Abstract
Atypical hemolytic uremic syndrome or the non-diarrheal form of hemolytic uremic syndrome is a rare disorder typically classified as familial or sporadic. Recent literature has suggested that approximately 50% of patients have mutations in Factor H (CFH), Factor I (CFI), or membrane cofactor protein (CD46). Importantly, results of renal transplantation in patients with mutations in either CFH or CFI are dismal with recurrent disease leading to graft loss occurring in the majority of cases. We describe an adult renal transplant recipient who developed recurrent hemolytic uremic syndrome one month after her transplant. Bidirectional sequencing of CFH, CFI and CD46 confirmed that the patient was heterozygous for a novel missense mutation, p.Tyr369Ser, in CFI. This report reemphasizes the importance of screening patients with atypical hemolytic uremic syndrome for mutations in these genes prior to renal transplantation and demonstrates the challenges in the management of these patients.
Keywords: kidney transplant, factor I, HUS, recurrent
Hemolytic uremic syndrome (HUS) was first described by Gasser in 1955 when he observed five children who presented with the triad of acute renal failure, microangiopathic hemolytic anemia, and thrombocytopenia.1 Recent literature has identified the non-diarrheal or non-Shiga toxin producing Escherichia coli form otherwise known as atypical HUS (aHUS) to be a disease predominantly related to complement regulatory protein deficiency.2 Other sporadic factors predisposing to aHUS include diverse disorders with a common physiopathologic thread – endothelial cell injury (Table 1). In patients with the familial form, mutations in one of three complement component regulatory proteins - complement factor H (CFH), membrane cofactor protein (CD46), complement factor I (CFI) or Factor B (CFB), a component of the complement activation cascade, have been identified.3,4 The first three proteins are important regulators of the activity of the alternative pathway (AP) of complement activation, while CFB is an essential component of the C3 convertase of the AP.5,6 Because the AP is constitutively active, tight control is essential. In aHUS with mutations in circulating complement regulatory proteins CFH and CFI disease recurrence and graft failure is very high.7–9 We report a novel missense mutation in the serine protease domain of CFI, with recurrent and aggressive HUS following renal transplantation, highlighting the importance of pre-transplant genetic testing in all patients with aHUS.
Table 1.
Conditions predisposing to atypical hemolytic uremic syndrome
| Complement regulatory defects |
| Factor H mutations |
| Factor I mutations |
| MCP mutations |
| Factor H autoantibodies |
| Factor B mutations |
| Bacterial |
| Streptococcus Pneumoniae |
| Viruses |
| HIV |
| Drugs |
| Chemotherapy (mitomycin C, gemcitabine, cisplatin, bleomycin) |
| Quinine |
| Calcineurin inhibitors (cyclosporine, tacrolimus) |
| Bevacizumab |
| Oral contraceptives |
| Illicit drugs (cocaine, heroin, ecstasy) |
| Systemic disease |
| Systemic lupus erythematosus |
| Scleroderma |
| Anti-phospholipid antibody |
| Pregnancy |
| Cancer |
| Hepatocellular carcinoma |
| Prostate adenocarcinoma |
| Gastric adenocarcinoma |
| Combined methylmalonic aciduria and homocystinuria |
Adapted from Kavanagh, et al. Br Med Bull 2006;77–78:5–22. Reprinted with permission from Oxford University Press.
Methods
Mutation Screening and Sequence Analysis of CFH, CFI and CD46
DNA was isolated from whole blood using established techniques and used as a template for amplification of coding and adjacent intronic regions of CFH, CFI and CD46. In general, polymerase chain reaction (PCR) conditions for each exon included amplification for 35 cycles of 30 seconds each at 94°C denaturing, 61°C annealing and 70°C extension. Primers used for CFI amplification are shown in Table 2. Product generation was verified by agarose gel electrophoresis and amplicons were then bi-directionally sequenced. Sequence analysis was completed using Sequencer 4.8. Nucleotide changes were analyzed for possible functional and structural importance using the ConSeq (http://conseq.bioinfo.tau.ac.il/), a web server for the identification of structurally and functionally important residues in protein sequences. Any putative pathological changes were screened against 200 normal controls.
Table 2.
Primers used for IF amplification
| Primer Name | Primer Sequence | Location with reference to NG_007569 |
Bp |
|---|---|---|---|
| IF Ex1 F | CAAAAGTACAAAGCTCTTTAGGAGG | 4883 – 4907 | 329 |
| IF Ex1 R | TTGCTGACTATAGAGTGGCATTG | 5211 – 5189 | |
| IF Ex2 F | TTGAAGCCACCAGACAACAC | 40028 – 40047 | 573 |
| IF Ex2 R | GGCAACCCCTGATTTGTTTAG | 40600 – 40580 | |
| IF Ex3 F | CGTAAAATGATTGCTTACTATTACTTG | 42145 – 42171 | 431 |
| IF Ex3 R | TGATGCACATAGTTAATTTTCTTAGG | 42575 – 42550 | |
| IF Ex4 F | CTTGCCCAAGCTGTAACTCC | 45165 – 45184 | 396 |
| IF Ex4 R | AACGAGGCATCAATCATTTG | 45560 – 45541 | |
| IF Ex5–6 F | TCCATAAGCCAGGTTTGACC | 46161 – 46180 | 698 |
| IF Ex5–6 R | CCATAGTTCTGCAAATGCCC | 46858 – 46839 | |
| IF Ex7 F | AAAACAGAAATAAGGTGCAATGG | 49090 – 49112 | 274 |
| IF Ex7 R | CTAAACTAATGTTCAGGCTGGG | 49363 – 49342 | |
| IF Ex8 F | GGGAGGATAAGTTTTAAGGCAG | 54370 – 54391 | 252 |
| IF Ex8 R | CCAAAACTACTTGTTGCTTGAATC | 54621 – 54598 | |
| IF Ex9–10 F | TCCAGCCTGTCTTGTACTGG | 57264 – 57283 | 630 |
| IF Ex9–10 R | CTTTGCAATTAATATAGTGGAGTTTG | 57893 – 57868 | |
| IF Ex11 F | AAGCATTTACAAAATTCTGGGG | 60357 – 60378 | 539 |
| IF Ex11 R | GGCTGGATGTTTACTTTTCTGG | 60895 – 60874 | |
| IF Ex12 F | CACCCTTTCATAATCCCAATG | 64296 – 64316 | 312 |
| IF Ex12 R | CCAAATGGAGGGAATTAGGG | 64607 – 64588 | |
| IF Ex13 F | AAGGAGAGCCCATGCTATTG | 65745 – 65764 | 496 |
| IF Ex13 R | GGCATAAACTCTGTGGAGACC | 66240 – 66220 |
Case Report
A healthy 26-year-old white woman presented in May 2004 with fevers, vomiting, and diarrhea. Three weeks prior she noticed bruising over her extremities followed by upper respiratory tract symptoms with sore throat and rhinnorhea. Her husband had similar symptoms and was diagnosed with group A beta-hemolytic Streptococcal pharyngitis.
On presentation the patient’s labs included a hemoglobin level of 6.7 g/dL, platelet count of 19,000/mm3, BUN 75 mg/dL (convert to mmol/L, multiply by 0.357) and creatinine 4.6 mg/dL (convert to µmol/L, multiply by 88.4). Urinalysis showed 3+ protein and 3 + blood, with greater than 100 red blood cells (RBCs)/hpf, and granular casts. Evidence of hemolysis was demonstrated by a haptoglobin level <6mg/dL, lactate dehydrogenase (LDH) 1635 U/L, and significant fragmented red cells on peripheral blood smear. Stool for Escherichia coli O157:H7, Salmonella, Campylobacter, and Shigella species were negative. Other relevant tests were C3 of 58 mg/dL (normal range 88–201) and C4 of 20 (normal range 16–47). Her A Disintegrin And Metalloproteinase with a ThromboSpondin type 1 motif, member 13 (ADAMTS13) activity was 97% (normal range ≥67) and inhibitor <0.4 units (normal range ≤0.4). A renal biopsy revealed diffuse capillary fibrin deposition and segmental staining for fibrin and IgM along the glomerular basement membrane consistent with thrombotic microangiopathy (TMA). She was discharged with daily plasmapheresis followed by every-other-day treatment for a total of 41 treatments. She was re-admitted six weeks later and commenced on hemodialysis. In the weeks leading to her transplant her LDH was elevated at 1073 U/L and at 883 U/L the week prior to transplant.
In December 2004, six months after initiating hemodialysis, the patient received a haploidentical living-related kidney transplant from her father and underwent bilateral native nephrectomies for refractory hypertension and to minimize the risk of recurrent disease.10,11 She received alemtuzumab, dexamethasone and intravenous immunoglobulin (IVIG) as induction. Her creatinine was 1.1 mg/dL on discharge and maintenance immunosuppression consisted of prednisone, mycophenolate mofetil, and tacrolimus. One month later, the patient was re-admitted with a serum creatinine of 1.9 mg/dL and biopsy showed recurrent TMA (Figure 1). Peritubular capillaries were negative for C4d deposition and single antigen bead assay using a multiplex Luminex® (Austin, TX) was negative for donor specific antibody.
Figure 1.
(A) –PAS-stained allograft biopsy (400×) reveals intracapillary fibrin thrombi (arrowhead), moderate increase in mesangial matrix and cellularity and global reduplication of glomerular capillary walls. A small capillary (top left corner) also shows a luminal fibrin thrombus. (B) PAMM Silver stained section (400×) shows another glomerulus with moderate increase in mesangial matrix and cellularity, and highlights the splitting of glomerular capillary walls. Luminal fibrin thrombi (arrowhead) are also noted.
Rituximab (375 mg/1.73 m2 BSA/dose × 4) and IVIG therapy (100 mg/Kg/dose) were reinstituted,9,12 and she continued plasma exchange 2–3 times per week for one month. Her tacrolimus was changed to sirolimus and follow-up creatinine stabilized to 1.4 mg/dL. Over the next several months, despite continued treatment, her renal function deteriorated with ongoing evidence of hemolysis. Late in the course of the disease while still on plasma infusions, further testing showed a quantitative Factor H level of 244.9 ug/mL (normal range 160–412). She resumed hemodialysis in October 2006 and subsequently underwent a transplant nephrectomy. The explanted graft showed changes consistent with chronic glomerulopathy and persistent TMA.
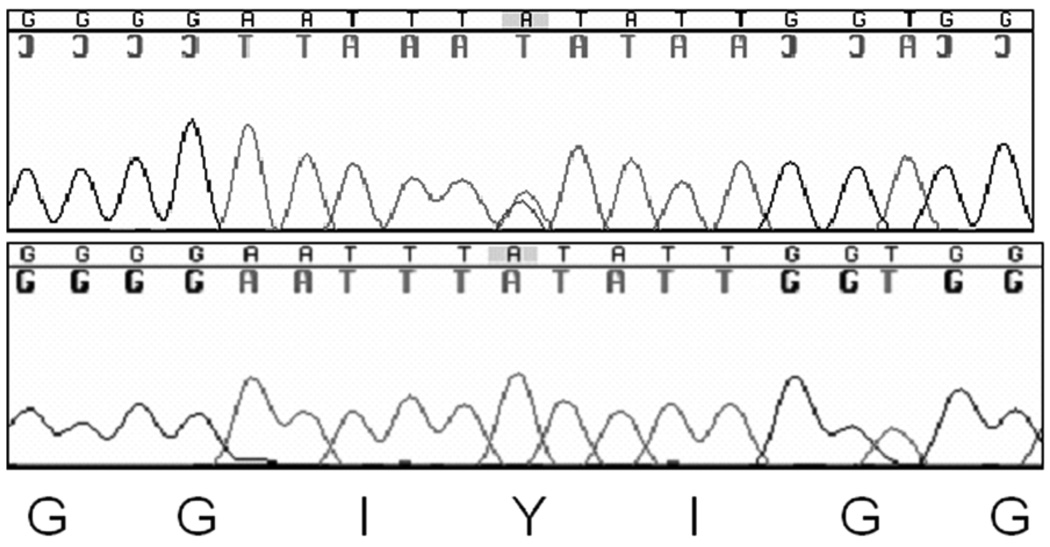
Given the rapid recurrence of HUS in the renal allograft, CFH, CD46 and Factor I were directly sequenced from genomic DNA isolated from peripheral blood. No mutations were found within the coding regions for CFH or CD46. A heterozygous missense mutation which changes a tyrosine to a serine residue was identified in the serine protease domain of CFI (Figure 2). This mutation, which has not been previously reported, is within a functional domain and is conserved across species with a conseq score of 6 indicating that it is likely to be functional. It is located in the 38kDa light chain in the serine protease domain that is linked by a disulfide bond to the noncatalytic heavy chain of Factor I (Figure 3).
Figure 2.
A heterozygous missense mutation c.1106A>C, p.Tyr369Ser lies in the serine protease domain of Factor I.
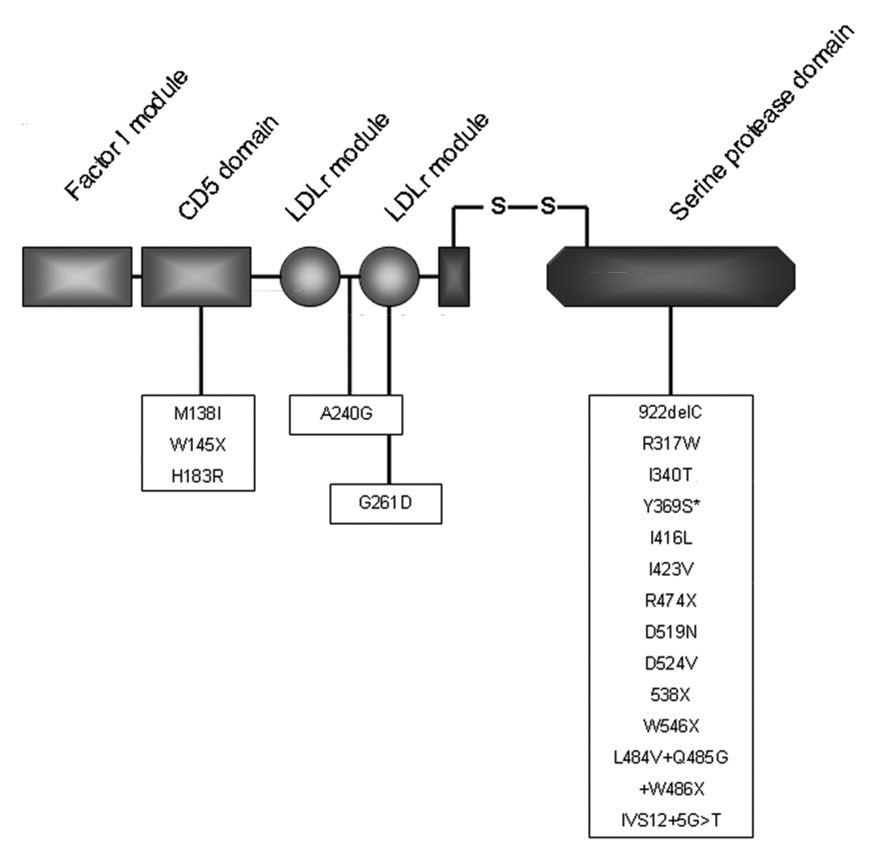
Figure 3.
Mutations in complement factor I associated with aHUS. Novel mutation Y369S noted with asterisk * Used with full permission from Oxford University Press
Discussion
Atypical HUS is a life threatening disorder with up to 50% of affected persons progressing to end-stage renal failure and 25% dying during the acute phase.3 In our case, the patient presented with renal failure and intravascular hemolysis and thrombocytopenia, consistent with the diagnosis of aHUS. Identification of a missense mutation in CFI provided genetic confirmation of the diagnosis of aHUS and is consistent with the clinical course of the disease in this patient.
CFI deficiency was first discovered in a patient who had increased susceptibility to infection.13 Accelerated inactivation of C3 to C3b was noted, and in vitro studies later showed that this was due to insufficient complement regulatory factor I.13 CFI is located on chromosome 4q25 and encodes an 88kDa serine protease that is predominantly synthesized in the liver.14 This protein regulates AP activity in the presence of a cofactor, which can be CFH, C4bBP, CD46 or CR1.15 The frequency of aHUS-causing CFI mutations is reported to be 3–12%, and these mutations are classified as either Type I mutations (decrease the amount of circulating CFI) or Type II mutations (normal serum levels of abnormally functioning CFI).3,16 Most mutations in the serine protease domain of CFI cause impairment of C3b and C4b cofactor activity and are probably the commonest form of type II mutations.17 Although factor I levels were not measured in our patient, we suspect that the identified mutation affects CFI function. Some other described mutations, especially those in CFI heavy chain do not appear to affect CFI level or activity and the pathophysiological basis for disease in these instances is unclear.15,17
Atypical HUS is more frequently associated with mutations in CFH and this association was first recognized 10 years ago when a candidate gene approach was used to identify two different mutations in CFH in patients with aHUS.18 Approximately 15–30% of cases of aHUS are associated with mutations in CFH.4 Interestingly, CFH is immediately upstream of five other genes in the CFH family, CFHR1–5 (Complement Factor H-Related 1–5). A deletion in CFHR1 and CFHR3 has been associated with anti-CFH antibodies which leads to an acquired form of functional CFH deficiency and aHUS.19
Kidney transplantation in patients with aHUS is associated with a high rate of disease recurrence. Outcome is mutation dependent, with approximately 80% of patients with CFH mutations developing recurrence, 100% of patients with CFI mutations, and only approximately 10% of patients with CD46 mutations.3 Both CFH and CFI are largely made in the liver, while CD46 is expressed locally within the kidney. As a result, a donor kidney with normal CD46 transplanted into a patient with CD46-related aHUS replaces the genetic defect within the kidney and presumably accounts for the reduced disease recurrence. In the case of CFH or CFI mutations, in contrast, haploinsufficiency continues, as the native liver is the primary source of CFH and CFI, and explains the frequent disease recurrence.3
Living-related renal transplantation is controversial. Prior to genetic testing for aHUS, multiple studies showed recurrence of disease in the recipient together with the appearance of de novo aHUS in the donor.20–24 On this basis, if a living-related donor transplantation is considered, the donor and recipient should have complete molecular testing of CFH, CFI, CD46 and CFB. In the absence of mutations in these genes or if a deletion of CFHR1 and CFHR3 is found, screening should include circulating autoantibodies for CFH and peripheral blood mononuclear cell CD46 expression.25 Recently, in an attempt to restore full synthesis of functional complement factors, combined liver-kidney transplantation has been attempted with mixed results. 8,26,27
In summary, we present a patient with aHUS who underwent an unsuccessful renal transplantation. The patient carries a novel mutation in CFI. Our experience with this case emphasizes the challenges inherent in the management of patients with aHUS and reiterates the importance of comprehensive evaluation in all patients who present with this disease Until better treatment protocols are developed, the likelihood of successful renal transplantation in patients with CFH or CFI mutations is low.
Acknowledgment
The authors thank David Kavanagh, MD PhD for his expert opinion on Factor I mutation and use of his diagrams and Maria A. Abeleda, BS of Molecular Otolaryngology Research Laboratories (MORL) for designing the primers. This research was supported in part by Grant RO1DK074409 to RJHS.
Funding: This research was supported in part by Grant RO1DK074409 to RJHS.
References
- 1.Gasser C, Gautier E, Steck A, Siebenmann RE, Oechslin R. Hemolytic-uremic syndrome: bilateral necrosis of the renal cortex in acute acquired hemolytic anemia. Schweiz Med Wochenschr. 1955;85:905–909. [PubMed] [Google Scholar]
- 2.Atkinson JP, Liszewski MK, Richards A, Kavanagh D, Moulton EA. Hemolytic uremic syndrome. An example of insufficient complement regulation on self-tissue. Ann NY Acd Sci. 2005;1056:144–152. doi: 10.1196/annals.1352.032. [DOI] [PubMed] [Google Scholar]
- 3.Kavanagh D, Goodship TH, Richards A. Atypical haemolytic uraemic syndrome. Br Med Bull. 2006;77–78:5–22. doi: 10.1093/bmb/ldl004. [DOI] [PubMed] [Google Scholar]
- 4.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- 5.Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, et al. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci U S A. 2007;104:240–245. doi: 10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jokiranta TS, Zipfel PF, Fremeaux-Bacchi V, Taylor CM, Goodship TJ, Noris M. Where next with atypical hemolytic uremic syndrome? Mol Immunol. 2007;44:3889–3900. doi: 10.1016/j.molimm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Bresin E, Daina E, Noris M, Castelletti F, Stefanov R, Hill P, Goodship T, Remuzzi G. Outcome of renal transplantation in patients with non-Shiga toxin-associated haemolytic syndrome: prognostic significance of genetic background. Clin J Am Soc Nephrol. 2006;1:88–99. doi: 10.2215/CJN.00050505. [DOI] [PubMed] [Google Scholar]
- 8.Kavanagh D, Goodship TH. Membrane cofactor protein and factor I: mutations and transplantation. Semin Thromb Hemost. 2006;32:155–159. doi: 10.1055/s-2006-939771. [DOI] [PubMed] [Google Scholar]
- 9.Sellier-Leclerc AL, Fremeaux-Bacchi V, Dragon-Durey MA, et al. Differential impact of complement mutations on clinical characteristics in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2007;18:2392–2400. doi: 10.1681/ASN.2006080811. [DOI] [PubMed] [Google Scholar]
- 10.Artz MA, Steenbergen EJ, Hoitsma AJ, Monnens LA, Wetzels JF. Renal transplantation in patients with hemolytic uremic syndrome: High rate of recurrence and increased incidence of acute rejections. Transplantation. 2003;76:821. doi: 10.1097/01.TP.0000085083.74065.1B. [DOI] [PubMed] [Google Scholar]
- 11.Remuzzi G, Ruggenenti P, Colledan M, et al. Hemolytic uremic syndrome: a fatal outcome after kidney and liver transplantation performed to correct factor h gene mutation. Am J Transplant. 2005;5:1146–1150. doi: 10.1111/j.1600-6143.2005.00783.x. [DOI] [PubMed] [Google Scholar]
- 12.Ponticelli C, Banfi G. Thrombotic microangiopathy after kidney transplantation. Transpl Int. 2006;19:789–794. doi: 10.1111/j.1432-2277.2006.00354.x. [DOI] [PubMed] [Google Scholar]
- 13.Alper CA, Abramson N, Johnston RB, Jr, Jandl JH, Rosen FS. Increased susceptibility to infection associated with abnormalities of complement-mediated functions and of the third component of complement (C3) N Engl J Med. 1970;282:350–354. doi: 10.1056/nejm197002122820701. [DOI] [PubMed] [Google Scholar]
- 14.Geelen J, van den Dries K, Roos A, et al. A missense mutation in factor I (IF) predisposes to atypical haemolytic uraemic syndrome. Pediatr Nephrol. 2007;22:371–375. doi: 10.1007/s00467-006-0320-2. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson SC, Karpman D, Vaziri-Sani F, et al. A mutation in factor I that is associated with atypical hemolytic uremic syndrome does not affect the function of factor I in complement regulation. Mol Immunol. 2007;44:1835–1844. doi: 10.1016/j.molimm.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Kavanagh D, Goodship TH. Update on evaluating complement in hemolytic uremic syndrome. Curr Opin Nephrol Hypertens. 2007;16:565–571. doi: 10.1097/MNH.0b013e3282f0872f. [DOI] [PubMed] [Google Scholar]
- 17.Kavanagh D, Richards A, Noris M, Hauhart R, Liszewski MK, Karpman D, Goodship JA, Fremeaux-Bacchi V, Remuzzi G, Goodship TH, Atkinson JP. Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol. 2008;45:95–105. doi: 10.1016/j.molimm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Warwicker P, Goodship TH, Donne RL, et al. Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 19.Dragon-Durey MA, Loirat C, Cloarec S, et al. Anti-Factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16:555–563. doi: 10.1681/ASN.2004050380. [DOI] [PubMed] [Google Scholar]
- 20.Donne RL, Abbs I, Barany P, et al. Recurrence of hemolytic uremic syndrome after live related renal transplantation associated with subsequent de novo disease in the donor. Am J Kidney Dis. 2002;40:1–4. doi: 10.1053/ajkd.2002.36938. [DOI] [PubMed] [Google Scholar]
- 21.Conlon PJ, Brennan DC, Pfaf WW, et al. Renal transplantation in adults with thrombotic thrombocytopenic purpura/haemolytic-uraemic syndrome. Nephrol Dial Transplant. 1996;11:1810–1814. [PubMed] [Google Scholar]
- 22.Ducloux D, Rebibou JM, SemhounDucloux S, et al. Recurrence of hemolytic-uremic syndrome in renal transplant recipients- A meta-analysis. Transplantation. 1998;65:1405–1407. doi: 10.1097/00007890-199805270-00023. [DOI] [PubMed] [Google Scholar]
- 23.Lahlou A, Lang P, Charpentier B, et al. Hemolytic uremic syndrome. Recurrence after renal transplantation. Groupe Coopératif de l'Ile-de-France (GCIF) Medicine. 2000;79:90–102. doi: 10.1097/00005792-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Quan A, Sullivan EK, Alexander SR. Recurrence of hemolytic uremic syndrome after renal transplantation in children: A report of the North American Pediatric Renal Transplant Cooperative Study. Transplantation. 2001;72:742–745. doi: 10.1097/00007890-200108270-00033. [DOI] [PubMed] [Google Scholar]
- 25.Kavanagh D, Richards A, Fremeaux-Bacchi V, Noris M, Goodship T, Remuzzi G, Atkinson JP. Screening for complement system abnormalities in patients with atypical hemolytic uremic syndrome. Clin J Am Soc Nephrol. 2007;2:591–596. doi: 10.2215/CJN.03270906. [DOI] [PubMed] [Google Scholar]
- 26.Saland JM, Emre SH, Shneider BL, et al. Favorable long-term outcome after liver-kidney transplant for recurrent hemolytic uremic syndrome associated with a factor H mutation. Am J Transplant. 2006;6:1948–1952. doi: 10.1111/j.1600-6143.2006.01375.x. [DOI] [PubMed] [Google Scholar]
- 27.Jalanko H, Peltonen S, Koskinen A, et al. Successful liver-kidney transplantation in two children with aHUS caused by a mutation in complement factor H. Am J Transplant. 2008;8:216–221. doi: 10.1111/j.1600-6143.2007.02029.x. [DOI] [PubMed] [Google Scholar]