Abstract
We recently reported that, in a European-American (EA) sample, the interaction between two cannabinoid receptor 1 gene (CNR1) variants significantly increased risk for drug dependence (DD), including cocaine dependence (CD). The present study aimed to investigate directly the association between CNR1 and CD in four independent samples. Eight markers across the 45kb CNR1 region and four large samples, i.e., family-based European-American (EA) sample (n=734), case-control EA sample (n=862), family-based African-American (AA) sample (n=834) and case-control AA sample (n=619) were examined in the present study. We investigated the association of these markers with CD and cocaine-induced paranoia (CIP) in the EA family sample first, and then replicated positive results in the other three samples.
The interaction between two independent CNR1 variants, i.e., the G allele-containing genotypes of rs6454674 (SNP3^G+), and the T/T genotype of rs806368 (SNP8^T/T), significantly increased risk for CD in the EA family (PGEE=0.015) and EA case-control (Pregression=0.003) samples. EA subjects with SNP3^G+ and SNP8^T/T had higher risk to develop CD than those EA subjects with the other genotypes for these two SNPs (LR+=1.4). The SNP3^G-SNP8^T haplotype also showed significant association (P=0.018) with CD in the EA case-control sample. SNP8-containing haplotypes showed significant association with both CD (Pglobal=0.007) and CIP (Pglobal=0.003) in the EA family sample. In the AA family sample, SNP8^T/T significantly confered higher risk for CD (P=0.019).
We conclude that wo independent CNR1 variants have significant interaction effects on risk for CD in EAs; they may also have effects on risk for CD in AAs.
Keywords: Interaction, CNR1, cocaine dependence, cocaine induced paranoia
Introduction
Cocaine dependence (CD), which occurs commonly in the United States, is characterized by compulsive cocaine seeking and continued cocaine use despite health problems. Cocaine-dependent individuals are at high risk of relapse to heavy cocaine use even after a period of abstinence. Limited efficacy of prevention and treatment efforts for CD are attributable in part to the unclear etiology of CD. Genetic epidemiological studies support a genetic contribution to the risk for CD [discussed in (Gelernter et al 2005)]. However, to date, few genetic variants have been confirmed as contributing to this vulnerability.
The cannabinoid receptor 1 gene (CNR1) has emerged as a promising contributor to CD vulnerability. CNR1 is located in 6q14–15 and encodes a seven transmembrane signaling protein, i.e., the cannabinoid receptor 1 (CB1), which is preferentially distributed in the presynaptic membrane of neurons (Di Marzo et al 2004). Delta-9-tetrahydrocannabinol (THC), the main active ingredient in marijuana, is an exogenous ligand for CB1. Anandamide (AEA) and 2-arachidonoyl-glycerol (2-AG) are the two major known endogenous ligands for CB1. Both AEA and 2-AG act as retrograde messengers, moving backward across the synapse from the postsynaptic neuron, and binding to CB1 to depress neurotransmitter release either transiently or over a longer time course (Chevaleyre et al 2006; Lupica and Riegel 2005). Neurobiological studies show that CB1 is one of the most abundant neuromodulators in the mammalian brain, including neocortex, hippocampus, basal ganglia, cerebellum, striatum, and the ventral tegmental area (VTA) (Arnold 2005; Di Marzo et al 2004; Herkenham et al 1990; Mailleux and Vanderhaeghen 1992; Solinas et al 2008; Tsou et al 1998).
Supporting possible relevance to CD, many lines of evidence show that CB1 and its ligands are involved in the regulation of mesocorticolimbic dopamine (DA) reward pathways that project from VTA to nucleus accumbens (NAc), prefrontal cortex (PFC), amygdala and hippocampus. The mesocorticolimbic DA reward pathways are the main hypothesized pathways for addiction, including CD (Di Chiara and Imperato 1988; Koob 1992; Koob and Le Moal 2001; Koob and Le Moal 2008; Lupica and Riegel 2005; Nestler 2005; Tanda and Goldberg 2003). Although all classes of commonly abused substances activate the mesocorticolimbic system, the mechanisms of this activation differs across abusable substances. In contrast to opioids, nicotine, or alcohol, the main receptor for cocaine is the DA transporter, to which cocaine binds, resulting in the suppression of DA reuptake, with a subsequent increase in extracellular DA level (Hyman et al 2006). Some studies suggested that CB1 might not play a critical role in cocaine self-administration (Caille et al 2007; Cossu et al 2001), but the role of the DA system on the effects of cocaine and the modulatory role of the endocannibinoid system in DA reward pathways suggest that CB1 may be involved in the risk of developing CD.
Consistent with evidence of a potential shared mechanism of risk, marijuana dependence frequently co-occurs with CD (Miller et al 1990). Further, cocaine administration increases levels of anandamide in striatum. This effect decreases by pharmacological inhibition of DA-2-like receptors (Centonze et al 2004). Besides the reduction effect on anandamide release by pharmacological inhibition of D2-like receptors, administration of D2-like receptor agonists increased anandamide release (Giuffrida et al., 1999). Additionally, blockade of CB1 receptors can partially prevent the inhibitory effect of cocaine on GABA transmission (Centonze et al 2004). Blockade of CB1 can also decrease the DA signal induced by cocaine in NAc (Cheer et al 2007). Endocannabinoid system-mediated synaptic plasticity, e.g., long-term potentiation (eCB-LTP) and long-term depression (eCB-LTD), have been identified in brain (Carlson et al 2002; Chevaleyre et al 2006), and the endocannabinoid system is involved in the mediation of cocaine-induced LTD in midbrain DA neurons (Pan et al 2008). Animal studies further have shown that, after a prolonged withdrawal period, pretreatment with a CB1 antagonist (SR141716) attenuates cocaine relapse induced by exposure to cocaine or cocaine-related cues, while pretreatment with a CB1 agonist (HU210) precipitates a relapse to cocaine-seeking behavior (De Vries et al 2001). Finally, the motivation to seek cocaine is decreased in CB1 knockout mice compared to wild type mice (Soria et al 2005).
Recently, in an European American (EA) sample, we observed that the interaction between two variants of CNR1 significantly increased risk for drug dependence (DD), which included CD and/or OD (Zuo et al 2007). The present study further investigated the association between CNR1 and CD. Moreover, a related phenotype, i.e., cocaine induced paranoia (CIP), was included to increase the information gained from the present study (Gelernter et al 2005). Genetic variants in the dopamine transporter gene (DAT1) and the dopamine β-hydroxylase gene (DBH) were reported to confer genetic vulnerability to CIP (Cubells et al 2000; Gelernter et al 1994; Kalayasiri et al 2007). We have recently reported several additional associations based on a low-density genomewide association scan (Yu et al 2008).
Materials and Methods
Subjects
Four independent samples, i.e., the EA family sample, the EA case-control sample, the African American (AA) family sample and the AA case-control sample, were included in the present study. All subjects gave written informed consent as approved by the relevant Institutional Review Boards. The EA family sample (n=734) included 307 pedigrees (551 CD cases and 394 CIP cases). The AA family sample (n=834) included 316 pedigrees (664 CD cases, 432 CIP cases). The EA case-control sample (n=862) included 299 CD cases and 563 healthy controls. The AA case-control sample (n=619) included 406 CD cases and 213 healthy controls. Of the case-control samples, 175 EA CD cases, 403 EA controls, 136 AA CD cases and 48 AA controls were included in our initial study (Zuo et al 2007). Demographic characteristics are listed in Table 1.
Table 1.
Demographic characteristics of four samples
| Sample type | Characteristics | EAsa | AAsb |
|---|---|---|---|
| Family Sample | No. of pedigrees | 307 | 316 |
| No. of pedigrees with 1 CDc | 81 | 40 | |
| No. of pedigrees with 2 CD | 200 | 211 | |
| No. of pedigrees with 3 CD | 24 | 55 | |
| No. of pedigrees with 4 CD | 2 | 7 | |
| No. of nuclear families | 277 | 242 | |
| No. of total subjects | 734 | 834 | |
| % male | 51.0 | 41.0 | |
| Age (mean±SD) | 37.9±10.3 | 40.8±7.3 | |
| No. of total CD | 551 | 664 | |
| No. of CIPd | 394 | 432 | |
| Population Sample | No. of total CD (No. in initial studye) | 299 (175) | 406 (136) |
| No. of control (No. in initial study) | 563 (403) | 213 (48) | |
| % male CD | 68.6 | 63.1 | |
| Age of CD (mean±SD) | 38.1±8.7 | 40.9±8.1 | |
| % Male controls | 44.6 | 28.2 | |
| Age of controls (mean±SD) | 30.4±11.2 | 35.2±13.2 | |
European American (EAs);
African American (AAs);
cocaine dependence (CD);
cocaine induced paranoia (CIP);
subjects involved in our initial study.
All CD cases met lifetime DSM-III-R or DSM-IV criteria for CD (APA, 1987; 1994). Individuals with schizophrenia or another psychotic illness were excluded. CIP was defined as having a transient, unrealistic and self-limited paranoid experience, e.g., irrational intense suspicion of others. CIP occurs during cocaine use and typically resolves with abstinence. CIP was assessed only among individuals with a diagnosis of CD. The controls were screened to exclude Major Axis I disorders, including substance dependence (SD), schizophrenia, mood disorder, major anxiety disorder and more, using DSM-III-R, DSM-IV, and the Schedule for Affective Disorders and Schizophrenia (SADS) (Spitzer and Endicott 1975). The subjects were recruited from four sites: Yale University School of Medicine, University of Connecticut Health Center, McLean Hospital of Harvard Medical School, and Medical University of South Carolina.
Marker selection, genotyping and error checking
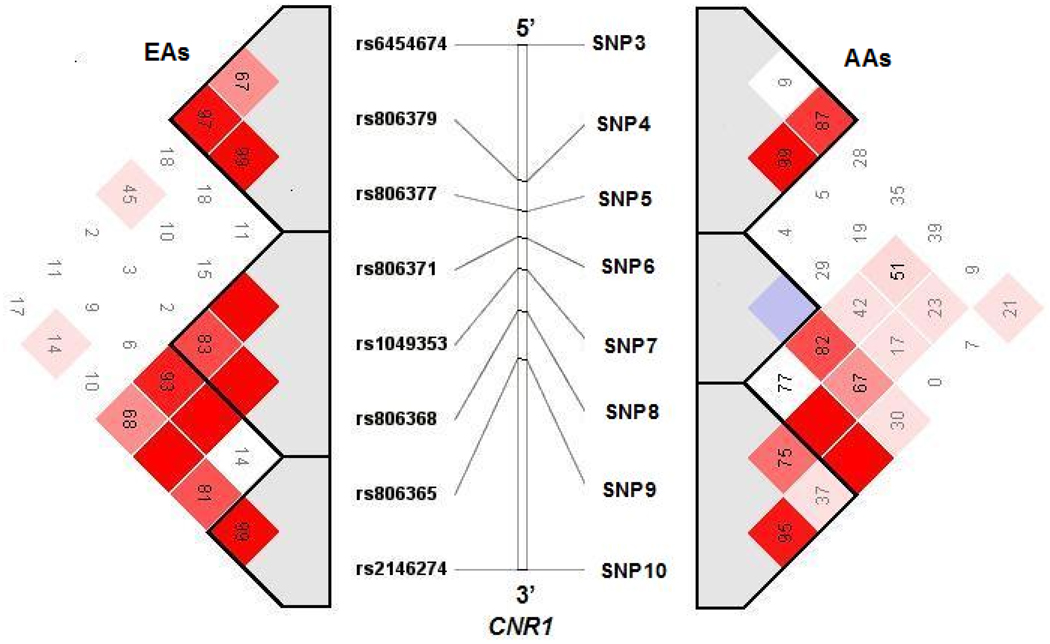
Eight markers across the 45kb region in CNR1 were selected (Figure 1). From 5’ to 3’ of CNR1, the 8 markers were numbered as SNPs 3–10 [matching their designation in our initial study (Zuo et al 2007)]. They were selected from the NCBI dbSNP database or from the Applied Biosystems SNP database. Marker selection was based on published data, minor allele frequency, information content, LD structure, region coverage, and potential function.
Figure 1. Pairwise Linkage disequilibrium (LD) of CNR1 markers in Family samples.
LD blocks were detected by haploview v3.32 and haplotype blocks were defined by solid spine of LD (Barrett et al., 2005). EAs and AAs represent European Americans and African Americans, respectively; the numbers inside the square are D’×100; the red squares indicate D’=1.
Genomic DNA was obtained from immortalized cell lines, blood or saliva. All SNPs were genotyped by a fluorogenic 5’ nuclease assay [Taqman technique; (Shi et al 1999)]. Mendelian inconsistencies in family genotype data were identified by implementing Pedcheck (O'Connell and Weeks 1998). One inconsistency was found and the genotype data for this entire pedigree were excluded from this study. Hardy-Weinberg Equilibrium (HWE) was tested using the program PowerMarker (Liu and Muse 2005).
Study design
Genetic populations were identified through Bayesian clustering using STRUCTURE (Gelernter et al 2005; Yang et al 2005). Because family-based association tests (FBAT) avoid confounding effects from model misspecification and population stratification or admixture (Laird et al 2000), we performed association tests in the EA family sample first, and then replicated positive results in the other three independent samples. To exclude the potential confounding effects on CD from other subtypes of comorbid SD, we adopted a one-by-one exclusion strategy described below.
Family based association test
The FBAT (family based association test) program was used to perform association tests in the family sample (Laird et al 2000). FBAT was developed on a principle similar to the transmission disequilibrium test (TDT), which controls for population stratification by comparing the alleles transmitted by parents to affected offspring with the nontransmitted alleles (Spielman et al 1993). The null hypothesis is “no linkage and no association” or “no association, in the presence of linkage”. The FBAT is a more general test that avoids confounding due to model misspecification as well as admixture or population stratification (http://www.biostat.harvard.edu/~fbat/default.html).
Genetic EA and AA ancestries for the family sample were assigned based on our earlier study (Gelernter et al 2005) and all of the analyses in the present study were performed in genetic EAs and AAs separately.
Haplotype associations were tested by the program HBAT in the FBAT package (Laird et al 2000). We adopted the global haplotype association test with Monte Carlo simulation, i.e., repeated random sampling, to compute an exact global p-value for multi-haplotype based association (http://www.biostat.harvard.edu/~fbat/default.html).
Case-control association test
Allelewise and genotypewise associations were tested using Fisher’s exact test by comparing allele and genotype frequency distributions between cases and controls. Haplotype Trend Regression (HTR) analysis was performed using Powermarker through permutation (permutation number was set as 100,000) by comparing the haplotype frequency distributions between cases and controls. The haplotype simulation association test was implemented using the program Haplo.score in the Haplo.stats package (http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm), with sex and age as covariates.
The case-control design is vulnerable to population stratification and admixture effects. To control for these effects, we also performed structured association analysis. First, we estimated the ancestry proportions for each unrelated subject, based on the information content of 37 AIMs using the program STRUCTURE (Pritchard et al 2000a). Parameters for burn-ins, iterations, and k were set as 100,000, 100,000 and 2, respectively. STRAT (Pritchard et al 2000b) was used to analyze allelewise and genotypewise associations between the candidate markers and CD by excluding the admixture effects. We performed 10,000 simulation iterations per marker.
Marker-marker interaction
Genotypes within pedigrees are correlated, The generalized estimating equations (GEE) method (Liang and Zeger 1986; Zeger and Liang 1986), an extension of the quasi-likelihood approach (Wedderburn 1974), is used to analyze correlated data, longitudinal data and family data (Chen et al 2004; Hanley et al 2003; Lange et al 2003). In the present study, GEE as implemented in SPSS v15 (SPSS Inc.), was implemented to model CD using a two-way interaction between the risk SNP and other SNPs of CNR1. The model applied a logit link function and Pearson chi-square estimating method, while adjusting for sex and age. The working correlation matrix was set as unstructured, and maximum iteration was set as 100,000. For GEE there is no prior assumption for data distribution, and since the number of unaffecteds in our family sample was limited, for GEE analysis, we increased the control sample size by pooling 403 EA controls and 48 AA controls from our case-control samples with the corresponding family samples.
Marker-marker interaction analyses in case-control sample were performed by mixed model backward logistic regression (Zuo et al 2007). The phenotypes served as the dependent variable and sex, age and genotypes were covariates.
Linkage disequilibrium
The dependence of allele frequencies at two loci is called linkage disequilibrium (LD) (Lonjou et al., 2003). D’(the value of LD normalized to the highest observable value between the two loci considered) values for pairwise LD of CNR1 markers were calculated and visualized through the program haploview v3.32 (Barrett et al 2005).
Results
Associations of CNR1 with CD in the EA family sample
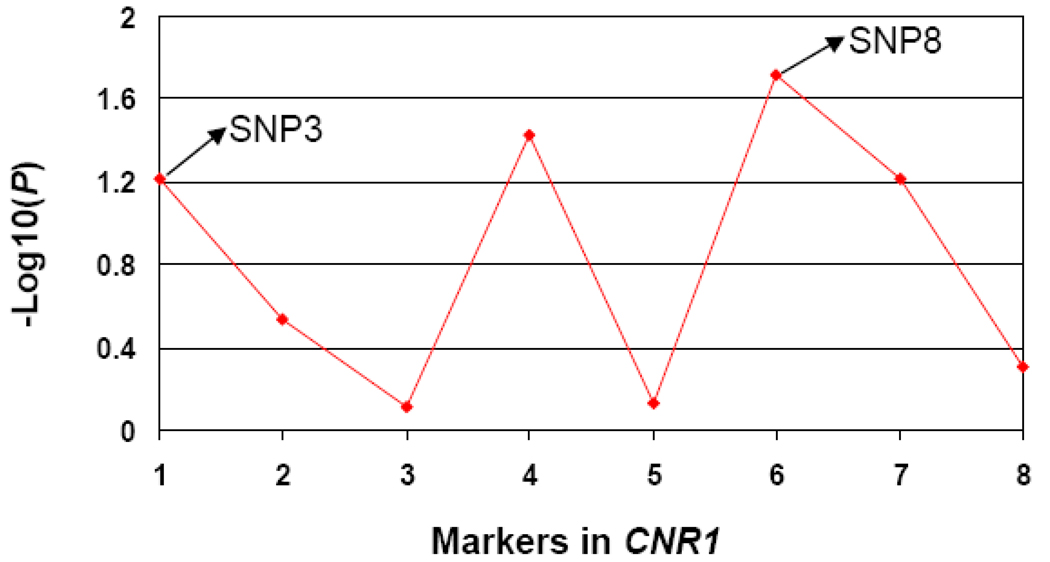
Associations between CD and the eight SNPs in CNR1 in the EA family sample are shown in Figure 2. rs806368 (SNP8) showed the most significant association signal (P=0.019). The second most significant association signal was for rs806371 (SNP6, P=0.038), which showed almost as high a δ peak as SNP8 in the fine-mapping analysis in our initial study (δ is a measure of LD used in fine-mapping analysis) (Zuo et al 2007). SNP6 is located within the SNP8-containing haplotype block and is correlated with SNP8 (D’=0.84, r2=0.49). Thus, the CD association signal at SNP6 is consistent with the association between SNP8 and CD, and also suggests that SNP6 may help to capture the association signal of SNP8 locus. The third most significant CD association signal was seen for rs6454674 (SNP3), though this did not reach statistical significance (P=0.060). No other association signal was observed.
Figure 2. Log10(P) values for the associations of cocaine dependence (CD) with 8 markers in CNR1 in EA family sample.
Markers 1 to 8 represent rs6454674 (SNP3), rs806379 (SNP4), rs806377 (SNP5), rs806371 (SNP6), rs1049353 (SNP7), rs806368 (SNP8), rs806365 (SNP9), and rs2146274 (SNP10), respectively.
In the global haplotype association test, the frequency distribution of SNP8-containing haplotypes (constructed from three SNPs within the haplotype block that contains SNP8) in CD cases differed significantly from controls (Pglobal=0.007, with α=0.017). Moving the haplotype window in the 5’ or 3’ direction in CNR1, this association signal decreased (data not shown). We failed to observe an association between SNP3-containing haplotypes and CD.
Based on the findings in our initial study and on the significant association signal observed here at SNP8, we performed an interaction analysis between SNP8^T/T and SNP3^G+ (G/G and G/T for SNP3), which conferred significantly higher risk for CD (P=0.015, Table 2). Additionally, exhaustively analyzing two-way marker-marker interactions showed that SNP8^T/T had no significant interaction with the other markers (except for SNP3) in CNR1 (P>0.05). As showed in Figure 1, SNP3 and SNP8 are neither in LD (D’= 0.026) nor correlated with each other (r2=0).
Table 2.
Interaction effects of SNP3^G+a and SNP8^T/T on CD in EA family sample and EA case-control sample
| Parameter | EA Family sample | E A Case-control sample | ||
|---|---|---|---|---|
| β | P value | β | P value | |
| Sex | 0.62 | 3.14×10−5 | 0.93 | 1.3×10−7 |
| Age | 0.10 | 5.29×10−16 | 0.07 | 4.2×10−16 |
| SNP3^G+ × SNP8^T/Tb | 0.29 | 0.015 | 0.42 | 0.003 |
G/G and G/T genotype for SNP3;
"SNP3^G+ × SNP8^T/T" denotes the interaction between G/G and G/T genotypes of SNP3 and T/T genotype of SNP8.
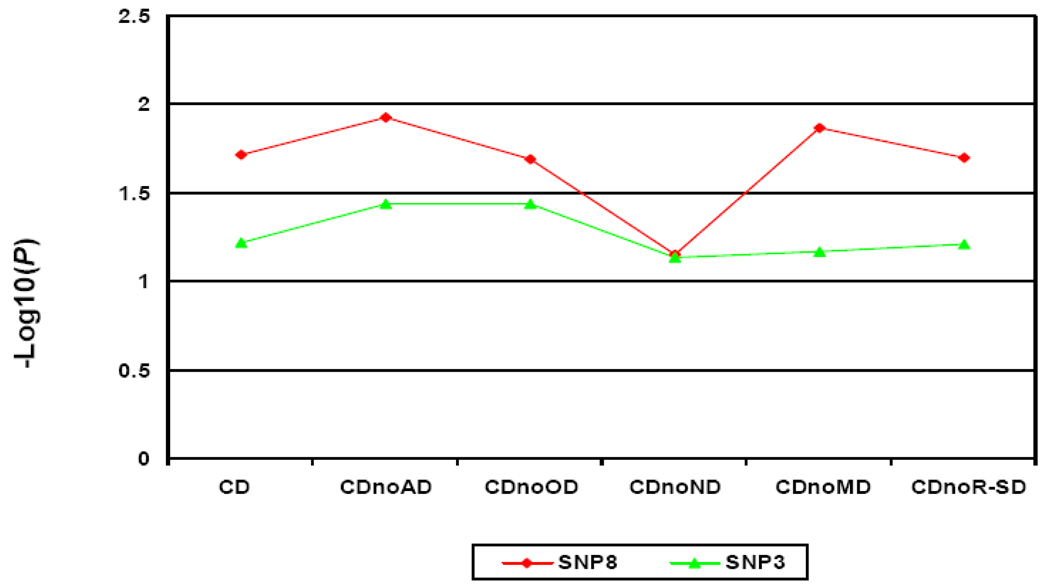
CD is highly comorbid with other SD (Kranzler et al 2008). In our family sample, the majority of CD cases were comorbid with another SD, including 77.0% with nicotine dependence (ND), 64.2% with opioid dependence (OD), 52.1% with alcohol dependence (AD), 35.9% with marijuana dependence (MD) and 26.0% with other types of SD (R-SD). Because of the high degree of comorbidity, it was not feasible to study a “CD-only” phenotype (4%, n=22) (nor is this phenotype important clinically, compared to comorbid phenotypes). Consequently, we excluded different types of comorbid SD one-by-one to control for the potential confounding effects from other SD (Figure 3). This one-by-one exclusion created five subgroups of CD, i.e. CDnoAD (CD without comorbid AD), CDnoOD (CD without comorbid OD) CDnoMD (CD without comorbid MD), CDnoND (CD without comorbid ND) and CDnoR-SD (CD without comorbid R-SD). After excluding the effects of AD, OD, MD and R-SD, the association signal remained significant for SNP8 (PCDnoAD=0.012; PCDnoOD=0.021; PCDnoMD=0.014; PCDnoR-SD=0.020). After excluding ND, the association signal for CDnoND decreased; a “trend-level possible” association signal remained (P=0.071). At SNP3, the “trend-level possible” association signals (P=0.060) remained or became significant after excluding the effect of AD (PCDnoAD=0.036), OD (PCDnoOD=0.036), ND (PCDnoND = 0.072), MD (PCDnoMD=0.068) or R-SD (PCDnoR-SD=0.062).
Figure 3. Log10(P) values of associations of SNP3 and SNP8 with six different CD subgroups in the EA family sample.
CDnoAD, CDnoOD, CDnoMD, CDnoND and CDnoR-SD represent CD without comorbid alcohol dependence, CD without comorbid opioid dependence, CD without comorbid marijuana dependence, CD without comorbid nicotine dependence and CD without comorbid rest type of substance dependence, respectively.
Associations of CNR1 with CIP in EA family sample
For CIP, nominally significant association signals were observed at two SNP8-linked loci, i.e., rs1049353 (SNP7) (P=0.015) and rs2146274 (SNP10) (P=0.010). SNP3 and SNP8 were not significantly associated with CIP (PSNP3=0.081, and PSNP8 = 0.071). In the global haplotype association test, the SNP8-containing haplotype showed a highly significant association with CIP (P=0.003), which remained significant after correcting for multiple testing (with α=0.017). These findings suggest that the SNP8 locus may also be important in CIP. However, we failed to observe an interaction effect between SNP3^G+ and SNP8^T/T on CIP.
Associations of CD with the positive variants identified in the EA family sample in the other three independent samples
The above EA family-based association tests suggested that SNP8 and the interaction between SNP8 and SNP3 were associated with risk for CD. We replicated these positive associations using the other three independent samples, i.e., an EA case-control sample, an AA case-control sample and an AA family sample.
In the EA case-control sample, SNP3 and SNP8 were in HWE in both cases and controls. In the AA case-control sample, SNP3 was in significant Hardy-Weinberg disequilibrium (HWD) in cases (P=7.0×10−4), and SNP8 was in nominal HWD in cases (P=0.025), but both were in HWE in controls (P>0.05), which may be an indication of marker-phenotype association (Luo et al 2006).
The allele and genotype frequencies of SNP3 and SNP8 in EA and AA case-control samples are listed in Table 3. In controls, we observed that the G allele frequency of SNP3 and T allele frequency of SNP8 in AAs were significantly higher than those in EAs (PSNP3=0.006, PSNP8= 4.5×10−10).
Table 3.
Allele and genotype frequencies of SNP3 and SNP8 in case-control samples
| SNP | rs# | Ethnicity | Allele or Genotype |
CD cases | Controls | P value | ||
|---|---|---|---|---|---|---|---|---|
| # | Freq | # | Freq | |||||
| SNP3 | rs6454674 | EA | G | 183 | 0.33 | 289 | 0.29 | - |
| T | 365 | 0.67 | 703 | 0.71 | ||||
| G/G | 33 | 0.12 | 46 | 0.09 | - | |||
| G/T | 117 | 0.43 | 197 | 0.40 | ||||
| T/T | 124 | 0.45 | 253 | 0.51 | ||||
| AA | G | 223 | 0.33 | 131 | 0.37 | - | ||
| T | 445 | 0.67 | 221 | 0.63 | ||||
| G/G | 51 | 0.15 | 20 | 0.11 | 0.003 | |||
| G/T | 121 | 0.36 | 91 | 0.52 | ||||
| T/T | 162 | 0.49 | 65 | 0.37 | ||||
| SNP8 | rs806368 | EA | T | 439 | 0.80 | 822 | 0.77 | - |
| C | 109 | 0.20 | 244 | 0.23 | ||||
| T/T | 178 | 0.65 | 318 | 0.60 | - | |||
| T/C | 83 | 0.30 | 186 | 0.35 | ||||
| C/C | 13 | 0.05 | 29 | 0.05 | ||||
| AA | T | 707 | 0.90 | 379 | 0.91 | - | ||
| C | 79 | 0.10 | 39 | 0.09 | ||||
| T/T | 322 | 0.82 | 174 | 0.83 | - | |||
| T/C | 63 | 0.16 | 31 | 0.15 | ||||
| C/C | 8 | 0.02 | 4 | 0.02 | ||||
“-“ represent p>0.05
In the single-marker-wise analysis in EAs, we failed to observe that allele and genotype frequency distribution for SNP3 or SNP8 was significant differenct from controls (Table 3). However, as expected, in the EA case-control sample, the interaction between SNP3^G+ and SNP8^T/T significantly increased risk for CD (P=0.003) while adjusting for sex and age. This is consistent with our initial study, which showed that this interaction significantly increased risk for DD (P=0.0002), including CD (Table 2). Through permutation, haplotype trend regression (HTR) analysis using the Powermarker program showed a trend-level possible association between CD and the global haplotypes composed of these two SNPs (Pglobal= 0.069). Haplotype simulation association tests using Haplo.stats showed that global haplotypes and the GT haplotype (constructed from SNP3^G and SNP8^T) displayed significant association signals (Pglobal=0.054, PGT=0.018), adjusting for sex and age.
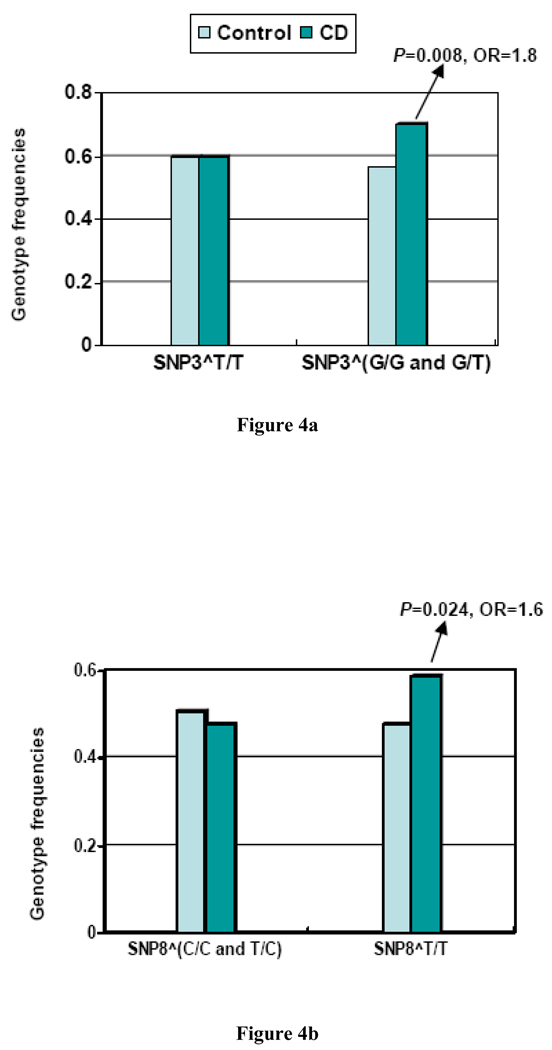
In subjects carrying the SNP3^G allele, the frequency of SNP8^T/T in CD cases was significantly higher than controls (P=0.008, OR=1.8, 95%CI: 1.1~ 2.9), while no significant difference was observed within subjects not carrying SNP3^G. On the other hand, in CD subjects carrying SNP8^T/T, the frequency of SNP3^G+ was nominally higher than in controls (P=0.024, OR=1.6, 95%CI: 1.0~2.4), but no difference was observed within subjects not carrying SNP8^T/T (P>0.05) (Table 4, Figure 4).
Table 4.
Associations of CD with SNPs 3 and 8 in EA family sample and EA case-control sample
| SNPs | Family sample |
Case-control sample |
|||||
|---|---|---|---|---|---|---|---|
| No. of informative families |
P | Genotype | CDs | Controls | P | OR | |
| SNP3 | 31 | 0.06 | G/G+G/T | 98 | 134 | 0.024a | 1.6 |
| T/T | 68 | 146 | |||||
| SNP8 | 33 | 0.019 | T/T | 98 | 134 | 0.008b | 1.8 |
| T/C+C/C | 41 | 102 | |||||
P value for SNP3^(G/G+G/T) vs. T/T in subjects carrying the SNP8^T/T genotype;
P value for SNP8^T/T vs. (T/C+C/C) in the subjects carrying the SNP3^(G/G or G/T).
Figure 4.
Figure 4a. SNP8^T/T genotype frequency difference between CD cases and controls in different genotype groups of SNP3 in EAs. SNP3^T/T and SNP3^(G/G or G/T) represent T/T genotype and G allele-containing genotype of SNP3, respectively
Figure 4b. SNP3^(G/G and G/T) genotype frequency difference between CD cases and controls in different genotype groups of SNP8 in EAs. SNP8^(C/C and T/C) and SNP8^T/T represent C allele containing genotype and T/T genotype of SNP8, respectively.
In the AA family sample, we observed that SNP8^T/T significantly increased risk for CD (P=0.019) while adjusting for sex and age. However, neither SNP3 nor the interaction between SNP3^G+ and SNP8^T/T was significantly associated with CD (P>0.05). In the haplotypewise analysis, no association between CD and SNP8-containing haplotypes was observed.
In the AA case-control sample, as shown in Table 3, the allele frequency distributions for both SNP3 and SNP8, and the genotype frequency distribution for SNP8 in cases, were not significantly different from controls. The genotype frequency distribution for SNP3 in cases was significantly different from controls, however the difference did not reach statistical significance while adjusted for sex and age, and corrected for multiple testing (P=0.028, α=0.025). We failed to observe an interaction between SNP3 and SNP8 having a significant effect on CD in the AA case-control sample.
Controlling for admixture effects on the association analysis in the above EA and AA case-control samples using the program STRAT did not change the results substantially (data not shown), suggesting that admixture effects in our samples did not contribute to the observed associations.
Discussion
In the present study, the interaction between two independent CNR1 variants, i.e., SNP3^G+ and SNP8^T/T, was associated with CD in an EA family sample. The association was replicable in an EA case-control sample. Sliding window haplotype analysis in the EA family sample showed that the haplotype constructed from SNP8 and its two neighboring SNPs displayed a higher association signal (P=0.007) than SNP8 alone, suggesting that SNP8 is in LD with a disease–influencing locus rather than being a risk locus itself. This SNP8-linked risk locus could be located either in the coding region of CNR1 directly affecting the function of the CB1 protein or in the 3’UTR regulating translational efficiency, mRNA stability, or polyadenylation signals. Prediction of miRNA target sites for CNR1 suggests that the 3’UTR region of CNR1 might be targeted by miRNAs affecting posttranscriptional CNR1 expression (http://www.targetscan.org). SNP8 was in complete LD with a well-studied polymorphism in CNR1, i.e., SNP7. This SNP is the Thr453Thr synonymous variant, located in the coding region of CNR1. Although it did not show association with CD, it showed nominal association with CIP. This SNP was reported to be associated with alcohol withdrawal delirium in an unrelated German population (Schmidt et al 2002).
Within the subjects with SNP8^T/T, we observed that SNP3^G+ nominally significantly increased the risk for CD (P=0.026). Although this relationship fails rigorous criteria for statistical significance after correcting for multiple tests, the association may not be a random false positive. Within the SNP3-containing haplotype block (Figure 1), a SNP, rs806380, which is correlated with SNP3 (r2=0.63), was reported to be associated with cannabis dependence symptoms in an adolescent Caucasian sample (Hopfer et al 2006); a TAG haplotype in the SNP3-containing haplotype block was reported to be associated with polysubstance abuse in an EA sample (Zhang et al 2004). Further, another SNP in this SNP3-containing haplotype block, i.e., rs2023239, was reported to be associated with CB1 binding in the prefrontal cortex (Hutchison et al 2008), supporting that this SNP3 locus may play a role in substance dependence. SNP3 per se could alter RNA secondary structure (IDT SciTools: http://www.idtdna.com/SciTools/SciTools.aspx). RAVEN [regulatory analysis of variation in enhancers; (Andersen et al 2008)] predicts that SNP3 is located within a conserved region (phastCon score>0.4) and may be a binding site for transcription factors and thereby influence the efficiency of CNR1 transcription.
SNP3 and SNP8 are located within the same gene rather than two different genes, so it could be argued that the interaction effect detected in the present study might simply reflect a haplotype effect which implicates a single risk variant effect for CD. But, as noted previously, SNP3 and SNP8 are neither in strong LD (D’= 0.026) nor correlated with each other (r2=0); both SNP3 and SNP8 may have functional implication, suggesting that the interaction is more likely to imply joint effects of two independent and functional variants at the locations of SNP3 and SNP8.
The associations observed in the present study seem unlikely to be spurious for a number of reasons. First, as noted above, the association signals were replicable between case-control and family based samples. Second, the association signals of SNP3 and SNP8 were observed across five phenotype groups that were defined based on comorbidity with CD, i.e., total CD, CDnoAD, CDnoOD, CDnoMD and CDnoR-SD (Figure 3). The consistency across these five groups not only argues against a false positive association but suggested that the association signals were from CD rather than from other types of SD. Third, another CD-related trait, CIP, also showed association with SNP8 in haplotypewise analysis (Pglobal=0.003), providing further evidence for an association between CNR1 and CD. Fourth, the association signals were consistent when we used different analytic approaches. Genotype frequency comparison analysis showed that the genotype frequency of SNP8^T/T in CD cases was significantly higher than in controls among subjects carrying SNP3^G; Logistic regression analysis showed that the interaction between SNP3^G+ and SNP8^T/T significantly increased the risk for CD; and haplotypewise analysis also consistently showed that the GT haplotype (constructed from SNP3^G and SNP8^T) increased risk for CD (P=0.018). Finally, as shown in Figure 2, both SNP8 and a SNP8-linked SNP, i.e., SNP6, showed nominally significant association signals, but the other SNPs did not, which also argues against a false positive association at the SNP8 locus.
There is considerable evidence for both shared and nonshared genetic factors between different forms of substance dependence. Our initial study tested case-control-based association between CNR1 and DD, which categorized CD cases and OD cases together, and alcohol dependence was also included in that initial study, but the specific association between CNR1 and CD (nEA=175, nAA =136) was not analyzed previously (Zuo et al 2007). The present study greatly increased the sample size of CD cases (nEA=299, nAA=406) and analyzed the specific association between CNR1 and CD. In the present study, we also tested for family-based associations between CNR1 and CD in two independent large samples, i.e., an EA family sample (n=734) and an AA family sample (n=834). The present study extended the initial study and had different study targets of interest.
There is only one prior study directly reporting an association between CD and an (AAT)n polymorphism in CNR1, a population-based association study conducted in a male African Caribbean sample (Ballon et al 2006). African Caribbeans are an admixed population (Benn-Torres et al 2008) and allele frequencies of CNR1 variants differ among different ancestral populations (Covault et al 2001; Herman et al 2006; Zhang et al 2004). Although an adjustment for admixture effects is required to avoid potential spurious associations, it was not included in that study. The (AAT)n polymorphism is located between SNP9 and SNP10, 7kb away from SNP10. SNP10 showed nominal association with CIP in the EA family sample in the present study. Thus, further work is warranted to investigate the association between this locus and CD.
CD is a complex non-Mendelian trait. The present study shows that the positive likelihood ratio (LR+) value for the subjects carrying SNP8^T/T to develop CD was 1.1, which shows that this variant of CNR1 exerts a minor effect on the vulnerability to CD. This could explain why single locus analysis of this SNP in the case-control sample did not show a significant association with CD. Alternatively, the interaction model may more accurately reflect the genetic effects underpinning CD. The present study showed that the LR+ value for interaction between SNP3^G+ and SNP8^T/T was 1.4, considerably higher than that for either SNP3^G+ or SNP8^T/T alone, showing that the interaction analysis is more powerful than the single locus analysis in detecting the association of CNR1 with CD.
Whether or not CD is included, substance abuse and/or dependence have been reported to be associated with CNR1 in some (Comings et al 1997; Ponce et al 2003; Schmidt et al 2002; Zhang et al 2004), but not all studies (Covault et al 2001; Heller et al 2001; Herman et al 2006; Li et al 2000). There are several potential explanations for these results. First, the inconsistent findings may result in part from a lack of phenotypic comparability across different studies. Substance dependence (SD) is a complex phenotype, which includes CD, OD, MD, AD, and others. This phenotypic heterogeneity could lead to different results. Second, inconsistent findings in relation to the association of CNR1 and substance use disorders may also result from differences in the populations studied, as well as from population stratification and admixture effects. Most previous studies based population membership on self-report, which may not be adequate to exclude potential population stratification effects. Third, some of the differences could derive from the use of different marker sets and analytic models. Finally, the inconsistent findings could have resulted from small sample sizes leading to inadequate statistical power in some studies.
In addition to the above positive findings in EAs, we observed that SNP8^T/T conferred significantly higher risk for CD in the AA family sample, which is consistent with the finding in EA samples. In AA case-control sample, we failed to observe any significant association for SNP3 and SNP8; we also failed to detect a significant role of the interaction between SNP3^G+ and SNP8^T/T on CD in AAs. The decay of linkage disequilibrium caused by more recombination events that have occurred in AAs could underlie this finding (see Figure 1). On the other hand, both the two SNPs showed HWD in cases and HWE in controls suggesting potential associations with CD in AAs too. A SNP8-containing haplotype (constructed from SNP8, SNP7 and a SNP between them) was reported to be associated with polysubstance abuse in AAs in another study (Zhang et al 2004), supporting a role for SNP8 in CD in AAs.
In conclusion, the present study provides evidence in support of an interaction model of the role of CNR1 in the risk for CD in EAs. SNP8 locus may be implicated in CIP as well. CNR1 could also play a role in the risk for CD in AAs. The two loci could ultimately be relevant to research on the prevention and treatment of CD. Further studies are warranted to replicate the findings in the present study, to locate the causal variants around SNP3 and SNP8 that contribute to risk of CD and to uncover the mechanism by which these two independent loci modulate that risk.
Acknowledgement
Ann Marie Lacobelle and Greg Kay provided excellent technical assistance. This work was supported in part by National Institute on Drug Abuse (NIDA) grants R01-DA12849, R01-DA12690, K24-DA15105 (J. Gelernter), and K24-DA022288 (R. Weiss), and K01-DA029643 (L. Zuo); National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R01-AA11330, R01-AA016015, P50-AA12870, P50-AA03510, and K24-AA13736 (H. Kranzler); National Center for Research Resources (NCRR) grant M01-RR06192; and by Alcoholic Beverage Medical Research Foundation (ABMRF) grant award R06932 (X. Luo); U.S. Department of Veterans Affairs (the VA Medical Research Program, VA Alcohol Research Center, and the VA Connecticut–Massachusetts Mental Illness Research, Education and Clinical Center [MIRECC]; and the VA Research Enhancement Award Program [REAP] research center).
Footnotes
Conflict of Interest Statement
Dr. Kranzler has received financial support or compensation for the following: related to consultation on grant reviews for the National Institutes of Health and related to academic lectures and editorial functions in various scientific venues; and has been a paid consultant to Alkermes, Inc., Ortho-McNeil Pharmaceuticals, Thomson Healthcare, Sanofi-Aventis, Lundbeck, Forest Pharmaceuticals, elbion NV, Bristol-Myers Squibb Co., and Solvay Pharmaceuticals. He has received research support from Ortho-McNeil Pharmaceuticals and Bristol-Myers Squibb Co. Dr. Weiss has received financial support or compensation for the following: related to consultation on grant reviews for the National Institutes of Health and related to academic lectures and editorial functions in various scientific venues; and has been a paid consultant to Eli Lilly Co., Novartis Pharmaceuticals, Titan Pharmaceuticals and AstraZeneca Pharmaceuticals. Dr Gelernter has received financial support or compensation for the following: related to consultation for Columbia University, the Thailand Center for Excellence for Life Sciences (TCELS), the University of CT Health Center, NIH, related to grant reviews for the National Institutes of Health; and related to academic lectures and editorial functions in various scientific venues (including from ACNP). The other authors declare that they have not received any compensation for research or professional service that could be perceived as constituting a potential conflict of interest.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders, third edition, revised. Washington, DC: American Psychiatric Press; 1987. [Google Scholar]
- American Psychiatric Association. (1994) Diagnostic and statistical manual of mental disorders, fourth edition. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P, Lenhard B, et al. In silico detection of sequence variations modifying transcriptional regulation. PLoS computational biology. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JC. The role of endocannabinoid transmission in cocaine addiction. Pharmacol Biochem Behav. 2005;81:396–406. doi: 10.1016/j.pbb.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Ballon N, Leroy S, Roy C, Bourdel MC, Charles-Nicolas A, Krebs MO, et al. (AAT)n repeat in the cannabinoid receptor gene (CNR1): association with cocaine addiction in an African-Caribbean population. The pharmacogenomics journal. 2006;6:126–130. doi: 10.1038/sj.tpj.6500352. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benn-Torres J, Bonilla C, Robbins CM, Waterman L, Moses TY, Hernandez W, et al. Admixture and population stratification in African Caribbean populations. Annals of human genetics. 2008;72:90–98. doi: 10.1111/j.1469-1809.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- Caille S, Alvarez-Jaimes L, Polis I, Stouffer DG, Parsons LH. Specific alterations of extracellular endocannabinoid levels in the nucleus accumbens by ethanol, heroin, and cocaine self-administration. J Neurosci. 2007;27:3695–3702. doi: 10.1523/JNEUROSCI.4403-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson G, Wang Y, Alger BE. Endocannabinoids facilitate the induction of LTP in the hippocampus. Nature neuroscience. 2002;5:723–724. doi: 10.1038/nn879. [DOI] [PubMed] [Google Scholar]
- Centonze D, Battista N, Rossi S, Mercuri NB, Finazzi-Agro A, Bernardi G, et al. A critical interaction between dopamine D2 receptors and endocannabinoids mediates the effects of cocaine on striatal gabaergic Transmission. Neuropsychopharmacology. 2004;29:1488–1497. doi: 10.1038/sj.npp.1300458. [DOI] [PubMed] [Google Scholar]
- Cheer JF, Wassum KM, Sombers LA, Heien ML, Ariansen JL, Aragona BJ, et al. Phasic dopamine release evoked by abused substances requires cannabinoid receptor activation. J Neurosci. 2007;27:791–795. doi: 10.1523/JNEUROSCI.4152-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Broman KW, Liang KY. Quantitative trait linkage analysis by generalized estimating equations: unification of variance components and Haseman-Elston regression. Genetic epidemiology. 2004;26:265–272. doi: 10.1002/gepi.10315. [DOI] [PubMed] [Google Scholar]
- Chevaleyre V, Takahashi KA, Castillo PE. Endocannabinoid-mediated synaptic plasticity in the CNS. Annual review of neuroscience. 2006;29:37–76. doi: 10.1146/annurev.neuro.29.051605.112834. [DOI] [PubMed] [Google Scholar]
- Comings DE, Muhleman D, Gade R, Johnson P, Verde R, Saucier G, et al. Cannabinoid receptor gene (CNR1): association with i.v. drug use. Molecular psychiatry. 1997;2:161–168. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- Cossu G, Ledent C, Fattore L, Imperato A, Bohme GA, Parmentier M, et al. Cannabinoid CB1 receptor knockout mice fail to self-administer morphine but not other drugs of abuse. Behavioural brain research. 2001;118:61–65. doi: 10.1016/s0166-4328(00)00311-9. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Kranzler H. Association study of cannabinoid receptor gene (CNR1) alleles and drug dependence. Molecular psychiatry. 2001;6:501–502. doi: 10.1038/sj.mp.4000925. [DOI] [PubMed] [Google Scholar]
- Cubells JF, Kranzler HR, McCance-Katz E, Anderson GM, Malison RT, Price LH, et al. A haplotype at the DBH locus, associated with low plasma dopamine beta-hydroxylase activity, also associates with cocaine-induced paranoia. Molecular psychiatry. 2000;5:56–63. doi: 10.1038/sj.mp.4000657. [DOI] [PubMed] [Google Scholar]
- De Vries TJ, Shaham Y, Homberg JR, Crombag H, Schuurman K, Dieben J, et al. A cannabinoid mechanism in relapse to cocaine seeking. Nature medicine. 2001;7:1151–1154. doi: 10.1038/nm1001-1151. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V, Bifulco M, De Petrocellis L. The endocannabinoid system and its therapeutic exploitation. Nature reviews. 2004;3:771–784. doi: 10.1038/nrd1495. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Satel SL, Rao PA. Genetic association between dopamine transporter protein alleles and cocaine-induced paranoia. Neuropsychopharmacology. 1994;11:195–200. doi: 10.1038/sj.npp.1380106. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Giuffrida A, Parsons LH, Kerr TM, Rodríguez de Fonseca F, Navarro M, Piomelli D. Dopamine activation of endogenous cannabinoid signaling in dorsal striatum. Nat Neurosci. 1999;2(4):358–363. doi: 10.1038/7268. [DOI] [PubMed] [Google Scholar]
- Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. American journal of epidemiology. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- Heller D, Schneider U, Seifert J, Cimander KF, Stuhrmann M. The cannabinoid receptor gene (CNR1) is not affected in German i.v. drug users. Addiction biology. 2001;6:183–187. doi: 10.1080/13556210020040271. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, de Costa BR, et al. Cannabinoid receptor localization in brain. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J. Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:499–503. doi: 10.1002/ajmg.b.30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Young SE, Purcell S, Crowley TJ, Stallings MC, Corley RP, et al. Cannabis receptor haplotype associated with fewer cannabis dependence symptoms in adolescents. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:895–901. doi: 10.1002/ajmg.b.30378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison KE, Haughey H, Niculescu M, Schacht J, Kaiser A, Stitzel J, et al. The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Archives of general psychiatry. 2008;65:841–850. doi: 10.1001/archpsyc.65.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annual review of neuroscience. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Kalayasiri R, Sughondhabirom A, Gueorguieva R, Coric V, Lynch WJ, Lappalainen J, et al. Dopamine beta-hydroxylase gene (DbetaH) −1021C-->T influences self-reported paranoia during cocaine self-administration. Biological psychiatry. 2007;61:1310–1313. doi: 10.1016/j.biopsych.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Koob GF. Drugs of abuse: anatomy, pharmacology and function of reward pathways. Trends in pharmacological sciences. 1992;13:177–184. doi: 10.1016/0165-6147(92)90060-j. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology. 2001;24:97–129. doi: 10.1016/S0893-133X(00)00195-0. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Addiction and the brain antireward system. Annual review of psychology. 2008;59:29–53. doi: 10.1146/annurev.psych.59.103006.093548. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Wilcox M, Weiss RD, Brady K, Hesselbrock V, Rounsaville B, et al. The validity of cocaine dependence subtypes. Addictive behaviors. 2008;33:41–53. doi: 10.1016/j.addbeh.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genetic epidemiology. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics (Oxford, England) 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- Li T, Liu X, Zhu ZH, Zhao J, Hu X, Ball DM, et al. No association between (AAT)n repeats in the cannabinoid receptor gene (CNR1) and heroin abuse in a Chinese population. Molecular psychiatry. 2000;5:128–130. doi: 10.1038/sj.mp.4000670. [DOI] [PubMed] [Google Scholar]
- Liang K, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics (Oxford, England) 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lonjou C, Zhang W, Collins A, Tapper WJ, Elahi E, Maniatis N, Morton NE. Linkage disequilibrium in human populations. Proc Natl Acad Sci U S A. 2003;100(10):6069–6074. doi: 10.1073/pnas.1031521100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Lupica CR, Riegel AC. Endocannabinoid release from midbrain dopamine neurons: a potential substrate for cannabinoid receptor antagonist treatment of addiction. Neuropharmacology. 2005;48:1105–1116. doi: 10.1016/j.neuropharm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Mailleux P, Vanderhaeghen JJ. Distribution of neuronal cannabinoid receptor in the adult rat brain: a comparative receptor binding radioautography and in situ hybridization histochemistry. Neuroscience. 1992;48:655–668. doi: 10.1016/0306-4522(92)90409-u. [DOI] [PubMed] [Google Scholar]
- Miller NS, Klahr AL, Gold MS, Sweeney K, Cocores JA. The prevalence of marijuana (cannabis) use and dependence in cocaine dependence. New York state journal of medicine. 1990;90:491–492. [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nature neuroscience. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American journal of human genetics. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Hillard CJ, Liu QS. Endocannabinoid signaling mediates cocaine-induced inhibitory synaptic plasticity in midbrain dopamine neurons. J Neurosci. 2008;28:1385–1397. doi: 10.1523/JNEUROSCI.4033-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce G, Hoenicka J, Rubio G, Ampuero I, Jimenez-Arriero MA, Rodriguez-Jimenez R, et al. Association between cannabinoid receptor gene (CNR1) and childhood attention deficit/hyperactivity disorder in Spanish male alcoholic patients. Molecular psychiatry. 2003;8:466–467. doi: 10.1038/sj.mp.4001278. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000a;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. American journal of human genetics. 2000b;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, Wendel B, et al. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug and alcohol dependence. 2002;65:221–224. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA. High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol. 1999;52:295–299. doi: 10.1136/mp.52.5.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas M, Goldberg SR, Piomelli D. The endocannabinoid system in brain reward processes. British journal of pharmacology. 2008;154:369–383. doi: 10.1038/bjp.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Mendizabal V, Tourino C, Robledo P, Ledent C, Parmentier M, et al. Lack of CB1 cannabinoid receptor impairs cocaine self-administration. Neuropsychopharmacology. 2005;30:1670–1680. doi: 10.1038/sj.npp.1300707. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) American journal of human genetics. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Spitzer R, Endicott J. Schedule for affective disorders and schizophrenia: Lifetime version. New York: New York Biometrics Research Division, New York State Psychiatric Institute; 1975. [Google Scholar]
- Tanda G, Goldberg SR. Cannabinoids: reward, dependence, and underlying neurochemical mechanisms--a review of recent preclinical data. Psychopharmacology. 2003;169:115–134. doi: 10.1007/s00213-003-1485-z. [DOI] [PubMed] [Google Scholar]
- Tsou K, Brown S, Sanudo-Pena MC, Mackie K, Walker JM. Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience. 1998;83:393–411. doi: 10.1016/s0306-4522(97)00436-3. [DOI] [PubMed] [Google Scholar]
- Wedderburn R. Quasi-likelihood functions, generalized linear models, and the Gauss-Newton method. Biometrika. 1974;61:439–447. [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genetic epidemiology. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Yu Y, Kranzler HR, Panhuysen C, Weiss RD, Poling J, Farrer LA, et al. Substance dependence low-density whole genome association study in two distinct American populations. Human genetics. 2008;123:495–506. doi: 10.1007/s00439-008-0501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- Zhang PW, Ishiguro H, Ohtsuki T, Hess J, Carillo F, Walther D, et al. Human cannabinoid receptor 1: 5' exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Molecular psychiatry. 2004;9:916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biological psychiatry. 2007;62:616–626. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]