito Aedes triseriatus. J Med Entomol. 1972;9:125–127. doi: 10.1093/jmedent/9.2.125. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wondji C, Hemingway J, Ranson H. Identification and analysis of Single nucleotide polymorphisms (SNPs) in the mosquito Anopheles funestus, malaria vector. BMC Genomics. 2007;8:5. doi: 10.1186/1471-2164-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring J, Chandler LJ, Oray CT, McGaw MM, Blair CD, Beaty BJ. Short report: Diapause, transovarial transmission, and filial infection rates in geographic strains of La Crosse virus-infected Aedes triseriatus. Am J Trop Med Hyg. 1998;58:587–588. doi: 10.4269/ajtmh.1998.58.587. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-Statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
- Wu G, Chai JJ, Suber TL, Wu JW, Du CY, Wang XD, Shi YG. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.