Summary
The critical initial step in V(D)J recombination, binding of RAG1 and RAG2 to recombination signal sequences flanking antigen receptor V, D, and J gene segments, has not previously been characterized in vivo. Here we demonstrate that RAG protein binding occurs in a highly focal manner to a small region of active chromatin encompassing Igκ and Tcrα J gene segments and Igh and Tcrβ J and J-proximal D gene segments. Formation of these small RAG-bound regions, which we refer to as recombination centers, occurs in a developmental stage- and lineage-specific manner. Each RAG protein is independently capable of specific binding within recombination centers. While RAG1 binding was detected only at regions containing recombination signal sequences, RAG2 binds at thousands of sites in the genome containing histone 3 trimethylated at lysine 4. We propose that recombination centers coordinate V(D)J recombination by providing discrete sites within which gene segments are captured for recombination.
Introduction
The vertebrate adaptive immune system recognizes antigens through the use of a highly diverse repertoire of immunoglobulins (Igs) and T cell receptors (Tcrs) expressed on the surface of mature B and T lymphocytes. The genes encoding Ig and TCR polypeptides are assembled during lymphocyte development by V(D)J recombination, a site-specific recombination reaction named for the arrays of V (variable), D (diversity), and J (joining) gene segments that are its substrates. The mechanisms that regulate V(D)J recombination are of considerable interest not only because of the central role the reaction plays in adaptive immunity and lymphocyte development, but also because of the strong connection that has emerged between aberrant V(D)J recombination, genomic instability, and the development of lymphoid malignancies (Lieber et al., 2006; Mills et al., 2003).
V(D)J recombination is initiated when the proteins encoded by the recombination activating genes RAG1 and RAG2, probably together with high mobility group protein HMGB1 or HMGB2, bind to recombination signal sequences (RSSs) that flank V, D, and J gene segments (Swanson, 2004) (Fig. 1A). The RAG proteins then introduce DNA double strand breaks between the RSSs and gene segments, and the reaction is completed by DNA end processing and ligation mediated by non-homologous end joining repair factors (Gellert, 2002). RSSs consist of conserved heptamer and nonamer elements separated by a spacer whose length is either 12 or 23 bps (12RSS or 23RSS, respectively). Efficient recombination requires a 12RSS and 23RSS, a restriction known as the 12/23 rule. DNA cleavage occurs by a two step mechanism involving nicking of one strand followed by attack on the other strand by the liberated 3′ hydroxyl group to generate hairpin sealed coding ends and blunt signal ends (Fig. 1B) (McBlane et al., 1995). Biochemical experiments suggest a “capture” model for the initiation of V(D)J recombination in which RAG1/RAG2/HMGB1/2 first bind to one RSS and then capture a second RSS lacking bound RAG proteins to form the synaptic or paired complex (Fig. 1A) (Jones and Gellert, 2002; Mundy et al., 2002).
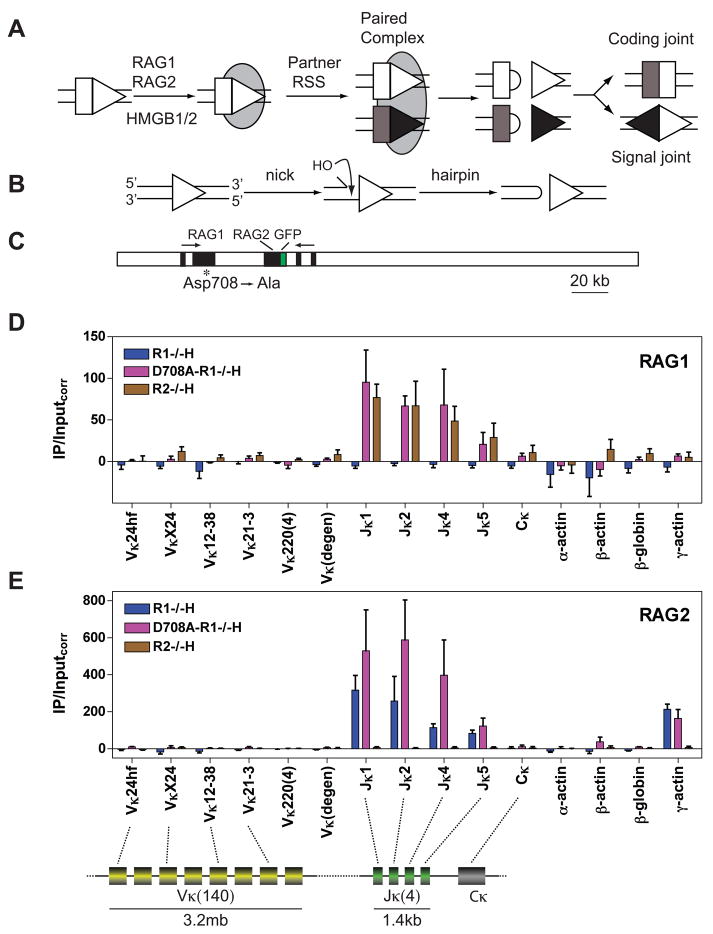
Figure 1. RAG binding to the Igκ locus in pre-B cells.
(A) Steps in V(D)J recombination. RAG1-RAG2-HMGB1/2 complex is represented as a gray oval, RSSs as triangles, and coding segments as rectangles.
(B) DNA cleavage by the RAG proteins.
(C) The D708A BAC. RAG1 and RAG2 exons are represented as black boxes and the direction of transcription is indicated with arrows. The RAG2 locus expresses green fluorescence protein (GFP) instead of RAG2. A mutation introduced into Rag1 (asterisk) changes the codon for Asp708 to an alanine codon.
(D-E) Binding of RAG1 (D) or RAG2 (E) was assessed by ChIP in primary CD19+ bone marrow B-lineage cells from Rag1−/− B1-8i Igh knockin (R1−/−H), D708A transgene-positive Rag1−/− B1-8i Igh knockin (D708A-R1−/−H), and Rag2−/− B1-8i Igh knockin (R2−/−H) mice. DNA recovery in immunoprecipitates was measured by qPCR using primers that detect four individual Vκ gene segments, four closely related Vκ gene segments (Vκ220), a group of about 40 Vκ gene segments (Vκ(degen)) (Curry et al., 2005), the four individual functional Jκ gene segments, the constant region exon (Cκ), and four non-antigen receptor genes as indicated below each graph and in the schematic diagram of the Igκ locus at the bottom of the figure. The diagram indicates only the relative locations of the various Igκ gene segments and is not drawn to scale. IP/Inputcorr values have been corrected for background and normalized to the input signal as described in Experimental Procedures, with bars indicating the mean of three independent experiments and error bars representing the SEM. See also Figs. S1–S4.
RAG1 plays direct roles in both RSS binding and DNA cleavage. It contains domains that interact with the nonamer and heptamer (De and Rodgers, 2004; Swanson, 2004) as well as three acidic amino acids (D600, D708, and E962) that coordinate divalent metal ions and are essential for DNA cleavage (Fugmann et al., 2000; Kim et al., 1999; Landree et al., 1999). The functions of RAG2 are less well understood. It interacts with RAG1, enhances the specificity and affinity of RSS binding, and is essential for DNA cleavage. While RAG2 has no detectable DNA binding activity by itself (Swanson, 2004), it contains a plant homeodomain (PHD) finger that recognizes histone H3 trimethylated at lysine 4 (H3K4me3) (Liu et al., 2007; Matthews et al., 2007). This interaction is important for V(D)J recombination in vivo (Liu et al., 2007; Matthews et al., 2007) and stimulates the cleavage activity of the RAG proteins in vitro (Shimazaki et al., 2009).
V(D)J recombination is tightly regulated in a lineage- and developmental stage-specific manner (Cobb et al., 2006). Current evidence indicates that the reaction is controlled primarily at the step of RAG-mediated DNA cleavage, and that this in turn is controlled by three general mechanisms: restriction of RAG expression to developing lymphocytes, regulation of the physical accessibility of substrate RSSs to RAG binding through the modulation of chromatin structure, and regulation of synapsis through the control of long range chromosome conformation (Cobb et al., 2006; Jhunjhunwala et al., 2008; Jung et al., 2006; Krangel, 2007).
Many experiments have helped to establish a tight link between the ability of a gene segment to participate in V(D)J recombination and an “open” or “accessible” chromatin configuration, as reflected by transcription, activating histone modifications, nuclease sensitivity, and the movement of loci away from repressive nuclear compartments (Cobb et al., 2006; Jung et al., 2006; Krangel, 2007). Importantly, when isolated lymphocyte nuclei were incubated with the RAG proteins, RSS cleavage occurred in a lineage- and developmental stage-appropriate manner, directly linking chromatin structure to the ability of the RAG proteins to initiate V(D)J recombination (Stanhope-Baker et al., 1996). Positioning an RSS within a nucleosome strongly inhibits cleavage by the RAG proteins in vitro (Golding et al., 1999; Kwon et al., 1998). The current model is that cis-acting accessibility control elements, such as enhancers and promoters, recruit chromatin modifying and remodeling enzymes and RNA polymerase II, which act together to free RSSs from repressive associations with chromatin factors and allow RAG binding.
Given the importance of RSS recognition by the RAG proteins in the initiation of V(D)J recombination, it is remarkable that the pattern of binding of RAG1 and RAG2 to antigen receptor loci remains unknown. Patterns of recombination, RSS cleavage, and chromatin accessibility have provided indirect information as to where the RAG proteins are likely to be bound, but these measures are inadequate because RSS binding need not result in DNA cleavage and because it is unknown whether current measures of accessibility adequately capture the features required for RAG binding. This has left numerous fundamental questions concerning the regulation of V(D)J recombination unanswered: Is RAG binding to the RSS a tightly regulated step? Can the initial binding event occur at either type of gene segment involved, or do the RAG proteins consistently bind one type of gene segment first? In large arrays of similar gene segments, do the RAG proteins bind uniformly across the array or in an uneven or focal manner?
Here, we use chromatin immunoprecipitation (ChIP) to demonstrate that RAG binding is tightly regulated during lymphocyte development and focuses on a small region encompassing J (and where present, J-proximal D) gene segments in the Igh, Igκ, Tcrβ, and Tcrα loci. These regions, which we refer to as a recombination centers (Jung et al., 2006), are rich in activating histone modifications and RNA polymerase II. RAG2 binds very broadly in the genome at sites with substantial levels of H3K4me3, while RAG1 binding is more tightly restricted and likely requires direct interaction with the RSS. Surprisingly, in most loci examined, each RAG protein exhibits its specific binding pattern in the absence of the other, suggesting the possibility of several distinct pathways for the recruitment of the RAG proteins into recombination centers. We propose that recombination centers are specialized sites of high local RAG concentration that facilitate RSS binding and synapsis and help regulate recombination order, fidelity, and perhaps allelic exclusion during V(D)J recombination.
Results
Tools for the detection of RAG protein binding in vivo by ChIP
Wild type (WT) developing lymphocytes contain a heterogeneous mixture of V(D)J recombination events and byproducts that could complicate the interpretation of ChIP data involving antigen receptor loci. To avoid this problem, we generated transgenic mice expressing an Asp708→Ala active site mutant RAG1 protein that interacts with RAG2 and binds DNA normally but lacks catalytic activity (Fugmann et al., 2000; Kim et al., 1999; Landree et al., 1999). The D708A RAG1 mutation was engineered into a bacterial artificial chromosome (Fig. 1C) previously demonstrated to accurately recapitulate the endogenous pattern of RAG expression in transgenic mice (Yu et al., 1999). One founder line (copy number of ≈ 4) with an appropriate pattern of transgene expression (data not shown) was selected for further analysis.
The D708A transgene was bred onto a Rag1−/− background to generate D708A-R1−/− mice, which exhibit blocks at the pro-B and pro-T stages of lymphocyte development equivalent to those of R1−/− mice (Fig. S1A, C). To allow analysis of pre-B and pre-T cells, D708A-R1−/− mice were bred with mice harboring a functionally rearranged B1-8i Igh allele (Sonoda et al., 1997) or the 2B4 Tcrβ transgene (Berg et al., 1989) to generate D708A-R1−/− H or D708A-R1−/− β mice, respectively. These mice exhibit developmental arrests at the pre-B and pre-T stages of development, respectively, equivalent to those observed in Rag1−/− x B1-8i (R1−/− H) and Rag1−/− x 2B4 (R1−/− β) mice (Fig. S1B, D), and express D708A RAG1 and RAG2 protein in thymocytes and bone marrow B lineage cells (Fig. S2A, B). These data demonstrate that the D708A transgene expresses RAG1 but does not support lymphocyte development, presumably because it does not support V(D)J recombination. To confirm this, pre-B and pre-T cells from D708A-R1−/− H and D708A-R1−/− β mice, respectively, were examined for Igκ and Tcrα locus rearrangements by PCR. As expected, no rearrangements of either locus could be detected (Figs. S2C, D). Therefore, D708A-R1−/− mice provide a system in which RAG protein binding to Ig and Tcr loci can be assessed in the absence of V(D)J recombination.
ChIP experiments were performed with polyclonal rabbit α-RAG1 or α-RAG2 antibodies (Leu and Schatz, 1995), and with antibodies that recognize acetylated H3 at lysines 9 and 14, H3K4me3, and RNA polymerase (RNAP) II. Immunoprecipitated and input chromatin samples were analyzed by qPCR using primers (Table S1) that amplify a region adjacent to or spanning the RSS of antigen receptor gene segments as well as various other genomic regions. The amount of DNA immunoprecipitated with specific antibodies was corrected for the amount recovered with control rabbit IgG, yielding corrected IP/input values (IP/Inputcorr) (see Experimental Procedures).
The Igκ locus
The Igκ locus contains ≈ 140 Vκ gene segments and 4 functional Jκ gene segments (Fig. 1, bottom) with initial Igκ rearrangements showing a bias in favor of Jκ1 (Victor et al., 1994). RAG binding to Igκ was analyzed in CD19+ bone marrow B lineage cells (almost entirely pre-B cells) purified from D708A-R1−/− H, R1−/− H, and Rag2-deficient x B1-8i (R2−/− H) mice. The RAG proteins were found to associate with the J portion of the locus, with strongest binding observed at the Jκ1, 2, and 4 gene segments (Fig. 1D, E, pink bars). In contrast, little or no binding could be detected to numerous different Vκ gene segments. The specificity of the ChIP assays was demonstrated by the lack of signal obtained at any region from R1−/− H cells with the α-RAG1 antibody (Fig. 1D, blue bars) or from R2−/− H cells with the α-RAG2 antibody (Fig. 1E, brown bars) In addition, no binding was detected to Cκ or several control loci (α-actin, β-globin, and β-actin) (Fig. 1D, E).
Interestingly, this pattern of binding was recapitulated in cells expressing only RAG1 or only RAG2: binding of WT RAG1 to Jκ but not Vκ gene segments was observed in R2−/− H cells (Fig. 1D, brown bars), and binding of RAG2 to Jκ but not Vκ gene segments was observed in R1−/− H cells (Fig. 1E, blue bars). These data indicate that each RAG protein has a mechanism for specific localization within chromatin.
While D708A RAG1 is known to recapitulate the DNA binding behavior of WT RAG1 in biochemical assays, it was important to verify that this is also the case in vivo. RAG binding to Igκ was assessed in a v-Abl transformed pre-B cell line with inducible expression of WT RAG1 and RAG2 (data not shown), and in pre-B cells purified from WT mice (Fig. S3A-C). In both cases, WT RAG1 binds strongly to Jκ but not Vκ gene segments. These data argue that D708A RAG1 retains normal DNA binding activity in lymphocytes and that D708A-R1−/− mice provide a suitable tool for the analysis of RAG binding in vivo.
Additional ChIP experiments revealed that the region of Igκ to which the RAG proteins bind corresponds to a domain of highly active chromatin. Levels of H3K4me3, H3 acetylation, and association with RNAP II were much higher at Jκ than at Vκ (Fig. S4). This is consistent with the well documented correlation between an accessible chromatin configuration, transcription, and V(D)J recombination, and raises the possibility that certain features of the chromatin in such open domains facilitate the binding of the RAG proteins to RSSs.
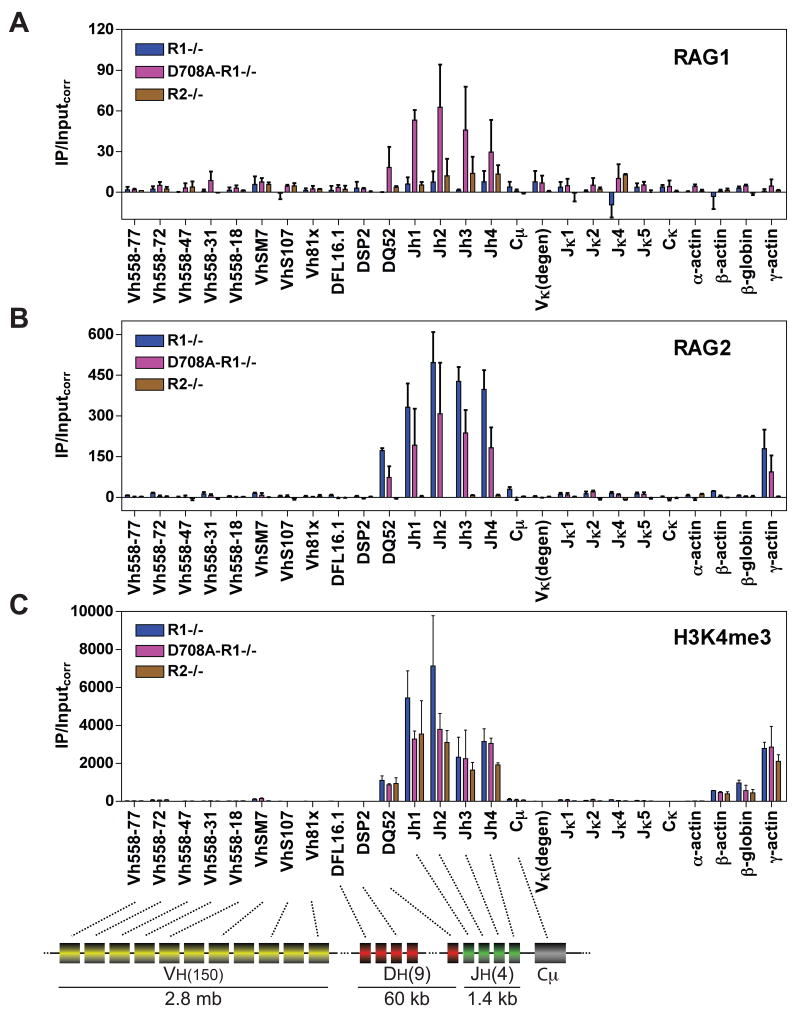
Igκ rearrangement is strongly upregulated during the pro-B to pre-B cell transition, but it had not previously been possible to determine whether this is regulated at the level of RAG binding. When we assessed RAG binding in CD19+ bone marrow pro-B cells purified from D708A-R1−/−, R1−/−, and R2−/− mice (Fig. 2), we found that, indeed, RAG1 and RAG2 binding is greatly upregulated at Jκ gene segments in pre-B cells as compared to pro-B cells (compare Figs. 1D, E with 2A, B), as is H3K4me3 (Fig. 2C), H3 acetylation, and association with RNAP II (data not shown). Hence, the onset of efficient Igκ rearrangement corresponds to a dramatic increase in measures of accessibility and RAG protein binding at Jκ gene segments, arguing that RAG binding is an important regulated step in the initiation of Igκ recombination.
Figure 2. RAG binding to the Igh and Igκ loci in pro-B cells.
(A-C) Binding of RAG1 (A), RAG2 (B), or levels of H3K4me3 (C) were assessed by ChIP in primary CD19+ bone marrow B-lineage cells from Rag1−/− (R1−/−), D708A transgene-positive Rag1−/− (D708A-R1−/−), and Rag2−/− (R2−/−) mice at the gene segments or regions indicated. Data are the average of 2 independent experiments and are presented as in Fig. 1. Cμ, Igμ constant region. See also Figs. S1, S3, and S5.
In summary, the Igκ locus exhibits developmentally regulated focal binding of the RAG proteins to a small, highly transcribed domain that contains the Jκ gene segments and their flanking 23RSSs.
The Igh locus
The Igh locus of C57BL/6 mice consists of ≈ 150 V gene segments, 9 functional D gene segments, and 4 functional J gene segments (Fig. 2, diagram) (Jung et al., 2006; Ye, 2004), with only 700 bp separating the 3′ most D gene segment (DQ52) from Jh1. In pro-B cells, RAG1 and RAG2 binding was observed in a small domain encompassing the Jh elements and closely linked DQ52 gene segment, but could not be detected at Vh gene segments, a DSP2 gene segment, or the frequently recombined DFL16.1 gene segment (Fig. 2A, B). A nearly identical pattern of binding was observed upon induction of expression of D708A RAG1 and RAG2 in the v-abl transformed cell line D345 (data not shown). RAG2 binding was easily detected in the absence of RAG1, in a pattern identical to that observed in the presence of RAG1 (Fig. 2B). However, RAG1 binding in R2−/− cells was not clearly distinguishable from the background established by R1−/− cells and was much lower than that observed in the presence of RAG2 (Fig. 2A). This suggests that RAG2 facilitates RAG1 recruitment/retention at Igh. As was observed at the Igκ locus, RAG binding correlates tightly with high levels of H3K4me3 (Fig. 2C), H3 acetylation, and RNAP II occupancy (data not shown).
The Tcrα locus
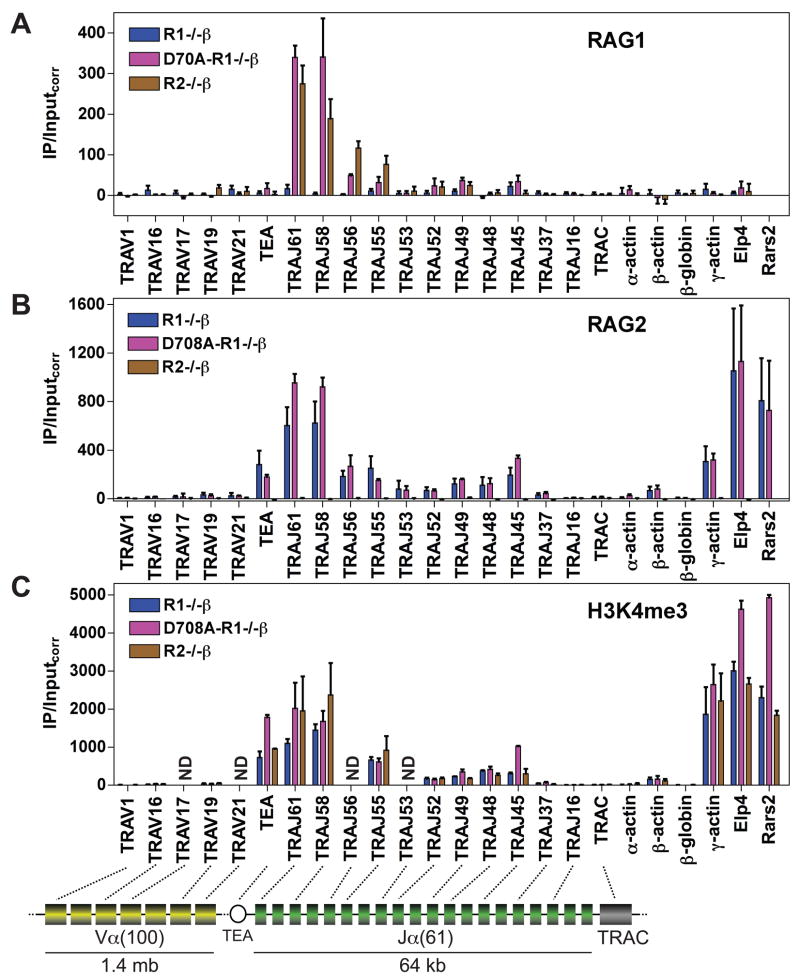
The Tcrα locus contains ≈ 100 Vα (TRAV) gene segments and 61 Jα (TRAJ) gene segments (Fig. 3, diagram). Initial rearrangements involve preferential use of more 3′ Vα gene segments and focus on the most 5′ Jα gene segments because of the strong T early-α (TEA) promoter that lies 2 kb upstream of TRAJ61 (Krangel, 2007).
Figure 3. RAG binding to the Tcrα locus in pre-T cells.
(A-C) Binding of RAG1 (A), RAG2 (B), or levels of H3K4me3 (C) were assessed by ChIP in primary thymocytes from Rag1−/− 2B4 Tcrβ transgenic (R1−/−β), D708A transgene-positive Rag1−/− 2B4 Tcrβ transgenic (D708A-R1−/−β), and Rag2−/− 2B4 Tcrβ transgenic (R2−/−β) mice at the gene segments or regions indicated. Data are presented as in Fig. 1 and are the average of three (RAG1 and H3K4me3) or four (RAG2) independent experiments except for TRAJ56 and TRAJ53, which were analyzed twice for RAG1 and RAG2. ND, not done; TRAC, Tcrα constant region; TEA, TEA promoter. See also Figs. S1 and S3.
RAG protein binding to Tcrα was analyzed in total thymocytes (almost entirely CD4+CD8+ pre-T cells; Fig. S1D) from D708A-R1−/− β, R1−/− β, and R2−/− β mice. Focal binding of both RAG1 and RAG2 to the 5′ portion of the Jα cluster was observed, with the strongest association seen at the most 5′ TRAJ gene segments analyzed (TRAJ61 and TRAJ58) (Fig. 3A, B). Binding was also detected in the region spanning TRAJ56 to TRAJ45 but not at more 3′ TRAJ gene segments, Cα (TRAC), or the five Vα gene segments tested. Each RAG protein, when expressed alone, was able to recapitulate the binding pattern observed when the two RAG proteins were coexpressed (Fig. 3A, B). And as observed at Ig loci, RAG binding occurred in a chromatin region of high H3K4me3 (Fig. 3C), H3 acetylation, and RNAP II occupancy (data not shown).
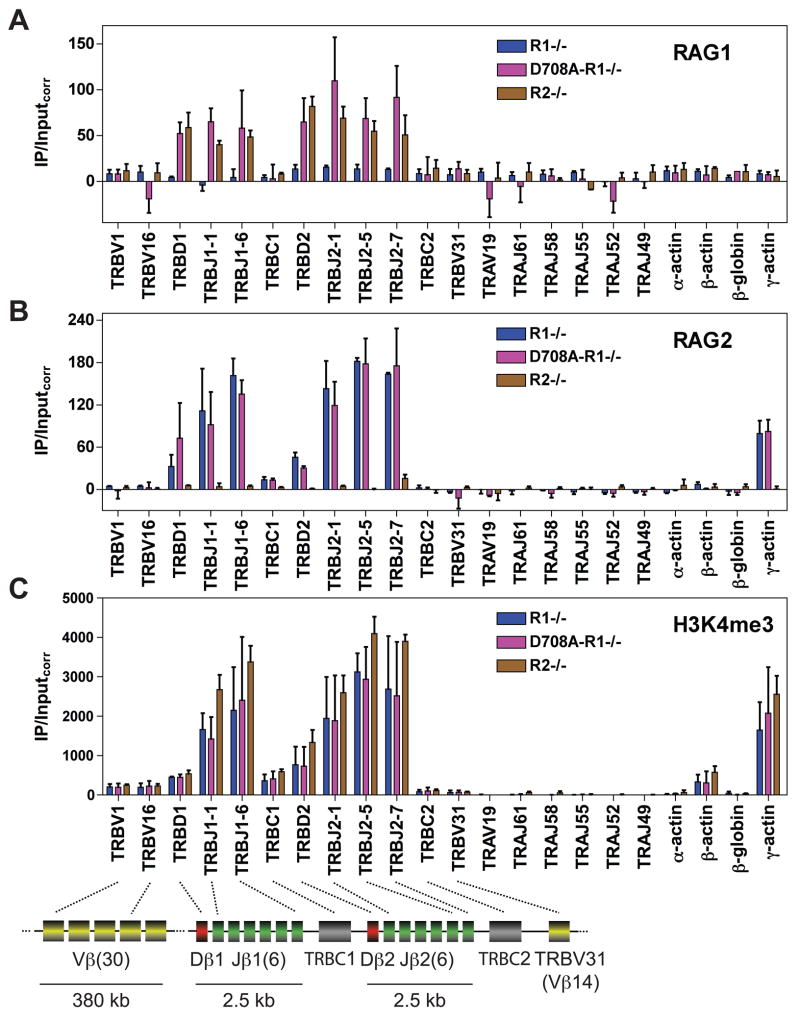
Recombination of Tcrα does not occur in pro-T cells despite high level Rag1 and Rag2 expression and efficient recombination of Tcrβ. Is this regulated at the level of RAG binding? When we analyzed total thymocytes (almost entirely CD4−CD8− pro-T cells; Fig. S1C) from D708A-R1−/−, R1−/−, and R2−/− mice, no binding of RAG was observed to any of the Tcrα gene segments analyzed, including TRAJ61 and TRAJ58 (Fig. 4A, B). These regions have very low levels of H3K4me3 (Fig. 4C), H3-acetylation, and RNAP II association (data not shown) in pro-T cells. We conclude that developmental regulation of locus accessibility and RAG binding represents one mechanism by which Tcrα locus recombination is prevented in pro-T cells.
Figure 4. RAG binding to the Tcrβ and Tcrα loci in pro-T cells.
(A-C) Binding of RAG1 (A), RAG2 (B), or levels of H3K4me3 (C) were assessed by ChIP in primary thymocytes from Rag1−/− (R1−/−), D708A transgene-positive Rag1−/− (D708A-R1−/−), and Rag2−/− (R2−/−) mice at the gene segments or regions indicated. Data are the average of three (RAG1) or two (RAG2 and H3K4me3) independent experiments and are presented as in Fig. 1. TRBC1 and TRBC2, Tcrβ constant regions 1 and 2; TRBD1 (Dβ1) and TRBD2 (Dβ2), red rectangles. See also Figs. S1, S3, and S5.
In summary, in pre-T but not pro-T cells the RAG proteins bind to a discrete portion of the Tcrα locus containing 5′ Jα gene segments. These Jα gene segments correspond closely to those involved in initial Tcrα rearrangements.
The Tcrβ locus
The Tcrβ locus contains 31 Vβ (TRBV) gene segments and two D-J-Cβ clusters, each containing one Dβ (TRBD) and six functional Jβ (TRBJ) gene segments, with only ≈650 bp separating Dβ from the nearest Jβ gene segment. In pro-T cells, binding of RAG1 and RAG2 was detected at both D-Jβ clusters, but not at TRBC1, TRBC2, or the three Vβ gene segments assayed (Fig. 4A, B, pink bars). This binding pattern was recapitulated in cells expressing either RAG1 or RAG2 alone (Fig. 4A, brown bars; Fig. 4B, blue bars). As with the other antigen receptor loci analyzed, RAG binding to Tcrβ correlates strongly with high levels of H3K4me3 (Fig. 4C), H3-acetylation and RNAP II association (data not shown). We conclude that in all four antigen receptor loci examined, RAG binding occurs detectably in a very small, highly accessible region containing J (and where present, J-proximal D) gene segments.
Persistent RAG binding to Igh in pre-B cells and Tcrβ in pre-T cells
Recombination of the Igh and Tcrβ loci is regulated to ensure that each B or T lymphocyte expresses Igh or Tcrβ protein, respectively, from only a single allele. Allelic exclusion at these loci is thought to be mediated by a feedback mechanism in which assembly of a productive VDJ allele leads to inhibition of further Igh or Tcrβ V-to-DJ recombination (Cobb et al., 2006; Jung et al., 2006; Krangel, 2007). To test whether allelic exclusion might be mediated in part by a failure of the RAG proteins to reassemble robust recombination centers on the Igh and Tcrβ loci in pre-B and pre-T cells, we examined RAG binding to Igh and Tcrβ in these cells.
The results were similar to those obtained in pro-B and pro-T cells: strong binding of RAG1 and RAG2 at the DQ52-Jh region in pre-B cells (Fig. S5A, B) and the Dβ-Jβ regions in pre-T cells (Fig. S5C, D), with little or no signal observed at DFL16.1 or any Vh or Vβ gene segment. We also detect RAG binding to the J and proximal D but not V portions of Igh and Tcrβ in purified WT pre-B and pre-T cells, respectively (see below, and data not shown), arguing that these results are not simply a consequence of the accelerated development caused by providing a functional Igh or Tcrβ allele in the germline. Levels of H3-acetylation and RNAP II remain high in the DQ52-Jh region in pre-B cells and in the Dβ-Jβ regions in pre-T cells (data not shown). Together, our results indicate that RAG1 and RAG2 efficiently associate with the J/proximal D regions of the Igh and Tcrβ loci in pre-B/pre-T cells. This supports the current model that feedback inhibition of Igh and Tcrβ recombination operates primarily on Vh and Vβ gene segments, respectively, and is consistent with previous data demonstrating that the D-J portions of these loci are competent for recombination in pre-B/pre-T cells ((Jung et al., 2006; Whitehurst et al., 1999) and references therein).
Lineage specificity of RAG binding
Tcr loci recombine in developing T but not developing B cells while the reciprocal is true for recombination of Ig loci, with the exception that Igh D-to-J recombination can be detected in thymocytes (Jung et al., 2006). We find that RAG binding exhibits a similar lineage specificity: RAG1 and RAG2 do not bind detectably to Dβ, Jβ, or Jα gene segments in WT pre-B cells, or to Vκ or Jκ gene segments in WT pre-T cells, while there appears to be a low level of binding in the vicinity of Jh1 in pre-T cells (Fig. S3D, E). These results indicate that the RAG proteins bind in a lineage-specific manner, consistent with previous data demonstrating tissue-specific RAG cleavage activity in isolated nuclei (Stanhope-Baker et al., 1996).
The RSS contributes to RAG recruitment
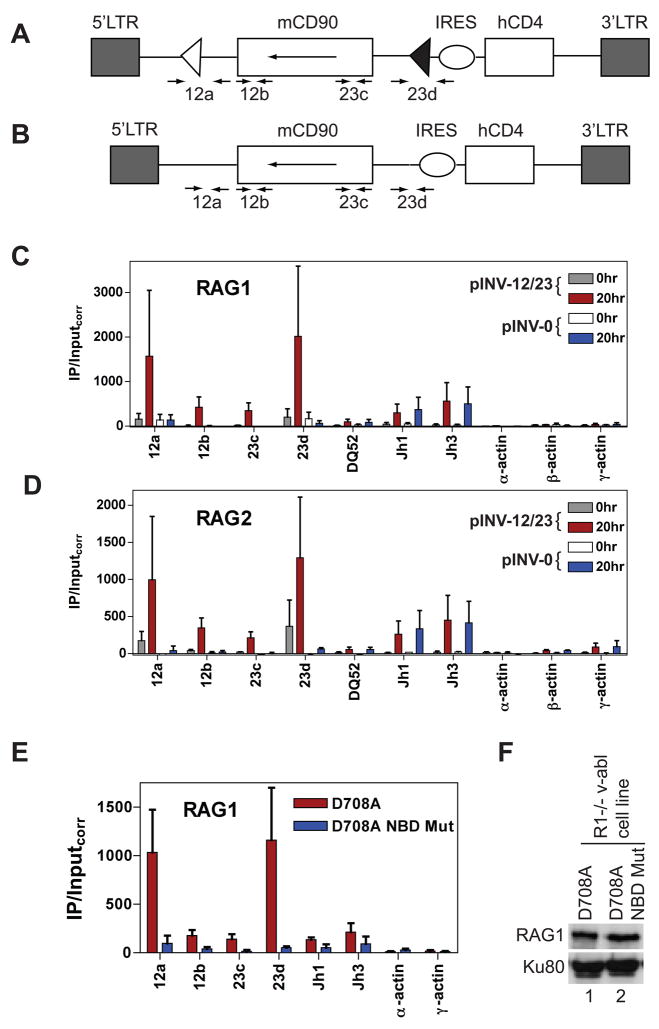
To explore the role of the RSS in RAG recruitment, we created retroviral recombination substrates containing (pINV-12/23) or lacking (pINV-0) a 12RSS and a 23RSS (Fig. 5A, B). We introduced these substrates into a pro-B cell line that expresses D708A RAG1 and RAG2 and performed ChIP on polyclonal populations of infected cells. After induction of RAG expression for 20 hours, much stronger binding of D708A RAG1 and RAG2 was detected to pINV-12/23 than to pINV-0 (Fig. 5C, D). Similar results were obtained with a second group of independently derived populations of infected cells (data not shown). Equivalent RAG binding was observed at the endogenous Jh1 and Jh3 gene segments in pINV-12/23 and pINV-0 infected cells (Fig. 5C, D), suggesting equivalent induction of RAG binding activity in the two cell populations. This and Western blot (Fig. S6A), substrate expression (Fig. S6B) and histone modification (Fig. S6C) data strongly argue that the large difference in RAG binding observed between pINV-12/23 and pINV-0 is not due to differences in RAG expression or binding activity, or in substrate accessibility. We conclude that the RSSs are required for strong binding of RAG1 and RAG2 to the recombination substrate under these assay conditions. Consistent with an important role for the RSS, mutation of RAG1 at three residues (R391A, R393A, and R402A) known to make critical contacts with the nonamer (Yin et al., 2009) dramatically reduced recruitment of D708A RAG1 to the RSSs of pINV-12/23 (Fig. 5E, F).
Figure 5. The RSS and nonamer binding domain are important for RAG binding.
(A and B) The pINV-12/23 (A) and pINV-0 (B) recombination substrates. The 5′ and 3′ long terminal repeats (LTR), mouse CD90 gene (mCD90) encoding Thy1.1, and human CD4 gene (hCD4) are indicated as rectangles, the internal ribosome entry site (IRES) as an oval, and the 12RSS and 23RSS as white and black triangles, respectively. The mCD90 gene lies in opposite transcriptional orientation (long arrow) to that of transcription originating in the 5′LTR. The positions of PCR primers are indicated with short arrows. Primer pairs 12a and 23d span the 12RSS and 23RSS respectively in pINV-12/23, and generate 109 bp (12a) and 61 bp (23d) smaller products with pINV-0 than with pINV-12/23 due to deletion of the RSSs. Primer pairs 12b and 23c lie 69 bp 3′ of the 12RSS and 91 bp 5′ the 23RSS, respectively. Not drawn to scale.
(C-D) Binding of RAG1 (C) or RAG2 (D) were assessed by ChIP in the D345 v-abl transformed cell line infected with pINV-12/23 (gray and red bars) or pINV-0 (white and blue bars) either prior to RAG induction (0hr) or after 20 hours of RAG induction (20hr) at the gene segments or regions indicated. Data are the average of two independent experiments and are presented as in Fig. 1. See also Fig. S6.
(E) Binding of RAG1 was assessed in a R1−/− cell line containing pINV-12/23 and infected with a retrovirus expressing D708A RAG1 (red bars) or nonamer binding domain (NBD) mutant D708A RAG1 (blue bars) and a linked blastocydin resistance gene. Data were collected from blastocydin-resistant clones 20 hours after treatment with STI-571 and are the average of two independent experiments. Similar data were obtained using a second set of independently derived clones (data not shown).
(F) Western blot of D708A RAG1 and NBD-mutant D708A RAG1 proteins in infected R1−/− v-abl cells. A separate blot of the same protein extracts was probed for Ku80 to confirm equal loading.
RAG2 binds throughout the genome at sites of H3K4me3
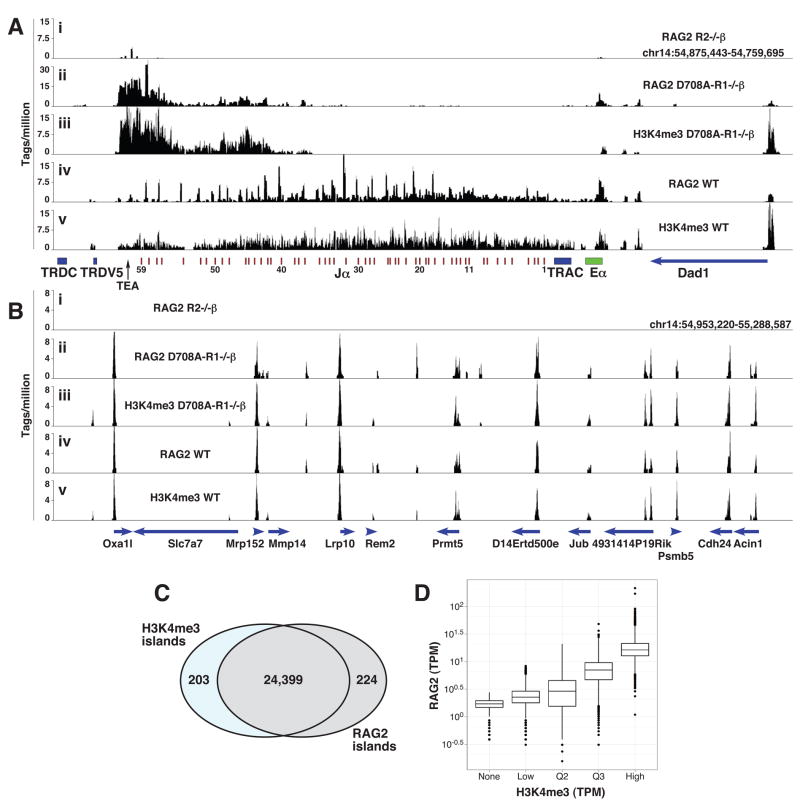
Our qPCR-ChIP analyses revealed several examples of RAG2 binding to sites with substantial H3K4me3 but lacking RSSs: the γ-actin promoter in pro-B, pre-B, pro-T, and pre-T cells (Figs. 1–4), and the TEA, Elp4 and Rars2 promoters in pre-T cells (Fig. 3). To examine this on a genome-wide basis, we performed ChIP-sequence (ChIP-seq) analysis of RAG2 binding and H3K4me3 in total thymocytes from WT and D708A-R1−/− β mice (and control R2−/− β thymocytes for the RAG2 antibody). At least 7 million sequence tags were mapped to the mouse genome in each ChIP-seq experiment (Table S2). The results reveal an extraordinary spatial and quantitative correlation between RAG2 binding and H3K4me3 (Fig. 6). RAG2 and H3K4me3 were each detected at over 24,000 peaks or “islands” (see Supplemental Methods), with a greater than 99% overlap in the locations of these islands (Fig. 6C). Islands containing higher levels of H3K4me3 tended to have higher levels of RAG2 binding (Fig. 6D). Analysis of the germline Tcrα locus in D708A-R1−/− β mice revealed RAG2 binding at the 5′ end of the Jα cluster (Fig. 6A, panel ii) in a pattern very similar to that observed by qPCR-ChIP (Fig. 3B). In contrast, RAG2 binding is detected across the entire Jα cluster in WT thymocytes (Fig. 6A, panel iv), presumably because these cells contain a complex mixture of different V-Jα rearrangements, with each Vα promoter conferring accessibility to a small region of downstream Jα gene segments (Hawwari and Krangel, 2007). Consistent with our qPCR-ChIP analysis, little RAG2 binding or H3K4me3 is observed at Vα gene segments (data not shown) relative to Jα. Overall, these data demonstrate that RAG2 is recruited to chromatin throughout the genome in a pattern that mirrors that of H3K4me3. Consistent with the idea that this is mediated by an interaction between the RAG2 PHD domain and H3K4me3, we find that a mutation in the PHD domain of RAG2 (W453A) previously demonstrated to abrogate H3K4me3 binding (Liu et al., 2007; Matthews et al., 2007) greatly reduces association of RAG2 with multiple genes containing H3K4me3 (Fig. S7).
Figure 6. ChIP-seq analysis of RAG2 binding in thymocytes.
(A and B) The number of sequence tags per million mapped tags is plotted in 100 bp windows across a region spanning the Jα gene segments (A) or a gene rich region on chromosome 14 (B). Immunoprecipitations were performed with anti-RAG2 (panels i, ii, and iv) or anti-H3K4me3 (panels iii and v) antibodies using total thymocytes from Rag2−/− 2B4 Tcrβ transgenic (R2−/−β) (panel i), D708A transgene-positive Rag1−/− 2B4 Tcrβ transgenic (D708A-R1−/−β) (panels ii and iii), or WT (panels iv and v) mice. The locations of genes and gene segments are indicated below the X-axes, with the direction of transcription indicated with arrowheads. Eα, Tcrα enhancer; TRDC, Tcrδ constant region.
(C) Venn diagram showing spatial overlap between RAG2 and H3K4me3 islands using ChIP-seq data pooled from WT and D708A-R1−/−β thymocytes.
(D) Combined H3K4me3 islands were separated into 4 groups of equal size (Low, Q2, Q3, High) according to the numbers of reads mapped to the island in the H3K4me3 Chip-Seq experiment; a fifth group of RAG2 islands lacking H3K4me3 reads was also defined (none). The numbers of reads in tags per million aligned reads (tpm) for these five groups in the Rag2 Chip-Seq is shown for each of the groups as a box and whisker plot on a logarithmic scale. See also Fig. S7 and Table S2.
Discussion
No direct assessment of RAG protein binding to antigen receptor loci has previously been performed, leaving important gaps in our understanding of the initiation of V(D)J recombination. Here, we demonstrate that RAG1 and RAG2 associate with a small region encompassing some or all of the J gene segments in the Igκ and Tcrα loci and J and J-proximal D gene segments in the Igh and Tcrβ loci. These RAG-bound regions assemble in a tightly regulated, lineage- and developmental stage-specific manner. Our data provide insights into how the RAG proteins are recruited to antigen receptor loci and have implications for the organizing principles that govern assembly of these large loci as well as for mechanisms that might contribute to aberrant V(D)J recombination and the development of lymphoid tumors.
Initial RSS binding and the ordered assembly of synaptic complexes
Given biochemical evidence for a “capture” model of RAG-mediated DNA cleavage (Jones and Gellert, 2002; Mundy et al., 2002), it is of considerable importance to understand how binding of the first RSS and capture of the second RSS are orchestrated in vivo. Two general scenarios can be envisioned: the binding and capture steps are carefully regulated and occur in a particular order, or the process is not ordered and either partner can be bound first. In the absence of a method to assess RAG binding in vivo, it has been difficult to distinguish between these scenarios.
Based on the detection of nicks at 12RSSs but not 23RSSs in vivo (Curry et al., 2005), it was proposed that V(D)J recombination is initiated by RAG binding to a 12RSS followed by capture of a 23RSS (“12RSS first” model). However, when we directly assess RAG binding in vivo, we do not see a correlation with a particular type of RSS: we detect binding to Jh and Jκ (23RSSs) and to Jβ and Jα (12RSSs), but not to V and distal Dh gene segments, including Vκ and DFL16.1, which are flanked by 12RSSs. One way to reconcile our findings with the 12RSS first model is to propose that most of the RAG-chromatin complexes we detect are synaptic complexes, but there are reasons for doubting that this is the case. First, formation of the stable 12RSS/23RSS synaptic complex requires both RAG1 and RAG2 (Swanson, 2004), and yet cells expressing only RAG1 or only RAG2 largely recapitulate the binding patterns observed in cells expressing both RAG proteins. Second, the pattern of binding observed at the Igh locus cannot readily be explained by invoking synaptic complexes. In this locus, the majority of D-J rearrangement events involve the four Jh gene segments and just two of the nine Dh gene segments (DQ52 and DFL16.1) (Chakraborty et al., 2007). While we observe relatively strong RAG binding at Jh gene segments, binding is weaker at DQ52 and not detected at all at DFL16.1 (Figs. 2 and S5). Were synaptic complexes the principal entity detected by our assay, ChIP signals should have been at least as strong at DQ52 and DFL16.1 as at the Jh gene segments. Synaptic complexes can likely be detected by the ChIP assay, but this appears to occur sufficiently infrequently with any given partner that such binding is below the detection limits of the assay.
We favor the idea that the discrete regions of RAG binding that we observe at J and closely linked Dh or Dβ gene segments are the primary sites of RAG recruitment and RSS binding in each antigen receptor locus. These regions exhibit much higher levels of H3 acetylation, H3K4me3, and bound RNAP II than other portions of the loci, creating favorable conditions for RAG protein recruitment. We propose that initial RSS engagement during V(D)J recombination is not restricted to 12RSSs, but rather occurs with whatever RSSs are found within recombination centers. It remains to be determined why nicking was detected at Vκ but not Jκ gene segments in a v-abl transformed pre-B cell line (Curry et al., 2005). One possibility is that initial engagement of the RAG proteins with chromatin-associated Jκ 23RSSs yields a complex that is not competent for catalysis and that most Vκ nicking occurs within the paired complex. We cannot rule out the possibility of heterogeneous RAG binding in individual cells to a small number of gene segments that make up large (e.g., Vκ) gene segment arrays.
Pathways of Recruitment of RAG1 and RAG2 to DNA
We find a very tight correlation between RAG2 binding and H3K4me3 at all loci and in all cell types examined. Together with the results of previous studies (Liu et al., 2007; Matthews et al., 2007), our data strongly argue that the major factor controlling the association of RAG2 with chromatin is the interaction of its PHD domain with H3K4me3. Elevated levels of H3K4me3 are found in the 5′ portions of active genes and are strongly associated with active or poised RNAP II (Barski et al., 2007; Ruthenburg et al., 2007). Hence, our data indicate that RAG2 associates with many different transcriptionally active regions of the genome. We previously estimated that thymocytes contain, on average, thousands of molecules each of RAG1 and RAG2, with RAG2 in excess over RAG1 (Leu and Schatz, 1995). This, together with the results of our RAG2 ChIP-seq analysis, argues that RAG2 binds to many different sites in the genome of each RAG-expressing lymphocyte. The functional significance of this is unknown: possibilities include a regulatory function in V(D)J recombination (e.g., sequestration of RAG2) or an influence on transcriptional activity, histone modifications, or chromatin structure of non-antigen receptor loci.
Importantly, binding of RAG2 to the non-antigen receptor loci we have examined is not accompanied by detectable binding of RAG1. This suggests that RAG2 need not exist in a stable complex with RAG1 and that developing lymphocytes contain a substantial pool of chromatin-associated RAG2 that is not in complex with RAG1. While this RAG2 by itself would pose no threat to genome integrity, even inefficient recruitment of RAG1 to non-antigen receptor loci by H3K4me3-bound RAG2 would create a risk of aberrant DNA nicks or double strand breaks, particularly at nearby cryptic RSSs, non-B form DNA structures (Raghavan et al., 2004), or DNA mismatches (such as might be created by the activation induced deaminase) (Tsai et al., 2008). RAG2 binding to H3K4me3 increases the catalytic activity of the RAG complex and hence might enhance this risk (Shimazaki et al., 2009), as might the recently described interaction of RAG1 with histone H3 (Grazini et al., 2010). Poor solubility of RAG1 and its tight association with as yet unknown elements of the nucleus have made it difficult to determine the extent to which RAG1 and RAG2 exist in RAG1-RAG2 complexes in vivo (Leu and Schatz, 1995).
RAG1 binding is more restricted than that of RAG2, being detected only in regions that display the two hallmark features of recombination centers: i) highly active chromatin and ii) the presence of arrays of RSSs. The recombination centers defined here contain multiple, closely spaced RSSs, and it remains to be determined whether the number and/or spacing of the RSSs is important for stable recruitment of the RAG proteins. With the exception of the Igh locus, RAG1 does not require RAG2 for binding and it is likely that direct interactions with the RSS play a critical role in RAG1 recruitment to recombination centers. Our findings that RSSs and an intact nonamer binding domain are required for RAG1 binding to a chromosomal recombination substrate (Fig. 5) strongly support this notion. The finding that RAG1-bound RSSs invariably reside in transcribed chromatin marked with activating histone modifications likely reflects the need to free RSSs from repressive interactions with nucleosomes. The ability of RAG1 to bind the RSS in the absence of RAG2 is consistent with the hypothesis that RAG1 evolved from the transposase gene of Transib transposons, which contain terminal inverted repeats resembling RSSs but no RAG2 homologue (Kapitonov and Jurka, 2005).
Our data support the possibility that there are three pathways by which the RAG proteins are recruited to DNA: i) binding of RAG2 to H3K4me3 followed by recruitment of RAG1; ii) binding of RAG1 to the RSS followed by recruitment of RAG2; and iii) binding of a pre-formed RAG1-RAG2 complex to the RSS and/or H3K4me3. A variety of previous findings (Swanson, 2004) suggest that pathway iii is likely to play a significant role in resting and G1 phase cells when RAG1 and RAG2 are co-expressed. The availability of two binding elements (H3K4me3 and the RSS) likely enhances the efficiency with which the RAG proteins are recruited to and retained in recombination centers. In this regard, it is noteworthy that RAG1 binding to Igh, unlike the other antigen receptor loci examined, is dependent on the presence of RAG2 (Figs. 2A and S5A). It remains to be determined what feature of the Igh locus is responsible for this difference. Weak binding of RAG2 to pINV-0 in the D345 cell line (Fig. 5D) represents a situation in which the RSS (presumably together with RAG1) facilitates RAG2 recruitment. Because RAG1 and RAG2 interact with one another, it is plausible that each RAG protein can increase the efficiency and rapidity of binding of the other in ways that are not captured in our analyses of primary lymphocyte populations.
RAG2 is highly unstable in the S, G2, and M phases of the cell cycle (Jiang et al., 2005) and hence developing lymphocytes have periods during which RAG1 is present in the absence of RAG2. We propose that as RAG2 levels rise at the beginning of G1, RAG1 is already localized to recombination centers and facilitates the recruitment of RAG2 (pathway ii above).
Recombination Centers and the Regulation of V(D)J Recombination
Based on our findings and those of others, we propose the following model for the initiation of V(D)J recombination (Fig. 7). Transcriptional control elements work in concert to create a chromatin domain encompassing J (and proximal D) gene segments that is marked by high levels of RNAP II and activating histone modifications. RAG2-H3K4me3 and RAG-RSS interactions lead to efficient and stable recruitment of the RAG proteins into this domain, creating the recombination center. RAG proteins in the recombination center then capture a partner RSS that is RAG-free. Capture might occur through simple diffusive interactions in the case of short range recombination events (e.g., D-to-J recombination), but likely requires large scale reorganization of the chromatin fiber for long range recombination events involving V gene segments (Jhunjhunwala et al., 2008). Based on recent findings from the Murre and Alt laboratories (Bassing et al., 2008; Jhunjhunwala et al., 2008; Ranganath et al., 2008; Wu et al., 2007), we envision that numerous gene segments compete for stable capture by the recombination center in a process that typically involves multiple nonproductive encounters. Formation of the stable synaptic complex activates coordinate double strand DNA cleavage by the RAG proteins, completing the first phase of the recombination reaction.
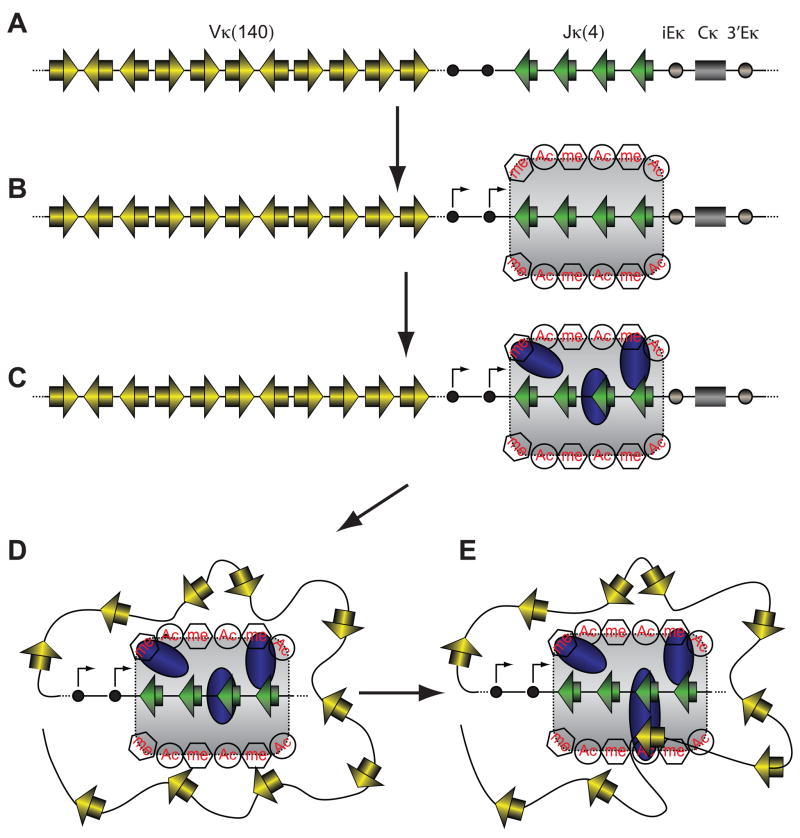
Figure 7. Recombination center model for V(D)J recombination.
(A) Schematic depiction of the Igκ locus showing V and J gene segments (yellow and green rectangles, respectively), 12RSSs and 23RSSs (yellow and green triangles, respectively), germline promoters (black circles), the intronic (iEκ) and 3′ (3′Eκ) enhancers (gray circles) and the constant region (gray rectangle).
(B) Germline promoters and enhancers cooperate to create a domain (gray shaded area) with high levels of germline transcription (arrows) and activating histone modifications including histone acetylation (Ac) and H3K4me3 (me) encompassing the Jκ gene segments.
(C) The RAG proteins (blue ovals) are recruited into this domain by virtue of direct RAG-RSS interactions and binding of the PHD domain of RAG2 to H3K4me3, forming the recombination center. No attempt is made to distinguish between the RAG1-RAG2 complex and the individual RAG proteins.
(D) Large scale reorganization of the chromatin fiber brings V gene segments into close proximity of the recombination center, where they compete for capture by the RAG proteins in the recombination center.
(E) One V gene segment is stably captured in the recombination center in a synaptic complex (large blue oval) containing RAG1, RAG2, the Vκ and Jκ RSSs, and probably HMGB1/2. Within this complex, the RAG proteins introduce double strand breaks between the gene segments and their flanking RSSs (not shown), completing the first phase of V(D)J recombination.
Focal recruitment of the RAG proteins into recombination centers provides an appealing mechanism for regulating the initiation of V(D)J recombination and limiting the possible dangerous outcomes of the reaction. Restricting initial RAG binding to a small fraction of the gene segments within each locus should reduce the generation of potentially recombinogenic DNA nicks (Lee et al., 2004). In the Igh locus, the absence of stable RAG binding to 5′ D gene segments should help prevent direct V-to-D recombination involving these D segments. Perhaps most importantly, recombination centers provide a means of coordinating V(D)J recombination events on a single allele or even between alleles, for example by helping to prevent multiple simultaneous recombination events in a single lymphocyte. Consistent with this idea, RAG-dependent, developmentally-regulated, homologous pairing of Ig alleles has recently been described and suggested to contribute to allelic exclusion of Ig recombination (Hewitt et al., 2009); the recombination center provides an attractive mediator of such pairing and of crosstalk between the two alleles. And given the ability of the RAG proteins to channel DNA double strand breaks into the non-homologous end joining repair pathway (Corneo et al., 2007; Cui and Meek, 2007) and interact (perhaps indirectly) with the Ku proteins (Raval et al., 2008), we speculate that recombination centers also serve as sites for the recruitment and organization of DNA damage response and repair factors, similar to ideas recently proposed by others (Matthews and Oettinger, 2009).
Chromosomal translocations between antigen receptor loci and oncogenes are a hallmark of lymphocytic leukemias and lymphomas and many are thought to arise from V(D)J recombination between a bone fide RSS and a cryptic RSS near an oncogene (Lieber et al., 2006; Marculescu et al., 2006). We propose that in addition to capturing partner gene segments for antigen receptor gene assembly, the RAG proteins in recombination centers sporadically capture and cleave cryptic RSSs in non-antigen receptor loci, leading occasionally to large chromosomal rearrangements or translocations. In keeping with this idea, for translocation events involving the loci we have analyzed, the participating antigen receptor gene segments are almost invariably located within predicted human recombination centers ((Marculescu et al., 2006) and references therein). It is therefore plausible that recombination centers are an important contributor both to the proper regulation of V(D)J recombination and to aberrant recombination events that underlie the generation of B and T cell malignancies.
Experimental Procedures
Mice and Cells
The HG BAC (Yu et al., 1999) was modified to alter the mouse RAG1 nucleotide sequence GAT encoding aspartate 708 to GCG encoding alanine by recombineering in bacteria (see Supplemental Data) and was used to generate transgenic mice on a C57BL/6-SJL background by standard techniques. D708A BAC transgenic mice were bred with C57BL/6 mice and subsequently intercrossed with Rag1-deficient, B1-8i Igh knockin, or 2B4 Tcrβ transgenic mice on a C57BL/6 background, or Rag2-deficient mice on a mixed 129/C57BL/6 background. B cell precursors were enriched from bone marrow using CD19 magnetic beads (Miltenyi Biotec) (final purity > 90%). The R1−/−, R2−/−, and D345 pro-B cell lines were prepared by infection of bone marrow of R1−/−, R2−/−, or D708A-R1−/− Bcl2 transgenic mice with the pMSCV-v-abl retrovirus as described (Bredemeyer et al., 2006). Where indicated, v-abl-transformed cell lines were induced to express RAG with 3 μM STI-571 (Novartis) as described (Bredemeyer et al., 2006). All animal procedures have been approved by the Institutional Animal Care and Use Committee of Yale University.
Chromatin Immunoprecipitation
RAG1- and RAG2-specific rabbit polyclonal antibodies were raised to RAG1 aa 56-123 and RAG2 aa 70-516, respectively (Leu and Schatz, 1995). ChIP was also performed with antibodies for H3K4me3 (#07-473 or #07-745), acetylated H3 (#06-599), normal rabbit IgG (#12-370) (Millipore), and RNAP II (#sc-899X; Santa Cruz Biotechnology). The ChIP procedure is described in detail in Supplemental Data. Briefly, cells were crosslinked with 1% HCHO, quenched with 0.125 M glycine, washed and resuspended in RIPA buffer (10 mM Tris pH 7.4, 1 mM EDTA, 1% tritonX-100, 0.1% sodium deoxycholate, 0.1% SDS) containing 0.8 M NaCl and sonicated using a water bath sonicator (Diagenode) to obtain DNA of ≈ 300–500 bp. After pre-clearing of the chromatin, an aliquot (5 × 105 cell equivalents) was set aside as the input sample. Chromatin from 5 × 106 cells was then incubated with specific antibody or normal rabbit IgG (Millipore) and immune complexes were pulled down with Protein A agarose beads (Millipore). After reversal of crosslinks and purification of the DNA, duplicate Taqman qPCR reactions were performed with Qiagen HotStart Taq with a 3000 XP thermocycler (Stratagene). Table S1 lists the primers and probes used. Input samples were diluted so that IP and input samples would give ≈ equal qPCR signals if 1% of the region of interest were present in the IP sample. Using standard curves generated for each region analyzed in each experiment, the amount of DNA recovered in immunoprecipitates and the input chromatin was calculated. IP/Inputcorr was then calculated as ((IPsp – IPrIg)/Input) × 1000, where IPsp and IPrig are the amount of DNA recovered in IPs with the specific antibody and rabbit IgG, respectively.
The ChIP-Seq procedure and analysis is described in Supplemental Data.
Supplementary Material
Acknowledgments
The authors thank S. Desiderio, R. Sen and R. Subrahmanyam for lentiviral vectors expressing WT and W453A RAG2, M. Nussenzweig for providing the HG BAC, B. Sleckman and A. Bredemeyer for providing cell lines and technical advice, S. Zhou for help with the analysis of mice, J. Duke for analysis of genome-wide H3K4me3 data, the Yale Transgenic Mouse Service for generating transgenic mice, members of the Schatz lab for helpful discussions, the Yale Immunobiology Cell Sorting Facility for cell sorting, and M. Krangel, M. Schlissel, and B. Sleckman for critical reading of the manuscript. This work was supported in part by Public Health Service grant AI32524 to D. G. S. and in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. D. G. S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Whitlow S, Mostoslovasky R, Yang-Iott K, Ranganath S, Alt FW. Vbeta cluster sequences reduce the frequency of primary Vbeta2 and Vbeta14 rearrangements. Eur J Immunol. 2008;38:2564–2572. doi: 10.1002/eji.200838347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg LJ, Pullen AM, Fazekas de St Groth B, Mathis D, Benoist C, Davis MM. Antigen/MHC-specific T cells are preferentially exported from the thymus in the presence of their MHC ligand. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the D-H-C-mu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- Cui X, Meek K. Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc Natl Acad Sci USA. 2007;104:17046–17051. doi: 10.1073/pnas.0610928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: Evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- De P, Rodgers KK. Putting the pieces together: identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazini U, Zanardi F, Citterio E, Casola S, Goding CR, McBlane F. The RING Domain of RAG1 Ubiquitylates Histone H3: A Novel Activity in Chromatin-Mediated Regulation of V(D)J Joining. Mol Cell. 2010;37:282–293. doi: 10.1016/j.molcel.2009.12.035. [DOI] [PubMed] [Google Scholar]
- Hawwari A, Krangel MS. Role for rearranged variable gene segments in directing secondary T cell receptor alpha recombination. Proc Natl Acad Sci USA. 2007;104:903–907. doi: 10.1073/pnas.0608248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Chang FC, Ross AE, Lee JY, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and Control of V(D)J Recombination at the Immunoglobulin Heavy Chain Locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel MS. T cell development: better living through chromatin. Nat Immunol. 2007;8:687–694. doi: 10.1038/ni1484. [DOI] [PubMed] [Google Scholar]
- Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Leu TM, Schatz DG. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol Cell Biol. 1995;15:5657–5670. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Yu KF, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marculescu R, Vanura K, Montpellier B, Roulland S, Le T, Navarro JM, Jager U, McBlane F, Nadel B. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair (Amst) 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AGW, Oettinger MA. RAG: a recombinase diversified. Nat Immunol. 2009;10:817–821. doi: 10.1038/ni.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- Mundy CL, Patenge N, Matthews AGW, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- Ranganath S, Carpenter AC, Gleason M, Shaw AC, Bassing CH, Alt FW. Productive coupling of accessible Vbeta14 segments and DJbeta complexes determines the frequency of Vbeta14 rearrangement. J Immunol. 2008;180:2339–2346. doi: 10.4049/jimmunol.180.4.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucl Acids Res. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. H3K4me3 Stimulates the V(D)J RAG Complex for Both Nicking and Hairpinning in trans in Addition to Tethering in cis: Implications for Translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Pewznerjung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vivo. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- Tsai AG, Lu HH, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human Chromosomal Translocations at CpG Sites and a Theoretical Basis for Their Lineage and Stage Specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor KD, Vu K, Feeney AJ. Limited functional diversity in kappa light chains - junctional sequences from CD43(+)B220(+) early B cell progenitors resemble those from peripheral B cells. J Immunol. 1994;152:3467–3475. [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- Wu C, Ranganath S, Gleason M, Woodman BB, Borjeson TM, Alt FW, Bassing CH. Restriction of endogenous T cell antigen receptor beta rearrangements to Vbeta14 through selective recombination signal sequence modifications. Proc Natl Acad Sci USA. 2007;104:4002–4007. doi: 10.1073/pnas.0700081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. The immunoglobulin IGHD gene locus in C57BL/6 mice. Immunogenetics. 2004;56:399–404. doi: 10.1007/s00251-004-0712-z. [DOI] [PubMed] [Google Scholar]
- Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Misulovin Z, Suh H, Hardy RR, Jankovic M, Yannoutsos N, Nussenzweig MC. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.