Abstract
A leaf develops from a few cells that grow, divide, and differentiate to form a complex organ that is precisely positioned relative to its neighbors. How cells communicate to achieve such coordinated growth and development is the focus of this review. We discuss (1) how the stem cells within the shoot meristem gain competence to form organs, (2) what determines the positioning and initiation of new organs, and (3) how the new organ attains its characteristic shape and polarity. Special emphasis is given to the recent integration of mathematics and physics in the study of leaf development.
FROM FUNCTION TO FORM
During the evolution of multicellularity, plants adopted a division of labor whereby some organs produce energy and others consume it. The leaf is essentially a solar panel that uses photosynthetic cells to convert carbon dioxide and water into sugars and oxygen and efficiently supplies these products to heterotrophic cells. What is the optimal form of a leaf? How would an engineer design a functional leaf? To maximize light capture, we arrange our photosynthetic cells in a flat, thin structure comparable to a solar panel. Such a design also puts light capture in close proximity to the substrates, water and CO2. A system of pipes is needed to transport water in and the sugars out, and the same pipes could be used to give the structure mechanical support. CO2 could enter through pores, which are preferentially on the side away from the sun. Finally, when we combine these light-harvesting structures into a superstructure, we need to avoid self-shading and maintain mechanical stability.
We have now defined a basic light-capturing/energy-converting structure from a design perspective. A flat, thin, yet flexible structure (the leaf blade) that is supported by a robust internal network of pipes (the veins), whose upper surface is specialized for light capture and whose lower surface facilitates gas exchange (through stomata). These structures are then organized into a higher-order structure (the shoot). The above analogy is useful when thinking about form and function, yet it misses a major point: a leaf is not built like a bridge, a building, or a solar panel. Instead, it develops from a few cells that grow, divide, and differentiate to form a complex organ that is precisely positioned relative to its neighbors. Unlike the building of a bridge, organ development requires continuous communication between cells in space and time.
Cell fate can depend on a cell's lineage as well as its position within a tissue. In plants, position plays a major role, which implies communication between cells, and such communication requires signaling compounds that can move from cell to cell. The mobile signaling mechanisms that carry out cell-to-cell communication during early leaf development are the focus of this review. These mechanisms include receptor-ligand signaling, hormone dynamics, small RNA gradients, and the potential of mechanical forces. We will also discuss the importance of formal mathematical and computational modeling as tools to deal with the ever increasing complexity of the experimental data (previously reviewed in Heisler and Jönsson, 2007; Lewis, 2008; Jönsson and Krupinski, 2010). The review is divided into three parts: (1) how the leaf founder cells in the shoot apical meristem are maintained and how they lose their stem cell character and gain competence for organ formation, (2) what determines the initiation and positioning of new organs, and (3) how the new organ attains its characteristic three-dimensional shape and polarity. These three topics are essential steps in the early growth and development of leaves and illustrate the many ways that mobile signals coordinate development.
FROM STEM CELLS TO DAUGHTER CELLS: THE SHOOT MERISTEM
The growth and development of aerial organs originates at the shoot apical meristem, which is situated at the tip of the stem (Figure 1). Its key functions are to maintain itself as a source of cells and to generate cells that are competent to differentiate into stems, leaves, and flowers. The organization of the meristem is established during embryogenesis and is maintained throughout the plant's life cycle. The balance between the stem cell population and the differentiating daughter cells is essential to the iterative production of new leaves.
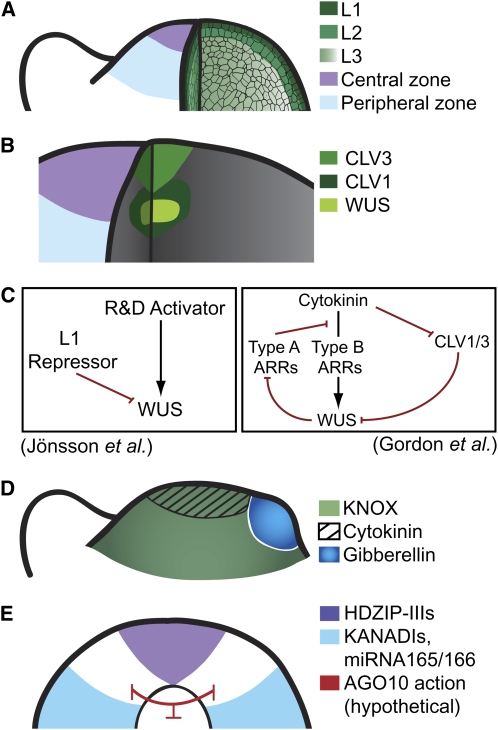
Figure 1.
The Shoot Apical Meristem: A Source for New Leaves.
(A) The organization of the meristem in terms of functional zones and cell layers.
(B) Expression domains of the basic WUS/CLV regulatory loop components.
(C) The role of cytokinin signaling and putative R and D in the spatial regulation of WUS, as described by Gordon et al. (2009) and Jönsson et al. (2005). Red lines illustrate negative interactions, while black arrows indicate a positive one. ARRs are authentic response regulators.
(D) KNOX expression within the meristem and areas of high cytokinin and gibberellin (relative to each other).
(E) The interaction between HD-ZIPIIIs and KANs with respect to meristem maintenance. HD-ZIPIII expression demarcates the top of the peripheral zone and KAN expression the lower boundary. Included is a diagrammatic representation of hypothetical ways for ZWILLE/PINHEAD/AGO10 (red lines) to affect this boundary.
In this section, we will examine how the meristem is organized and how it serves as a template for organ formation. We will discuss (1) the relative positioning and establishment of the stem cells and organizing center, (2) the maintenance and subsequent loss of meristematic potential, and (3) the potential role of small RNAs in meristem maintenance.
The Organization of the Shoot Apical Meristem
The meristem can be divided into functional zones based upon developmental potentials, molecular markers, and rates of cell division (Szymkowiak and Sussex, 1996; Kwiatkowska, 2008). The central zone comprises the stem cells and the organizing center and contains cells that remain undifferentiated. The peripheral zone surrounds the central zone, and its cells are competent to form organs, even though not all of its cells share that eventual fate. The rib zone lies below the central zone and gives rise to the central tissues of the stem. Although cell cycle times of individual cells within the meristem vary widely, it is generally accepted that the cells in the central zone divide more slowly than those in the peripheral zone (Kwiatkowska and Dumais, 2003; Reddy et al., 2004; Traas and Bohn-Courseau, 2005).
The meristem can also be divided into independent cell layers that are characterized by the orientation of cell division and molecular markers (Figures 1A and 1B). Within the central zone of the meristem, the outermost layers (L1 and L2 in dicots) divide anticlinally (perpendicular to the surface) and therefore remain separate from each other and the underlying cell layer (L3). The L3 layer undergoes both peri- and anticlinal divisions (as such it is not really a layer). The L1 gives rise to the plant's epidermis. The strict anticlinal orientation of cell divisions in the L1 is maintained during leaf development and results in a leaf epidermis that has increased as much as a millionfold in surface area but essentially remains a single cell layer in depth. Once the leaf has been initiated, both the L2 and L3 contribute to the organ body, but the proportions of their individual contributions are quite variable.
Stem Cell Identity: Receptor-Ligand Signaling
Stem cells are present in the center of the L1, L2, and L3 layers. Clonal analysis has demonstrated that there are only one to three cells per layer that act as true stem cells (Stewart and Dermen, 1970). These stem cells are centered within a group of cells marked by the expression of CLAVATA3 (CLV3) and other genes (Fletcher et al., 1999; Yadav et al., 2009). This region is subtended by the organizing center, which is marked by expression of WUSCHEL (WUS). The maintenance of the stem cell population is regulated by WUS/CLV signaling (Figure 1B). Mutants of WUS rapidly consume their stem cells, whereas in clv mutants, the stem cells overproliferate; the wus clv double mutants have a wus phenotype (Laux et al., 1996; Clark et al., 1997; Kayes and Clark, 1998; Fletcher et al., 1999; Jeong et al., 1999). In addition, CLV3 represses WUS (Schoof et al., 2000). These phenotypes can be explained by a regulatory loop in which WUS promotes stem cell fate, whereas CLV represses WUS (Otsuga et al., 2001; Dinneny and Benfey, 2008; Gray et al., 2008; Bleckman and Simon, 2009). The WUS/CLV loop serves as a mechanism to maintain a relatively constant number of cells in each of the domains of the shoot apical meristem and thus allows an indeterminate number of organs.
There are likely two mobile signals in this system: CLV3 and a hypothetical WUS-triggered signal. CLV3 is active as a 13–amino acid arabinosylated peptide that diffuses until it binds to its receptors: the LRR receptor kinase CLV1, the receptor-like kinase CLV2, and the Ser-Thr kinase CORYNE (Rojo et al., 2002; Lenhard and Laux, 2003; Kondo et al., 2006; Muller et al., 2008; Ohyama et al., 2009). These three proteins form two types of CLV3 receptor complexes: one complex comprises a CLV1 homodimer, and the other comprises a tetramer consisting of two CLV2 and two CORYNE proteins (Muller et al., 2008; Bleckmann et al., 2010). The interaction between CLV3 and either receptor complex then signals the inhibition of WUS expression. As to the second mobile signal, it is hypothesized that WUS causes a non-cell-autonomous signal to move to the stem cells and trigger the expression of CLV3, setting up feedback regulation between WUS and CLV3, which serves to delineate the relative position of the stem cells and organizing center (Schoof et al., 2000; Brand et al., 2002). As a point of interest, Suzaki et al. (2006) demonstrated that CLV function in rice (Oryza sativa) has been apportioned; one CLV3- like protein, FLORAL ORGAN NUMBER2 (FON2), is responsible for CLV function within reproductive meristems, while another CLV3-like protein, FON2-LIKE CLE PROTEIN1, provides this function within vegetative meristems. This demonstrates the conservation of the WUS/CLV mechanism and also shows that specialization of the process that has occurred, reminding us that a single species may provide invaluable information about development but may also shortchange the potential diversity of the mechanism.
Two questions dealing with the self-organization and reorganization of the WUS/CLV domains are hard to answer through conceptual models of the genetic circuitry. (1) How does it maintain its position in the center of the meristem? And (2) since perturbation of the stem cells can be overcome by respecification of neighboring cells, how can we explain its regeneration from those more differentiated neighbors? Traditional conceptual genetic models are inadequate to describe the increasingly complex genetic circuitry, especially when feedback loops are involved. A useful way to describe such systems is through mathematical and computational models; such models can integrate experimental data, discover missing parameters and generate hypotheses. This can then inspire new experiments and further refine hypotheses. Quantitative descriptions of regulatory networks have benefited the study of plant development in many ways, several of which will be discussed in this review.
A two-dimensional model of the shoot apex by Jönsson et al. (2005) recapitulates the position of the WUS domain simply using a spatially uniform activator and a slowly diffusing L1-derived repressor (Figure 1C). The model depicts a transverse section through the meristem at the level of the organizing center. In theory, the extension of the model into the longitudinal dimension requires only the addition of a repressive signal from the stem or rib zone (Jönsson et al., 2005). This is an elegant demonstration of how a simple computational model can recapitulate a central feature of meristem organization. The model raises several new questions for biologists: What is the identity of the L1-derived repressor? What is the repressive signal originating from the rib zone? Recent work by Tucker et al. (2008) suggests that the rib zone expressed gene ZWILLE/PINHEAD/AGO10 plays a role in stem cell maintenance, possibly via its role in small RNA processing. This will be discussed further in a later section.
Experimental data show that the meristem is able to recover from injury by respecification of adjacent cells into meristem cells (Pilkington, 1929; Sussex, 1964; Reinhardt et al., 2003; de Reuille et al., 2006; Reddy et al., 2007). The simple activator model described above is not able to predict WUS reestablishment after simulated destruction. This leads to the second question: How can we explain the regeneration of the organizing center? Jönsson et al. (2005) address this by replacing the uniform activator by a Reaction-Diffusion (R and D) activator/inhibitor system. R and D models (Meinhardt, 1982) provide a theoretical basis for de novo pattern generation in biological systems; the simplest case consists of a slowly diffusing activator and a more rapidly diffusing inhibitor. The activator is autocatalytic and also activates the inhibitor; the inhibitor in turn inhibits the activator. This system can yield stable patterns of the activator in space and/or time. More details on R and D patterning mechanisms can be found in Kondo (2002). Basically, a dynamic patterning mechanism is realized through an activator of WUS that does not only rely on a strict expression pattern but whose activity and localization is instead determined through an R and D model. This model is able to recapitulate the experimental data; when the organizing center is removed by simulated ablations, the model predicts the recruitment of adjacent cells into new organizing center foci. Although there is no implicit evidence as to the molecular identity of any of the mechanistic components, such computational models suggest possible requirements for correct WUS positioning and help biologists in designing experiments aimed at uncovering the molecular identity of the postulated activators and inhibitors.
Gordon et al. (2009) add a number of important molecular components to the system. They show that the highest level of cytokinin signaling, as visualized by the cytokinin signaling reporter PTCS:GFP, colocalizes with WUS expression. This suggests that cytokinin signaling is an upstream activator of WUS. Gordon et al. (2009) present a model in which the positive cytokinin effectors (Type B ARRs) cause upregulation of WUS directly and indirectly (by suppressing CLV1/3), while the negative effectors (Type A ARRs) provide negative feedback upon the Type B ARRs (Figure 1C). In addition, WUS negatively regulates the Type A ARRs that sets up an overall positive feedback loop between cytokinin signaling and WUS levels. In this model, the activation and repression of WUS (whether direct or indirect) are based upon the static cytokinin-responsive PTCS:GFP expression domain. It will be interesting to determine how the computational model (Jönsson et al., 2005) and the molecular pathways (Gordon et al., 2009) could fit together. Could the theoretical R and D mechanism of WUS positioning be upstream of cytokinin signaling, or could components of the cytokinin system be mobile and participate directly in an R and D mechanism?
The WUS/CLV signaling mechanism for meristem maintenance and the still-unresolved patterning mechanism illustrate the increasing complexity of our knowledge of regulatory mechanisms and demonstrate the usefulness of integrating computational and mathematical modeling into developmental biology. Both traditional developmental biology and computational modeling together feed forward into new questions and hopefully new answers as well.
Maintaining Apical-Basal Boundaries in the Meristem
While the above section deals with establishing radial positioning of the central zone, the apical-basal boundaries are also important. The cells of the meristem exhibit a developmental partitioning: the stem cells are at the very tip, followed by the peripheral zone within which cells are competent for organ formation, and below are cells that no longer form organs. How is this apical-basal pattern of increasing differentiation maintained? The KANADI (KAN) transcription factors are key regulators during embryogenesis with respect to the formation of cotyledons and the first true leaves: ectopic expression in the central zone completely abolishes stem cell function, whereas a kan triple mutant extends organogenic potential to cells below the peripheral zone (Izhaki and Bowman, 2007). As we will see in the section on organ polarity, KAN function is antagonistic to that of the class III homeodomain zipper (HD-ZIPIII) proteins. HD-ZIPIIIs are normally expressed in the central zone, while the KANs are excluded from this tissue and expressed below the peripheral zone (Figure 1E). Consistent with these data, hd-zipIII triple mutants fail to maintain a meristem; this meristem loss is suppressed by the addition of triple kan mutations, indicating that the hd-zipIII meristem consumption is due to an expansion of KAN expression into the central zone and/or vasculature (Izhaki and Bowman, 2007). While these data relate mostly to the embryo, this concept plausibly may be extended to the vegetative and reproductive meristems.
What sets up these apical-basal expression patterns? There are interesting indications that small RNAs, negative regulators of their target genes, are involved. The HD-ZIPIIIs mRNAs are degraded through their interaction with microRNA165/166 (miR165/166). Mutations in the Arabidopsis thaliana PINHEAD/ZWILLE/ARGONAUTE10 (hereafter referred to as AGO10) gene cause elevated levels of miR165/166. This indicates that AGO10 is somehow responsible for the downregulation of miR165/166 (and thus indirectly the upregulation of HD-ZIPIIIs) in the developing meristem (Figure 1E). In ago10 embryos, the meristem is established normally, based on SHOOTMERISTEMLESS (STM) expression, but is not maintained, yielding seedlings that produce few aerial organs and exhibit terminal differentiation of the shoot meristem. These phenotypes indicate that AGO10 is required to maintain the meristem. The ARGONAUTE (AGO) family is integral to small RNA pathways in plants where they act to mediate small RNA activity. In plants, different AGO proteins recruit different classes of small RNAs and thus provide a certain specificity (for a comparison of AGO1 and AGO10 activities, see Mallory et al., 2009). Tucker et al. (2008) showed that complementation of the ago10 mutant phenotype requires AGO10 expression within the vasculature. Thus, AGO10 within the vasculature is required to restrict miR165/166 from being active in the meristem above it (Figure 1E). All indications point to the involvement of a nonautonomous factor, but it remains to be seen what this factor might be. For example, it is possible that AGO10 mediates the production of a mobile signal, conceivably a small RNA. Alternatively, AGO10 might bind miR165/166 and prevent it from moving into the meristem via the vasculature. Insights into the role of AGO10 are eagerly awaited.
Stem Cell Maintenance: Hormone Regulation via KNOX Genes
The identity and position of the stem cells within the meristem are maintained during growth, but at the same time the stem cell derivatives (transit amplifying cells) in the peripheral zone must acquire competence to undergo organogenesis. The maintenance and loss of stem cell fate is controlled by the KNOTTED LIKE HOMEOBOX (KNOX) transcription factors. Mutants in the Arabidopsis KNOX gene STM have no meristem, indicating that KNOX transcription factors repress differentiation (Barton and Poethig, 1993; Kerstetter et al., 1997). KNOX expression marks cells within the central and peripheral zones but is absent from sites of new organ formation (Figure 1D) (Jackson et al., 1994; Long et al., 1996), and the loss of KNOX expression is a prerequisite for organ initiation.
Two important downstream targets of the KNOX transcription factors are the phytohormones gibberellin and cytokinin. Cytokinins positively regulate cell division, while gibberellins positively regulate cell elongation, which is usually associated with differentiation (Veit, 2009). KNOX proteins regulate the balance between gibberellin and cytokinin; for example, STM upregulates genes encoding cytokinin biosynthesis enzymes, downregulates genes encoding GA inactivating enzymes, and upregulates genes encoding gibberellin inactivating enzymes (Jasinski et al., 2005; Bolduc and Hake, 2009). A high cytokinin/low gibberellin ratio is thought to be important for preventing cell differentiation and thus maintaining stem cell fate; this is informative if one recalls the work of Gordon et al. (2009), indicating an importance of cytokinin in WUS expression. The absence of KNOX expression at incipient organ sites correlates with a reversal in the cytokinin/gibberellin ratio, the consequence of which may be cell differentiation. Thus, the interplay between the KNOX transcription factors and the cytokinin/gibberellin ratio is an important part of the switch toward organogenic competence (Figure 1D; see also Shani et al., 2006; Veit, 2009). Given the close coincidence of KNOX expression and the site of cytokinin activity, and the sharp boundaries obtained between emerging organs and the meristem, it seems unlikely that cytokinin and gibberellin are mobile within the meristem. While we have a fairly detailed molecular understanding of the regulation of cytokinin and gibberellins by KNOX proteins, the downstream integration of such signals into developmental processes remains an interesting subject of future research. As detailed later in this review, the phytohormone auxin is also important as a regulator of KNOX expression and organ positioning.
The organization of the shoot meristem and the dynamics of stem cells and daughter cells are integral to setting the stage for early leaf development. Within this section, we examined how small mobile ligands, hormones, and small RNAs may all contribute to the patterning of the meristem; however, as we will see, each of these mechanisms is reutilized during the steps of early leaf development that follow.
FROM ONE LEAF TO THE NEXT: PHYLLOTAXIS
In contrast with the stem cells in the central zone, their daughter cells in the peripheral zone are competent to differentiate into lateral organs; however, not all of the daughter cells form new organs. What then determines where the new organs will form within this field of competent cells? The pattern of organs about the meristem is termed phyllotaxis. The most prevalent phyllotactic pattern in nature is spiral, but often decussate, alternate, and whorled phyllotaxis are observed (Figure 2A). What virtually all phyllotactic patterns have in common is that new organs tend to form as far away as possible from previously initiated organs (known historically as Hofmeister's Rule, after Hofmeister, 1868).
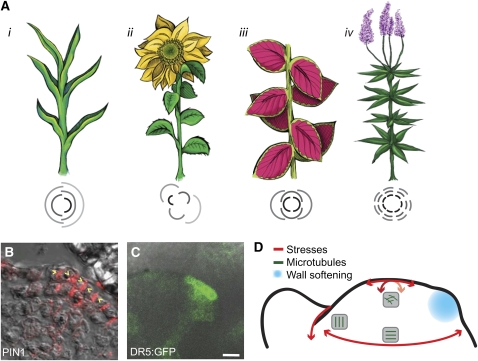
Figure 2.
Phyllotaxis: the Relative Positioning of New Leaves.
(A) Four major phyllotactic patterns depicted as plants and as top view line diagrams showing relative leaf positions: (i) distichous with a divergence angle of 180° (maize), (ii) spiral with an angle of ∼137.5° (sunflower), (iii) decussate with pairs of leaves at 90° (Solenostemon sp), and (iv) whorled with three or more leaves originating from the same node (Veronicastrum virginicum). In line diagrams, lighter gray indicates older leaves.
(B) Immunolocalization of PIN1 protein in an Arabidopsis vegetative meristem. PIN1 exhibits polar localization within the L1 (direction indicated by yellow arrow heads). (Image provided by E.R. Pesce and C. Kuhlemeier.)
(C) Expression of the auxin responsive reporter DR5:GFP in an Arabidopsis inflorescence meristem. Bar = 10 μm. (Reproduced from Smith et al. [2006].)
(D) Tissue mechanics and components within the meristem as illustrated by major axis of stress (red arrows) and the coincident microtublue organization (green lines in gray boxes), as described by Hamant et al. (2008). Predicted wall softening (expansin activity and pectin modification) at the site of a new leaf in blue.
In terms of biological relevance, Hofmeister's Rule is thought to be advantageous for efficient light capture; however, different phyllotactic patterns yield similar light interception efficiencies (Valladares and Brites, 2004). Thus, light capture does not explain why spiral patterns are so prevalent in nature or under which environmental conditions distichous or decussate leaf arrangements might have a selective advantage. Alternatively, efficient packing of primordia at the meristem may serve a protective role by shielding them from environmental stresses. From a structural perspective, regular phyllotactic patterns may balance the forces that leaves exert on the stem. Endress and Doyle (2007) propose that spiral positioning of floral organs promotes adaptive radiations. Given the highly uncertain nature of the “whys” of phyllotactic patterns, let us return to the mechanistic question: How is new leaf (primordia) positioning determined? This will be the main focus of this section.
Hofmeister's Rule can easily be explained by assuming that each primordium is the source of an inhibitor that decreases in strength with time and/or distance. The new organ then arises at the site of lowest inhibition within the peripheral zone. Fairly simple mathematical models based on mutual inhibition can recreate almost any observed phyllotactic pattern (Douady and Couder, 1992; Smith et al., 2006). But what is the molecular nature of the phyllotactic patterning mechanism? As mentioned in the previous section, new organs coincide with sites of downregulated KNOX gene expression, but how are these sites determined? The meristem is a radially symmetric structure, and the initiation of lateral organs in a phyllotactic pattern implies the breaking of radial symmetry. The earliest indication of this newly emerging pattern is the establishment of auxin maxima at the position of presumptive primordia. Over the last few years, it has become clear that phyllotactic patterning is set up by a positive feedback loop between auxin and its transporter, PIN1. Within this section, we will examine how the auxin/PIN feedback loop contributes to (1) positioning of new organs and (2) transitions between phyllotactic patterns. We will also examine the possible role of tissue mechanics in reinforcing the auxin/PIN patterning mechanism.
Positioning of New Organs: Auxin Dynamics
Auxin is a small, mobile phytohormone important for many aspects of plant growth and development. In the case of phyllotactic patterning, the ability of auxin to move between cells, so-called polar auxin transport, makes it special and capable of acting as a signaling molecule. Inactivation of polar auxin transport, either using inhibitors or by mutations in the gene encoding the PIN1 auxin exporter, abolishes organ formation and yields stems without organs, so called naked pins. Application of microdroplets of auxin to naked pins induces organ initiation at the site of application, indicating that artificially induced auxin maxima are sufficient to trigger organ initiation (Reinhardt et al., 2000). Importantly, PIN1 displays a coordinated polar subcellular localization in the cells of the L1, indicating that directed transport within the L1 alone may be sufficient to provide maxima and thus positional information (Figures 2B and 2C; (Reinhardt et al., 2003; de Reuille et al., 2006; Jönsson et al., 2006; Smith and Bayer, 2009). It should also be noted that while auxin dynamics in the L1 alone may position organs, it is likely that the underlying tissue layers also play a role in subsequent development, as is the case in organ outgrowth and tissue differentiation (Bayer et al., 2009).
The experimental data show that auxin transport somehow directs the positioning and initiation of primordia, but fail to give a clue about the quantitative aspects of phyllotaxis: How does auxin space primordia, how does it determine divergence angles, and how does it condition transitions between distinct stable patterns, such as decussate (pairs of organs at 90°) and Fibonacci spirals (divergence angle ∼137.5°)? Again, we have an instance where developmental biology has benefited from mathematics and computational models.
Modeling has shown that the observed pattern of PIN1 localization in the L1 layer of the meristem accurately predicts the observed positions of the auxin maxima (de Reuille et al., 2006). But how is the pattern of PIN1 polarization established in the first place? This relates to a classical question in developmental biology: How can a pattern form de novo from a field of equivalent cells? In the previous section describing WUS positioning, we described R and D (reaction diffusion) models that are capable of de novo pattern formation. Unlike R and D, auxin-based patterning acts by a mechanism that does not rely on diffusion but on active transport. All auxin-based computational models of phyllotaxis exploit the concept of a positive feedback loop between auxin and its exporter PIN1.
One type of model proposes that PIN1 protein within a cell is preferentially allocated toward the neighboring cell with highest auxin concentration (“up the gradient” model). PINs have been shown to cycle rapidly between the plasma membrane and endosomal compartments, and this intracellular trafficking of PIN is controlled by auxin. Auxin inhibits endocytosis and thereby stabilizes PINs at the plasma membrane (Paciorek et al., 2005). Thus, if a cell is exposed to a gradient of auxin, PIN1 will polarize toward the side of the cell exposed to the highest concentration and thereby direct transport up the auxin concentration gradient. In the other type of models, it is not the concentration but the flux of auxin through a cell, which promotes the subcellular localization of PINs and thereby enhances auxin transport, comparable to water carving a channel when it flows down a soft terrain. This type of “with the flux” or “canalization” model is traditionally invoked to explain vein formation. For detailed discussions of auxin-based modeling, see Heisler and Jönsson (2007), Kramer (2008), and Smith and Bayer (2009).
While computational modeling shows that a positive feedback loop between auxin and PIN is a plausible patterning mechanism, it does not tell us how cells perceive their own auxin status relative to their neighbors; is it the difference in auxin concentration or the direction of auxin flux that is measured? Or is an unknown intercellular signaling molecule involved? Again, we can see the advantages gained using a combination of traditional developmental biology and computational modeling, both in hypothesis testing and generation of new hypotheses.
We can also link the mechanism by which KNOX genes are downregulated at sites of new organ formation to auxin maxima. Auxin has been shown to negatively regulate the KNOX genes; maize (Zea mays) apices grown in the presence of an auxin transport inhibitor fail to exhibit downregulation of KNOX genes within the meristem (Scanlon, 2003). In addition, a maize mutant ectopically expressing several KNOX genes also shows decreased polar auxin transport, and the KNOX gene BREVIPEDICELLUS is regulated by auxin (Hay et al., 2006). These studies point to a role for auxin in downregulating KNOX expression and provide a functional link between maxima and loss of stem cell identity (Barkoulas et al., 2007).
Phyllotactic Transitions: Hormone Dynamics and Meristem Geometry
Phyllotactic patterns can be disrupted by experimental interference, but, within limits, they will quickly recover and reestablish the original arrangement. On the other hand, transitions between patterns, for instance, from decussate to spiral, occur frequently during the life of a single plant, indicating that developmental switches can override the self-correction mechanism. Several mathematical theories attribute different observed patterns to the properties of meristem size and primordia size (Hofmeister, 1868; Douady and Couder, 1992). Intuitively this makes sense, as mentioned above, since more or less available space will alter the PIN1/auxin positive feedback dynamics distribution. It should be pointed out that the parameters of meristem and primordial size do not necessarily refer to physical size, but instead may refer to some chemical size or size of influence that may or may not be bounded by what we observe physically.
Recent work in maize indicates that feedback between auxin and cytokinin may also be important for phyllotactic patterning. The maize mutant aberrant phyllotaxy1 (abph1) displays, as its name suggests, aberrant phyllotaxis; maize normally exhibits distichous patterning (alternate organs at 180°), but the abph1 mutant displays decussate patterning (paired organs at 90°). ABPH1 encodes a cytokinin-inducible type A response regulator, a protein involved in cytokinin signaling. Based on the role of cytokinin in meristem maintenance and Hofmeister's Rule, it is tempting to hypothesize that the change of patterning in abph1 phyllotaxis might be due to an increase in meristem size; however, the true mechanism is more complex. Lee et al. (2009) have recently demonstrated that while ABPH1 in maize may function to negatively regulate cytokinin signaling and therefore regulate meristem size, it also appears to positively affect auxin and PIN1 expression themselves. This is in line with modeling efforts on one- and two-dimensional surfaces that show that reduction of active auxin transport relative to diffusion will increase the spacing between auxin peaks. These results suggest that the interplay between cytokinins, meristem size, and auxin/PIN1 determined patterning are intimately linked through ABPH1.
Wall Mechanics: Pattern Reinforcement via Mechanical Properties
While auxin dynamics alone can dictate phyllotactic patterns in a computer simulation, biology is much more complex than a model. Traditionally, another theory of phyllotactic patterning has had its proponents as well: mechanical buckling. Recent experimental and mathematical results indicate that both auxin dynamics and mechanics may be important for phyllotaxis.
Mechanical phyllotactic theories postulate that differences in tension between the L1 surface layer and the inner tissues lead to tissue buckling (Green, 1999; Shipman and Newell, 2004; Dumais, 2007). In addition, existing primordia may constrain the pattern of new outgrowths at the meristem and thereby set up positioning of the next primordia. Again the size of both primordia and meristem would be contributing factors. Differential tension and compression within the meristem has been experimentally suggested for the developing sunflower (Helianthus annuus) capitulum (Dumais and Steele, 2000). Savaldi-Goldstein et al. (2007) demonstrated that the epidermis both promotes and restricts shoot growth by providing a nonautonomous signal to the ground tissues. This highlights the unique identity of the L1, which might also include different mechanical properties.
Work regarding cell wall modifications in the meristem may indicate that cell wall changes, and thus changes in tissue mechanics, may be important for patterning. Local application or gene induction of expansin, a protein that regulates wall extensibility in vitro, induces primordia at aberrant positions (Fleming et al., 1997; Reinhardt et al., 1998; Pien et al., 2001). Recently, Peaucelle et al. (2008) demonstrated that alterations in pectin structures within the meristem affect organ outgrowth; transgenic alterations in pectin structure were sufficient to abolish organ outgrowth at the meristem, although it is unclear whether phyllotaxis was affected. These two studies link cell wall structural modifications (predicted wall softening; Figure 2D) to meristem structure and primordia formation, perhaps through mechanics.
Hamant et al. (2008) explored the relationship between microtubules and stress forces within the Arabidopsis inflorescence meristem; due to the connection between microtubule orientation and cellulose microfiber orientation, this may provide insight on tissue mechanics (Baskin, 2005). The authors present a physically based model of tensile forces within the L1 that accurately predicts the orientation of microtubules within cells during growth and external manipulation. Microtubules align along the axes of principal stress, indicating that cells within the meristem respond to stress vectors by reinforcing their wall strength along the appropriate axis (Figure 2D). When the microtubules were depolymerized using the drug oryzalin, the meristem turned into a balloon-like structure, not unlike a collection of soap bubbles (Corson et al., 2009). However, in the presence of oryzalin, growth and organ formation continued, at least in the short term. The authors concluded that mechanics is required for specific morphogenetic events, such as tissue folding, but that it acts largely independently from auxin-mediated organogenesis. In the light of these and other results, it is likely that patterning is set up via an auxin/PIN patterning generator and that the resulting pattern is stabilized by mechanics.
A recent mathematical treatment by Newell et al. (2008) combines both the auxin chemical theory and the mechanical buckling theory and in doing so highlights the similarities in the mathematical basis behind both theories. The authors indicate that situations may exist where the two processes are strongly linked (both are important for patterning), but also situations where they are weakly linked such that one may take precedence over the other. These current studies open up a new area for phyllotactic theory involving both auxin and mechanics, perhaps in different measure depending on the species, and provide many new avenues of research. The emerging picture of phyllotaxis is one of a process requiring both auxin dynamics and biomechanical changes to allow for proper organ outgrowth and feedback into new organ positioning (Figures 2B to 2D). Future studies of the feedback between auxin and cell mechanics within the meristem will provide valuable information toward understanding phyllotaxis.
We now have an idea of how the meristem is structured and the importance of its proper organization for plant development and how auxin and changes in cell wall mechanics may explain the positions of new organs within the peripheral zone. We also have an idea of the complex interactions between plant hormones with respect to meristem size and, thus, organ positioning and how the maintenance and loss of stem cell identity via KNOX genes is required for proper organogenesis and plant function. But once a position is determined for a new leaf, how does it grow out and become the flat leaf that is required?
FROM TOP TO BOTTOM: LEAF POLARITY AND DETERMINING WHAT IS UP
In most plants, the two surfaces of the leaf are specialized, with the photosynthesizing light harvesting cells concentrated at the upper side and gas-exchanging cells at the lower side. This leaf polarity is also evident in the differential distribution of trichomes and stomata and in the position of cell types in the vascular bundles (Figure 3A). The up-down polarity of an organ is formally known as adaxial-abaxial (ad-ab) polarity, where adaxial refers to the side of the organ that originated closest to the meristem and abaxial the opposite (Figure 3A). Most of the described polarity mutants fall into distinct classes of highly conserved transcription factors (for a recent review, see Husbands et al., 2009). Generally speaking, the HD-ZIPIIIs and the AS1/AS2 complex promote adaxial identity; whereas KANADIs and the auxin-dependent ARF3/ARF4 promote abaxial identity. All of these factors have other functions in addition to their roles in leaf polarity; for instance, the KANADIs are also involved in specifying the lower boundary of the shoot apical meristem, as mentioned in a previous section. Intriguingly, although the polarity-determining factors are highly conserved, their contributions to organ polarity differ between species; to use an analogy, it is as if the same jigsaw puzzle pieces could be put together in different ways to produce the same final picture. Within this section, we will (1) discuss the roles of key transcription factors in polarity and (2) the role of small RNAs in patterning said transcription factors.
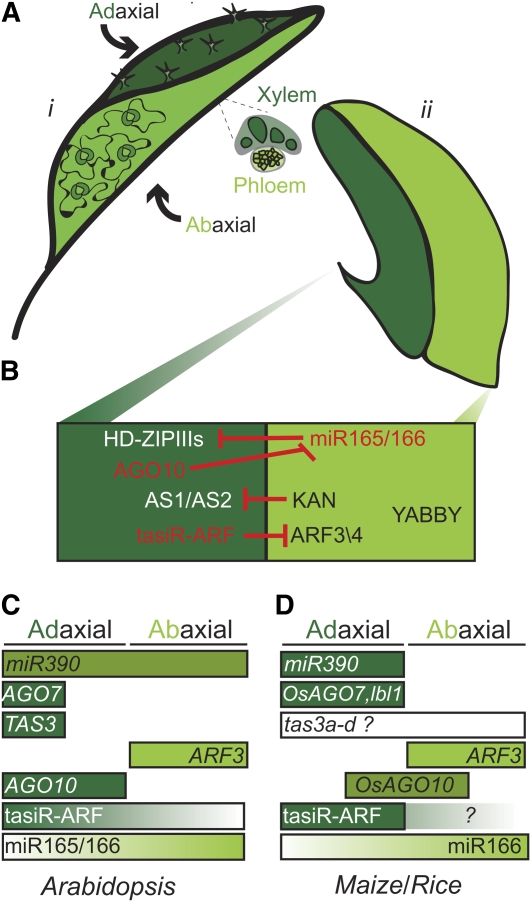
Figure 3.
Polarity: Determining What Is Up.
(A) An illustration of (i) a mature leaf showing differences in adaxial (dark green) and abaxial (light green) traits and (ii) a young primordium.
(B) Key regulators for organ polarity in Arabidopsis. HD-ZIPIIIs, KAN, ARF, AGO10, tasiR-ARF, microRNA165/166 (miR165/166), and ASYMMETRIC LEAVES1 (AS1).
(C) A schematic diagram showing the localization of important players in the smallRNA-polarity regulatory pathway in Arabidopsis. MicroRNA390 (miR390), AGO7, and TRANS-ACTING SIRNA3 (TAS3).
(D) As in (C) except showing the important players in maize and/or rice (Os). Maize TAS3 (tas3a-d) expression pattern is unknown, maize tasiR-ARF expression is low, and it is unclear if it shows a gradient. OsAGO7 is expressed adaxially (Nagasaki et al., 2007). Maize ARF3 and OsARF3 are expressed abaixially (Itoh et al., 2008). OsAGO10 is expressed in the vasculature of young leaves (Nishimura et al. 2002).
Transcription Factors Involved in Polarity
Phantastica (phan) in Antirrhinum majus was the first polarity mutant described (Waites and Hudson, 1995); the PHAN gene encodes a Myb-type transcription factor. In phan mutants, the leaf has a rod-like shape with abaxial characters. Thus, adaxial characters are lost and replaced by abaxial markers. Moreover, the leaf blade is missing. This suggests (1) that adaxial and abaxial cell fates are mutually exclusive, and (2) that the outgrowth of the flat leaf blade and ad-ab polarity are mechanistically linked. phan mutations have similar phenotypes in tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum; Kim et al., 2003; McHale and Koning, 2004), but in the homologous maize rs2 and Arabidopsis as1 mutants, the leaves develop with essentially normal polarity (Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000). This highlights the fact that mutations in homologous genes can cause different phenotypes depending on the species (see Kidner and Timmermans, 2007).
The HD-ZIPIII transcription factor family also plays a major role in polarity. The HD-ZIPIII proteins PHAVOLUTA (PHV), PHABULOSA (PHB), and REVOLUTA (REV) specify adaxial cell fate (McConnell et al., 2001; Emery et al., 2003; Itoh et al., 2008), and wild type HD-ZIPIII genes are expressed on the adaxial side (Figure 3B). Interestingly in semidominant gain-of-function mutants, which exhibit adaxialization, the expression domain expands throughout the leaf (McConnell et al., 2001; Emery et al., 2003); the mutations in these alleles are located in a small part of the sequence and cause the HD-ZIPIII mRNA to become resistant to cleavage by miR165/166.
The abaxial counterparts to the HD-ZIPIIIs are the KAN and AUXIN RESPONSE FACTOR (ARF) families of transcription factors (Kerstetter et al., 2001; Eshed et al., 2004; Pekker et al., 2005; Candela et al., 2008). KAN genes are expressed on the abaxial side, complementary to the adaxial PHV/PHB/REV domain (Figure 3B). Loss-of-function kan mutants are adaxialized and display expanded expression of the HD-ZIPIIIs, a demonstration at the molecular level of the mutual exclusivity of adaxial and abaxial cell fates. Similarly, ubiquitous overexpression of KAN1 and KAN2 causes abaxialization. As mentioned in a previous section, both the KANs and HD-ZIPIIIs have an effect on the meristem; hd-zipIII triple loss-of-function mutants also lack an apical meristem, a phenotype that is mirrored by the overexpression of KAN. Conversely, the gain-of-function hd-zipIII mutants develop extra axillary meristems and an enlarged apical meristem. These data indicate that HD-ZIPIIIs or the exclusion of KANs is required for meristem identity, and they point to a tight connection between the patterning of meristems and leaves.
The auxin response factors ARF3 and ARF4 act partially downstream of KAN. While ARF transcription is induced all over the leaf, as was shown for ARF3, the ARF proteins are restricted to the abaxial side by a small RNA–based mechanism we will discuss in the next section (Figures 3B to 3D) (Pekker et al., 2005). The presence of ARFs in the polarity chain further suggests a role for auxin in ad-ab patterning. Could auxin gradients be regulating both polarity and phyllotactic patterning? As we have seen in the section on phyllotaxis, auxin maxima position the leaf primordia within the peripheral zone of the meristem. Moreover, during leaf outgrowth, auxin flows through the epidermis toward the leaf tip, where it reverses direction and flows back through the midvein, ultimately being drained by the central vasculature of the stem. How the young leaf might maintain an ad-ab auxin gradient superimposed on such a “reverse fountain” flow of auxin is puzzling.
A final set of transcription factors that act relatively late during leaf development are the YABBYs. In Arabidopsis, the YABBYs are abaxially expressed, and mutation of multiple yab genes leads to formation of abaxialized organs (Stahle et al., 2009). However, abaxial localization is not conserved between species: in maize, YABBYs are expressed adaxially and in rice in a nonpolar pattern (Juarez et al., 2004a; Toriba et al., 2007). This may suggest that their function in polarity determination is not conserved between species. Instead, data suggest a shared function in blade outgrowth (Kidner and Timmermans, 2007).
What emerges from the previous section is that sets of transcription factors are expressed in mutually exclusive domains and that they specify either adaxial or abaxial cell fates (Figure 3B). However, this gives us no clue as to how these domains are set up in the first place. Neither HD-ZIPIIIs, KANs, nor any of the other polarity transcription factors are known to move between cells; thus, their differential localization must depend on other intercellularly mobile signals. Historically, the first evidence for a mobile signal came from ablation experiments by Ian Sussex (1951). When the incipient primordium was isolated from the meristem by a tangential incision, an abaxialized primordium resulted that is comparable to what is seen in phan mutants. More recent ablation experiments specifically show that an intact L1 layer is required for the maintenance of abaxial identity (Reinhardt et al., 2005). The interpretation of these experiments is that the ablation interrupts the movement of an L1-based adaxializing signal from the meristem into the leaf primordium (for an alternative view, see Efroni et al., 2010). The molecular identity of this Sussex signal remains unknown, and experimental evidence is eagerly awaited.
Regulation of Polarity by Small RNAs
The maize leafbladeless1 (lbl1) mutant has a strongly abaxialized phenotype; thus, the normal function of LBL1 is to specify adaxial identity (Figure 3D; Timmermans et al., 1998). Unlike previously described polarity determinants, LBL1 is not a transcription factor: it is a component of a specialized pathway involved in the generation of a unique class of small RNAs, the transacting short interfering RNAs (tasiRNAs) (Allen et al., 2005; Husbands et al., 2009). The tasiRNAs involved in polarity are generated from the TAS3 genes, which do not code for proteins and are functional as RNAs. They are targeted by miR390 for cleavage by AGO7/ZIPPY and converted into a long double-stranded RNA by LBL1 (SGS3 in Arabidopsis) and RDR6. This double-stranded RNA is then digested into precisely phased 21-bp fragments by DCL4. A subset of these tasiRNAs (the tasiR-ARFs) finally targets the ARF3 and ARF4 mRNAs for cleavage and degradation. The components of the miR390/tasiR-ARF pathway are highly conserved in plants; yet, there are important differences in expression and phenotypes between maize and Arabidopsis (Figures 3C and 3D) (Nogueira et al., 2007; Chitwood et al., 2009). In Arabidopsis, mutants in the tasiR-ARF pathway do not display the dramatic phenotypes seen in maize but have rather subtle effects, mostly leaf curling, which is interpreted as a consequence of underlying polarity defects.
In maize, the mature miR390 localizes to the adaxial side of the incipient primordium, where it triggers the production of tasiR-ARF small RNAs. These tasiR-ARFs then travel toward the abaxial side, providing a mobile component in ad-ab patterning. In Arabidopsis, miR390 also accumulates outside the expression domains of its precursors and is ubiquitously present in the meristem and leaf primordia. However, its activity is restricted to the two most adaxial cell layers of the leaf because of the restricted expression of AGO7 and TAS3 (Figure 3C). Thus, as in maize, the tasiR-ARF small RNAs are produced adaxially and travel toward the abaxial side, thereby creating a gradient of expression. This gradient results in a precisely defined abaxial expression of ARF3/4 (Figures 3B and 3C).
MiR166, the regulator of the HD-ZIPII family, was shown in maize to exist in a gradient with its maximum below the abaxial side (Juarez et al., 2004b). It is conceptually possible that miR166 is mobile. However, the more conservative alternative is that its gradient is formed in response to another, preexisting, gradient. Whether the Arabidopsis miR165/166 also forms a gradient is not known.
The general picture that emerges is that mobile tasiR-ARF small RNAs form a gradient toward the abaxial side and miR166 forms an opposing gradient. These two gradients might give rise to mutually exclusive domains of the ARF3/4 and HD-ZIPIII transcription factors, respectively (Figure 3B). How gradients of mobile factors are converted into discrete domains of nonmobile transcription factors has been the subject of much discussion (for an insightful recent review see, Lewis, 2008). Theoretical work shows that opposing gradients of small RNAs would be well suited for this purpose (Levine et al., 2007). However, the patterning scenario sketched above is far from complete. What sets up the earliest polar determinants? Could it be the enigmatic Sussex signal? Or is leaf polarity a manifestation of the apical-basal polarity in the meristem?
CONCLUSIONS AND PROSPECTS
In this review, we focused on the early phase of leaf development, from the origin of leaf primordia in the shoot apical meristem, through organ positioning, to the establishment of leaf polarity. Subsequent leaf development is reviewed by Efroni et al. (2010).
Mobile Signals: Old and New
In the past, major insights into leaf development have been achieved through the characterization of mutants with specific developmental defects. Transcription factors in particular have been prime targets for developmental genetics given their hierarchical position in many signaling cascades; however, with a few notable exceptions, transcription factors are cell autonomous. As such, they can relay information from mobile signals into cellular responses but are unable themselves to coordinate differentiation across complex tissues.
The existence of mobile signals was first inferred from ingenious experiments performed decades ago, and for an increasing number, the molecular identity and mode of action are being revealed. Most prominent among them is the small peptide CLV3, the ligand for a receptor-like kinase complex that interacts with WUS to regulate stem cell identity. Recent computational models have also provided new clues toward understanding WUS positioning within the meristem, although the identity of predicted mobile factors remains to be seen.
Hormones are quintessential mobile regulators of development. Auxin is obviously mobile and involved in almost every developmental process in plants (the only exception to date seems to be the induction of flowering). Curiously, the action of cytokinin and gibberellin in the switch to organogenic competence appears to be strictly confined to their sites of synthesis (Jasinski et al., 2005; Yanai et al., 2005; but see Gordon et al., 2009). For example, in the meristem, the sharp coincidence of KNOX expression and high cytokinin/low gibberellin levels provides compelling evidence for this idea given their recently described interactions (Jasinski et al., 2005). Thus, in the specific context of the shoot meristem, these two hormones do not seem to act as mobile signals. By contrast, alteration of brassinsteroid or its perception has non-cell-autonomous effects (Savaldi-Goldstein et al., 2007).
Small RNAs represent a new class of mobile developmental signals. The recently described mobility of miRNA390 and tasiARF RNAs are essential, and we are only just beginning to understand their significance for intercellular signaling with respect to development. It is likely that other small RNAs play a mobile role in development, and the high specificity of nucleic acid base pairing seems ideal for exclusive relationships between spatially separated components of a gene regulatory network.
The Role of Mechanics in Tissue Patterning
The Sleeping Beauty of the phyllotaxis story is mechanics. Mechanical theories of development were en vogue in the early 1900s (Thompson, 1942) but lost their luster with the advance of biochemistry and genetics. Since mechanics operates at the tissue and organ levels, it has clear non-cell-autonomous properties and the potential to coordinate growth and development. A problematic characteristic of mechanics has been that it does not yield easily to genetic dissection. Because mechanics involves turgor, wall properties, cytoskeletal architecture, and growth in general, relevant genes tend to encode structural proteins and enzymes of primary metabolism, and mutations in such housekeeping genes likely affect many different processes. The theory and technology are now poised to define the roles of mechanics in plant development. Today, it may appear unlikely that mechanical forces are primary regulators of development. However, the realization is growing that chemical signaling is constrained and potentially stabilized by the mechanical properties of a tissue. What forces are created when a leaf bulges out from the meristem, and how do those forces feedback on the chemical signaling machinery? What determines final leaf shape? Surely development involves intricate chemical signaling, but it is undeniable that morphogenesis must also obey basic rules of geometry and physics as well (Coen et al., 2004).
The Integration of Mathematics and Computer Science
When the number of known molecular components in developmental pathway was limited, simple diagrams were adequate to visualize their interactions. However, the flood of data generated in the genomics revolution has had an impact on how we construct developmental models: As we add more and more components, which may be connected through feedback and non-linear interactions, it becomes more and more difficult to devise simple conceptual models.
Throughout this review, important contributions of mathematical and computational modeling to plant development have been highlighted. Such models can test hypotheses, make quantitative predictions about interactions between components, and identify missing factors. The best models are based on a combination of experimental evidence and biologically plausible assumptions. As phrased by Salazar et al. (2009), “Analysis of the models confirms our understanding of [a developmental process] in some areas. Specific failures of the models in other areas predict new regulatory interactions or components that can be tested by molecular experimentation.” Computational models cannot guarantee biological relevance; they simply show that the underlying assumptions do or do not reproduce the observed patterns. A good example is phyllotaxis, for which quantitative models have been constructed based on three entirely different mechanisms: R and D, mechanical buckling, and active auxin transport. In fact, the latter two mechanisms can be represented by mathematically similar sets of equations (Newell et al., 2008). It is the experimental observations of PIN1 and auxin that tip the balance toward an auxin-transport model, while buckling has not yet garnered much experimental support.
Beyond Arabidopsis
Like most recent reviews on plant development, this one prominently features Arabidopsis. The implicit assumption is that what is true for Arabidopsis will be true for a redwood tree, and up until now, we have often been staggered by how true this is. For one example, the KNOX genes, first discovered in maize, have functional homologs in many other plants. As another example, the FLORICAULA gene in Antirrhinum regulates the switch from vegetative to inflorescence meristem identity (Coen et al., 1990), and the Arabidopsis homolog LEAFY is highly conserved and has essentially the same function (Weigel et al., 1992). Also, auxin regulates phyllotaxis in a similar manner in both tomato and Arabidopsis. It would be easy to extend this list, but it is also important to note the exceptions. As we have seen, the same molecular building blocks regulate organ polarity in Arabidopsis, maize, tobacco, and Antirrhinum; however, mutations in the individual components can have drastically different phenotypes depending on the species. This signifies more than trivial differences in redundancy and may rather reflect different evolutionary trajectories. Thanks to the groundwork done in Arabidopsis, we are now beginning to understand the evolutionary diversity of leaf development at the molecular level.
Acknowledgments
We thank Rüdiger Simon, Thomas Laux, Richard Smith, Henrik Jönsson, Marja Timmermans, and Yuval Eshed for their insightful comments on the manuscript and Nina Smith for expert phyllotactic illustrations. Work from the authors’ laboratory was supported by grants from the Swiss National Science Foundation and SystemsX.ch, the Swiss Initiative in Systems Biology. S.B. is supported by the International Research Fellowship Program from the U.S. National Science Foundation (Award OISE-0853105).
References
- Allen E., Xie Z.X., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Galinha C., Grigg S.P., Tsiantis M. (2007). From genes to shape: Regulatory interactions in leaf development. Curr. Opin. Plant Biol. 10: 660–666 [DOI] [PubMed] [Google Scholar]
- Barton M.K., Poethig R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Baskin T. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Smith R.S., Mandel T., Nakayama N., Sauer M., Prusinkiewicz P., Kuhlemeier C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckman A., Simon R. (2009). Interdomain signaling in stem cell maintenance of plant shoot meristems. Mol. Cells 27: 615–620 [DOI] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A.M., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Grunewald M., Hobe M., Simon R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Candela H., Johnston R., Gerhold A., Foster T., Hake S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20: 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Coen E., Rolland-Lagan A.G., Matthews M., Bangham J.A., Prusinkiewicz P. (2004). The genetics of geometry. Proc. Natl. Acad. Sci. USA 101: 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula - A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Corson F., Hamant O., Bohn S., Traas J., Boudaoud A., Couder Y. (2009). Turning a plant tissue into a living cell froth through isotropic growth. Proc. Natl. Acad. Sci. USA 106: 8453–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuille P.B., Bohn-Courseau I., Ljung K., Morin H., Carraro N., Godin C., Traas J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Benfey P.N. (2008). Plant stem cell niches: Standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Douady S., Couder Y. (1992). Phyllotaxis as a physical self-organized growth process. Phys. Rev. Lett. 68: 2098–2101 [DOI] [PubMed] [Google Scholar]
- Dumais J. (2007). Can mechanics control pattern formation in plants? Curr. Opin. Plant Biol. 10: 58–62 [DOI] [PubMed] [Google Scholar]
- Dumais J., Steele C.R. (2000). New evidence for the role of mechanical forces in the shoot apical meristem. J. Plant Growth Regul. 19: 7–18 [DOI] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves—A critical review. Plant Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class IIIHD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Endress P.K., Doyle J.A. (2007). Floral phyllotaxis in basal angiosperms: development and evolution. Curr. Opin. Plant Biol. 10: 52–57 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Fleming A.J., McQueen-Mason S., Mandel T., Kuhlemeier C. (1997). Induction of leaf primordia by the cell wall protein Expansin. Science 276: 1415–1418 [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.E., Casson S., Hunt L. (2008). Intercellular peptide signals regulate plant meristematic cell fate decisions. Sci. Signal. 1: pe53. [DOI] [PubMed] [Google Scholar]
- Green P.B. (1999). Expression of pattern in plants: combining molecular and calculus-based biophysical paradigms. Am. J. Bot. 86: 1059–1076 [PubMed] [Google Scholar]
- Hamant O., Heisler M.G., Jonsson H., Krupinski P., Uyttewaal M., Bokov P., Corson F., Sahlin P., Boudaoud A., Meyerowitz E.M., Couder Y., Traas J. (2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Jönsson H. (2007). Modelling meristem development in plants. Curr. Opin. Plant Biol. 10: 92–97 [DOI] [PubMed] [Google Scholar]
- Hofmeister W. (1868). Allgemeine Morphologie der Gewachse. (Leipzig, Germany: Engelmann; ). [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C.P. (2009). Signals and prepatterns: New insights into organ polarity in plants. Genes Dev. 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J.I., Hibara K.I., Sato Y., Nagato Y. (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A., Bowman J.L. (2007). KANADI and Class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson H., Heisler M., Reddy G.V., Agrawal V., Gor V., Shapiro B.E., Mjolsness E., Meyerowitz E.M. (2005). Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics 21: i232–i240 [DOI] [PubMed] [Google Scholar]
- Jönsson H., Heisler M.G., Shapiro B.E., Meyerowitz E.M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson H., Krupinski P. (2010). Modeling plant growth and pattern formation. Curr. Opin. Plant Biol. 13: 5–11 [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. (2004b). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C.P. (2004a). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kayes J.M., Clark S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Timmermans M.C. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10: 13–20 [DOI] [PubMed] [Google Scholar]
- Kim M., McCormick S., Timmermans M., Sinha N. (2003). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 425: 102. [DOI] [PubMed] [Google Scholar]
- Kondo S. (2002). The reaction-diffusion system: a mechanism for autonomous pattern formation in the animal skin. Genes Cells 7: 535–541 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kramer E.M. (2008). Computer models of auxin transport: A review and commentary. J. Exp. Bot. 59: 45–53 [DOI] [PubMed] [Google Scholar]
- Kwiatkowska D. (2008). Flowering and apical meristem growth dynamics. J. Exp. Bot. 59: 187–201 [DOI] [PubMed] [Google Scholar]
- Kwiatkowska D., Dumais J. (2003). Growth and morphogenesis at the vegetative shoot apex of Anagallis arvensis L. J. Exp. Bot. 54: 1585–1595 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jurgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Johnston R., Yang Y., Gallavotti A., Kojima M., Travençolo B.A., Costa Lda F., Sakakibara H., Jackson D. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M., Laux T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130: 3163–3173 [DOI] [PubMed] [Google Scholar]
- Levine E., McHale P., Levine H. (2007). Small regulatory RNAs may sharpen spatial expression patterns. PLOS Comput. Biol. 3: e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. (2008). From signals to patterns: space, time, and mathematics in developmental biology. Science 322: 399–403 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Hinze A., Tucker M.R., Bouché N., Gasciolli V., Elmayan T., Lauressergues D., Jauvion V., Vaucheret H., Laux T. (2009). Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 5: e1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McHale N.A., Koning R.E. (2004). PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. Plant Cell 16: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H. (1982). Models of Biological Pattern Formation. (London: Academic Press; ). [Google Scholar]
- Muller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., Nagato Y., Sato Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell A.C., Shipman P.D., Sun Z. (2008). Phyllotaxis as an example of the symbiosis of mechanical forces and biochemical processes in living tissue. Plant Signal. Behav. 3: 586–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A., Ito M., Kamiya N., Sato Y., Matsuoka M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30: 189–201 [DOI] [PubMed] [Google Scholar]
- Nogueira F.T.S., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C.P. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazimalova E., Ruthardt N., Petrasek J., Stierhof Y.-D., Kleine-Vehn J., Morris D.A., Emans N., Jurgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18: 1943–1948 [DOI] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Wyrzykowska J., McQueen-Mason S., Smart C., Fleming A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington M. (1929). The regeneration of the stem apex. New Phytol. 28: 37–53 [Google Scholar]
- Reddy G.V., Gordon S.P., Meyerowitz E.M. (2007). Unraveling developmental dynamics: Transient intervention and live imaging in plants. Nature 8: 491–501 [DOI] [PubMed] [Google Scholar]
- Reddy G.V., Heisler M.G., Ehrhardt D.W., Meyerowitz E.M. (2004). Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131: 4225–4237 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2003). Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2005). Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development 132: 15–26 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Wittwer F., Mandel T., Kuhlemeier C. (1998). Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 10: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Sharma V.K., Kovaleva V., Raikhel N.V., Fletcher J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar J.D., Saithong T., Brown P.E., Foreman J., Locke J.C.W., Halliday K.J., Carre I.A., Rand D.A., Millar A.J. (2009). Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139: 1170–1179 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Scanlon M.J. (2003). The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol. 133: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Shipman P.D., Newell A.C. (2004). Phyllotactic patterns on plants. Phys. Rev. Lett. 92: 168102.1–168102.4 [DOI] [PubMed] [Google Scholar]
- Smith R.S., Bayer E.M. (2009). Auxin transport-feedback models of patterning in plants. Plant Cell Environ. 32: 1258–1271 [DOI] [PubMed] [Google Scholar]
- Smith R.S., Kuhlemeier C., Prusinkiewicz P. (2006). Inhibition fields for phyllotactic pattern formation: A simulation study. Can. J. Bot. 84: 1635–1649 [Google Scholar]
- Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.N., Dermen H. (1970). Determination of number and mitotic activity of shoot apical initial cells by analysis of mericlinal chimeras. Am. J. Bot. 57: 816–826 [Google Scholar]
- Sussex I.M. (1951). Experiments on the cause of dorsiventrality in leaves. Nature 167: 651–652 [DOI] [PubMed] [Google Scholar]
- Sussex I.M. (1964). The permanence of meristems: Developmental organizers or reactors to exogenous stimuli? In Proceedings of Brookhaven Symposium in Biology, Miksche J.P., ed (Upton, NY: Brookhaven National Laboratory; ), pp. 1–12 [Google Scholar]
- Suzaki T., Toriba T., Fujimoto M., Tsutsumi N., Kitano H., Hirano H.-Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Szymkowiak E.J., Sussex I.M. (1996). What can chimeras tell us about plant development? Annu. Rev. Plant Physiol. 47: 351–376 [DOI] [PubMed] [Google Scholar]
- Thompson D.A.W. (1942). On Growth and Form: A New Edition. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Timmermans M.C.P., Hudson A., Becraft P.W., Nelson T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C.P., Schultes N.P., Jankovsky J.P., Nelson T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125: 2813–2823 [DOI] [PubMed] [Google Scholar]
- Toriba T., Harada K., Takamura A., Nakamura H., Ichikawa H., Suzaki T., Hirano H.Y. (2007). Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genomics 277: 457–468 [DOI] [PubMed] [Google Scholar]
- Traas J., Bohn-Courseau I. (2005). Cell proliferation patterns at the shoot apical meristem. Curr. Opin. Plant Biol. 8: 587–592 [DOI] [PubMed] [Google Scholar]
- Tsiantis M., Brown M.I.N., Skibinski G., Langdale J.A. (1999). Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol. 121: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Hinze A., Tucker E.J., Takada S., Jürgens G., Laux T. (2008). Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development 135: 2839–2843 [DOI] [PubMed] [Google Scholar]
- Valladares F., Brites D. (2004). Leaf phyllotaxis: Does it really affect light capture? Plant Ecol. 174: 11–17 [Google Scholar]
- Veit B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69: 397–408 [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Girke T., Pasala S., Xie M., Reddy G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]