This work describes the flowering time gene DAY NEUTRAL FLOWERING (DNF), which acts in the same flowering pathway as CONSTANS (CO). DNF is a membrane-bound E3 ligase that represses CO expression and plays an important role in maintaining low levels of CO expression in short days; it is thus essential for the ability of the Arabidopsis plant to have a different flowering response in long and short days.
Abstract
The photoperiodic response in Arabidopsis thaliana requires the precise regulation of CONSTANS (CO) expression in relation to the light period during the day. In short days (SDs) levels of CO expression are normally low during the light period, and this results in delayed flowering compared with long days (LDs) when CO expression rises to high levels before the end of the light period. We identified a novel flowering time gene called DAY NEUTRAL FLOWERING (DNF) that acts in the same flowering pathway as CO. DNF is a membrane-bound E3 ligase that represses CO expression and plays an important role in maintaining low levels of CO expression in SDs. The effect of DNF on the rhythm of CO expression is essential for the photoperiodic response of Arabidopsis, enabling it to have a different flowering response in LDs and SDs.
INTRODUCTION
Many plants regulate the timing of the transition from vegetative to reproductive growth to coincide with favorable seasons of the year. They are able to do this through their perception of, and response to, environmental signals such as temperature and photoperiod (Yanovsky and Kay, 2003; Michaels, 2009). These stimuli are perceived in different organs of the plant: vernalizing temperatures are detected in the shoot apical meristem, whereas photoperiod is detected in the leaves. Perception of an inducing photoperiod in the leaves results in the production of a systemic flowering signal that moves to the apex where it triggers flower development (Zeevaart, 1976). The identity of this mobile signal in Arabidopsis thaliana has been shown to include the FLOWERING LOCUS T (FT) protein (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007), the expression of which is principally regulated by the CONSTANS (CO) gene (Samach et al., 2000). Key questions still remain, however, regarding the control of both CO transcription and the stability/activity of the CO protein (reviewed in Imaizumi and Kay, 2006).
Arabidopsis is a facultative long-day plant in which long days (LDs) promote more rapid flowering than short days (SDs). Different flowering responses to changes in photoperiod are brought about through the interaction of light with the circadian clock–regulated rhythmic expression of CO. In SDs of 8 to 10 h, CO expression is low during the light period, whereas in LDs of 14 to 16 h, the level of CO expression rises toward the end of the day, and the coincidence of light with high levels of CO expression leads to the induction of FT and flowering (Suàrez-López et al., 2001). As evidence to support this model, it has been shown that flowering can be induced in SDs by constitutive overexpression of CO or by altering the rhythm of CO expression such that it is expressed at high levels during the light period of an SD (Onouchi et al., 2000; Roden et al., 2002; Yanovsky and Kay, 2002). In addition to transcriptional regulation, there is also regulation at the level of CO protein stability, which is affected by light signals acting through photoreceptors (Valverde et al., 2004). To generate the level of sensitivity required to distinguish between photoperiods that may only differ by a couple of hours, both the transcription of CO and CO protein stability have to be very tightly regulated.
Transcription of CO is known to be controlled by a number of factors, one of which is the circadian clock, which causes rhythmic oscillations in CO expression (Suàrez-López et al., 2001). GIGANTEA (GI) is also known to affect the expression of CO (Suàrez-López et al., 2001; Mizoguchi et al., 2005). GI has been shown to bind a transcriptional repressor of CO expression called CYCLING DOF FACTOR1 (CDF1). The stability of CDF1 is controlled by an F-box protein called FLAVIN BINDING, KELCH REPEAT, F-BOX 1 (FKF1) (Imaizumi et al., 2003, 2005; Sawa et al., 2007). FKF1 has also been shown to bind to GI in a blue light–dependent manner. This has led to the proposal of a model in which the CDF1 repressor bound to the CO promoter is bound by GI; binding of FKF1 to this complex later on in the day results in the degradation of CDF1, thus allowing CO expression to increase at the end of an LD (Sawa et al., 2007). It has recently been shown that other related DOF factors, CDF2, CDF3, and CDF5, act redundantly with CDF1 to repress CO expression and delay flowering and that CDF2 is also targeted for degradation by FKF1 (Fornara et al., 2009). Overexpression of GI in the fkf1 mutant still causes early flowering, indicating that GI is able to promote flowering independently of the FKF1-mediated degradation of the CDF proteins (Sawa et al., 2007); however, this has been shown to be due to partial redundancy between FKF1 and its close homologs ZEITLUPE and LOV kelch protein2 (Somers et al., 2000; Schultz et al., 2001; Fornara et al., 2009).
Interestingly, Fornara et al. (2009) also demonstrated that this whole layer of regulation of CO expression by GI and the CDF proteins can be removed without affecting the rhythm of CO expression or its response to photoperiod. In a quintuple mutant carrying the gi mutation combined with mutations in the four CDF genes (CDF1, 2, 3, and 5), flowering was responsive to photoperiod and the rhythm of CO expression in SDs and LDs was similar to the wild type but at slightly elevated levels. This means that other regulators of CO transcription must be generating this photoperiodic-responsive rhythm of CO expression and that other factors apart from GI are also able to induce CO transcription. The role of GI and CDFs 1, 2, 3, and 5 appears to be to modulate the amplitude of this underlying rhythm of CO expression.
Apart from the CDF proteins, one other transcriptional repressor of CO has been reported called RED AND FAR-RED INSENSITIVE2, which affects the expression of CO and FT and flowering, and this acts primarily in LD (Chen and Ni, 2006). In this article, we describe the identification of a repressor that regulates the rhythm of CO expression in SDs. This factor, called DAY NEUTRAL FLOWERING (DNF), is crucial in enabling Arabidopsis to distinguish between LDs and SDs, as loss of this repressor alters the rhythm of CO expression and the critical photoperiod for flowering with the result that Arabidopsis flowers at the same time in 8-h SDs as in 16-h LDs.
RESULTS
Isolation of the Early Flowering dnf Mutant
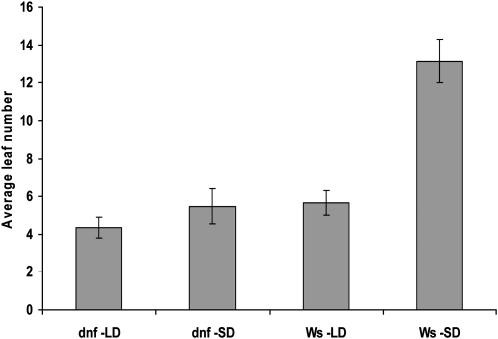
A mutant that flowered early in 8-h SDs was isolated from a screen of the Institut National de la Recherche Agronomique Versailles T-DNA knockout mutant population. The mutant is in the Wassilewskija (Ws) background and has been called day neutral flowering (dnf). The mutant is only affected in flowering time in one photoperiod, flowering early in SDs but at the same time as the wild type in LDs (Figure 1), indicating that the mutation affects the photoperiodic flowering pathway. As rosette leaf number is taken as a measure of flowering time, we checked that the dnf mutation did not affect leaf production. The rate of leaf development in the dnf mutant grown in SDs was shown to be the same as that in wild-type plants (see Supplemental Figure 1 online). The phenotype of the mutant resembles the wild type in all other aspects, suggesting that the mutation does not have any pleiotropic effects and specifically affects the flowering pathway.
Figure 1.
Flowering Times in LDs and SDs.
Average leaf number at flowering of Ws and dnf mutant plants in LDs (16 h light/8 h dark) and SDs (8 h light/16 h dark). Error bars show sd; n = 20 plants.
As the T-DNA carried a gene for phosphinothricin resistance (PPTR), following a backcross to Ws, the F2 population (∼500 lines) was analyzed for segregation of the early flowering phenotype with the PPTR gene. All early flowering lines were PPT resistant, suggesting linkage between the dnf mutation and the PPTR gene. A ratio of 1 early flowering (PPTR) to 2.7 late/intermediate flowering (PPTR) to 0.96 late flowering (PPTS) was obtained. The reason for the slightly skewed ratio is unknown, but a 4:1 ratio rather than a 3:1 ratio was observed for both the flowering phenotype and PPT resistance (of 492 plants in total, 92 were early flowering, while 400 were late/intermediate flowering, and 102 were PPTS, while 390 were PPTR). It is therefore possible that the dnf mutant may contain more than one T-DNA insertion affecting flowering time.
Isolation of the DNF Gene
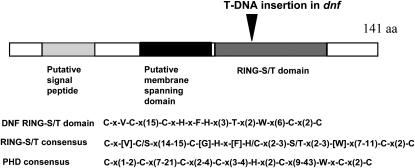
A fragment of the T-DNA sequence was used to probe a genomic library made from the dnf mutant to isolate clones containing a T-DNA insertion and flanking DNA sequences. Analysis of the flanking sequences of the clones obtained showed that the T-DNA insertion was located within the coding sequence of a putative RING finger domain gene, At3g19140. This gene encodes a small protein of 141 amino acids that according to The Arabidopsis Information Resource annotation is predicted to be localized in the endomembrane system. Using the bioinformatic protocols outlined by Emanuelsson et al. (2007), it was shown to have a predicted cleavable signal sequence at the N terminus followed by a transmembrane domain that is the typical structure of a class I membrane protein (Figure 2; von Heijen 1988). Type I membrane proteins are orientated such that the C-terminal part of the protein is in the cytoplasm. The C-terminal domain of DNF contains a consensus sequence of a RING-S/T domain, which is a modified RING finger domain (Stone et al., 2005). RING domains are present in E3 ubiquitin ligases that are involved in targeted protein degradation by the proteosome. Functional analysis of all predicted RING domain proteins in Arabidopsis found that of the predicted RING-S/T proteins tested, which included At3g19140 (DNF), none had detectable E3 ligase activity when assayed with Arabidopsis UBC8, UBC10, UBC11, UBC35, or UBC36 as the E2 conjugating enzyme (Stone et al., 2005). It is possible, however, that some or of all of them may function as E3 ligases specifically with one of the other E2s that were not tested, as Arabidopsis has 37 E2 conjugating enzymes.
Figure 2.
Predicted Domains of the DNF Protein.
Schematic of the DNF protein showing predicted domains and the site of the T-DNA insertion in the dnf mutant. The amino acid sequence of the DNF RING-S/T domain is illustrated together with the consensus sequences for RING-S/T and PHD domains.
PHD domains are closely related to RING finger domains and have a similar consensus sequence to the RING domain. PHD domains are protein–protein interaction domains typically involved in chromatin remodeling (Bienz, 2006); both EARLY BOLTING IN SHORT DAYS (EBS) and VERNALIZATION INSENSITIVE3 are examples of PHD domain proteins involved in the control of flowering time (Pineiro et al., 2003; Sung and Amasino 2004). In the case of DNF, however, the similarity to the PHD consensus breaks down after the Cys in position 3 (Figure 2), and so it is unlikely to act as a PHD domain protein. Apart from some sequence similarity to other proteins in the RING/PHD domain region, DNF does not show any homology to other plant proteins in the databases.
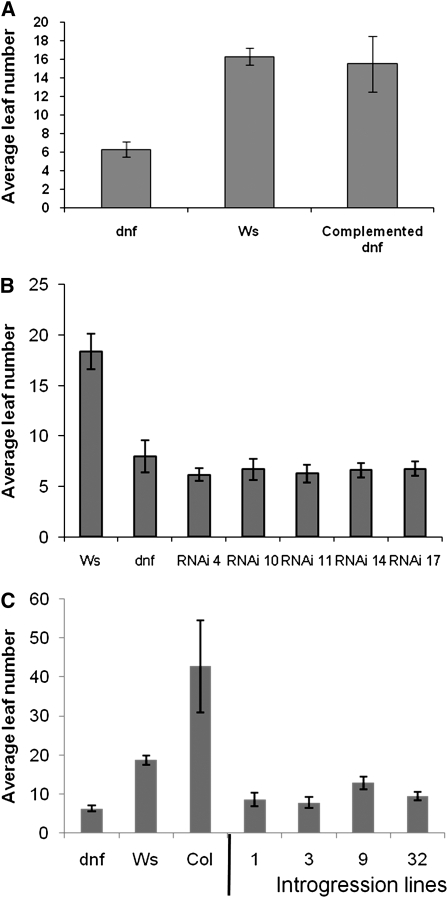
To confirm that the T-DNA insertion in At3g19140 is responsible for the early flowering phenotype of the dnf mutant in SDs, the wild-type DNF allele with 1.1 kb of upstream sequence was cloned from Ws genomic DNA and transformed into the dnf mutant to test for complementation. Figure 3A shows that the DNF transgene restores wild-type flowering to the dnf mutant; this complementation confirms that At3g19140 encodes the DNF gene. As only one line showing full complementation was obtained (the other lines were later flowering than dnf but were not completely restored to wild-type flowering), and as the complementation effect was unstable and frequently lost in subsequent generations, we also recreated the early flowering phenotype of the dnf mutant by downregulating the At3g19140 gene in wild-type plants through RNA interference (RNAi). The whole DNF coding sequence was used for the RNAi construct, as BLAST searches showed that no other Arabidopsis gene has any significant sequence similarity to DNF and, thus, the RNAi construct would target DNF specifically. Several RNAi lines were obtained that all exhibited early flowering to a similar extent as the original dnf mutant (Figure 3B), and this was stable in successive generations. Expression levels of DNF in the two RNAi lines (4 and 10) used in subsequent experiments was shown to be greatly reduced compared with Ws in SDs at ZT5 (see Supplemental Figure 2 online), a time point at which DNF expression levels are known to be high in Ws (see below). The complementation and RNAi results confirm that mutation of the At3g19140 gene results in early flowering.
Figure 3.
Flowering Time of a Complemented dnf Mutant Line, DNF RNAi Plants, and Col Introgression Lines in 8-h SDs.
(A) Flowering times of Ws, the dnf mutant, and a homozygous complemented mutant line (dnf mutant expressing the DNF transgene driven by DNF promoter sequences).
(B) Flowering times of several independent DNF RNAi lines (RNAi of the DNF gene in Ws) compared with the original dnf mutant and Ws plants.
(C) Leaf number at flowering of homozygous progeny from selfed plants derived from four rounds of backcrossing of the dnf mutant into Col. Flowering times of Ws, dnf, and Col are also shown. Error bars show sd; n = 12 to 15 plants.
To show that the early flowering in SD caused by the dnf mutation was not dependent upon the Ws genetic background (as Ws itself is early flowering compared with Columbia [Col] or Landsberg erecta [Ler] ecotypes), the dnf mutation was introgressed into the Col background through four backcrosses. After each backcross, lines containing the T-DNA insertion were selected based upon their resistance to PPT. Following four rounds of backcrossing, PPTR lines were selfed to produce a segregating population containing homozygous mutant lines. The progeny of these selfed lines were screened for flowering time and PPTR. All of the lines that showed 100% PPTR were also early flowering compared with wild-type Col (Figure 3C); these lines were genotyped to confirm that they were homozygous for the dnf mutation. Therefore, the dnf mutation can also cause early flowering in the Col ecotype and is not dependent upon the Ws genetic background.
A search for other mutant alleles of the DNF gene yielded only one line where a T-DNA insertion disrupts the DNF open reading frame (GABI-Kat line 857H08). Plants homozygous for this insertion line did not flower early in SDs as expected. However, this is probably because the position of the insertion is right at the 3′ end of the DNF gene, only 5 bp upstream from the TAG stop codon; thus, it is possible that functional DNF protein could still be produced in these plants. Analysis of DNF transcript levels in this GABI-Kat insertion line showed that DNF transcript levels were unaffected by the insertion and accumulated to the same level in SDs as in wild-type Col plants (see Supplemental Figure 2 online). DNF expression in the dnf mutant is not above background levels.
DNF Is an E3 Ligase
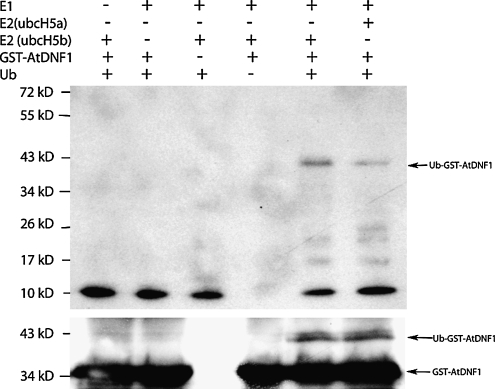
As the DNF protein contains a RING-S/T domain, we tested whether DNF had E3 ligase activity. We expressed and affinity purified DNF without the N-terminal putative signal peptide sequence as a glutathione S-transferase (GST) fusion from Escherichia coli (the complete DNF protein containing this sequence could not be resolubilized from the pellet following extraction). Ubiquitination activity was observed for the purified GST-DNF fusion protein in the presence of yeast E1 and the human E2 Hubc5b and to a lesser extent with human E2 Hubc5a (Figure 4, lanes 5 and 6). This ubiquitination was dependent upon the presence of the E1, E2, and GST-DNF, and the level of activity varied depending upon which E2 was present. DNF primarily directed ubiquitination of one major protein in the E. coli extract, and this was the DNF protein itself. This was shown by probing the immunoblot with a GST antibody, which bound to the GST tag of the expressed DNF protein (Figure 4, bottom panel); thus, DNF has autoubiquitination activity. DNF may also ubiquitinate other plant proteins that are not present in the E. coli extract; the fact that it does not ubiquitinate many E. coli proteins suggests that it may only target specific proteins for ubiquitination. Our findings contrast with those of Stone et al. (2005) who did not detect any ubiquitination activity when assaying the recombinant full-length protein together with a selection of Arabidopsis E2s; UBC8, UBC10, UBC11, UBC35, or UBC36.
Figure 4.
E3 Ubiquitin Ligase Activity of DNF.
GST-DNF was expressed and purified from E. coli and tested for ubiquitination activity in the presence of yeast E1, human E2 (Hubc5b or Hubc5a), and ubiquitin. The immunoblots were probed with anti-Ub antibodies (top panel) to detect ubiquitinated E. coli proteins. Anti-GST antibodies (bottom panel) were used to detect GST-DNF.
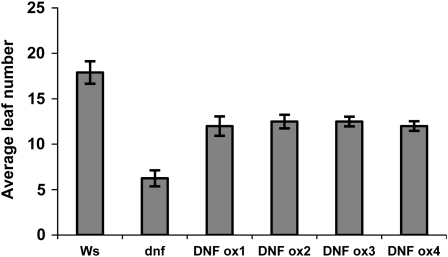
Overexpression of DNF
Downregulation or mutation of DNF causes early flowering in SDs; DNF must therefore be involved in the repression of flowering in SDs. We produced Ws plants overexpressing DNF to see whether this would cause the plants to be delayed in flowering. Interestingly, the overexpressing lines were all early flowering compared with the wild type but not as early as the dnf mutant (Figure 5). This is unlikely to be due to cosuppression, as RNA expression levels in the overexpressing lines were shown to be much higher than Ws (see Supplemental Figure 2 online). A similar observation was reported for overexpression of the floral repressor EBS, where the overexpressers had a similar early flowering phenotype as the ebs mutant (Pineiro et al., 2003). This was thought to be due to the disruption of the formation of complexes necessary for floral induction by either the mutation or by overexpression, which could cause sequestering of other proteins in the complex and prevent formation of fully active complexes.
Figure 5.
Flowering Times of DNF Overexpressers in SD.
Flowering time of 35S:DNF overexpressing lines compared with Ws and the dnf mutant in 8-h SDs. Error bars show sd; n = 12 plants.
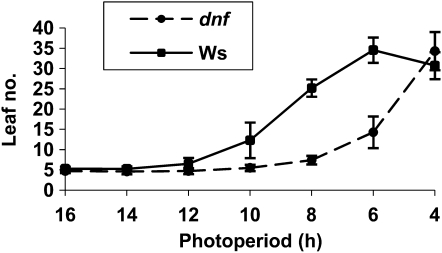
The dnf Mutant Has an Altered Critical Photoperiod
As the dnf mutant has an altered response to photoperiod, we tested whether this is reflected in an altered critical photoperiod for flowering. This was done using small purpose-built light boxes in which the fluorescent lights were timed to come on for 4, 6, 8, 10, 12, 14, or 16 h per day, so that we could define the critical photoperiod for flowering. While wild-type plants showed a delay in flowering time once the daylength was reduced to 10 h or less, flowering of the dnf mutant was only delayed once the daylength was reduced to 6 h or less (Figure 6). In very short photoperiods (4 h), the dnf mutant exhibited a wild-type late-flowering response. Thus, flowering time in the dnf mutant is only accelerated compared with the wild type in short photoperiods of between 4 and 10 h. The accelerated flowering in the mutant compared with the wild type means that DNF must act to repress flowering. The fact that the difference in flowering time between the mutant and wild-type plants is only observed when the daylength is somewhere between 4 and 10 h suggests that DNF only represses flowering between 4 and 10 h after dawn. At or before 4 h, or after 10 h, in the light DNF does not affect flowering because the mutant behaves as the wild type in photoperiods of these lengths.
Figure 6.
Critical Photoperiod of dnf and Ws.
Average leaf number at flowering of Ws and dnf mutant plants grown in photoperiods of different lengths ranging from 4 to 16 h of light. Error bars show sd; n = 12 plants.
DNF Acts in the Same Pathway as CO and GI and Downstream of the Circadian Clock
Defects in photoperception, or circadian timing, are known to affect flowering time (Yanovsky and Kay, 2003) and so the dnf mutant was analyzed for defects in light perception and/or in the function of the circadian clock. Hypocotyl elongation in red, far-red, and blue light was found to be normal (see Supplemental Figure 3 online), indicating that perception of these wavelengths of light is unaffected in the dnf mutant. Mutants that are defective in the perception of these wavelengths of light were included as controls to show that the light treatments used were appropriate to detect such defects in light perception. The circadian clock was analyzed by looking at CAB gene expression in continuous light. The phase of CAB gene expression in the dnf mutant upon transfer from light/dark cycles into continuous light was indistinguishable from the wild type (see Supplemental Figure 4 online). This suggests that the dnf mutation affects neither photoperception pathways nor the clock and that it acts downstream of these processes in the photoperiodic pathway to influence flowering time in SDs.
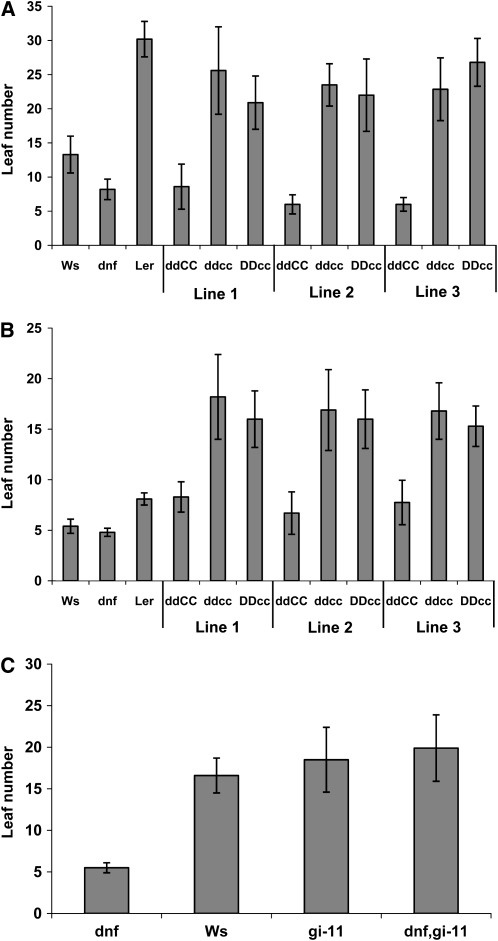
To investigate whether DNF is acting in the same pathway as CO to affect the photoperiodic flowering response, the dnf mutant (Ws) was crossed into the co-2 (Ler) mutant background. Due to technical difficulties, homozygous double mutant lines were not identified until the F4 generation. To make allowance for possible variation in flowering time caused by background flowering quantitative trait loci that would segregate after crossing Ler and Ws, three different homozygous dnf co-2 double mutant lines were analyzed together with their sibling lines that were only homozygous for the co-2 mutation but that carried the wild-type DNF allele. These plants were grown in both LDs and SDs and scored for flowering time. It should be noted that the late flowering phenotype caused by the co mutation is normally only observed in LDs (Putterill et al., 1995), and the effect of the dnf mutation is only observed in SDs. The dnf mutation caused early flowering in SDs in Ler plants that had the wild-type CO allele (ddCC). In SDs (as well as in LDs) the double mutant lines (ddcc), however, flowered as late as their siblings carrying just the co-2 mutation (DDcc) showing that the co-2 mutation is epistatic to the dnf mutation (Figures 7A and 7B). The late flowering of the double mutants in SDs means that functional CO protein is required for the early flowering phenotype of the dnf mutant in SD and, thus, that DNF and CO are acting in the same flowering control pathway.
Figure 7.
Flowering Times of Double Mutants.
(A) and (B) Average leaf number at flowering in SDs (A) and LDs (B) of three different homozygous dnf co-2 double mutant lines (ddcc) compared with their siblings that carry the wild-type DNF allele but are still homozygous for the co-2 mutation (DDcc), and those carrying the wild-type CO allele but homozygous for the dnf mutation (ddCC). Flowering times of Ws, the dnf mutant, and Ler are also shown for comparison. Error bars show sd; n = 15-20 plants.
(C) Average leaf number at flowering of Ws, dnf, gi-11, and dnf gi-11 double mutant plants in SDs. Error bars show sd; n = 10 plants.
The gi-11 mutant, which is in the Ws background, was used to cross with the dnf mutant. The dnf gi-11 homozygous double mutant flowered as late as the gi-11 mutant in SDs (Figure 7C). This shows that GI function is required for the dnf mutation to cause early flowering and that DNF is therefore also acting in the same flowering pathway as GI.
Localization and Expression of DNF
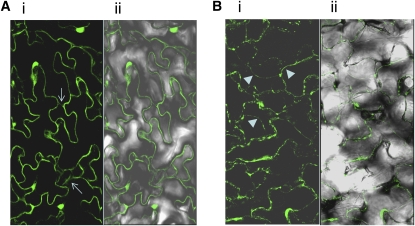
As DNF is predicted to be a type I membrane protein, we investigated the intracellular localization of a DNF–green fluorescent protein (GFP) fusion protein. GFP fluorescence was observed in the plasma membrane of leaf epidermal cells of plants transformed with a 35S:DNF-GFP construct, and there also appears to be evidence of the DNF:GFP protein in endomembrane structures within the cell (Figure 8A). The 4',6-diamidino-2-phenylindole staining indicates that the bright globular structures showing fluorescence are not nuclei but some other cellular compartment. Following plasmolysis of the leaf tissue, the GFP fluorescence is still observed in the plasma membrane, which has become detached from the cell wall (Figure 8B). In this case, the DNF:GFP protein does not seem to be as evenly distributed throughout the membrane as in nonplasmolyzed tissue.
Figure 8.
Intracellular Localization of DNF Protein.
Localization of DNF-GFP fusion protein in leaf epidermal cells of 35S-DNF-GFP plants before (A) and after plasmolysis (B). Panel (i), GFP fluorescence; panel (ii), GFP with transmitted light. Arrows in panel (Ai) indicate possible internal endomembrane structures within the cell containing the DNF:GFP protein. Arrowheads in panel (Bi) show where the plasma membrane has separated from the cell wall.
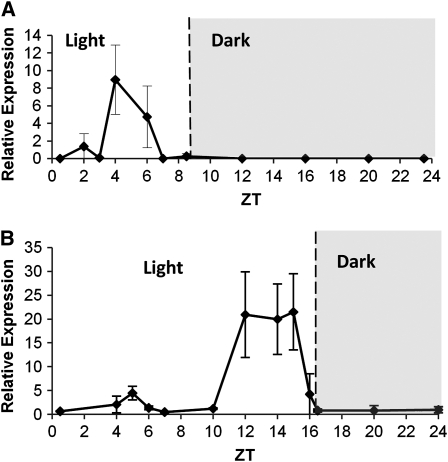
The expression of DNF was examined in wild-type and dnf plants in both SDs and LDs. Expression levels were very low (undetectable by RNA gel blots); therefore, real-time PCR was necessary for quantification. No expression was detectable in dnf mutant plants, indicating that it is probably a null mutation. Expression of DNF in wild-type plants was observed at very precise times of the day. In SDs, expression of DNF was observed in the period 4 to 6 h after dawn (ZT4-ZT6; Figure 9A). Up until ZT3, there is very little expression of DNF and expression levels had fallen to zero again by ZT7, suggesting that there is very tight regulation of its expression. Interestingly, the expression profile of DNF fits nicely with the critical photoperiod data that show that in the first 4 h of the day, there is no difference in flowering response between dnf and wild-type plants; only in SD photoperiods greater than 4 h is a difference in flowering time observed (i.e., just after the point when DNF expression is observed in wild-type plants). In LDs, the expression pattern is very different with a major peak in DNF expression occurring between ZT12 and ZT15 (Figure 9B); there is a minor peak in expression between ZT4 and ZT6 at the same time as in SDs, and the expression levels at this time of day are similar in LDs and SDs (see Supplemental Figure 5A online), but the induction at this time is small in comparison to the later peak. The reason why DNF is expressed so highly in LD when its absence in the dnf mutant has no effect on flowering in LD is unclear. The large second peak in expression at ZT12-ZT15 is not observed in SDs when the plants are in the dark, which indicates that light is required for DNF expression or that DNF expression may be repressed in the dark.
Figure 9.
Expression Pattern of DNF.
(A) Expression of DNF in wild-type plants in 8-h SDs.
(B) Expression of DNF in wild-type plants in 16-h LDs. Expression levels were determined by quantitative RT-PCR and are normalized to β-Actin. Data points represent an average of two experimental replicates each with three technical replicates. Error bars represent sd.
DNF expression was analyzed in different organs of the plant to examine where it is expressed. It was found to be expressed in leaves, stem, roots, and flowers with highest expression in rosette leaves (see Supplemental Figure 5B online). No obvious circadian regulation of DNF expression was observed when Ws plants were sampled for 3 d in continuous light following transfer from SD conditions (see Supplemental Figure 6 online). Diurnal peaks in expression are observed at ZT4 in both the first SD and following the dark period in the first subjective day as expected; however, in continuous light, DNF appears to be deregulated and expressed at continuously high levels.
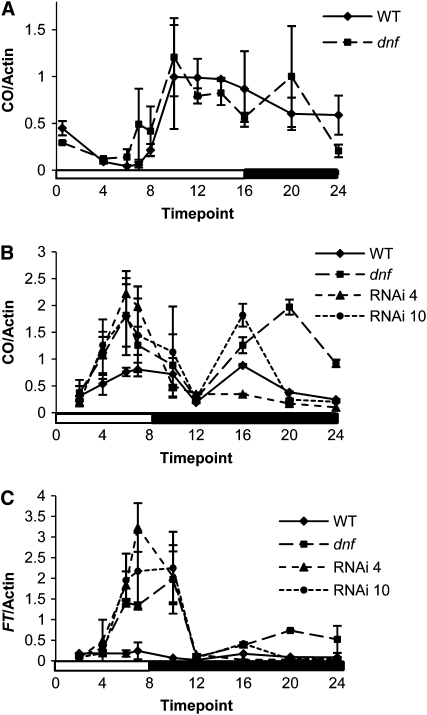
To address the question of how the dnf mutation affects flowering time only in SDs, the expression of CO and FT was analyzed in the dnf mutant compared with the wild type. In the dnf mutant, CO expression is the same as the wild type in LDs (Figure 10A) consistent with the lack of effect of the dnf mutation on flowering in LDs. In SDs, however, the expression of CO is altered such that it starts to rise by 4 h after dawn and is expressed at high levels before the end of an 8-h SD (Figure 10B). The usual nighttime peak of CO expression is also observed. The elevated levels of CO transcript in the light before the end of the SD must result in elevated CO protein levels because induction of FT is also observed before the end of the SD in the dnf mutant. The induction of FT expression follows that of CO and occurs between 4 and 6 h after dawn (Figure 10C). CO and FT expression was also induced in SD in the DNF RNAi lines, and this occurred at the same time as in the dnf mutant, demonstrating that the altered expression pattern of CO and FT is due to the dnf mutation and not due to some second site mutation in the dnf mutant. The induction of CO and, therefore, FT in SD explains the early flowering phenotype of the dnf mutant and the RNAi lines in SDs. The role of DNF must therefore be to prevent the expression of CO during the light period of an SD, thus enabling the plant to prevent flowering and continue vegetative growth in SDs.
Figure 10.
Expression of CO and FT in the dnf Mutant.
(A) Expression of CO in Ws and the dnf mutant in 16-h LDs.
(B) Expression of CO in Ws, dnf, and DNF RNAi lines 4 and 10 in 8-h SDs.
(C) Expression of FT in Ws, dnf, and DNF RNAi lines 4 and 10 in 8-h SDs.
Expression levels were determined by quantitative RT-PCR and are normalized to β-Actin, but as different standard curves were used for the LD and SD analysis, the levels between experiments cannot be compared. White and black bars represent light and dark periods, respectively. Data points represent an average of two experimental replicates each with three technical replicates. Error bars represent sd.
As GI is known to affect the expression of CO, the expression of GI in the dnf mutant was also investigated. The expression of GI in the dnf mutant over a SD (8 h) time course was found to be very similar to its expression in wild-type plants, with expression increasing around ZT4 to peak before the end of the SD before falling to low levels in the dark (see Supplemental Figure 7 online; Fowler et al., 1999). The fact that the dnf mutation causes alterations in both CO and FT expression without significantly affecting the expression of GI indicates that DNF acts upstream of CO but not of GI in the photoperiodic pathway.
DISCUSSION
DNF is a novel flowering time gene that encodes a repressor of flowering; this is demonstrated by the fact that the dnf mutation causes early flowering in SDs when flowering of wild-type plants is normally delayed. In 8-h SD conditions, the dnf mutant flowers as early as the wild type, and dnf plants flower in 16-h LD conditions. The fact that it is induced to flower as much in 8-h photoperiods as it is in 16-h photoperiods indicates that it has lost the repression of flowering normally present in 8-h SDs. The dnf mutant exhibits an altered critical photoperiod, being induced to flower early in photoperiods as short as 6 h compared with the wild type, which requires longer 10-h photoperiods to attain the same level of induction (Figure 6). In 4-h photoperiods, the flowering of dnf is as late as the wild type, and this correlates to the fact that DNF is not expressed before ZT4 (Figure 9A) and therefore there will be no difference between dnf and the wild type up until this time of day. The absence of DNF expression in the dnf mutant from ZT4 onwards results in a lack of inhibition of CO expression; therefore, CO expression starts to increase in the mutant around ZT4 with significant levels of expression by ZT6 (Figure 10B). The high levels of CO expression at ZT6 in the mutant results in the induction of FT at this time; thus, early flowering of the dnf mutant is able to occur in SD photoperiods as short as 6 h.
DNF is expressed between ZT4 and ZT6, and the difference in CO expression between the dnf mutant and wild-type plants is observed between ZT4 and ZT7; DNF must therefore prevent the induction of CO specifically between ZT4 and ZT7. After ZT7, when DNF expression in wild-type plants has fallen to low levels, CO expression is no longer repressed and transcripts start to accumulate as the photoperiod becomes increasingly longer and more inductive. In 16-h LDs, CO expression starts to increase earlier in the dnf mutant than in the wild type, but overall the expression profiles of CO in the dnf mutant and the wild type later on in the day are very similar (Figure 10A). The reason for this may be that other mechanisms (such as the degradation of CDF proteins by FKF1) are also acting to increase CO expression toward the end of an LD, and this may mask the effect of the dnf mutation. This probably explains why there is no significant effect of the loss of the DNF repressor in the dnf mutant on flowering time in LDs. The high level of expression of DNF at the end of an LD is curious given that it does not act to repress CO expression at this time, it may be that an interacting cofactor that is also required for the DNF-mediated repression of CO expression is missing at this time of day.
DNF is thus an important regulator of the rhythm of CO expression, but it is not acting through the GI/FKF1/CDF regulatory mechanism to modulate the amplitude of the rhythm because the effect of the dnf mutation on CO expression in SDs and LDs is different to the constitutively high levels of CO expression observed in the cdf1-R cdf2-1 cdf3-1 cdf5-1 quadruple mutant (cf. Figure 10 and Fornara et al., 2009). Without the DNF-mediated repression of CO transcription between ZT4 and ZT7, the rhythm of CO expression would start to increase after ZT4, and Arabidopsis would not able to distinguish between LDs and SDs (except if the SD was 4 h or less); the specific timing of DNF expression is thus crucial in establishing a photoperiodic flowering response.
The mechanism by which DNF represses CO transcription is unknown, but it could be through the ubiquitin/proteosome degradation pathway. DNF contains a RING-S/T domain, and we have shown that it has E3 ligase activity. DNF may specifically target an activator of CO transcription for degradation at specific times of the day (between ZT4 and ZT7). As the levels of the GI protein, which is known to promote CO expression, have been shown to be high at that time of day (David et al., 2006) DNF cannot be degrading GI, although it could be targeting another transcriptional activator protein that may interact with GI to induce CO expression. DNF is a membrane-bound E3 ligase. The auto-ubiquitination of DNF could be a mechanism by which it recycles and regulates the amount of DNF protein present in the membrane and cytosol; such a mechanism is known to occur in yeast and humans (Platta et al., 2007, 2009).
In summary, we have shown that DNF affects the rhythm of CO expression, particularly between ZT4 and ZT7, and that this regulation is involved in determining the critical photoperiod of the flowering response in Arabidopsis, as without it, Arabidopsis plants flower early even in days with photoperiods as short as 6 h.
METHODS
Plant Growth Conditions
All Arabidopsis thaliana seed, including the T-DNA mutant population and the mutants co-2 (Koornneef et al., 1991), phyA-1 (Whitelam et al., 1993), phyB-1, and cry2 (originally called hy3 and hy4, respectively; Koornneef et al., 1980), was obtained from the Nottingham Arabidopsis Stock Centre (NASC). This is apart from the GABI-Kat line 857H08, which was obtained from Bernd Weisshaar at Bielefeld University, Germany, and the gi-11 mutant (Richardson et al., 1998), which was obtained from Jo Putterill, University of Aukland, New Zealand.
Unless otherwise stated, plants were grown in Levingtons F2 compost containing six parts compost, one part sand, and one part vermiculite. Seeds were stratified in the dark at 4°C for 2 d to achieve uniform germination before being transferred to Sanyo MLR-350 growth cabinets and grown at 22°C in either SDs or LDs. SDs consisted of 8 h white light (100 μmol m−2 s−1) followed by 16 h darkness; LDs consisted of 16 h of white light followed by 8 h darkness. Lighting was supplied by BriteGro F36WT8 fluorescent lamps (Sylvania). Critical photoperiod experiments were performed in small purpose-built light boxes when the fluorescent lights (50 μmol m−2 s−1) were timed to come on for 4, 6, 8, 10, 12, 14, or 16 h per day. Flowering time was scored as the number of rosette leaves when the plant had developed a bolt of 1 cm. The variation in flowering times observed between different experiments is probably due to the growth cabinets not maintaining exactly the same temperatures; the variation observed within an experiment is much less.
Hypocotyl Elongation Assay
Seeds were sterilized in 20% bleach, washed five times in sterile water, and then pipetted onto 0.7% agarose plates. The plates were then transferred to a Percival growth cabinet (CLF plant Climatics model 1-3LEDDLL3). The seedlings were grown for 4 d at 22°C under continuous single fluence light provided by LEDs, red (2.5 μmol m−2 s−1), far red (0.1 μmol m−2 s−1), blue (0.4 μmol m−2 s−1), irradiances were measured using an EPP2000 fiber optic spectrometer (StellarNet UK). Seedlings were also grown in the dark as a control. The length of the hypocotyl was then measured.
Complementation of the dnf Mutant
Primers O3p5 and O3p9 (see Supplemental Table 1 online) were used to PCR a 1.6-kb fragment consisting of the full-length wild-type allele of the DNF gene plus 1.1 kb of upstream sequence from Ws genomic DNA using KOD Hot Start proofreading DNA polymerase (Novagen). This was cloned into the SmaI site of pUC 18, sequence verified, and then subcloned into pGVPT hygromycin transformation vector (Becker et al., 1992) using the HindIII and SstI sites. This construct was electroporated into Agrobacterium tumefaciens GV3101, and dnf mutant plants were transformed by the floral dip method (Clough and Bent, 1998). Transformed seed were selected on hygromycin plates (20 μg/mL).
Overexpression and RNAi of DNF
The coding sequence of the DNF gene was amplified by PCR using O3attB1 and O3attB2 primers (see Supplemental Table 1 online) and KOD DNA polymerase. The fragment obtained was cloned into the Gateway pDONR 207 vector using BP clonase and sequence verified. The insert was then transferred into the Gateway CaMV 35S overexpression vector pB2GW7 and the RNAi vector pB7GWIWG2 (http://www.psb.ugent.be/gateway/index.php) by the LR reaction. These constructs were transformed into GV3101 and then by floral dip into wild-type Ws plants. Transformed plants were selected by spraying young plants with BASTA (0.02% Challenge; BAYER).
GFP Constructs
The cauliflower mosaic virus 35S and DNF promoters (P35S and PDNF, respectively), the DNF coding sequence (without the stop codon), and the GFP coding sequence were PCR amplified using the following primers (for = forward; rev = reverse): P35Sfor, P35Srev, PDNFfor, PDNFrev, DNFfor1, DNFrev1, EGFPfor, and EGFPrev (see Supplemental Table 1 online).
The PCR fragments were subcloned into pBluescript vector using the restriction sites present in the primer sequences. The fragments were sequence verified. The GFP coding sequence fragment was subcloned behind the DNF coding sequence, and the cauliflower mosaic virus 35S or DNF promoter fragments were cloned in front of the DNF-GFP fusion protein sequence. The whole promoter fusion protein sequence was then subcloned into the BIB-HYG transformation vector using the HindIII and SacI restriction sites. The constructs were transformed into wild-type Ws plants by floral dip and transformants selected on hygromycin plates (20 μg/mL).
Expression Analysis
Quantitative real-time PCR was used to detect the levels of CO, FT, GI, CAB, and DNF mRNA abundance. Plants were grown in either SDs or LDs, and samples from four plants were harvested at the 4/5 leaf stage and pooled for each RNA extraction. Five micrograms of total RNA was DNase treated with 1 μL DNase (Roche) and made up to a total of 9 μL with MilliQ water. The RNA samples were then incubated at 37°C for 1 h before inactivating the DNase at 75°C for 10 min. The RNA samples were then used to synthesize cDNA using the Super Script first-strand synthesis system for RT-PCR (Invitrogen) following the manufacturer's instructions. Real-time PCR assays were performed using a Taqman machine (ABI Prism 7900HT; Applied Biosystems). Each reaction contained 0.4 μM of the forward and reverse primers (see below), 6 μL of diethylpyrocarbonate-treated water, and 7.5 μL of Applied Biosystems SYBR GREEN PCR 2× Master Mix, with the exception of DNF primers, where the concentration was reduced to 0.2 μM. Triplicate reactions were run for each sample.
The cycling parameters consisted of 95°C for 10 min, followed by 50 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 1 min. The raw data were analyzed using the default settings of the software for determining both the threshold value and baseline. In each assay, a standard curve for the primer set was generated using 10-fold serial dilutions of a cDNA sample where expression was known or expected to be high. Reactions were optimized so that efficiencies were equal to 100% ± 10%. Melt curve analyses were performed to show that only a single product was being amplified in each reaction. ABI prism software version SDS2.1 was used to analyze the assay results. Duplicate RNA samples were assayed for each time point (i.e., eight leaf samples per time point), and each real-time PCR assay for each RNA sample had three technical replicates. The expression levels of β-Actin were used to normalize the expression of the target genes between samples.
Primer sequences were DNFfor2, DNFrev2, Actinfor, Actinrev, FTfor, FTrev, COfor, COrev, GIfor, GIrev, CABfor, and CABrev (see Supplemental Table 1 online).
Ubiquitination Assay
A DNF clone lacking the first 39 N-terminal amino acids containing the putative signal peptide sequence was cloned into the pGEX-4T-1 vector (Amersham Pharmacia Biotech) to produce an in-frame fusion with the GST tag. All recombinant fusion proteins were retained mostly in the insoluble fraction of Escherichia coli strain BL21 (DE3) pLysS; the insoluble fraction was solubilized and dialyzed according to the protein refolding kit (Novagen), and the soluble protein was used for in vitro ubiquitination assays.
In vitro ubiquitination assays were performed as described previously (Hardtke et al., 2002). Each reaction (50 μL final volume) contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM ATP, 0.2 mM DTT, 10 mM phosphocreatine, 0.1 unit of creatine kinase (Sigma-Aldrich), 2 μg purified His-ubiquitin, 50 ng of yeast E1 (Biomol), 150 ng E2 UbcH5b or UbcH5a (Biomol), and 1 μg of refolded GST-DNF. The reactions were incubated at 37°C for 3 h and stopped by adding 4× SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 20% [v/v] glycerin, 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) at 100°C for 5 min and analyzed by SDS-PAGE electrophoresis followed by immunoblotting.
Immunoblotting
Immunoblots were performed with mouse monoclonal anti-Ub antibodies (Roche) and rabbit anti-GST antibodies (Novagen). The primary antibodies were used at 1:5000 dilution, and the secondary horseradish peroxidase–conjugated secondary antibodies were used at a 1:20,000 dilution. Amersham ECL-plus protein gel blotting chemiluminescence detection kits were used to detect levels of horseradish peroxidase and develop the blots on light-sensitive autoradiograph films.
Confocal Microscopy
Sections of Arabidopsis leaves were mounted for microscopy observation in water under glass cover slips. Plasmolyzed leaf samples were prepared by immersing them in 0.8 M mannitol for 20 min. The leaves were examined using an Olympus confocal fluoview IX70 laser microscope. The argon laser excitation wavelength was 488 nm, and EGFP emission was detected with the filter set for fluorescein isothiocyanate (505 to 530 nm). The fluorescence of the images was assessed using the Olympus fluoview software.
Mutant Crosses and Introgression
The dnf mutant was always used as the male parent in the crosses so that F1 progeny from successful crosses could be selected for on their resistance to PPT. For introgression of the dnf mutation into Col, progeny from the cross and from each of the subsequent rounds of backcrossing were selected for PPT resistance. After four rounds of back crossing, PPTR plants were selfed and lines homozygous for the dnf mutation were selected.
Genotyping the dnf mutation was done in a single PCR reaction using three PCR primers: DNFF and DNFR designed to the DNF gene each side of the T-DNA insertion site in the dnf mutant, and the RBR primer designed to the right border of the T-DNA (see Supplemental Table 1 online).
DNFF and DNFR amplify a fragment 178 bp from the DNF gene that does not contain the T-DNA insertion, whereas DNFR and RBR amplify a fragment 482 bp from the mutated dnf gene containing the T-DNA insertion; the size of the T-DNA insertion prevents amplification from DNFF and DNFR primers in the mutant. Homozygous dnf mutant lines only produce the 482-bp fragment, homozygous DNF lines only the 178-bp fragment, while heterozygous T-DNA lines will amplify both fragments.
Genotyping the co-2 mutation was done by PCR amplifying the region containing the position of the single base change (Putterill et al., 1995) (using primers CO-Span 2F and CO-Span 2R) and sequencing the fragments obtained. Plants homozygous for the co-2 mutation possess an A at that position, whereas wild-type plants posses a G, and heterozygous plants have a mix of G and A at that position.
Genotyping the gi-11 mutation was also done by PCR using primers designed to the 5′ deleted region of the GI gene in the gi-11 mutant (Fowler et al., 1999): GI-For6 and GI-Rev5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3G19140 (DNF), At3G18780 (β-Actin), At5G15840 (CO), At1G22770 (GI), and At1G29930 (CAB).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Rate of Leaf Production in dnf and Ws Plants.
Supplemental Figure 2. DNF Expression in RNAi and Overexpressing Lines, and the GABI-Kat Insertion Line in 8-h SDs.
Supplemental Figure 3. Hypocotyl Elongation of Ws and dnf Mutant Plants in Different Light Qualities.
Supplemental Figure 4. Analysis of Circadian CAB Expression.
Supplemental Figure 5. DNF Expression at ZT5 in LDs and SDs and DNF Expression at ZT5 in SDs in Different Tissues.
Supplemental Figure 6. DNF Expression after Transfer from One SD to Continuous Light.
Supplemental Figure 7. GI Expression in dnf and Ws Plants.
Supplemental Table 1. Primers Used in This Study.
Acknowledgments
We thank the Station de Génétique et d'Amélioration des Plantes, Institut National de la Recherche Agronomique, and the NASC for the production and distribution of the T-DNA mutant population. We also thank Linda Brown for technical assistance and Jo Putterill for the gi-11 seed. This work was funded by the Biotechnology and Biological Science Research Council.
References
- Becker D., Kemper E., Schell J., Masterson R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bienz M. (2006). The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31: 35–40 [DOI] [PubMed] [Google Scholar]
- Chen M., Ni M. (2006). RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J. 46: 823–833 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- David K.M., Armbruster U., Tama N., Putterill J. (2006). Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 580: 1193–1197 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C.S., Gissot L., Sauerbrunn N., Ruhl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Okamoto H., Stoop-Myer C., Deng X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30: 385–394 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Kay S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Rolff E., Spruit C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100: 147–160 [Google Scholar]
- Mathieu J., Warthmann N., Kuttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Michaels S.D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wright L., Fujiwara S., Cremer F., Lee K., Onouchi H., Mouradov A., Fowler S., Kamada H., Putterill J., Coupland G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H., Igeno M.I., Perilleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro M., Gomez-Mena C., Schaffer R., Martinez-Zapater J.M., Coupland G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15: 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Baumer B.E., Schlee D., Girzalsky W., Erdmann R. (2009). Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein inport. Mol. Cell. Biol. 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., Erdmann R. (2007). Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Richardson K., Fowler S., Pullen C., Skelton C., Morris B., Putterill J. (1998). T-DNA tagging of a flowering time gene and improved gene transfer by in planta transformation of Arabidopsis. Aust. J. Plant Physiol. 25: 125–130 [Google Scholar]
- Roden L.C., Song H.R., Jackson S., Morris K., Carre I.A. (2002). Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 13313–13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz T.F., Kiyosue T., Yanovsky M., Wada M., Kay S.A. (2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Schultz T.F., Milnamow M., Kay S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Hauksdottir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suàrez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- von Heijen G. (1988). Transcending the impenetrable: how proteins come to terms with membranes. Biochim. Biophys. Acta 947: 307–333 [DOI] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.C., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2003). Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 4: 265–275 [DOI] [PubMed] [Google Scholar]
- Zeevaart J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27: 321–348 [Google Scholar]