This work analyzes the surprising result that Nicotiana benthamiana transformed with RNA-dependent RNA polymerase 1 from Nicotiana tabacum (Nt-RDR1) is hypersusceptible to several viruses. It provides evidence supporting a dual role for RDR1 in contributing to salicylic acid–mediated antiviral defense at the same time as it suppresses RDR6-mediated antiviral RNA silencing.
Abstract
Endogenous eukaryotic RNA-dependent RNA polymerases (RDRs) produce double-stranded RNA intermediates in diverse processes of small RNA synthesis in RNA silencing pathways. RDR6 is required in plants for posttranscriptional gene silencing induced by sense transgenes (S-PTGS) and has an important role in amplification of antiviral silencing. Whereas RDR1 is also involved in antiviral defense in plants, this does not necessarily proceed through triggering silencing. In this study, we show that Nicotiana benthamiana transformed with RDR1 from Nicotiana tabacum (Nt-RDR1 plants) exhibits hypersusceptibility to Plum pox potyvirus and other viruses, resembling RDR6-silenced (RDR6i) N. benthamiana. Analysis of transient induction of RNA silencing in N. benthamiana Nt-RDR1 and RDR6i plants revealed that Nt-RDR1 possesses silencing suppression activity. We found that Nt-RDR1 does not interfere with RDR6-dependent siRNA accumulation but turns out to suppress RDR6-dependent S-PTGS. Our results, together with previously published data, suggest that RDR1 might have a dual role, contributing, on one hand, to salicylic acid–mediated antiviral defense, and suppressing, on the other hand, the RDR6-mediated antiviral RNA silencing. We propose a scenario in which the natural loss-of-function variant of RDR1 in N. benthamiana may be the outcome of selective pressure to maintain a high RDR6-dependent antiviral defense, which would be required to face the hypersensitivity of this plant to a large number of viruses.
INTRODUCTION
Most eukaryotes possess an RNA silencing system that is mediated by small RNAs of 21 to 24 nucleotides in length. RNA silencing provides sequence-specific degradation or translational repression of target RNA and eliminates RNAs that are sensed as aberrant through direct recognition or formation of double-stranded RNA (dsRNA) (Baulcombe, 2004, 2005). In plants as well as the nematode Caenorhabditis elegans, RNA silencing, which is initiated with small amounts of initiator dsRNA, involves an amplification process that requires RNA-directed RNA polymerases (RDRs) for persistent silencing (Baulcombe, 2007). In plants, the initiator dsRNAs are processed by Dicer-like proteins to produce small interfering RNAs (siRNAs). These primary siRNAs bind to the Argonaute (AGO)-containing RNA-induced silencing complex (RISC) that cleaves targeted RNA. The cleaved RNA then recruits RDR to generate more dsRNA from regions surrounding the primary trigger sequence, producing secondary siRNAs (Axtell et al., 2006). One of the six Arabidopsis thaliana RDRs, At-RDR6, has been shown to be indispensable in transitive RNA silencing for producing endogenous trans-acting siRNAs (tasiRNAs) from a cleaved RNA (Allen et al., 2005; Yoshikawa et al., 2005). RDR6 was also found to be essential for sense transgene-induced posttranscriptional gene silencing (S-PTGS), but it is not required for inverted repeat dsRNA-induced PTGS (IR-PTGS) (Dalmay et al., 2000; Mourrain et al., 2000; Vance and Vaucheret, 2001; Vaistij et al., 2002; Parizotto et al., 2004).
In addition to transitive silencing of single-stranded RNA, plant RDR6 is also required for spreading silencing through the plant. This occurs via amplification of secondary siRNAs and their movement from the original site of silencing via cell-to-cell and systemic transport (Voinnet et al., 2000; Klahre et al., 2002; Himber et al., 2003; Voinnet, 2005). This RDR6-dependent amplification and systemic spread of silencing is a crucial step toward achieving an efficient antiviral defense response in plants. In many cases, restriction of virus movement, rather than inability of the virus to replicate within cells, is responsible for host range limitations and such is the case with RNA silencing–mediated plant defense (Baulcombe, 2004; Alamillo et al., 2006). At-RDR6 is implicated in defense against Cucumber mosaic cucumovirus (CMV) (Dalmay et al., 2000; Mourrain et al., 2000). The RDR6 homolog in Nicotiana benthamiana (Nb-RDR6) has been determined to have effects on the silencing signal during Potato potexvirus X (PVX) infection, such that the systemic spread of PVX is limited (Schwach et al., 2005).
RDR1 was found to be involved in separate but overlapping viral resistance and PTGS mechanisms in plants. At-RDR1 and Nt-RDR1 (the Nicotiana tabacum ortholog of At-RDR1) both are elicited by salicylic acid (SA) treatment and virus infection, and they influence susceptibility to Tobacco mosaic tobamovirus (TMV) and Tobacco rattle virus (TRV) in Arabidopsis (Yu et al., 2003) and to TMV and PVX in tobacco (Xie et al., 2001). Although it has been suggested that At-RDR1's role in the activation of antiviral defenses is not through triggering virus-induced gene silencing (VIGS) (Yu et al., 2003), a recent study using siRNA deep sequencing revealed that At-RDR1 plays an important role in the biogenesis of TMV siRNAs (Qi et al., 2009). At-RDR1–dependent production of CMV siRNAs has also been found when the silencing suppressor of CMV is absent (Diaz-Pendon et al., 2007).
N. benthamiana has an RDR1 gene that contains a 72-nucleotide insert with consecutive in-frame stop codons in the 5′ half of the RDR1 open reading frame. This natural loss-of-function mutation was named Nb-RdRP1m (also called Nb-RDR1m) (Yang et al., 2004). N. benthamiana transformed with an SA-inducible RDR1 gene from Medicago truncatula (Mt-RDR1) showed enhanced resistance to tobamoviruses (e.g., TMV, Turnip vein-clearing virus [TVCV], and Sunn hemp mosaic virus [SHMV]) but not to other viruses, such as CMV and PVX (Yang et al., 2004). It has been suggested that the extreme susceptibility of N. benthamiana to a wide range of viruses (van Dijk and Huismans, 1987; van Dijk et al., 1987; Dawson and Hilf, 1992) is because it lacks an SA- and virus-inducible RDR1 (Yang et al., 2004).
In this study, N. benthamiana transformed with RDR1 from N. tabacum (Nt-RDR1 plants) exhibited hypersusceptibility to Plum pox potyvirus (PPV) in systemically infected leaves, resembling PPV-infected RDR6-silenced N. benthamiana (RDR6i) plants. Analysis of transient induction of RNA silencing in N. benthamiana wild-type, Nt-RDR1, and RDR6i plants revealed that Nt-RDR1 (1) possesses silencing suppressor activity and (2) suppresses RDR6-dependent S-PTGS, but (3) does not interfere with RDR6-dependent siRNA and tasiRNA accumulation. Our results suggest that RDR6-dependent antiviral PTGS may be negatively regulated by functional RDR1 and that the natural variant of RDR1 in N. benthamiana may be the outcome of the pressure of its extreme susceptibility to certain viruses (van Dijk and Huismans, 1987; van Dijk et al., 1987; Dawson and Hilf, 1992) to accomplish the RDR6-dependent antiviral defense successfully. The various possible mechanisms by which Mt-RDR1 (Yang et al., 2004) and Nt-RDR1 (this study) mediate viral resistance in N. benthamiana are discussed, as are the different effects of RDR1 in N. tabacum and N. benthamiana.
RESULTS
Constitutive or Induced Expression of N. tabacum RDR1 in N. benthamiana Results in Hypersusceptibility to PPV Infection
N. benthamiana containing the natural loss-of-function mutation Nb-RDR1m (Yang et al., 2004) was transformed with an RDR1 ortholog from N. tabacum (Nt-RDR1) fused to a myc-tag and expressed under the control of the 35S promoter. Nt-RDR1 is 95 and 93% identical in nucleotide and amino acid sequence, respectively, to Nb-RDR1m, not including the 72-nucleotide insertion sequence (Yang et al., 2004). The accumulation of myc-NtRDR1 protein in T1 progeny plants derived from eight independent primary transformants that contained one or two copies of the 35S-Myc-NtRDR1 DNA insert (see Supplemental Figure 1 online) was detected with anti-c-myc antibody in six of the eight lines (Figure 1A). The accumulation of myc-NtRDR1 RNA was detected in all eight lines at similar levels (Figure 2D). Hereafter, the protein-expressing lines (lines 1 to 6) are referred to as Nt-RDR1 plants, whereas the non-protein-expressing lines (lines 7 and 8) are referred to as Nt-RDR1(-) plants. All transgenic 35S-NtRDR1 lines appeared normal by visual inspection.
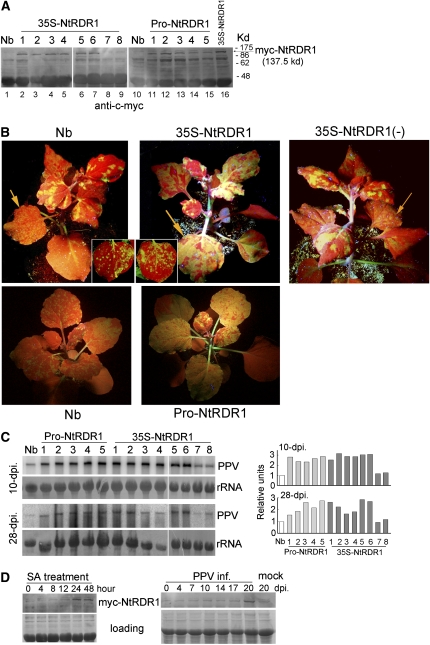
Figure 1.
Analysis of PPV Infection in the Transgenic N. benthamiana Plants Expressing Nt-RDR1.
(A) Protein immunoblot detection of myc-NtRDR1 with anti-c-myc antibody in pools of six T1 plants of different 35S-NtRDR1 and Pro-NtRDR1 transgenic lines. Wild-type N. benthamiana (Nb) was used as negative control.
(B) Wild-type control (Nb) or transgenic plants were inoculated with GFP-tagged PPV. Photographs were taken from line 1 and line 7 of 35S-NtRDR1 [each containing one copy of the 35S-Myc-NtRDR1 DNA insert, see Supplemental Figure 1 online, even though line 7, labeled 35S-NtRDR1(-), does not express the transgenic protein] at 25 DAI (top panels) and line 1 of Pro-NtRDR1 at 18 DAI (bottom panels) under UV light to show the GFP expression associated with PPV infection in systemically infected leaves. Local infected leaves were photographed at 4 DAI (insets in top panels).
(C) PPV RNA accumulation was examined at similar layers of systemically infected leaves of Nb, Pro-NtRDR1, and 35S-NtRDR1 collected at 10 and 28 DAI. Pools of six plants were analyzed for each plant line. Hybridization was done with a 32P-labeled PPV antisense cDNA probe. Methylene blue–stained rRNA is shown as loading control. Quantification of PPV-GFP RNA relative to total RNA is shown at the right part of the panel. The value of Nb was arbitrarily designated as 1.
(D) Protein immunoblot analysis with anti-c-myc antibody of myc-NtRDR1 induction in Pro-NtRDR1 plants after SA treatment or PPV infection. Pools of four plants were analyzed for each time point. The result of representative line 3 was shown. Coomassie blue-stained total proteins are shown as loading controls.
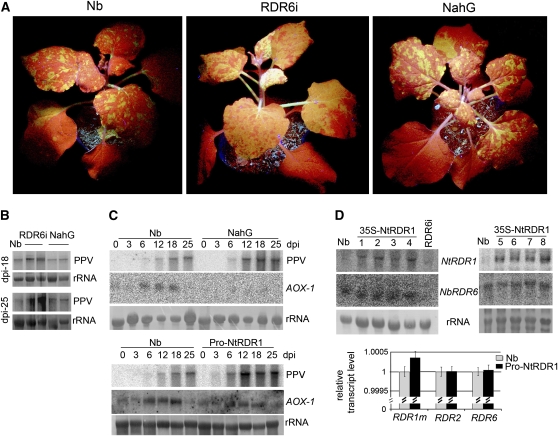
Figure 2.
Analysis of PPV Infection in RDR6-Silenced (RDR6i) and SA-Deficient (NahG) Transgenic N. benthamiana Plants.
(A) GFP-tagged PPV was inoculated to wild-type (Nb), RDR6i, and NahG plants, and photographs were taken at 18 DAI under UV light to show the GFP expression associated with PPV infection in systemically infected leaves.
(B) PPV-GFP RNA accumulation was examined at similar layers of systemic infected leaves of Nb, RDR6i, and NahG plants collected at 18 and 25 DAI. The duplicated lanes are two pools of six plants. Hybridization was done as described in Figure 1C.
(C) Kinetics of induction of AOX-1 in response to PPV infection. Systemically infected leaves of Nb, NahG, and Pro-NtRDR1 transgenic plants infected with PPV-GFP were collected at different days after inoculation. Pools of four plants were analyzed for each time point. The blot was probed for accumulation of PPV RNA and AOX-1 transcripts using 32P-labeled specific cDNAs. Methylene blue–stained rRNA is shown as loading control.
(D) Detection of RDR1m, RDR2, and RDR6 mRNA accumulation in Nb, 35S-NtRDR1, and RDR6i plants. The RNA gel blots were probed for accumulation of transgene Nt-RDR1 and endogenous RDR6 transcripts using 32P-labeled specific transcribed RNA. Pools of four plants were analyzed for each plant line. Methylene blue–stained rRNA is shown as loading control. Quantitative real-time RT-PCR for the assessment of RDR1m, RDR2, and RDR6 transcript levels in Nb and Pro-NtRDR1 plants were made on pools of four plants each. Error bars represent standard deviations for three replicates. Relative transcript levels were calculated by the ΔΔC(t) method (Livak and Schmittgen, 2001) using GAPDH transcripts (Schwach et al., 2005) as the internal standard.
To determine the effect of Nt-RDR1 expression in virus infection, seven T1 plants from each transgenic line were inoculated with PPV and green fluorescent protein (GFP)-tagged PPV, a recombinant PPV encoding GFP (Alamillo et al., 2006). Wild-type N. benthamiana and vector-transformed plants were used as controls. Similar striking chlorosis and curling disease symptoms were observed in systemically infected leaves of both wild-type and vector-transformed plants infected with PPV or PPV-GFP, confirming that expression of the GFP marker within the viral genome did not modify the outcome of the infection (see Supplemental Figure 2 online) (Alamillo et al., 2006). Systemic symptoms occurred mainly in the four leaves above the inoculated ones but also reached leaves further up, where virus accumulation was confirmed by direct observation of GFP fluorescence (Figure 1B) and by RNA gel blot analysis (Figure 1C). To our surprise, there was not much difference between the inoculated leaves of the wild-type plants and those of the transgenic Nt-RDR1 plants (Figure 1B, inset). However, all Nt-RDR1 plants appeared to have stronger GFP fluorescence in the noninoculated upper leaves (Figure 1B) with higher levels of viral RNA [about 3 and 2 times that in the wild-type and Nt-RDR1(-) plants at 10 d after inoculation [DAI] and 28 DAI, respectively] (Figure 1C). The GFP fluorescence in the lower systemically infected leaves of the Nt-RDR1 plants was diffuse to agglomerate, whereas in wild-type and Nt-RDR1(-) leaves at the same level, it was in small speckles (Figure 1B, arrow). The upper systemically infected leaves of Nt-RDR1 plants were almost fully GFP fluorescent, while wild-type and Nt-RDR1(-) leaves emerged with only a few areas of fluorescence (Figure 1B). The same results were obtained in three independent experiments. These results ran counter to our initial idea of using Nt-RDR1 in N. benthamiana to gain protection against PPV infection.
To determine whether the increased susceptibility to PPV infection was due to the constitutive expression of Nt-RDR1 from the 35S promoter, the endogenous promoter from the natural defective variant Nb-RDR1m was cloned to replace the 35S promoter. Five individual transgenic lines (Pro-NtRDR1) were obtained. All PPV-GFP–infected Pro-NtRDR1 plants (six plants for each line in each assay, repeated more than three times) showed less severe symptoms in newly emerged leaves and less green fluorescence compared with the infected 35S-NtRDR1 plants but considerably stronger symptoms and fluorescence than the infected wild-type plants (Figure 1B). Viral RNA accumulation was also about 2 to 3 times higher than that in wild-type plants (Figure 1C). Protein immunoblots showed that, regardless of the promoter, all transgenic lines constitutively accumulated myc-NtRDR1 protein to comparable levels (Figure 1A). To validate the Nb-RDR1m endogenous promoter, which was shown to be induced by SA (Yang et al., 2004), the induction of myc-NtRDR1 in Pro-NtRDR1 plants was examined by application of SA and PPV infection. Figure 1D shows that myc-NtRDR1 expression of representative line 3 was stimulated by SA treatment, reaching a maximum of ∼2.5-fold at 24 h post-treatment (hpt), and it was maintained at a high level up to 48 hpt. Myc-NtRDR1 was increased ∼3.8-fold in upper noninoculated PPV-infected leaves compared with leaves at the same level in mock-inoculated plants at 20 DAI. There was a slight induction of myc-NtRDR1 at 8 hpt in 35S-NtRDR1 plants in parallel SA treatment and no induction with PPV infection assays (see Supplemental Figure 3 online). These results suggest that the cloned Nb-RDR1m promoter was correct and functional.
Thus, expression of Nt-RDR1 by either the 35S or the Nb-RDR1m promoter in N. benthamiana enhanced systemic spread of PPV-GFP, and this hypersusceptibility operated at the Nt-RDR1 protein, rather than RNA, level.
The Hypersusceptibility to PPV-GFP in Nt-RDR1 Plants Resembles That in RDR6i but Contrasts with the Normal Susceptibility to PPV of NahG N. benthamiana Plants
To investigate whether the hypersusceptibility to PPV-GFP in NtRDR1 plants results from interference with host RNA silencing and/or SA-mediated defense pathway(s) that are known to restrict the systemic spread of PPV in tobacco (Alamillo et al., 2006), we performed the following experiments.
Nb-RDR6 has been shown to be required for the cell to amplify the primary VIGS-derived signal to block systemic virus spread (Schwach et al., 2005). RDR6i N. benthamiana plants, in which Nb-RDR6 has been silenced by an RNA interference (RNAi) hairpin construct (Schwach et al., 2005), were inoculated with PPV-GFP. Inoculation was also performed in wild-type plants as controls. Extensive green fluorescence and more severe symptoms were observed in the systemically infected leaves in RDR6i plants (Figure 2A) compared with those in the infected wild-type plants, which displayed a few fluorescent areas (Figure 2A), whereas there was no significant difference in the GFP fluorescence of the inoculated leaves. Elevated viral RNA levels, up to 4 to 6 times higher than those of infected wild-type plants at 25 DAI, were detected in RDR6i plants (Figure 2B). These results indicate that RDR6 inhibits PPV systemic spread.
Next, to examine whether the SA-mediated host defense inhibits PPV systemic spread, N. benthamiana was transformed with bacterial NahG, which encodes the SA-degrading enzyme salicylate hydroxylase (Gaffney et al., 1993). Primary transformants were confirmed by RNA gel blot analysis, indicating the accumulation of NahG transcripts (see Supplemental Figure 4 online). Six T1 progeny plants from two individual lines were inoculated with PPV-GFP. There was no significant difference in symptoms or GFP fluorescence between the infected wild-type and NahG plants (Figure 2A). Expression of the alternative oxidase gene AOX-1, which is known to depend on SA accumulation, was transiently induced in response to PPV-GFP infection in wild-type plants but not in the NahG plants (Figure 2C), confirming that the NahG transgenic plants were SA deficient. Similar infection phenotypes and viral RNA levels (Figures 2A and 2B) in the wild-type and NahG plants suggested that the effect of SA-mediated defenses on PPV infection in N. benthamiana, if it exists, is minor. Furthermore, we examined the induction of AOX-1 in response to PPV-GFP infection in Pro-NtRDR1 plants. The kinetics of AOX-1 RNA induction is comparable to those observed in wild-type N. benthamiana in the parallel infection, suggesting that overexpression of Nt-RDR1 does not appreciably perturb the SA-dependent defense response in N. benthamiana (Figure 2C).
Taken together, our results argue against the possibility that the hypersusceptibility to PPV-GFP in NtRDR1 plants was due to disturbance of an SA-mediated defense pathway. However, the hypersusceptibility to PPV-GFP in RDR6i resembled the phenotype of PPV-GFP–infected NtRDR1 plants, suggesting that the RDR6-based VIGS antiviral resistance might be inhibited by the expression of Nt-RDR1 in N. benthamiana.
Overexpression of Nt-RDR1 in N. benthamiana Did Not Cause a General Cosuppression of Endogenous Nb-RDR Genes
To examine whether the hypersusceptibility to PPV-GFP infection in NtRDR1 plants, which resembles that of RDR6i plants, results from silencing of Nb-RDR6 transcripts by expression of the Nt-RDR1 gene (the two RDR nucleotide sequences share 50.1% similarity), Nb-RDR6 mRNA levels were examined. There were no obvious differences in Nb-RDR6 mRNA levels between the wild-type and NtRDR1 transgenic plants (Figure 2D). Quantitative real-time RT-PCR was also performed to examine Nb-RDR1m, Nb-RDR6, and Nb-RDR2 transcript levels; similar transcript levels were obtained for the three genes in wild-type and Nt-RDR1 plants (Figure 2D). Taken together with the accumulation of myc-NtRDR1 protein in the Nt-RDR1 transgenic plants (Figure 1A), our results indicate that, if the hypersusceptibility to PPV of the plants overexpressing Nt-RDR1 is caused by interference with an RDR6-mediated antiviral resistance mechanism, this interference was not due to suppression of Nb-RDR6 transcripts.
Nt-RDR1 Delayed Transient Induction of GFP Silencing
The hypersusceptibility to PPV and the possibility of subverting RDR6-based VIGS antiviral resistance in Nt-RDR1 plants prompted us to examine the silencing suppression activity of Nt-RDR1. First, an Agrobacterium tumefaciens coinfiltration assay was performed. In this system, an Agrobacterium strain expressing 35S-GFP that is infiltrated to GFP-transgenic N. benthamiana plants (line 16c) induces silencing of the GFP transgene, resulting in red fluorescence, unless the plants are coinfiltrated with another Agrobacterium strain expressing an RNA silencing suppressor (Brigneti et al., 1998). 35S-GFP and 35S-NtRDR1, which encoded a GFP and an Nt-RDR1 expression cassette, respectively, were coinfiltrated to leaves of the 16c GFP plants (Brigneti et al., 1998), leading to transient expression of both genes in the infiltrated regions. Coinfiltrations of 35S-GFP with an empty vector or with 35S-p19 (which encodes a viral silencing suppressor from a tombusvirus) were also performed as controls.
Induction of GFP silencing was evidenced by red fluorescence in the areas coinfiltrated with 35S-GFP and the empty vector at 4 d post agroinfiltration (dpa) (Figure 3A), whereas patches coinfiltrated with 35S-GFP and 35S-NtRDR1 showed slight enhancement of GFP fluorescence at this time point (Figure 3A). Coinfiltration of 35S-GFP with 35S-p19 maintained strong GFP fluorescence in infiltrated leaves (Figure 3A) to 9 to 10 dpa. Finally, systemic spread of GFP silencing was observed in all the samples, including plants coinfiltrated with 35S-GFP in combination with the empty vector or 35S-NtRDR1, except for the samples coinfiltrated with 35S-GFP and 35S-p19. The GFP silencing was confirmed by RNA gel blot analysis (Figure 3B) in which degradation of GFP mRNA was detected together with accumulation of GFP-specific siRNAs in the samples coinfiltrated with 35S-GFP and vector. Accumulation of GFP-specific siRNAs was also detected in the 35S-GFP/35S-NtRDR1 coinfiltrated sample (Figure 3B). This result suggested that Nt-RDR1 might function as a suppressor of RNA silencing in this coinfiltration assay, although its activity was not as strong as that of p19.
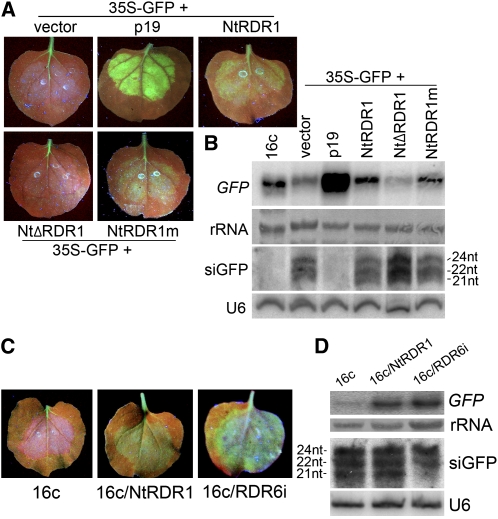
Figure 3.
Analysis of the Effect of Nt-RDR1 on Transient Induction of Silencing in GFP-Transgenic 16c N. benthamianaPlants.
(A) Leaves of 16c GFP-transgenic plants were coinfiltrated with 35S-GFP and either empty vector, 35S-p19, 35S-myc-NtRDR1, 35S-NtΔRDR1, or 35S-NtRDR1m. Photographs were taken under UV light at 4 dpa.
(B) RNA gel blot analysis of GFP mRNA and GFP-derived siRNA accumulation. 32P-labeled GFP DNA or RNA probes were used. Methylene blue–stained rRNA and U6 RNA hybridization are shown as loading controls.
(C) Leaves of 16c, 16c/NtRDR1, and 16c/RDR6i plants infiltrated with 35S-GFP. Photographs were taken under UV light at 4 dpa.
(D) RNA gel blot analysis of GFP mRNA and GFP-derived siRNAs accumulation as described in (B).
The eukaryotic RDRs share a conserved motif DXDGD (X represents any amino acid), which was shown to be at the catalytic active site (Iyer et al., 2003). To test whether the Nt-RDR1's catalytic activity is required for its silencing suppression activity, the Nt-RDR1 DLDGD sequence was replaced with VMVEV by site-directed mutagenesis, giving 35S-NtRDR1m. In addition, 35S-NtΔRDR1 was constructed by replacing the start codon of Nt-RDR1 with a stop codon. Coinfiltration of 35S-GFP with 35S-NtRDR1m showed slight enhancement of GFP fluorescence and mRNA accumulation at 4 dpa as found for 35S-NtRDR1 (Figures 3A and 3B); however, induction of GFP silencing was observed in the areas coinfiltrated with 35S-GFP/35S-NtΔRDR1 at this time point (Figures 3A and 3B). GFP-specific siRNAs were also detected in 35S-GFP/35S-NtRDR1m and 35S-GFP/35S-NtΔRDR1 coinfiltrated samples (Figure 3B). These results suggest that the Nt-RDR1 protein, but not RNA, possesses silencing suppression activity, and the catalytic activity of the Nt-RDR1 is not required for its suppression function.
Next, we examined the possible silencing suppression activity of constitutively expressed Nt-RDR1 in 16c/NtRDR1 transgenic plants. The 16c/NtRDR1 line was generated by crossing 16c and 35S-NtRDR1 (line 1) plants. The expression of the GFP and Nt-RDR1 transgenes in progeny plants was verified by green fluorescence and Nt-RDR1 protein accumulation, respectively. The 16c/RDR6i plants (Schwach et al., 2005) were also used in the following assays. Agrobacteriumcontaining 35S-GFP was infiltrated to 16c, 16c/NtRDR1, and 16c/RDR6i plants. Induction of GFP silencing was observed in the infiltrated areas in 16c plants at 4 dpa (Figure 3C), while infiltrated leaves of 16c/NtRDR1 and 16c/RDR6i plants maintained a strong GFP fluorescence at this time (Figure 3C). RNA gel blot analysis also showed higher levels of GFP mRNA in infiltrated leaves of 16c/NtRDR1 and 16c/RDR6i plants than those in infiltrated leaves of 16c plants (Figure 3D). Interestingly, GFP-specific siRNAs of 21, 22, and 24 nucleotides were detected in silenced 16c and nonsilenced 16c/NtRDR1 leaves, whereas only 22- and 24-nucleotide GFP-specific siRNAs were detected in 16c/RDR6i leaves at 4 dpa (Figure 3D). These results suggest that accumulation of the 21-nucleotide siRNAs and induction of GFP silencing were RDR6 dependent, and the constitutive expression of Nt-RDR1 in N. benthamiana did not reduce the accumulation of the 21-nucleotide siRNAs but did suppress induction of GFP silencing.
All these results indicate that, either transiently or constitutively expressed, Nt-RDR1 might function as a silencing suppressor. This further supports the idea that the hypersusceptibility to PPV infection in NtRDR1 plants is not due to the cosuppression of endogenous gene(s), such as Nb-RDR6, but rather to a direct role of the Nt-RDR1 transgene.
Nt-RDR1 Suppressed Sense PTGS but Did Not Interfere with RDR6-Dependent Secondary siRNA Synthesis
Nb-RDR6 has an effect on blockage of virus systemic spread (Schwach et al., 2005), and At-RDR6 has been shown to be required for secondary siRNA synthesis, a process involved in transitive amplification of RNA silencing (Vaistij et al., 2002; Parizotto et al., 2004). To investigate further the possible molecular basis of suppression of silencing by Nt-RDR1 in N. benthamiana, we examined whether Nt-RDR1 was able to interfere with RDR6-dependent secondary siRNA synthesis. A hairpin construct, named 35S-Fi, containing only a central part of the GFP target sequence (symbolized as “F” because it represents the middle of GFP) was created to induce IR-PTGS.
The 16c, 16c/Nt-RDR1, and 16c/RDR6i plants were infiltrated with 35S-Fi to induce silencing of the GFP transgene. Induction of GFP silencing was observed as red areas in infiltrated leaves of all three genotypes at 4 dpa, and similar levels and profiles of F-specific siRNAs (including all sizes of 21-, 22- and 24-nucleotide siRNAs) were detected (Figure 4A, lanes 1 to 3). The detection of 21-nucleotide siRNAs and induction of GFP silencing in 16c/RDR6i samples infiltrated with 35S-Fi, together with the above result that there were no 21-nucleotide siRNAs detected and no GFP silencing induced upon infiltration of full-length sense GFP transgene 35S-GFP in 16c/RDR6i plant samples (Figure 3D), suggests that the synthesis of 21-nucleotide siRNAs is important for induction of PTGS. Our result is also consistent with a previous finding that IR-PTGS is RDR6 independent (Beclin et al., 2002). Neither 5′ (upstream) nor 3′ (downstream) secondary siRNAs of F (referred to as G and P siRNAs, respectively) were detected in the three plant genotypes in this assay (Figure 4A, lanes 1 to 3). GFP silencing spread to upper noninfiltrated leaves at 7 dpa in all infiltrated plants (Figure 4B). In 16c/RDR6i plants, however, the GFP silencing spread only to one or two upper leaves, and red fluorescence was displayed as speckles or was restricted near veins. By contrast, GFP silencing spread throughout 16c and 16c/Nt-RDR1 plants, suggesting that RDR6-dependent responses to the systemic signal that silence the GFP transgene in upper leaves were not inhibited by Nt-RDR1 in 35S-NtRDR1 plants.
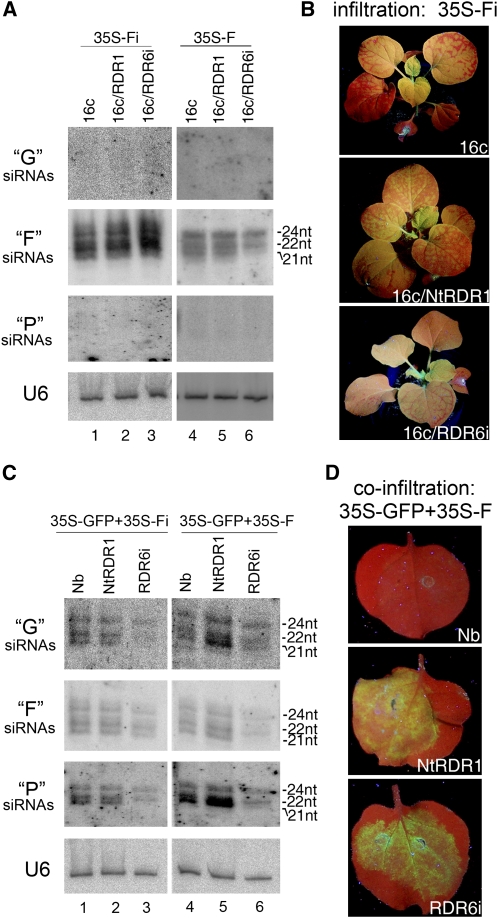
Figure 4.
Analysis of the Effect of Nt-RDR1 on S-PTGS and Secondary siRNA Synthesis.
(A) and (B) Induction of silencing of the GFP transgene in 16c, 16c/NtRDR1, and 16c/RDR6i plants. (A) shows an RNA gel blot analysis of G-, F-, and P-derived siRNA accumulation in 16c, 16c/NtRDR1, and 16c/RDR6i plants infiltrated with 35S-Fi or 35S-F at 4 dpa. 32P-labeled RNA probes specific for each corresponding GFP portion were used. U6 RNA hybridization is shown as loading control. (B) shows the systemic spread to the whole plant of GFP silencing induced by transient expression of 35S-Fi in 16c and 16c/NtRDR1 plants and the limited silencing spread to one or two upper leaves in a 16c/RDR6i plant. Photographs were taken under UV light at 2 weeks after agroinfiltration.
(C) and (D) Induction of silencing of transiently expressed GFP in Nb, Nt-RDR1, and RDR6i plants. (C) shows an RNA gel blot analysis of G-, F-, and P-derived siRNA accumulation in plants coinfiltrated with 35S-GFP and 35S-Fi or 35S-F as described in (A). (D) shows leaves of Nb, Nt-RDR1, and RDR6i plants coinfiltrated with 35S-GFP and 35S-F. Photographs were taken under UV light at 4 dpa.
We next constructed 35S-F, which expresses the F sense transcript to induce RDR6-dependent sense (S)-PTGS, and used it to infiltrate 16c, 16c/Nt-RDR1, and 16c/RDR6i leaves. GFP silencing was observed at 4 dpa in infiltrated 16c leaves but not in those of either 16c/Nt-RDR1 or 16c/RDR6i. Similar to the induction of GFP silencing by infiltration of 35S-GFP, F-specific siRNAs were detected in silenced 16c and nonsilenced 16c/Nt-RDR1 infiltrated leaf samples, and only 22- and 24-nucleotide F-specific siRNAs were detected in 16c/RDR6i infiltrated leaves (Figure 4A, lanes 4 to 6). Consistent with the above assay of full-length GFP induction (Figure 3D), Nt-RDR1 suppressed RDR6-dependent S-PTGS but did not block the synthesis of the 21-nucleotide siRNAs. G and P secondary siRNAs could not be detected by RNA gel blot analysis at high enough levels to distinguish differences among the three genotypes of plants (Figure 4A, lanes 4 to 6).
Therefore, induction of silencing by transient expression of 35S-GFP coinfiltrated with 35S-Fi or 35S-F was performed to examine further secondary siRNA accumulation. 35S-GFP/35S-Fi coinfiltration caused rapid silencing of GFP, and there were no differences in F-specific siRNA accumulation among wild-type, Nt-RDR1, and RDR6i plants (Figure 4C, lanes 1 to 3). The transitive RNA silencing of transiently expressed GFP was evidenced by detection of both G and P specific secondary siRNAs in infiltrated wild-type and Nt-RDR1 leaves (Figure 4C, lanes 1 and 2). The 21- and 22-nucleotide species accumulated at high levels, and 24-nucleotide secondary siRNAs were detected at slightly lower levels in these plants. Reduced amount of 22-nucleotide and no 21-nucleotide G or P specific secondary siRNAs were found in infiltrated RDR6i plants (Figure 4C, lane 3). This result suggests that the RDR6-dependent accumulation of 21-nucleotide secondary siRNAs induced by primary IR-PTGS was not disturbed by constitutive expression of Nt-RDR1 in N. benthamiana.
35S-GFP/35S-F coinfiltration caused GFP silencing in wild-type plants at 4 dpa. However, GFP silencing was blocked when 35S-GFP/35S-F were coinfiltrated to Nt-RDR1 or RDR6i plants (Figure 4D), although the intensity of green fluorescence was slightly lower in Nt-RDR1 than in RDR6i leaves, consistent with the GFP mRNA accumulation detected by RNA gel blot analysis (see Supplemental Figure 5 online). There were no obvious differences in F-specific siRNA accumulation between wild-type and NtRDR1 plants, but reduced amounts of 22- and 24-nucleotidet and no 21-nucleotide siRNAs were found in infiltrated RDR6i plants (Figure 4C, lanes 4 to 6). This result further confirmed that Nt-RDR1 suppresses RDR6-dependent S-PTGS but does not block siRNA accumulation. Both G and P secondary siRNAs accumulated at higher levels in Nt-RDR1 than in wild-type plants, especially the 21-nucleotide P siRNAs (Figure 4C, lanes 4 and 5). However, the high levels of secondary G and P siRNAs did not enhance GFP silencing in NtRDR1 plants, consistent with the above idea that Nt-RDR1 interferes with RDR6-dependent S-PTGS at a level other than siRNA accumulation.
Nt-RDR1 Suppressed Synthetic tasiRNA-Mediated Silencing but Did Not Interfere with RDR6-Dependent syn-tasiRNA Synthesis
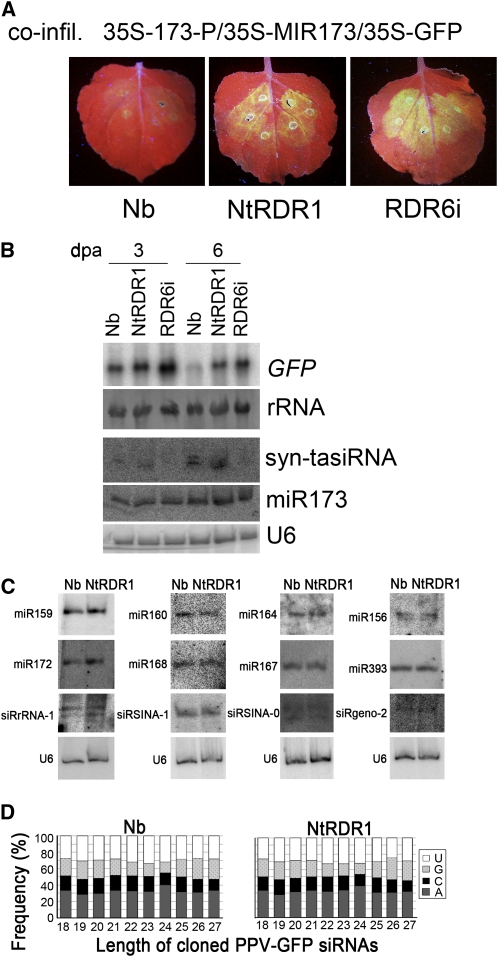
In Arabidopsis, the production of tasiRNAs, which are generated from TAS-derived transcripts after these are cleaved at a microRNA (miRNA) target site, is fully dependent on RDR6 (Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005). The technology for production of artificial (synthetic) syn-tasiRNAs in N. benthamiana mediated by miR173-guided cleavage has been recently developed (Montgomery et al., 2008; Felippes and Weigel, 2009). To examine whether Nt-RDR1 interferes with RDR6-dependent syn-tasiRNAs, we constructed 35S-173-P, in which an miR-173 cleavage site was inserted upstream of the 3′ part of the GFP sequence, P, and 35S-MIR173, which produces active miR173 (Montgomery et al., 2008). Coinfiltration of 35S-173-P/35S-MIR173/35S-GFP in wild-type, Nt-RDR1, and RDR6i plants was conducted and GFP fluorescence was monitored. In coinfiltrated wild-type plants, GFP fluorescence was low at 3 dpa (Figure 5A), and no GFP fluorescence was observed at 6 dpa. There was no great difference in GFP fluorescence between coinfiltrated Nt-RDR1 and RDR6i plants, which was clearly higher than that of wild-type plants (Figure 5A). RNA gel blot analysis confirmed the expression of miR173 in all three genotypes (Figure 5B). Similar accumulation levels of 21-nucleotide syn-tasiRNA were detected in wild-type and NtRDR1 plants at 3 and 6 dpa (Figure 5B), indicating that miR173-guided cleavage and syn-tasiRNA production occurred efficiently in Nt-RDR1 plants. However, the syn-tasiRNA–mediated GFP silencing was inhibited in Nt-RDR1 plants, consistent with the fact that the syn-tasiRNA–specific probe detected 22-nucleotide siRNAs only in the coinfiltrated wild-type plants at 6 dpa (Figure 5B). No 21- or 22-nucleotide syn-tasiRNA were detected in the coinfiltrated RDR6i plants (Figure 5B), consistent with the requirement of RDR6 for tasiRNA biogenesis. Our results indicate that Nt-RDR1 interferes with tasiRNA activity without disturbance of its synthesis.
Figure 5.
Analysis of the Effect of Nt-RDR1 on Small RNA Accumulation and syn-tasiRNA–Mediated Silencing.
(A) and (B) Induction of silencing of transiently expressed GFP in Nb, Nt-RDR1, and RDR6i plants coinfiltrated with 35S-173-P/35S-MIR173/35S-GFP. Photographs were taken under UV light at 3 dpa (A). (B) shows RNA gel blot analysis of GFP transcripts, syn-tasiRNAs, and miR173 in Nb, Nt-RDR1, and RDR6i plants at 3 and 6 dpa. 32P-labeled GFP DNA or oligodeoxynucleotide probes specific for syn-tasiRNA and miR173 were used, respectively. Methylene blue–stained rRNA and U6 RNA hybridization are shown as loading controls.
(C) RNA gel blot analysis of endogenous small RNAs in Nb and Nt-RDR1 plants. Oligodeoxynucleotide probes specific for each small RNA were used. U6 RNA hybridization is shown as a loading control.
(D) Analysis of vsiRNAs in PPV-GFP–infected Nb and Nt-RDR1 plants. The frequency of each nucleotide at the 5′ end of vsiRNAs of different lengths is indicated.
Overexpression of Nt-RDR1 in N. benthamiana Did Not Alter Accumulation of Endogenous Small RNAs
To examine whether the populations of endogenous small RNAs are altered in Nt-RDR1 plants, the accumulation levels of some conserved plant miRNAs and siRNAs with different nucleotides at the 5′ end were assessed by RNA gel blot analysis. Figure 5C shows that there were no obvious differences for any of the miRNAs/siRNAs tested, suggesting that the expression of Nt-RDR1 in N. benthamiana does not disturb the biogenesis and stability of the endogenous small RNAs. Sequence analysis of viral siRNAs (vsiRNAs) showed that the ratio of the different nucleotides at their 5′ ends are similar in wild-type and Nt-RDR1 plants infected with PPV-GFP (Figure 5D), although total amount of vsiRNAs was higher in PPV-GFP–infected NtRDR1 plants. Taken together with the fact that transiently expressed miR173 is active in syn-tasiRNA production in Nt-RDR1 plants (Figure 5B), these data argue against the possibility that hypersusceptibility to PPV-GFP infection in Nt-RDR1 plants could be due to saturation of AGO proteins by enhanced levels of endogenous small RNAs, which might result in out-competition for RISC loading of vsiRNAs or in the disturbance of the 5′ nucleotide-related selection of vsiRNAs by AGO proteins (Mi et al., 2008).
The Response of Nt-RDR1 Plants to Challenge by Other Viruses
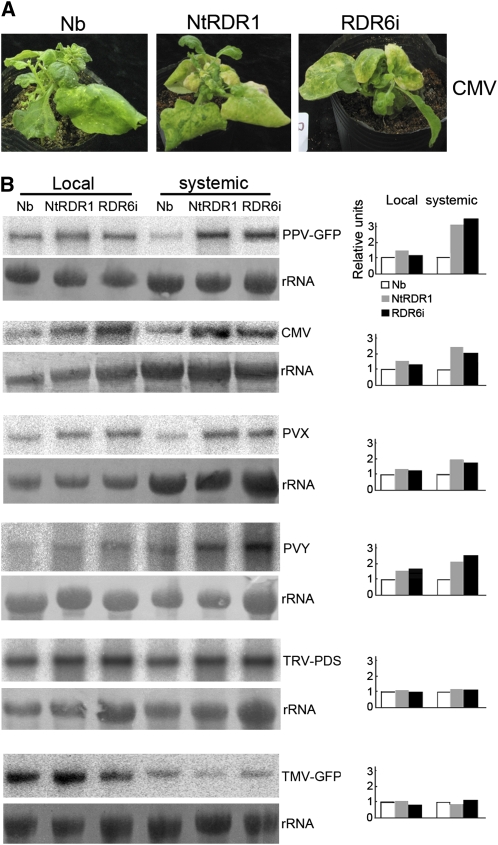
N. benthamiana expressing an RDR1 ortholog from M. truncatula (Mt-RdRP1), which has 62% identity with Nb-RDR1m and is induced by SA treatment (Yang et al., 2004), displayed enhanced resistance to several tobamoviruses (e.g., TMV, TVCV, and SHMV) but not to viruses outside of the Tobamovirus genus (e.g., CMV and PVX) (Yang et al., 2004). To test the virus susceptibility spectrum in 35S-NtRDR1 plants, we inoculated Nt-RDR1, RDR6i, and wild-type N. benthamiana plants with CMV (SD-CMV strain), PVX, PVY, TRV-PDS, and two tobamoviruses (TMV-GFP and Tomato mosaic virus [ToMV]). Symptoms of locally inoculated and systemically infected leaves were monitored, and infected leaves were collected for viral RNA analysis.
Infection with SD-CMV caused more severe systemic symptoms in the NtRDR1 and RDR6i plants than in wild-type plants, especially in newly developed systemically infected leaves (20 DAI). This virus caused leaf mosaic and curling in wild-type plants, but it caused a strong aberrant, narrow, asymmetric phenotype in both Nt-RDR1 and RDR6i plants, which rapidly withered and became yellow (Figure 6A), consistent with viral RNA levels that were about twofold higher in both Nt-RDR1 and RDR6i plants than in wild-type plants (Figure 6B). PVX-infected RDR6i and NtRDR1 plants were more stunted, and the development of new leaves was delayed relative to the infected wild-type plants (see Supplemental Figure 7 online). PVX RNA analysis showed that virus accumulated at higher levels in Nt-RDR1 and RDR6i plants (∼1.8-fold) than in wild-type plants (Figure 6B). The systemically PVY-infected leaves of RDR6i and Nt-RDR1 plants were more crimped and curled down relative to the infected wild-type plants (see Supplemental Figure 7 online). PVY RNA analysis also showed that virus accumulated at higher levels in Nt-RDR1 and RDR6i plants (about twofold) than in wild-type plants (Figure 6B). TRV-PDS, a recombinant TMV encoding PDS sequence (Liu et al., 2002), infection caused photobleaching symptoms in systemically infected leaves resulting from silencing of the endogenous PDS gene. The photobleaching phenotype appeared to similar extents in the Nt-RDR1 and RDR6i plants as in the wild-type plants (see Supplemental Figure 7 online), which correlated with similar viral RNA accumulation in plants of all three genotypes (Figure 6B). Similar to TRV-PDS infection, there were no obvious differences in systemic symptoms (see Supplemental Figure 7 online) or in viral RNA accumulation among the three genotypes infected with TMV-GFP (Figure 6B). ToMV infection caused rapid death in all three genotypes, and no viral RNA analysis was conducted. Taken together, except for the two tobamoviruses (TMV-GFP and ToMV), and TRV-PDS, which was shown not to hyperaccumulate in the absence of RDR6 (Schwach et al., 2005), Nt-RDR1 and RDR6i plants were more sensitive than wild-type N. benthamiana plants to all viruses tested in this study.
Figure 6.
Analysis of the Infection of Several Viruses in Wild-Type, Nt-RDR1, and RDR6i Plants.
(A) Symptoms of SD-CMV infection in wild-type (Nb), Nt-RDR1, and RDR6i plants. Photographs were taken at 30 DAI.
(B) RNA gel blot analysis of viral RNA accumulation in Nb, Nt-RDR1, and RDR6i plants infected with PPV-GFP, CMV, PVX, PVY, TRV-PDS, or TMV-GFP. Total RNA was extracted from inoculated and systemically infected leaves at 4 and 10 DAI. Blots were probed with 32P-labeled cDNAs specific for the corresponding virus. Methylene blue–stained rRNAs are shown as loading controls. Quantification of viral RNA relative to total RNA is shown at the right part of the panel. The value of Nb was arbitrarily designed as 1.
DISCUSSION
In this report, we have shown that N. benthamiana, a natural RDR1 mutant, expressing a tobacco RDR1 transgene under the control of either the 35S or the endogenous promoter of the mutant gene (Nb-RDR1m) exhibited hypersusceptibility to PPV and other viruses (CMV, PVX, and PVY) in systemically infected leaves (Figures 1 and 6) probably due to interference with the RDR6-mediated antiviral silencing pathway. In Nt-RDR1 plants, the expression of Nb-RDR1m, Nb-RDR2, and Nb-RDR6 and the induction by PPV infection of SA-dependent AOX-1 transcripts are comparable to those in wild-type N. benthamiana (Figures 2D and 2C), suggesting that expressing Nt-RDR1 neither causes cosuppression of Nb-RDR genes nor perturbs the SA-dependent defense pathway. The fact that enhanced PPV was evident only in Nt-RDR1 plants that accumulate high amount of transgene protein, regardless of transgene RNA accumulation levels, indicates that Nt-RDR1 protein, rather than RNA, is responsible for the hypersusceptibility phenotype.
In GFP silencing assays, transiently expressed Nt-RDR1 exhibited silencing suppression activity, although it was weak compared with the strong viral suppressor p19 (Figures 3A and 3B). Mutation of the conserved DXDGD motif did not compromise the silencing suppression activity of Nt-RDR1, suggesting that RNA polymerase activity was not required for the ability to suppress silencing of this protein (Figure 3A). The silencing suppression activity of Nt-RDR1 was also evident when 35S-GFP was infiltrated into 16c/Nt-RDR1 transgenic plants (Figures 3C and 3D). Using 16c/RDR6i plants, we confirmed that RDR6 was required for sense transgene-induced GFP 21-nucleotide siRNA accumulation and GFP silencing (Figure 3D). In 16c/Nt-RDR1 plants, in which sense silencing was suppressed, the GFP 21-nucleotide siRNAs accumulated normally. These data, together with the detection of syn-tasiRNA accumulation in Nt-RDR1 plants in which the induction of GFP silencing by miR173/syn-tasiRNA was suppressed (Figures 5A and 5B), suggest that suppression of silencing by Nt-RDR1 acts downstream of RDR6-dependent siRNA production. Both the 5′ (G)- and 3′ (P)-specific 21-nucleotide secondary siRNAs derived from GFP RNA coexpressed with a sense F RNA accumulated strongly in transgenic plants constitutively expressing Nt-RDR1 (Figure 4C), further supporting that RDR6-dependent synthesis of siRNA was not inhibited by Nt-RDR1 in N. benthamiana. The higher levels of G and P secondary siRNAs of transiently expressed GFP in Nt-RDR1 plants than in wild-type plants (Figure 4C) may result simply from the higher level of GFP transcripts in Nt-RDR1 plants due to the silencing suppression activity of Nt-RDR1. However, a contribution of Nt-RDR1 to transitive spreading of siRNA synthesis via recognition of compromised transcripts (for instance, the 5′ and 3′ cleaved G and F RNA fragments) to produce more G and F dsRNAs cannot be ruled out.
It was intriguing that high levels of secondary siRNAs (Figure 4C) fail in silencing of target transcripts. Similarly, in viral infection assays, virus-derived siRNAs and viral genomic RNAs accumulated to higher levels in Nt-RDR1 N. benthamiana than in wild-type plants (Figure 6B). Increases in both small RNAs and target transcripts in transgenic plants expressing a viral suppressor of RNA silencing (for example, HC-Pro protein encoded by potyviruses) have been reported (Llave et al., 2000; Kasschau et al., 2003). The silencing suppression activity of Nt-RDR1 expressed in N. benthamiana is likely due to its specific biochemical properties. In general, the biological specificity of RDR function is proposed to require interacting factors assembled in complexes termed as RDRCs (Motamedi et al., 2004; Duchaine et al., 2006; Aoki et al., 2007; Lee et al., 2009). For example, the C. elegans RDR family protein RRF-1 copurified with the putative helicase DRH-1, and RRF-3 was copurified as one of several proteins associated with DCR-1 (Duchaine et al., 2006; Aoki et al., 2007). Assembly of distinct RDRCs could be responsible for specific siRNA biogenesis and effector pathways. For instance, plant RDR6 recruits cleaved transcripts targeted by specific miRNAs to synthesize dsRNAs and then 21-nucleotide tasiRNAs associated with silencing amplification (Allen et al., 2005; Yoshikawa et al., 2005), while RDR2 recruits transcripts generated by plant-specific RNA polymerase IV to produce dsRNAs and then 24-nucleotide repeat-associated siRNAs associated with RNA-directed DNA methylation and histone modification (Xie et al., 2004; Huettel et al., 2006; Chinnusamy and Zhu, 2009). The similar accumulation levels of endogenous small RNAs in Nt-RDR1 and wild-type plants (Figure 5C) suggest that expressing Nt-RDR1 in N. benthamiana is not likely to cause saturation of AGO proteins by endogenous small RNAs in Nt-RDR1 plants. Increases in both small RNAs and target transcripts in Nt-RDR1 transgenic plants suggest that the suppression activity of Nt-RDR1 acts downstream of siRNA synthesis, and it may involve assembly of RISC-like complexes that disturb AGO1-containing RISC activity, which appears to be required for RDR6-dependent silencing but not IR-PTGS in Arabidopsis and N. benthamiana (Beclin et al., 2002; Jones-Rhoades et al., 2006). The idea that one RDR-related process might inhibit the function of another RDR pathway, as observed in this study, is supported by the discovery of the negative regulation of RNA silencing in C. elegans, in which the pathways for exogenous siRNA (exo-siRNA) and endogenous siRNA (endo-siRNA) use separate RDR homologs (RRF-1 and RRF-3, respectively) while sharing other factors, including DCR-1 (Duchaine et al., 2006; Lee et al., 2006). The finding that loss of RRF-3 causes C. elegans to be hyperactive in the RRF1-related exo-siRNA pathway (Simmer et al., 2002; Asikainen et al., 2007) suggests a competition between the exogenous and endogenous RNAi pathways for shared rate-limiting factors.
RDR1 in tobacco and N. benthamiana are 95 and 93% identical in nucleotide and amino acid sequence, respectively (Yang et al., 2004). We therefore speculate that if RDR1 in N. benthamiana was functional, it probably would interfere with RDR6-dependent silencing as Nt-RDR1 does, especially the RDR6-dependent antiviral silencing amplification and maintenance (persistence), since viral multiplication in cells was likely to be closer to transient agroinfiltration-mediated RNA production than to constitutive transgene expression. We therefore extrapolate that the absence of a functional RDR1 is averting a negative modulation of RDR6-dependent antiviral PTGS in N. benthamiana.
N. benthamiana expressing Mt-RDR1 displays enhanced resistance to tobamoviruses (e.g., TMV, TVCV, and SHMV) but not to other viruses (e.g., CMV and PVX) (Yang et al., 2004). Our Nt-RDR1 plants displayed no obvious differences in susceptibility to two different tobamoviruses, but they were hypersusceptible to species outside of the Tobamovirus group (e.g., PPV, CMV, PVX, and PVY) (Figure 6). The lower sequence identity between Nb-RDR1m and Mt-RDR1 (62%) than between Nb-RDR1m and Nt-RDR1 (95%) might explain the different susceptibility to viruses of Mt,RDR1 and Nt,RDR1 plants because of different specific biochemical properties of the RDRs. At-RDR1 is 66% identical at the nucleotide level to Nt-RDR1 and shows no silencing suppressor activity in N. benthamiana (see Supplemental Figure 6 online), and it has been shown to play an important role in biogenesis of TMV siRNAs (Qi et al., 2009). Therefore, sequence specificity might be responsible for the lack of suppression of antiviral silencing of Mt-RDR1 expressed in N. benthamiana. By contrast, an increase in siRNAs precursors caused by Mt-RDR1 recognizing some aberrant RNA (e.g., derived from TMV but not from CMV and PVX) might enhance anti-TMV defenses in N. benthamiana.
An alternative but not mutually exclusive possibility is that the anti-TMV defense response of Mt-RDR1 expressed in N. benthamiana probably activates the SA-related defense pathway upon virus infection. The pivotal anti-TMV pathway in plants might rely more on an SA resistance pathway, for example, the N gene–mediated signal transduction cascade induced by TMV infection that leads to induction of the hypersensitive response and restriction of virus spread in tobacco (Whitham et al., 1994; Padgett et al., 1997; Abbink et al., 1998; Erickson et al., 1999). TMV infection, SA, and biologically active SA analogs can induce Nt-RDR1 activity in tobacco (Xie et al., 2001). Moreover, TMV-infected Nt-RDR1–deficient transgenic tobacco plants accumulated higher levels of viral RNA and developed more severe symptoms compared with wild-type tobacco (Xie et al., 2001). However, SA treatment inhibited TMV RNA accumulation equally well in wild-type tobacco and RDR1-deficient plants (Xie et al., 2001), suggesting that SA has both RDR1-dependent and RDR1-independent anti-TMV effects in tobacco. Nevertheless, all these results suggest that Nt-RDR1 plays an important role in anti-TMV defenses likely associated with SA-mediated defense pathways in tobacco. However, unlike Mt-RDR1, the ability of Nt-RDR1 to suppress the RDR6-mediated antiviral silencing appears to compensate for its SA-related anti-TMV activity in N. benthamiana, since TMV infection appears not to be restricted in Nt-RDR1 transgenic N. benthamiana plants (Figure 6).
The fact that downregulation of Nt-RDR1 in transgenic tobacco enhanced susceptibility to PVX and PVY (Xie et al., 2001; Rakhshandehroo et al., 2009) contrasts with our result that PVX and PVY accumulation and symptoms were enhanced in N. benthamiana expressing Nt-RDR1 compared with wild-type N. benthamiana (Figure 6). These results suggest that PVX and PVY are restricted in tobacco by an efficient RDR1- and SA-mediated defense pathway, but this antiviral mechanism was not able to compensate for the suppression of the RDR6-mediated antiviral silencing in the Nt-RDR1–expressing N. benthamiana plants. Similarly, both SA-mediated defenses and the RNA silencing pathway could act together to limit PPV infection in tobacco, although the possible role of Nt-RDR1 in anti-PPV protection has not been assessed (Alamillo et al., 2006). The fact that PPV infection was not enhanced in SA-deficient NahG transgenic N. benthamiana plants (Figure 2) suggests that the SA-mediated anti-PPV pathway in N. benthamiana might not be as important as it is in tobacco. This appears not to be exclusively the consequence of the lack of a functional RDR1 gene because the transgenic N. benthamiana plants expressing Nt-RDR1 were comparable to the wild type in SA-dependent gene (AOX-1) induction (Figure 2C) and hypersusceptible to PPV infection, similar to the RDR6i N. benthamiana plants. Thus, RDR6-mediated antiviral silencing, which could be counteracted by RDR1 silencing suppression activity, might be the main anti-PPV defense in N. benthamiana. Our results, together with previously published data, suggest that RDR1 might have a dual role, contributing, on one hand, to SA-mediated antiviral defense, and suppressing, on the other hand, the RDR6-mediated antiviral RNA silencing.
It has been reported that TRV vsiRNAs in single loss-of-function rdr mutants were not substantially different from those in wild-type Arabidopsis and that their amounts were only slightly diminished in rdr1 rdr2 and rdr2 rdr6 mutants; however, they were strongly reduced in the rdr1 rdr2 rdr6 triple mutant compared with wild-type Arabidopsis (Donaire et al., 2008), suggesting that At-RDR1, 2 and 6 redundantly act on TRV siRNA biogenesis. Recently, using the same rdr mutants, Wang et al. (2010) found some target specificity for RDR1 and RDR6 in amplification of CMV-derived siRNAs. We cannot rule out a role of Nt-RDR1 in vsiRNA amplification in N. benthamiana in accordance with the current thought that host RDRs act on viral RNA substrates to trigger VIGS; however, our data suggest that another level of inter-RDR regulation exists that appears to occur downstream of siRNA biogenesis.
It has been suggested that the natural loss-of-function variant of RDR1 in N. benthamiana would be the cause of the extreme susceptibility of this species to virus infection (Yang et al., 2004). By contrast, our results that expression of a functional RDR1 enhances the susceptibility of N. benthamiana to a number of viruses from different families suggest that the natural dysfunctional variant of RDR1 in N. benthamiana might be the outcome of coevolution during a long-term host-virus arms race and strong selection pressure for a fully active RDR6-mediated antiviral system, as a consequence of the hypersensitivity of N. benthamiana to many viruses.
METHODS
Plant Material and Growth Conditions
The 16c GFP-transgenic Nicotiana benthamiana, RDR6i, and 16c/RDR6i N. benthamiana were described previously (Brigneti et al., 1998; Schwach et al., 2005). All transgenic plants and wild-type N. benthamiana were grown in glass house at 25°C and 16-h-light/8-h-dark cycles.
Plasmid Constructs
For the 35S-myc-NtRDR1 construction, according to the sequence of Nt-RDR1, three DNA fragments were amplified with three pair of primers, Nt-RDR1-F 5′/Nt-RDR1-F 3′, Nt-RDR1-M 5′/Nt-RDR1-M 3′ and Nt-RDR1-B 5′/Nt-RDR1-B 3′, and inserted into pGEM-T vector (Promega), giving pGEM-NtRDR1(F), pGEM-NtRDR1(M), and pGEM-NtRDR1(B). The EcoRV-SacI fragment of pGEM-NtRDR1(B) was inserted to EcoRV-SacI–digested pGEM-NtRDR1(M) to give pGEM-NtRDR1(M-B). The SacI-ScaI fragment of pGEM-NtRDR1(M-B) was then inserted into SacI-ScaI–digested pGEM-NtRDR1(F) to get pGEM-NtRDR1, a full-length clone of Nt-RDR1. The NcoI-SacI fragment of pGEM-NtRDR1 was inserted into pBA002 (Kost et al., 1998) digested with SmaI and SacI to obtain a myc-tagged Nt-RDR1 sequence. Myc-NtRDR1 was then inserted into pCAMBIA1300-221 digested with XbaI and SacI, giving the final clone pCAMBIA-35S-myc-NtRDR1. pCAMBIA1300-221 was constructed by inserting a fragment of HindIII-EcoRI from pBI221 (Clontech) into HindIII-EcoRI–digested pCAMBIA1300.
For Pro-myc-NtRDR1 construction, a fragment upstream of the ATG in Nb-RDR1m (2144 bp) was obtained by PCR amplification with primers Pro-Nb-RDR1m 5′/Pro-Nb-RDR1m 3′ to substitute for the 35S promoter in pCAMBIA1-35S-myc-NtRDR1 digested with HindIII and SmaI, giving pCAMBIA1-Pro-myc-NtRDR1.
pCAMBIA-F (35S-F) and pCAMBIA-Fi (35S-Fi) were constructed by PCR amplification of the middle part of GFP with primers gFp 5′ and gFp 3′ and cloned into pCAMBIA1300-221 digested with XbaI and SacI to obtain pCAMBIA-F. The F PCR fragment was cloned into the RNAi intermediate vector pSK-int (Guo et al., 2003) digested with ApaI and HindIII, giving pSK-F-int, then the F PCR fragment was cloned into XbaI-SacI–digested pSK-F-int to give pSK-Fi. The NcoI-SacI fragment containing the F hairpin sequence was inserted into pCAMBIA1300-221 digested with SmaI and SacI, giving the RNAi construct 35S-Fi.
pCAMBIA-MIR173 (35S-MIR173) and pCAMBIA-173-P (35S-173-P) were constructed by PCR amplification of the Arabidopsis MIR173 precursor sequence with primers MIR173 5′ and MIR173 3′, and of the 3′ part of the GFP sequence, P, of pSK-mGFP with primers 173-P 5′ and 173-P 3′ . The PCR fragments were cloned inter pGEM-Teasy vector getting pGEM-MIR173 and pGEM-173-P, respectively. The XbaI-SacI fragments containing MIR173 or 173-P were inserted into pCAMBIA1300-221 digested with XbaI and SacI to obtain pCAMBIA-MIR173 and pCAMBIA-173-P, respectively.
35S-NtRDR1m was constructed by amplification of DNA fragments from pCAMBIA-1300-myc-NtRDR1 with primers Nt-RDR1m-F 5′/Nt-RDR1m F 3′ and Nt-RDR1m-B 5′/Nt-RDR1m-B 3′. The PCR fragments were then annealed and the recovered product was used as template in next PCR amplification, using primers Nt-RDR1m-F 5′/Nt-RDR1m-B 3′ to get a fragment containing the mutant sequence, resulting in changing the conserved domain DLDGD to VMVEV, and was cloned into pGEM-Teasy vector, getting pGEM-NtRDR1m and confirmed by sequencing. The NsiI-NcoI of pGEM-NtRDR1m was inserted into pCAMBIA-1300-myc-NtRDR1 digested with NsiI and NcoI to get the final construct 35S-NtRDR1m.
35S-NtΔRDR1 was constructed by amplification of PCD fragment from pCAMBIA-1300-myc-NtRDR1 with primers Nt-RDR1-ΔATP 5′ and Nt-RDR1-ΔATP 3′, and the fragment was cloned into pGEM-Teasy and confirmed by sequencing. The fragment of XbaI-NsiI was used to replace the fragment of pCAMBIA-1300-myc-NtRDR1 digested with XbaI and NsiI, getting the final construct 35S-ΔRDR1.
35S-p19 construct was previously described (Liu et al., 2009).
NahG was amplified from transgenic Arabidopsis thaliana (van Wees and Glazebrook, 2003) with primers NahG 5′/NahG 3′, then the PCR fragment was inserted into pGEM-Teasy vector, making pGEM-Teasy-NahG. A 1.3-kb XbaI-SacI fragment from pGEM-Teasy-NahG was inserted into XbaI and SacI digested pCAMBIA1300-221, making pCMBIA1300-NahG.
At-RDR1 was amplified by reverse transcript PCR using SuperScript III reverse transcriptase following the manufacturer's protocol (Invitrogen) and LA Taq (Takara), with primers At-RDR1 5′/At-RDR1 3′. The PCR product was inserted into pGEM-Teasy vector. The BanHI/klenow-XbaI fragment from pGEM-Teasy-AtRDR1 was then inserted into pCAMBIA1300-221 digested with SacI/T4 and XbaI.
All primer sequences are listed in Supplemental Table 1 online.
Plant Transformation
After 2 d of culture on coculture medium (Murashige and Skoog medium containing 1.2 mg/L 6-benzylaminopurine and 0.1 mg/L a-naphthalene acetic acid [NAA]), sterilized leaf disks were cocultured with Agrobacterium tumefaciens carrying different constructs and were transferred to coculture medium for 2 d in the dark. Potential transformants were selected with 40 mg/L hygromycin, 1 mg/L 6-benzylaminopurine, and 0.1 g/L NAA. Regenerated shoots were rooted on Murashige and Skoog medium, adding 0.1 g/L NAA and 40 mg/L hygromycin. Rooted plants were transferred to soil that was kept wet and were grown with a 16-h-light/8-h-dark cycle.
Virus Inoculation and Transient Expression
Plants for virus inoculation were grown in a glasshouse with 16-h light and 8-h dark at 25°C. For sap inoculation of PPV-GFP (Alamillo et al., 2006), CMV (Du et al., 2007), PVX, and PVY (Schwach et al., 2005), 1 g of virus-infected leaves was ground in 1 mL of 5mM phosphate buffer, pH 7.2, in mortar prechilled on ice. Plants were inoculated with sap prepared freshly by rubbing mechanically. For PPV or PPV-GFP Agrobacterium infiltration, N. benthamiana plants were inoculated by infiltration with culture of AgrobacteriumEHA105 carrying PPV or PPV-GFP at OD 0.5. For TRV-PDS inoculation, a mixture of AgrobacteriumGV2260 carrying pTRV1 and pTRV-PDS at OD 0.8 at a ratio of 1:1 was used. Agrobacterium GV2260 carrying TMV-GFP at OD 0.5 (Liu et al., 2002) was used for inoculation. For transient expression, AgrobacteriumEHA105 carrying the indicated DNA construct was cultured and adjusted to OD 0.8 for infiltration. For coinfiltration of 35S-GFP and 35S-F, cultures of Agrobacteriumcarrying 35S-GFP and Agrobacteriumcarrying 35S-F were adjusted to OD 0.8 and 0.4, respectively, and equal volume mixed was used. For coinfiltratation of 35S-GFP with 35S-Fi, the ODs of Agrobacteriumcultures were 0.8 and 0.1, respectively, and equal volume mixed was used. For coinfiltration of 35S-GFP with 35S-NtRDR1 or 35S-p19, the ODs of Agrobacteriumcultures were 0.8 and 1.2, respectively, and equal volume mixed was used. For coinfiltration of 35S-173-P/35S-MIR173/35S-GFP, the ODs of Agrobacterium cultures were 0.4, 1.6, and 0.8, respectively, and equal volume mixed was used.
SA Treatment
Pro-Nt-RDR1 plants were sprayed with a 2 mM SA solution at the eight-leaf stage (Yang et al., 2004), and water treatment was used as the control.
GFP Imaging
GFP fluorescence in plants was photographed under UV light using a Kodak Easy Share DX7440 digital camera and a B-100AP longwave-UV lamp (Ultra-Violet Products).
Nucleic Acid Extraction and RNA Gel Blotting
For preparation of viral genomic RNA, total RNA was isolated from inoculated plants by a hot-phenol method (Fernandez et al., 1997). Other RNA was extracted from plants by Trizol. For hybridization, total RNA was separated on 1.2% agarose gels containing 6% formaldehyde and transferred to a nylon N+ membrane and UV cross-linked. The membrane was then stained with methylene blue solution (0.5 n NaAc, pH 5.2, and 0.04% methylene blue) for several minutes and rinsed with distilled water until the bands clearly visible. For detection of viral RNA and GFP mRNA, DNA fragments were amplified by PCR with respective primers (see Supplemental Table 1 online) and labeled with 32P using the Rediprime II System (Amersham). Hybridization was performed in Church-gilbert buffer at 65°C overnight. For Nt-RDR1 and Nb-RDR6 detection, DNA fragments corresponding to 1211 to 2539 of Nt-RDR1 and 114 to 1975 of Nb-RDR6 were inserted into the pGEM-Teasy vector (Promega) for transcription of RNA probes using the Maxiscript kit (Ambion). Hybridization was performed in buffer containing 50% formamide, 6× SSPE, 5× Denhardt's, and 1% SDS at 65°C overnight. Signal intensity was quantified using ImageQuant TL software (GE). For detection of siRNAs, 30 μg total RNA was separated on 15% polyacrylamide-8 M urea gels. After electrophoresis, the RNA was electroblotted onto Hybond N+ membranes with a semidry transfer cell with 1× TBE and fixed by UV cross-linking. DNA fragments corresponding to three regions of GFP (referred to in the text as G, F, and P) were cloned into the pGEM-Teasy vector for transcription of RNA probes using the MAXIscript kit (Ambion). [γ-32p]ATP labeled oligodeoxynucleotide probes specific for miR173, syn-tasiRNA, U6 control, and endogenous small RNAs (miR-159, miR160, miR164, miR156g, miR172e, miR168, miR167, miR393, siRrRNA-1, siRSINA-1, siRSINA-0, and siRgeno-2). All probe sequences are listed in Supplemental Table 1 online.
DNA Extraction and Blotting
Genomic DNA was extracted from the fully expand leaves of eight-leaf stage plants via the CTAB method (Stewart and Via, 1993). Thirty micrograms of DNA was digested in 300 μL of reaction buffer-K (Takara), 2.5 mM spermidine, and 200 units of HindIII at 37°C for 6 h. After precipitation, the digested DNA was separated on a 0.9% agarose gel and transferred onto Hybond-N+ membrane (Amersham) by capillary equipment with 20× SSC (3 n NaCl and 0.3 n trisodium citrate). After prehybridization, hybridization was performed in Church-Gilbert buffer (7% [w/v] SDS, 0.5 M phosphate buffer, pH 7.2, and 10 mM EDTA) (Church and Gilbert, 1984) at 65°C overnight with [α-32P]-labeled (Perkin-Elmer) DNA probe using the Rediprime II System (Amersham). The DNA probe was a 568-bp SacI-HindIII fragment from pGEM-Teasy-NtRDR1. The membrane was washed with 2× SSC and 0.1% SDS twice at 65°C, each time 20 min, then washed with 1× SSC and 0.1% SDS for 20 min.
Protein Extraction and Immunoblotting
Plant leaves were ground to fine powder in liquid nitrogen for protein extraction with buffer EB (50 mM sodium phosphate, pH 7.0, 200 mM NaCl, 10 mM MgCl, 10% glycerol, adding one minicocktail tablets/10 mL solution) at a ratio of 100 mg powder/100 μL EB and centrifuged at 13,000g for 30 min at 4°C. The supernatant was transferred into a new tube. Proteins were separated in a 10% polyacrylamide gel and transferred onto polyvinylidene fluoride membrane with a semidry transfer cell (Bio-Rad). The membrane was blocked with 5% BSA in TBS-T at 4°C overnight and then incubated with the primary antibody c-Myc (9E10) (Santa Cruz Biotechnology) for 3 h at room temperature and washed with TBS-T three times, each time for 5 min. After 0.5 h incubation with the AP-labeled secondary antibody (Beijing Dingguo), the membrane was washed thoroughly, and signals were detected with BCIP/NBT (Calbiochem). For Coomassie Brilliant Blue staining, the protein gel was immersed in Coomassie Brilliant Blue R 250 solution (0.1% w/v Coomassie Brilliant Blue R 250, 25% isopropyl alcohol, and 10% acetic acid) overnight with gentle shaking. The stained gel was then treated with solution (10% acetic acid and 5% ethanol) twice, each time for 30 min with gentle shaking.
vsiRNA Cloning and Analysis
Small RNA were extracted from PPV-GFP–infected Nb- and Nt-RDR1 at 20 DAI. Small RNA fractions were purified and cloned as described previously (Du et al., 2007). Analysis of the frequency of each nucleotide was performed using Microsoft Office Excel 2007.
Real-Time RT-PCR
RDR1m, RDR2, and RDR6 transcript levels in N. benthamiana and Pro-NtRDR1 plants were analyzed by quantitative real-time RT-PCR with a PTC-200 Thermal Cycler (MJ Research/Bio-Rad). Total RNA was extracted from pools of four plants at the six-leaf stage for each plant line using RNzol regent (Tiangen) and treated with DNaseI (Takara) and exacted with phenol-chloroform. Synthesis of cDNA from 4 μg of treated total RNA was performed with random hexanucleotides (Takara) and Primescript reverse transcriptase (Takara). Reaction without template was included as controls. For the quantitative real-time PCR, cDNA corresponding to 200 ng of total RNA was used in 20-μL reactions using SYBR green real-time PCR master mix (Toyobo) according to the manufacturer's instructions. Primers for RDR2, RDR6, and internal standard GAPDH were as described (Schwach et al., 2005). Primers realtime-Nb-RDR1m 5′/realtime-Nb-RDRm 3′ (see Supplemental Table 1 online) were used for Nb-RDR1m amplification (amplicon length of 221 bp).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Nt-RDR1 (AJ011576), Nb-RDR1m (AY574374), At-RDR1 (AY148431), Nb-RDR2 (AY722009), Nb-RDR6 (AY722008), MIR173 (NR_022764), NahG (X83926), pBI221 (AF502128), and pCAMBIA1300 (AF234296).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Detection of Copy Numbers of the Transgene 35S-Myc-NtRDR1 in Eight Independent Primary Transformants.
Supplemental Figure 2. Similar Symptom Phenotype in PPV and GFP-Tagged PPV-Infected Wild-Type N. benthamiana.
Supplemental Figure 3. No Induction of myc-NtRDR1 with PPV Infection in 35S-NtRDR1 Plants.
Supplemental Figure 4. Verification of NahG Transgenic N. benthamiana
Supplemental Figure 5. RNA Gel Blot Analysis of GFP mRNA Accumulation in 35S-GFP/35S-F Coinfiltration in Nb, Nt-RDR1, or RDR6i Plants.
Supplemental Figure 6. Analysis of the Effect of At-RDR1 on Transient Induction of Silencing in GFP-Transgenic 16c N. benthamiana Plants.
Supplemental Figure 7. Symptoms of SD-CMV, PVX, PVY, TRV-PDS, and TMV-GFP Infection in the Wild Type (Nb), Nt-RDR1, and RDR6i.
Supplemental Table 1. Primer and Probe Sequences Used in This Study.
Acknowledgments
We thank David Baulcombe for RDR6i and 16c/RDR6i seeds and Yule Liu for TRV-PDS, TMV-GFP, and ToMV viruses. This research was supported by grants from the National Science Foundation of China (Grants 90919010, 30525004, and 30700049) and the Ministry of Science and Technology (Grant 2006CB101601 and 2006CB101900).
References
- Abbink E.M., Tjernberg P.A., Bol J.F., Linthors J.M. (1998). Tobacco mosaic virus helicase domain induces necrosis in N gene-carrying tobacco in the absence of virus replication. Mol. Plant Microbe Interact. 11: 1242–1246 [Google Scholar]
- Alamillo J.M., Saenz P., Garcia J.A. (2006). Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 48: 217–227 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H. (2007). In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26: 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S., Storvik M., Lakso M., Wong G. (2007). Whole genome microarray analysis of C. elegans rrf-3 and eri-1 mutants. FEBS Lett. 581: 5050–5054 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2005). RNA silencing. Trends Biochem. Sci. 30: 290–293 [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (2007). Molecular biology. Amplified silencing. Science 315: 199–200 [DOI] [PubMed] [Google Scholar]
- Beclin C., Boutet S., Waterhouse P., Vaucheret H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W.X., Ji L.H., Ding S.W., Baulcombe D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chinnusamy V., Zhu J.K. (2009). RNA-directed DNA methylation and demethylation in plants. Sci. China C Life Sci. 52: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dawson W.O., Hilf M.E. (1992). Host range determination of plant viruses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 527–555 [Google Scholar]
- Diaz-Pendon J.A., Li F., Li W.X., Ding S.W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L., Barajas D., Martinez-Garcia B., Martinez-Priego L., Pagan I., Llave C. (2008). Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82: 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q.S., Duan C.G., Zhang Z.H., Fang Y.Y., Fang R.X., Xie Q., Guo H.S. (2007). DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 81: 9142–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Yates J.R., III, Mello C.C. (2006). Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Erickson F.L., Holzberg S., Calderon-Urrea A., Handley V., Axtell M., Corr C., Baker B. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18: 67–75 [DOI] [PubMed] [Google Scholar]
- Felippes F.F., Weigel D. (2009). Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 10: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Guo H.S., Saenz P., Simon-Buela L., Gomez de Cedron M., Garcia J.A. (1997). The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25: 4474–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T., Friedrich L., Vernooij B., Negrotto D., Nye G., Uknes S., Ward E., Kessmann H., Ryals J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Guo H.S., Fei J.F., Xie Q., Chua N.H. (2003). A chemical-regulated inducible RNAi system in plants. Plant J. 34: 383–392 [DOI] [PubMed] [Google Scholar]
- Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O. (2003). Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J., Matzke M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E.V., Aravind L. (2003). Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Klahre U., Crete P., Leuenberger S.A., Iglesias V.A., Meins F., Jr (2002). High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA 99: 11981–11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Lee R.C., Hammell C.M., Ambros V. (2006). Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.R., Talsky K.B., Collins K. (2009). A single RNA-dependent RNA polymerase assembles with mutually exclusive nucleotidyl transferase subunits to direct different pathways of small RNA biogenesis. RNA 15: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2009). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. in press; [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau K.D., Carrington J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97: 13401–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. (2004). Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Mourrain P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Padgett H.S., Watanabe Y., Beachy R.N. (1997). Identification of the TMV replicase sequence that activates the N gene-mediated hypersensitive response. Mol. Plant Microbe Interact. 11: 709–715 [Google Scholar]
- Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bao F.S., Xie Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One 4: e4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo F., Takeshita M., Squires J., Palukaitis P. (2009). The influence of RNA-dependent RNA polymerase 1 on potato virus Y infection and on other antiviral response genes. Mol. Plant Microbe Interact. 22: 1312–1318 [DOI] [PubMed] [Google Scholar]
- Schwach F., Vaistij F.E., Jones L., Baulcombe D.C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S.P., Nonet M.L., Fire A., Ahringer J., Plasterk R.H. (2002). Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Stewart C.N., Jr., Via L.E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14: 748–750 [PubMed] [Google Scholar]
- Vaistij F.E., Jones L., Baulcombe D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V., Vaucheret H. (2001). RNA silencing in plants–Defense and counterdefense. Science 292: 2277–2280 [DOI] [PubMed] [Google Scholar]
- van Dijk A.A., Huismans H. (1987). The identification of factors capable of reversing the core-mediated inhibition of the bluetongue virus transcriptase. Onderstepoort J. Vet. Res. 54: 629–633 [PubMed] [Google Scholar]
- van Dijk P., van der Meer F.A., Piron P.G.M. (1987). Accessions of Australian Nicotiana species as indicator hosts in the diagnosis of plant virus diseases. Neth. J. Plant Pathol. 93: 73–85 [Google Scholar]
- van Wees S.C., Glazebrook J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33: 733–742 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 6: 206–220 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer C., Baulcombe D.C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Wang X.B., Wu Q., Ito T., Cillo F., Li W.X., Chen X., Yu J.L., Ding S.W. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S.P., Choi D., Hehl R., Corr C., Baker B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Xie Z., Fan B., Chen C., Chen Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 98: 6516–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.J., Carter S.A., Cole A.B., Cheng N.H., Nelson R.S. (2004). A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 101: 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Fan B., MacFarlane S.A., Chen Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16: 206–216 [DOI] [PubMed] [Google Scholar]