The Arabidopsis histidine kinase CKI1 is essential for female gametogenesis and is able to activate cytokinin signaling by unknown mechanisms. This study shows that CKI1 acts upstream of HISTIDINE PHOSPHOTRANSFER PROTEINS to regulate downstream signaling events in a cytokinin receptor-independent manner and demonstrates that CKI1-AHP signaling is essential for plant growth and development.
Abstract
Cytokinin signaling is mediated by a multiple-step phosphorelay. Key components of the phosphorelay consist of the histidine kinase (HK)-type receptors, histidine phosphotransfer proteins (HP), and response regulators (RRs). Whereas overexpression of a nonreceptor-type HK gene CYTOKININ-INDEPENDENT1 (CKI1) activates cytokinin signaling by an unknown mechanism, mutations in CKI1 cause female gametophytic lethality. However, the function of CKI1 in cytokinin signaling remains unclear. Here, we characterize a mutant allele, cki1-8, that can be transmitted through female gametophytes with low frequency (∼0.17%). We have recovered viable homozygous cki1-8 mutant plants that grow larger than wild-type plants, show defective megagametogenesis and rarely set enlarged seeds. We found that CKI1 acts upstream of AHP (Arabidopsis HP) genes, independently of cytokinin receptor genes. Consistently, an ahp1,2-2,3,4,5 quintuple mutant, which contains an ahp2-2 null mutant allele, exhibits severe defects in megagametogenesis, with a transmission efficiency of <3.45% through female gametophytes. Rarely recovered ahp1,2-2,3,4,5 quintuple mutants are seedling lethal. Finally, the female gametophytic lethal phenotype of cki1-5 (a null mutant) can be partially rescued by IPT8 or ARR1 (a type-B Arabidopsis RR) driven by a CKI1 promoter. These results define a genetic pathway consisting of CKI1, AHPs, and type-B ARRs in the regulation of female gametophyte development and vegetative growth.
INTRODUCTION
The plant phytohormone cytokinin is a key growth regulator involved in the regulation of a wide range of developmental processes, including root and shoot growth, photomorphogenesis, flowering timing, senescence, and seed development (Davies, 1995; Mok and Mok, 2001). It is generally believed that cytokinin executes these physiological activities by regulating cell division and cell differentiation through a two-component system (TCS) signaling pathway (Hwang et al., 2002; Heyl and Schmülling, 2003; Kakimoto, 2003; Ferreira and Kieber, 2005; Müller and Sheen, 2007). Key components of the TCS have been well defined by genetic and biochemical studies. In Arabidopsis thaliana, upon binding of cytokinin, three homologous Arabidopsis histidine kinases (AHK2, 3, and 4; AHK4 is also known as CYTOKININ RESPONSE1 [CRE1] or WOODEN LEG [WOL]) are activated by autophosphorylation at a conserved His residue. Subsequently, the phosphoryl group is transferred to Arabidopsis histidine phosphotransfer proteins (AHP1 through AHP5). AHP proteins were suggested to localize in the cytoplasm in the absence of cytokinin and, upon treatment with cytokinin, to translocate into the nucleus (Hwang and Sheen, 2001). However, a recent study demonstrated that AHP proteins maintained a constant nuclear/cytosolic distribution by balancing active transport and that this subcellular pattern was independent of cytokinin signaling (Punwani et al., 2010). Nevertheless, the nuclear-localized and phosphorylated AHPs presumably activate a class of MYB transcription factors, designated as type-B Arabidopsis response regulators (ARR). These type-B ARRs directly activate expression of type-A ARR genes that, in turn, negatively regulate type-B ARRs (Hwang et al., 2002; Heyl and Schmülling, 2003; Kakimoto, 2003; Ferreira and Kieber, 2005; Müller and Sheen, 2007; Punwani et al., 2010). CRE1 is a bifunctional protein exerting kinase or phosphatase activities on AHPs in the presence or absence of cytokinin, respectively (Mähönen et al., 2006a). In addition, a pseudophosphotransfer protein, AHP6, was characterized as a negative regulator of cytokinin signaling (Mähönen et al., 2006b). This TCS-based phosphorelay establishes the framework of cytokinin signaling.
Mutations in the cytokinin receptor genes (ahk2,3,4 triple mutant) (Inoue et al., 2001; Yamada et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006) and AHP genes (ahp1,2,3,4,5 quintuple mutant) (Suzuki et al., 2002; Hutchison et al., 2006) cause a pleiotropic phenotype, including abolished sensitivity to cytokinins, a reduced meristematic activity, severely inhibited growth of roots and shoots, as well as impaired reproductive development with reduced fertility. However, given the critical role of cytokinin in plant growth and development, it is somewhat unexpected that ahk2,3,4 triple mutants have little, if any, effects on gametogenesis and embryogenesis (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). In addition, an ahp1,2,3,4,5 quintuple mutant can set seeds with reduced fertility (Hutchison et al., 2006), further challenging a possibly essential role of cytokinin signaling in plant growth and development.
In addition to the three cytokinin receptors, an additional AHK gene, CYTOKININ INDEPENDENT1 (CKI1), has been implicated in cytokinin signaling (Kakimoto, 1996). CKI1 was identified as a positive regulator that was capable of promoting shoot regeneration from hypocotyl explants in the absence of cytokinin and therefore was proposed to act in the early steps of cytokinin signaling (Kakimoto, 1996). The observation that CKI1 induces a typical cytokinin response (Kakimoto, 1996; Hwang and Sheen, 2001; Zheng et al., 2006) and that CKI1 is able to dephosphorylate AHP1 and AHP2 (Nakamura et al., 1999) or phosphorylate AHP1, 2, 3, and 5 (Mähönen et al., 2006a) implies that CKI1 plays an important role in cytokinin signaling. In addition, CKI1 has been found to interact with AHP2 and AHP3 in a yeast two-hybrid assay (Urao et al., 2000). However, CKI1 is incapable of binding to cytokinins (Yamada et al., 2001), suggesting that CKI1 is unlikely to act as a receptor. Loss-of-function mutations in CKI1 impair megagametogenesis, indicating an essential role in female gametophyte development (Pischke et al., 2002; Hejátko et al., 2003). Analysis of CKI1 RNA interference transgenic plants suggests that a reduced CKI1 expression level causes defects in procambial cell maintenance in shoots (Hejátko et al., 2009). In spite of these progresses, the role of CKI1 in cytokinin signaling has long been questioned, largely owing to the lack of genetic evidence to link CKI1 to the TCS components. Given the essential role of CKI1 in female gametogenesis, it remains unclear if cytokinin signaling is directly involved in or is required for female gametophyte development. In addition, because the cki1 mutant exhibits female gametophytic lethality, the functions of CKI1 during postgametophytic growth and development remain unknown.
In this study, we present evidence that CKI1-activated signaling is dependent on AHP genes and that this regulatory scheme is essential for female gametophyte development. Moreover, CKI1 also plays an important role during vegetative growth by regulating cell expansion. We demonstrate that the CKI1-AHP–dependent activity in female gametophyte development can be functionally substituted by the activation of cytokinin receptor-mediated signaling events. These results reveal an essential role of TCS-mediated signaling in plant growth and development.
RESULTS
Characterization of cki1-7 and cki1-8 Mutants
We previously reported the partial characterization of two mutants, cki1-7 and cki1-8 (Zheng et al., 2006). In these two mutants, an estradiol-inducible activation tagging vector was inserted 994 bp (cki1-7) and 498 bp (cki1-8) upstream of the putative translation start codon of CKI1, respectively (Figure 1A). In these two mutants, expression of CKI1 was highly inducible by estradiol in a dose-dependent manner (see Supplemental Figure 1A online), which caused a typical cytokinin response, including the inhibition of shoot growth and primary root elongation, delayed leaf senescence induced by dark, and the induction of the cytokinin primary response genes ARR6 and ARR7 (see Supplemental Figures 1B to 1E online). The phenotypic strength is correlated with the CKI1 expression levels induced by different concentrations of estradiol (see Supplemental Figures 1B and 1C online), mimicking wild-type plants treated with different concentrations of cytokinin. These gain-of-function mutant phenotypes could be recapitulated by overexpression of a CKI1 or a CKI1-green fluorescent protein (GFP) transgene in wild-type plants, and the transgenic phenotype was correlated with the accumulation of CKI1-GFP fusion protein induced by estradiol (see Supplemental Figure 2 online).
Figure 1.
Expression of CKI1 in Wild-Type and cki1-8 Mutant Plants.
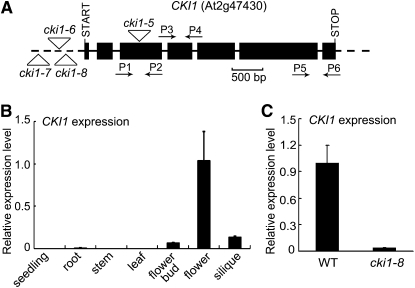
(A) A schematic map of the CKI1 gene and cki1 mutant alleles. Filled boxes and solid lines denote exons and introns, respectively. Untranscribed and untranslated regions are indicated as dashed lines. Putative translation start and stop sites, and positions of the T-DNA insertions in cki1 mutant alleles are shown. Relative positions and orientation of primers (P1 through P6; see Supplemental Table 2 online for sequences) used for qRT-PCR are shown.
(B) Analysis of expression of CKI1 in different organs and tissues of wild-type plants by qRT-PCR using primers P3 and P4 shown in (A). Data presented are mean values of three biological repeats with standard deviations. Relative expression level of CKI1 was normalized using ACTIN7 (At5g09810) as an internal control.
(C) CKI1 expression in wild-type and cki1-8 flowers analyzed by qRT-PCR as described in (B).
In addition to the gain-of-function mutant phenotype induced by estradiol as highlighted above, the T-DNA insertion in cki1-8 also caused severe abnormalities owing to the loss-of-function mutation (see below). Hereafter, we refer to mutant plants treated with estradiol as CKI1-OXi (for CKI1 overexpression inducible). Most overexpression experiments described below were performed using the cki1-7 mutant allele unless otherwise specified. On the other hand, plants grown under noninductive conditions are referred to as cki1-7 and cki1-8, which represent loss-of-function mutants by T-DNA insertional mutations.
Under normal growth conditions, cki1-7 plants had no detectable abnormalities. The position of the T-DNA insertion in cki1-8 was similar to that in cki1-6, which carried a T-DNA 587 bp upstream from the translation start codon and showed a female gametophytic lethal phenotype (Pischke et al., 2002). Consistent with this result, our initial attempts failed to recover cki1-8 plants homozygous for the T-DNA insertion. To analyze the cki1-8 phenotype more precisely, we performed reciprocal crosses between CKI1/cki1-8 and wild-type plants. In CKI1/cki1-8 plants, the transmission efficiency of the T-DNA insertion was 48.76% through male gametophytes (n = 252), suggesting that male gametogenesis was not affected by the cki1-8 mutation. However, the T-DNA insertion was transmitted through female gametophytes at an extremely low frequency in CKI1/cki1-8 plants (0.17%; n = 596). Note that cki1-6, which carried a T-DNA at a more distal location in the CKI1 promoter than cki1-8 (Figure 1A), did not appear to be capable of being transmitted through female gametophytes. This difference is most likely due to different sizes of F1 populations assayed (596 in cki1-8 and 173 in cki1-6) (Pischke et al., 2002). These results suggest that cki1-8 is not a null allele, and cki1-8 homozygous plants indeed showed residual expression of CKI1 (see below). Similar to the observation made in other cki1 mutant alleles (Pischke et al., 2002; Hejátko et al., 2003), confocal laser scanning microscopy (CLSM) revealed that ∼50% of ovules in CKI1/cki1-8 siliques showed various defects, including collapsed, degenerate, or multinucleated female gametophytes (see Supplemental Figure 3 online). Collectively, these results indicate that the cki1-8 mutation impairs female gametophyte development.
In correlation with the cki1 mutant phenotype, a quantitative RT-PCR (qRT-PCR) analysis revealed that CKI1 is predominantly expressed in flowers, followed by flower buds and siliques (Figure 1B). Expression of CKI1 was barely detectable in vegetative tissues by qRT-PCR (Figure 1B), although a recent study revealed CKI1 expression in specific tissues of stem vascular bundles (Hejátko et al., 2009). Whereas our qRT-PCR results are consistent with previous studies by in situ hybridization and reporter gene analyses (Pischke et al., 2002; Hejátko et al., 2003), we could not exclude the possibility that CKI1 is expressed in certain vegetative tissues at a lower level. To test this possibility, we employed a highly sensitive method to analyze CKI1 expression in wild-type seedlings. Poly(A) RNA was isolated and then used as a template for RT-PCR. Whereas expression of WUSCHEL (WUS), a meristem preferentially expressed gene (Mayer et al., 1998), was easily detected under the assay conditions, expression of CKI1 was undetectable (see Supplemental Figure 4 online). The RT-PCR products were then subjected to DNA gel blot analysis using a CKI1 cDNA fragment as a probe. Using a CKI1 cDNA clone as a positive control in the PCR–DNA gel blot analysis, we found that this method could detect 1 fmol CKI1 cDNA fragment added to the PCR reaction. However, no CKI1 expression could be detected in wild-type seedlings under the assay conditions (see Supplemental Figure 4 online), suggesting that the CKI1 expression level in vegetative tissues is under the detection limit of the current method.
Identification and Characterization of the cki1-8 Homozygous Mutant
Because the cki1-8 mutation can be transmitted through female gametophytes at low frequency, it should be possible to recover cki1-8 plants. Because of its defective female gametophyte development, a cki1-8 plant is expected to set few or no seeds. Therefore, we screened >1000 individual plants derived from self-pollinated CKI1/cki1-8 plants by visual inspection, from which we recovered a plant with significantly reduced fertility. Genotyping revealed that this plant was indeed homozygous for the T-DNA insertion at the CKI1 locus. We have been able to harvest ∼15 seeds from this plant, from which we subsequently amplified the cki1-8 mutant over several generations and have collected several thousand seeds for the experiments described below. Typically, a cki1-8 plant produced ∼10 to 40 seeds.
To evaluate the strength of the cki1-8 mutation, we analyzed CKI1 expression in wild-type and cki1-8 plants by qRT-PCR and in situ RNA hybridization. Compared with expression in the wild type, expression of CKI1 was 20-fold lower in cki1-8 flowers, as revealed by qRT-PCR (Figure 1C). In mature female gametophytes, CKI1 expression is confined mainly to the central cell nucleus and, to a lower level, to the egg cell nucleus (Pischke et al., 2002; Hejátko et al., 2003). However, we were unable to detect a similar signal by in situ RNA hybridization in cki1-8 ovules (see Supplemental Figures 5A to 5C online), indicating a lower CKI1 mRNA level in the mutant female gametophytes than in the wild-type gametophytes. These results indicate that cki1-8 is a weak mutant allele with residual CKI1 expression.
The cki1-8 Mutant Phenotype during Vegetative Growth
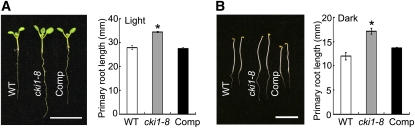
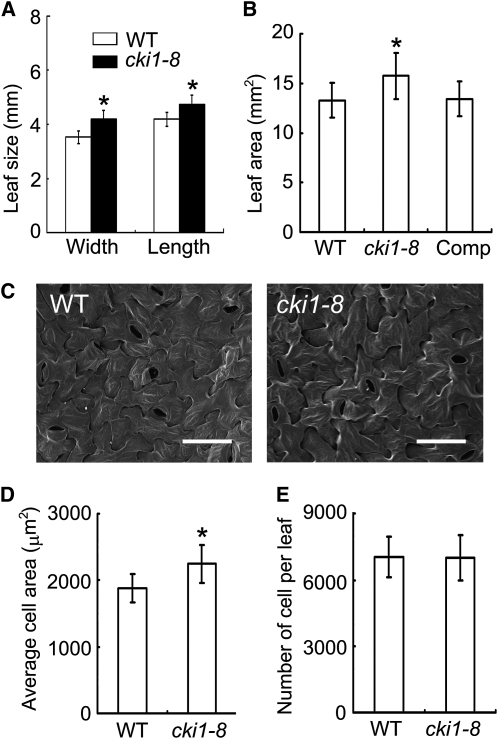
When germinated and grown under both continuous white light and in the dark, cki1-8 plants were larger than wild-type plants (Figures 2A and 2B). Quantitative analysis showed that cki1-8 had longer primary roots than wild-type seedlings under both the light and dark conditions (Figures 2A and 2B). True leaves were initiated in cki1-8 at a similar rate and timing as in wild-type seedlings. However, true leaves of cki1-8 plants were larger than those of wild-type plants, both in length and width (Figures 3A and 3B).
Figure 2.
The cki1-8 Mutant Phenotype.
(A) Left: Seven-day-old seedlings of the wild type, cki1-8, and cki1-8 carrying a CKI1:CKI1 transgene (Comp) germinated and grown under continuous white light. Right: Quantitative analysis of the primary root length of 7-d-old seedlings.
(B) Left: Five-day-old seedlings of wild-type, cki1-8, and cki1-8 plants carrying a CKI1:CKI1 transgene (Comp) germinated and grown in the dark for 5 d. Right: Quantitative analysis of the primary root length of 5-d-old seedlings.
Bar = 10 mm. Data presented are mean values of three independent experiments (n > 20 in each experiment) and standard deviations are given in the graphs. Asterisk indicates statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
[See online article for color version of this figure.]
Figure 3.
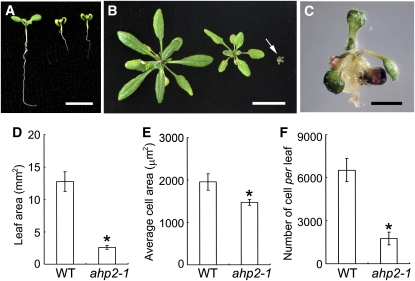
The Cellular Phenotype of the cki1-8 Leaves.
(A) Size comparison of wild-type and cki1-8 leaves. Data presented are mean values of two independent experiments (n > 20 in each experiment) and standard deviations are indicated by bars.
(B) The leaf blade area (in mm2) of wild-type and cki1-8 leaves. Data presented are mean values of two independent experiments (n > 15; collected from different seedlings), and standard deviations are indicated by bars.
(C) Scanning electron microscopy analysis of wild-type and cki1-8 leaves. Images show abaxial leaf surfaces. Bar = 50 μm.
(D) Average size of abaxial epidermal cells. Data presented are mean values of two independent experiments (nine and five leaves collected from different seedlings were measured in two experiments, respectively), and standard deviations are indicated by bars.
(E) The number of abaxial epidermal cells. Data presented are calculated by dividing leaf area (shown in [B]) with the average cell size (shown in [D]). Standard deviations are indicated by bars.
All data presented were obtained using the second true leaves derived from wild-type, cki1-8, and cki1-8 seedlings carrying a CKI1:CKI1 transgene (Comp; [B] only) germinated and grown under continuous white light for 14 d. Asterisks in (A), (B), and (D) indicate statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
To explore the cellular basis of the enlarged cki1-8 leaves, we analyzed the cell size and the number of cells in the mutant in comparison with wild-type leaves. Cells in cki1-8 leaves were larger than those in wild-type leaves (Figures 3C and 3D). However, similar cell numbers were observed in cki1-8 and wild-type leaves (Figure 3E). These results suggest that CKI1 may negatively regulate cell expansion in leaves. Note that our RT-PCR–DNA gel blot analysis was unable to detect CKI1 expression in wild-type seedlings (see Supplemental Figure 4 online). Whereas CKI1 may be expressed at an extremely low level in vegetative tissues, an alternative explanation could be that CKI1 protein synthesized in embryos and seeds, rather than CKI1 mRNA, remained active for a specific window during postgerminative growth.
A recent study revealed that CKI1 RNA interference transgenic plants showed defects in shoot procambial cell maintenance (Hejátko et al., 2009). In serial sections prepared from stems of cki1-8 plants (n = 5), vascular development appeared to be relatively normal compared with that of wild-type shoots (see Supplemental Figure 6 online; see also below). However, we could not exclude the possibility that analysis of additional samples or under certain assay conditions would be able to reveal specific phenotypes in cki1-8 during shoot vascular development. On the other hand, transgenic plants expressing an antisense CKI1 transgene were reported to show retarded vegetative growth under nutrition-restricted conditions (Glover et al., 2008). Under our assay conditions, no phenotype similar to that of the CKI1 antisense transgenic lines was observed in cki1-8 plants under both normal and nutrition-restricted conditions (Figures 2A and 2B; see Supplemental Figure 7 online).
The cki1-8 Mutant Phenotype during Reproductive Development
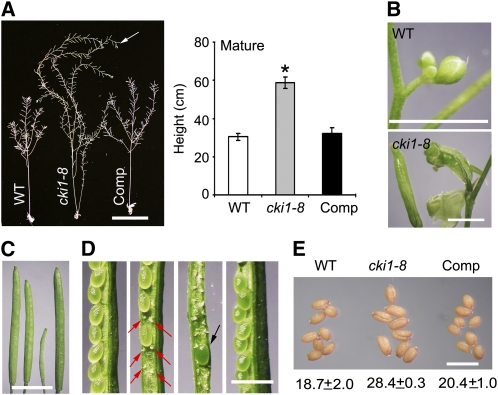
The cki1-8 plants could produce normal-looking flowers, of which both the number and the identity of the floral organs appeared to be normal (see Supplemental Figure 8A online). Scanning electron microscopy revealed that the initiation and development of the inflorescence meristem are normal (see Supplemental Figure 8B online). However, when the floral inflorescence meristem became inactive and flower production was terminated in wild-type plants, cki1-8 plants continued to grow and produced new flowers or flower-like structures for an additional 3 to 5 weeks, leading to a substantially increased size of the mutant plant (Figure 4A). Flowers at the inflorescence tips were often transformed into carpel-like structures (Figure 4B). It has been proposed that a signal derived from developing fruits triggers the arrest of proliferative activity at the inflorescence meristems, thereby ceasing flower production on all inflorescence branches. This global proliferative arrest of the inflorescence meristem does not occur in the male-sterile1 mutant or in wild-type plants when siliques are manually removed. The lack of the global proliferative arrest consequently results in an enlarged plant size and the formation of carpel-like structures in these plants (Hensel et al., 1994). Likewise, the cki1-8 mutant phenotype during the flowering stage is attributed to a similar mechanism, owing to the formation of abortive fruits (see below).
Figure 4.
The Reproductive Development Phenotype of the cki1-8 Mutant.
(A) Left: Ten-week-old plants of the wild type, cki1-8, and cki1-8 carrying a CKI1:CKI1 transgene (Comp) germinated and grown under continuous white light. Note that cki1-8 continues to produce flowers (indicated by an arrow). Bar = 10 cm. Right: Quantitative analysis of the height of 10-week-old plants (n = 8). Standard deviations are indicated by bars. Asterisk indicates statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
(B) Inflorescences derived from 8-week-old (wild type) or 10-week-old (cki1-8) plants. Bar = 2 mm.
(C) Siliques (from left to right) of the wild type, CKI1/cki1-8, cki1-8, and cki1-8 carrying a CKI1:CKI1 transgene. Bar = 5 mm.
(D) Seed development in siliques. From left to right: wild type, CKI1/cki1-8 (red arrows show abortive ovules), cki1-8 (a rarely set seed indicated by a black arrow), and cki1-8 carrying a CKI1:CKI1 transgene. Bar = 1 mm.
(E) Increased seed weight in cki1-8. Average weight (μg/seed; at least 500 seeds were analyzed in each genotype) and standard deviations are given below the image. Bar = 1 mm.
To explore if the cki1-8 reproductive phenotype is a secondary effect caused by defects in early development, we performed the following experiments. Wild-type and cki1-8 plants were germinated and grown in the presence of various concentrations of estradiol. Upon treatment with estradiol, the cki1-8 mutant phenotype was partially suppressed, and the seedling size was smaller than untreated cki1-8 and was comparable to wild-type seedlings (see Supplemental Figure 9A online). We found that treatment with 10 nM estradiol resulted in cki1-8 plants with a growth phenotype similar to that of wild-type seedlings under the assay conditions (see Supplemental Figures 9A and 9B online). A higher concentration of estradiol caused growth arrest in cki1-8 plants (see Supplemental Figure 9A online). We then transferred the partially rescued cki1-8 seedlings treated with 10 nM estradiol into soil and continued to grow these plants until maturation. Defective floral development was observed in these estradiol-treated cki1-8 plants, similar to that in untreated cki1-8 plants (see Supplemental Figure 9C online; Figure 4B). These results suggest that abnormal reproductive development in cki1-8 is not directly related to defects in early seedling development.
In cki1-8 flowers, pollen development was normal, as revealed by Alexander staining (see Supplemental Figure 8C online). However, >98% of ovules showed defective female gametophyte development, as determined by CLSM. Consequently, remarkably smaller siliques were formed in cki1-8 plants than in the wild type (Figure 4C), and these mutant siliques produced no seeds in most cases, or occasionally set seeds (Figure 4D). cki1-8 seeds were larger than wild-type seeds (Figure 4E), a phenotype similar to that observed in the ahk2,3,4 triple mutant (Riefler et al., 2006) and the ahp1,2,3,4,5 quintuple mutant (Hutchison et al., 2006) as well as of transgenic plants overexpressing cytokinin oxidase genes (Werner et al., 2003). This result suggests that CKI1 may play a similar role as cytokinin and the cytokinin signaling components in seed development. Taken together, these results suggest that, whereas flower development and male gametogenesis remain relatively normal, female gametogenesis and seed development are impaired in the cki1-8 mutant.
All of these developmental abnormalities of cki1-8 could be fully rescued by a CKI1:CKI1 transgene. In the 18 independent transgenic lines analyzed, we were able to recover fully fertile cki1-8 plants carrying a CKI1 transgene in all 18 transgenic lines, which showed normal growth and development throughout the life cycle (Figures 2A, 2B, 3B, 4A, and 4C to 4E). Similarly, 17 out of 21 analyzed transgenic lines carrying a CKI1:CKI1-GFP transgene were also able to fully rescue the cki1-8 mutant phenotype, indicating that the CKI1-GFP fusion protein is functional in planta.
cki1-8 Responds Normally to Cytokinin
Previous studies showed that overexpression of CKI1 mimicked cytokinin effects (Kakimoto, 1996; Hwang and Sheen, 2001; Zheng et al., 2006) (see Supplemental Figures 1 and 2 online). These observations prompted us to investigate the cytokinin response in cki1-8. Cytokinin inhibited primary root growth in the wild type, and this inhibitory effect was nearly unaltered in cki1-8 seedlings (see Supplemental Figure 10A online). Cytokinin specifically eliminates the protoxylem cell linage in root vascular tissues (Mähönen et al., 2006b). In the absence or presence of cytokinin, root protoxylem development in cki1-8 showed a pattern similar to that of the wild type (see Supplemental Figure 10B online). Moreover, cytokinin delayed dark-induced leaf senescence in wild-type and cki1-8 leaves by a similar degree (see Supplemental Figure 10C online). In a shoot formation assay, cki1-8 hypocotyl explants had a similar response to cytokinin as the wild type (see Supplemental Figure 10D online). Lastly, cytokinin-induced expression of ARR6 and ARR7 was comparable in the wild type and cki1-8 (see Supplemental Figure 10E online). Collectively, these results indicate that cki1-8 has a similar response to cytokinin as wild-type plants.
The CKI1-Induced Cytokinin Phenotype Is Independent of CRE1/WOL
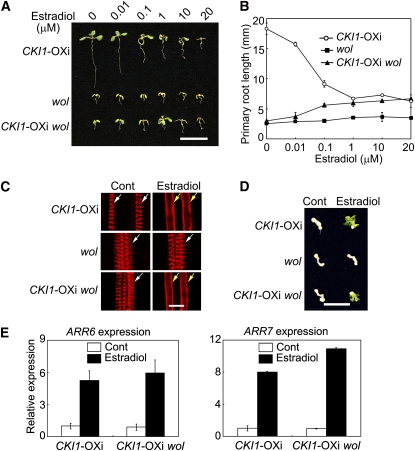
Because CKI1, a histidine kinase, is structurally and functionally similar to cytokinin receptors, we then asked if CKI1 could genetically interact with CRE1/WOL in the regulation of cytokinin signaling. The wol mutant protein is incapable of binding cytokinin, thus exerting a dominant-negative effect and resembling the ahk2,3,4 triple mutant phenotype (Mähönen et al., 2006a, 2006b). We generated a CKI1-OXi wol double mutant by crossing cki1-7 and wol and then analyzed the CKI1-induced cytokinin phenotype of the double mutant. The wol mutation inhibited seedling growth and primary root elongation, and this inhibitory effect was reduced in the CKI1-OXi wol double mutant in the presence of estradiol (Figures 5A and 5B). When grown under higher concentrations of estradiol, CKI1-OXi wol exhibited a phenotype similar to that of CKI1-OXi (Figures 5A and 5B). In particular, CKI1-OXi wol had longer primary roots than wol at all tested concentrations of estradiol (Figure 5B), indicating that overexpression of CKI1 is able to partially complement the wol phenotype in primary root growth.
Figure 5.
Overexpression of CKI1 Partially Rescues the wol Mutant Phenotype.
(A) Two-week-old seedlings germinated and grown in the presence of various concentrations of estradiol as indicated on the top of the panel. Bar = 10 mm.
(B) Primary root length of CKI1-OXi, wol, and CKI1-OXi wol seedlings germinated and grown as described in (A). Data presented are mean values of three independent experiments (n > 15 in each experiment) with standard deviations.
(C) Xylem development in CKI1-OXi, wol, and CKI1-OXi wol roots. Five-day-old seedlings were germinated and grown on germination medium (GM) containing DMSO (Control, Cont; dimethylsulfoxide, a solvent for estradiol) or 20 μM estradiol. The seedlings were stained with basic Fuchsin Red and then analyzed by confocal microscopy. White and yellow arrows denote protoxylem and metaxylem, respectively. Bar = 10 μm.
(D) Shoot regeneration assay of CKI1-OXi, wol, and CKI1-OXi wol hypocotyl explants treated with or without (Control, Cont) 20 μM estradiol. Bar = 10 mm.
(E) Expression of ARR6 and ARR7 as analyzed by qRT-PCR as described in Figure 1B. Total RNA prepared from 2-week-old seedlings treated with DMSO or estradiol (20 μM) for 12 h was used for qRT-PCR. Data presented are mean values of three biological repeats with standard deviations. Relative expression level of ARR6 and ARR7 was normalized using ACTIN7 (At5g09810) as an internal control.
All CKI1-OXi wol plants are in the cki1-7/cki1-7 background.
The wol mutation converts all cell files of the root vascular cylinder into protoxylem, whereas cytokinin exerts an opposite effect by inhibiting protoxylem differentiation (Scheres et al., 1995; Mähönen et al., 2000, 2006b). CKI1-OXi roots treated with estradiol showed a phenotype similar to that of wild-type plants treated with cytokinin (Figure 5C; see Supplemental Figure 11A online). When treated with cytokinin in the absence of estradiol, the cki1-7 wol double mutant showed a phenotype similar to that of wol; both mutants contained protoxylem but lacked metaxylem (see Supplemental Figure 11A online). However, when treated with estradiol, 76% of CKI1-OXi wol roots (n = 21) did not have protoxylem cell files but contained metaxylem (Figure 5C). Therefore, overexpression of CKI1 partially rescued the wol phenotype in xylem development.
In a shoot regeneration assay, while wol showed a reduced response to cytokinin (see Supplemental Figure 11B online), CKI1-OXi wol explants showed a similar phenotype as CKI1-OXi explants (Figure 5D). Moreover, expression of ARR6 and ARR7 was highly inducible in CKI1-OXi wol plants treated with estradiol (Figure 5E). Taken together, these results suggest that CKI1 is able to induce cytokinin responses independently of CRE1/WOL.
The CKI1-Induced Cytokinin Phenotype Is Dependent on AHP1-5
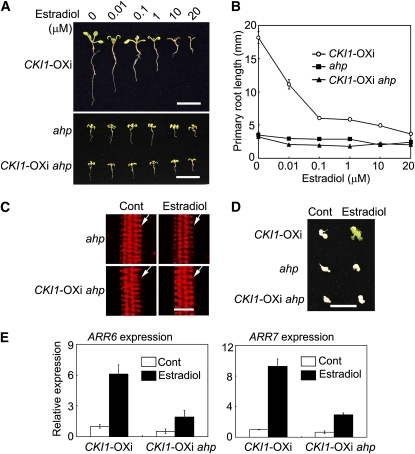
The data presented above indicated that CKI1 is capable of activating cytokinin signaling independently of CRE1/WOL. We next investigated if AHP genes are functionally required for CKI1 activity. Previous studies indicated that single ahp mutants did not have a detectable phenotype, and a quintuple mutant ahp1,2,3,4,5 showed various developmental defects and was insensitive to cytokinin (Hutchison et al., 2006). Therefore, we generated and analyzed a CKI1-OXi ahp1,2,3,4,5 hextuple mutant. In the presence of estradiol, the hextuple mutant showed a phenotype indistinguishable from that of the ahp1,2,3,4,5 mutant (Figures 6A and 6B), indicating that the CKI1-OXi gain-of-function phenotype requires AHP genes. In terms of xylem development, the estradiol-treated CKI1-OXi ahp1,2,3,4,5 hextuple mutant showed a phenotype similar to the ahp1,2,3,4,5 quintuple mutant, with overproliferated protoxylem and no differentiation of metaxylem (Figure 6C; see Supplemental Figure 11A online). This phenotype differs from the phenotype of CKI1-OXi roots, which lack the protoxylem cell lineage (Figure 5C). In contrast with CKI1-OXi, CKI1-OXi ahp1,2,3,4,5 explants were incapable of regenerating shoots when treated with estradiol (Figure 6D). Moreover, CKI1-induced expression of ARR6 and ARR7 was substantially reduced in the CKI1-OXi ahp1,2,3,4,5 hextuple mutant (Figure 6E). These results demonstrate that AHP genes are essential for CKI1-activated cytokinin signaling.
Figure 6.
CKI1-Mediated Signaling Is Dependent on AHP Genes.
(A) Two-week-old seedlings germinated and grown in the presence of various concentrations of estradiol. Note that ahp in (A) through (E) denotes the ahp1,2-1,3,4,5 quintuple mutation. Bar = 10 mm.
(B) Primary root length of CKI1-OXi, ahp1,2-1,3,4,5, and CKI1-OXi ahp1,2-1,3,4,5 seedlings germinated and grown as described in (A). Data presented are mean values of three independent experiments (n > 15 in each experiment) with standard deviations.
(C) Xylem development in ahp1,2-1,3,4,5 and CKI1-OXi ahp1,2-1,3,4,5 roots. See Figure 5C for technical details. All analyzed (31) CKI1-OXi ahp1,2-1,3,4,5 roots treated with estradiol show a phenotype similar to that of ahp1,2-1,3,4,5. White arrow denotes protoxylem. Bar = 10 μm.
(D) Shoot regeneration assay of CKI1-OXi, ahp1,2-1,3,4,5, and CKI1-OXi ahp1,2-1,3,4,5 hypocotyl explants treated with or without (Control, Cont) 20 μM estradiol. Bar = 10 mm.
(E) Expression of ARR6 and ARR7 as analyzed by qRT-PCR, as described in Figure 1B. Total RNA prepared from 2-week-old seedlings treated with DMSO or estradiol (20 μM) for 12 h was used for qRT-PCR. Data presented are mean values of three biological repeats with standard deviations. Relative expression level of ARR6 and ARR7 was normalized using ACTIN7 (At5g09810) as an internal control.
The Cytokinin Receptor Genes Are Not Required for Female Gametophyte Development
The data presented above indicate that CKI1 acts upstream of AHP to regulate cytokinin signaling in a cytokinin receptor-independent manner. Because gametophyte development appears to be unaffected in various cytokinin receptor mutants (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), we reasoned that the CKI1-AHP pathway, rather than the cytokinin receptor-AHP pathway, may be required for female gametogenesis. To test this hypothesis, we first analyzed female gametogenesis in wol and ahk2,3,4 mutants by CLSM analysis. No apparent abnormalities were observed in wol ovules and in ovules derived from ahk2,3/+,4 (the T-DNA insertions were heterozygous at the AHK3 locus) flowers that should produce 50% of ahk2,3,4 haploid female gametophytes (see Supplemental Figure 12 online). These results are consistent with the observation made in the genetic analysis of the ahk2/+,3,4 and ahk2,3/+,4 mutants, in which the T-DNA insertion at the AHK2 and AHK3 loci is segregated as a typical Mendelian trait in a 3:1 ratio, respectively (Nishimura et al., 2004). Taken together, these results indicate that gametophyte development is not affected by the receptor triple mutations.
AHP Genes Play a Critical Role in Female Gametophyte Development
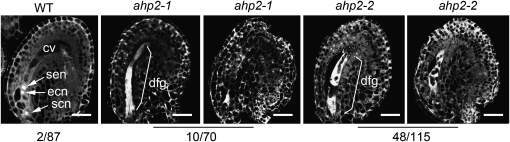
Given that CKI1 and AHP act in a linear pathway and that AHP genes are essential for CKI1 activity, one would expect that the ahp1,2,3,4,5 quintuple mutation may also cause abnormal development of female gametophytes. Among the analyzed ahp1,2,3,4,5 ovules, 38.2% (n = 178) showed severe defects in female gametophyte development at various stages, similar to those observed in cki1 mutants (see Supplemental Figure 13). In wild-type flowers 48 h after emasculation, most female gametophytes (98%) were at FG7 (female stage 7; the developmental stages were defined according to Christensen et al. [1997]). However, at the same stage, 37.3% (n = 110) of normal-looking female gametophytes in ahp1,2,3,4,5 siliques stayed at around FG5 to FG6, suggesting that the mutation also affects the progression of female gametogenesis. Moreover, in flowers derived from ahp1,2,3/+,4,5 plants (the T-DNA insertions were heterozygous at the AHP3 locus), 14.29% of ovules (n = 70) showed various defects, similar to those observed in ahp1,2,3,4,5 flowers (Figure 7).
Figure 7.
Female Gametophyte Development in the ahp1,2,3,4,5 Mutants.
CLSM of ovules derived from wild-type, ahp1,2-1,3,4,5/+ (ahp2-1), and ahp1,2-2,3/+,4,5 (ahp2-2) flowers, as indicated on the top of the images. Numbers below the images denote abnormal female gametophytes out of the examined ovules. cv, central cell vacuole; ecn, egg cell nucleus; dfg, degenerated female gametophyte; scn, synergid cell nucleus; sen, secondary nucleus. Bar = 25 μm.
Similar to cki1-8, no abnormality was observed in pollen development of the ahp1,2,3,4,5 mutant plants (see Supplemental Figure 14A online). Moreover, the number and identity of floral organs in the ahp1,2,3,4,5 quintuple mutant appeared to be normal (see Supplemental Figure 14B online). Again, similar to cki1-8, the ahp1,2,3,4,5 mutant plant produced enlarged seeds (Hutchison et al., 2006). Taken together, these results suggest that CKI1 and AHP genes function similarly in the regulation of reproductive development.
To analyze the transmission efficiency of T-DNA insertions through gametophytes in the ahp1,2,3,4,5 quintuple mutant, we constructed and characterized two different multiple mutants, ahp1,2,3/+,4,5 and ahp1,2,3,4,5/+. These two multiple mutants were reciprocally crossed to wild-type plants, and transmission efficiency of the T-DNA insertion at either AHP3 or AHP5 was analyzed in F1 plants. Unexpectedly, the transmission efficiency for both female and male gametophytes in ahp1,2,3,4,5 was only marginally altered compared with that of wild-type plants (Table 1). This result was not fully consistent with the CLSM analysis, in which 14.29% ahp1,2,3,4,5 ovules were found to be defective. This result raises the possibility that abnormal female gametophyte development in the ahp1,2,3,4,5 mutant might be caused by sporophytic defects. To clarify this question, we performed the following experiments.
Table 1.
Genetic Analysis of Transmission Efficiency of ahp1,2-1,3,4,5 and ahp1,2-2,3,4,5
| Parent |
F1 Genotype |
|||
| Female | Male | AHP/AHP | AHP/ahp | T-DNA TE (%) |
| Col-0 | ahp1,2-1,3/+,4,5 | 51 | 42 | 45.16 |
| Col-0 | ahp1,2-1,3,4,5/+ | 31 | 36 | 53.73 |
| Col-0 | ahp1,2-2,3/+,4,5 | 47 | 38 | 44.71 |
| Col-0 | ahp1,2-2,3,4,5/+ | 47 | 47 | 50.00 |
| ahp1,2-1,3/+,4,5 | Col-0 | 83 | 69 | 45.39 |
| ahp1,2-1,3,4,5/+ | Col-0 | 83 | 72 | 46.45 |
| ahp1,2-2,3/+,4,5 | Col-0 | 196 | 7 | 3.45 |
| ahp1,2-2,3,4,5/+ | Col-0 | 93 | 1 | 1.06 |
The ahp mutants used in crosses were heterozygous for the T-DNA insertion at AHP3 (ahp1,2,3/+,4,5) or AHP5 (ahp1,2,3,4,5/+) and were homozygous at other loci. Genotypes of both parents and the resulting progenies were determined by PCR analysis as described in Methods. Success of crossing was confirmed by genotyping F1 progenies using appropriate simple sequence length polymorphism markers (approximately three to five markers were used in each assay). Transmission efficiency (TE) is calculated as 100 × heterozygous/(heterozygous + wild type).
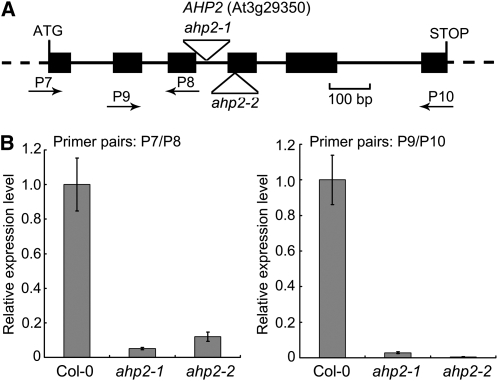
We noticed that the ahp2 allele used for the construction of the ahp1,2,3,4,5 quintuple mutant contained a T-DNA insertion in intron 3, which was not a null mutation, as it showed residual expression of AHP2 (Hutchison et al., 2006). Hereafter, we refer to this allele characterized by Hutchison et al. (2006) as ahp2-1. To more precisely define AHP function, we identified and characterized an additional mutant allele, ahp2-2 (SALK_019024), in which a T-DNA is inserted in exon 4 (Figure 8A). Whereas AHP2 expression was easily detectable in ahp2-1 by qRT-PCR analysis, as previously observed (Hutchison et al., 2006), essentially no AHP2 expression was detected in ahp2-2 (Figure 8B), indicating that ahp2-2 is a null mutant allele.
Figure 8.
Characterization of the ahp2-2 Mutant.
(A) A schematic map of the AHP2 gene and ahp2 mutant alleles. Filled boxes and solid lines denote exons and introns, respectively. Untranscribed and untranslated regions are indicated as dashed lines. Putative translation start and stop sites and positions of the T-DNA insertions in ahp2 mutant alleles are shown. P7 through P10 denote relative positions and orientation of primers used in qRT-PCR (see Supplemental Table 2 online for sequences).
(B) AHP2 expression in wild-type (Col-0), ahp1,2-1,3,4,5 (labeled as ahp2-1), and ahp2-2 seedlings (2 weeks old) analyzed by qRT-PCR using primer pairs as indicated in each graph. Data presented are mean values of three biological repeats with standard deviations. Relative expression level of AHP2 was normalized using ACTIN7 (At5g09810) as an internal control.
We used ahp2-2 to construct ahp multiple mutants and analyzed ahp1,2-2,3/+,4,5 and ahp1,2-2,3,4,5/+ in detail. No apparent developmental defects were observed in ahp1,2-2,3/+,4,5 and ahp1,2-2,3,4,5/+ plants except for defects in female gametophyte development (see below). These two multiple ahp mutants were reciprocally crossed to wild-type plants, respectively, and the transmission efficiency through female or male gametophytes was examined. We found that the T-DNA insertion at either the AHP3 or AHP5 locus was transmitted through male gametophytes with a transmission efficiency of ∼50% (Table 1), indicating that male gametophyte development is not affected by the ahp multiple mutations. However, the transmission efficiency of the T-DNA insertion through female gametophytes was dramatically reduced in both combinations (3.45 and 1.06% in ahp1,2-2,3/+,4,5 and ahp1,2-2,3,4,5/+, respectively; Table 1). Consistent with this result, CLSM revealed the presence of 41.74% (n = 115) abortive ovules in ahp1,2-2,3,4,5/+ flowers, a significantly higher phenotypic penetrance than that of the weaker allele ahp1,2-1,3/+,4,5 (14.29%; Figure 7). Taken together, these results indicate that the ahp1,2-2,3,4,5 quintuple mutation causes severe defects in female gametophyte development.
We noticed that T-DNA insertions were present in the introns of ahp1 and ahp4 in both ahp1,2-1,3,4,5 and ahp1,2-2,3,4,5 mutants. Although no full-length transcripts of AHP1 and AHP4 were detected by RT-PCR (Hutchison et al., 2006), one cannot exclude the possibility that either or both of these genes has residual expression. Indeed, we have been able to detect residual AHP4 expression in ahp1,2-1,3,4,5 seedlings using two different primer sets (see Supplemental Figure 15 online). This result indicates that residual AHP4 activities are present in both ahp1,2-1,3,4,5 and ahp1,2-2,3,4,5 mutants. Therefore, an ahp1,2,3,4,5 quintuple mutant carrying all five null mutations most likely has a stronger phenotype than ahp1,2-2,3,4,5.
The ahp1,2-2,3,4,5 Quintuple Mutant Is Seedling Lethal
To gain more insight into the function of AHPs during vegetative growth, we attempted to identify ahp1,2-2,3,4,5 quintuple mutant plants. In progeny obtained from self-pollinated ahp1,2-2,3/+,4,5 plants, 2.34% of seedlings (n = 598) showed a mutant phenotype, significantly lower than that obtained from ahp1,2-1,3/+,4,5 (22.00%; n = 559) and ahp1,2-1,3,4,5/+ (22.66%; n = 384) plants, consistent with the observation that ahp2-2 is a stronger mutant allele than ahp2-1. In progeny obtained from self-pollinated ahp1,2-2,3,4,5/+ plants, no mutants were recovered from 596 analyzed plants. This result is consistent with the observation that ahp1,2-2,3,4,5/+ plants show a lower transmission efficiency of T-DNA through female gametophytes than ahp1,2-2,3/+,4,5 plants (Table 1).
In wild-type-like progeny derived from self-pollinated ahp1,2-2,3/+,4,5 and ahp1,2-2,3,4,5/+ plants, genotyping revealed that ∼50% of plants were heterozygous for the T-DNA insertion at the AHP3 (50.00%; n = 58) or AHP5 (51.72%; n = 116) locus, and the remaining half were wild type at the AHP3 or AHP5 locus, respectively. No homozygous plants for the T-DNA insertion at the respective loci were recovered among the analyzed population, consistent with the gametophytic defective nature of the ahp1,2-2,3,4,5 quintuple mutation.
Compared with poorly developed ahp1,2-1,3,4,5 plants as previously described (Hutchison et al., 2006), the rarely recovered ahp1,2-2,3,4,5 mutant plants (derived from self-pollinated ahp1,2-2,3/+,4,5 plants) showed a stronger phenotype. When germinated and grown on Murashige and Skoog medium, both ahp1,2-1,3,4,5 and ahp1,2-2,3,4,5 mutants were significantly smaller than wild-type seedlings and were delayed in the initiation of true leaves (Figure 9A). Upon longer culture, the ahp1,2-1,3,4,5 mutant grew slowly and occasionally grew as wild-type seedlings. However, the growth and development of ahp1,2-2,3,4,5 seedlings were completely arrested, and these plants eventually died. Whereas ahp1,2-1,3,4,5 seedlings were able to grow and develop to maturity, albeit with reduced fertility, upon transfer to soil, ahp1,2-2,3,4,5 seedlings did not grow further and eventually died (Figures 9B and 9C). Therefore, the ahp1,2-2,3,4,5 mutation causes seedling lethality.
Figure 9.
Characterization of the ahp1,2,3,4,5 Quintuple Mutant.
(A) Seven-day-old seedlings of the wild type (Col-0; left), ahp1,2-1,3,4,5 (middle), and ahp1,2-2,3,4,5 (right). Bar = 5 mm.
(B) Four-week-old seedlings of the wild type (Col-0; left), ahp1,2-1,3,4,5 (middle), and ahp1,2-2,3,4,5 (right; denoted by an arrow). Bar = 20 mm.
(C) An enlarged view of an ahp1,2-2,3,4,5 seedling grown in soil for 4 weeks. Bar = 2 mm.
(D) to (F) The cellular phenotype of the ahp1,2-1,3,4,5 leaf. Leaves of the wild type and ahp1,2-1,3,4,5 (ahp2-1) were analyzed as described in Figure 3. Asterisks indicate statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
(D) The leaf blade area.
(E) Average size of abaxial epidermal cells.
(F) The number of abaxial epidermal cells.
AHP Genes Are Required for Cell Division and Cell Expansion
The ahp1,2-1,3,4,5 plants were smaller than wild-type plants (Figures 9A and 9B). Leaves of the quintuple mutant plants were substantially smaller than wild-type leaves (Figure 9D). At the cellular level, we found that both the cell size and the cell number are reduced in ahp1,2-1,3,4,5 leaves (Figures 9E and 9F). Notably, the cell number in ahp1,2-1,3,4,5 leaves was only 27.04% that of wild-type leaves (Figure 9F). The dramatically reduced cell number in ahp1,2-1,3,4,5 leaves is similar to that observed in ahk2,3,4 leaves (Nishimura et al., 2004). However, the cell size of ahk2,3,4 leaves was slightly increased compared with that of wild-type leaves (Nishimura et al., 2004), a phenotype similar to that of cki1-8 leaves (Figure 3D). Therefore, the AHP genes appear to play a more dominant role than the cytokinin receptor genes and CKI1, being involved in the regulation of both cell division and cell expansion during plant growth and development.
ARR1 Is Able to Partially Rescue the cki1-5 Mutant Phenotype
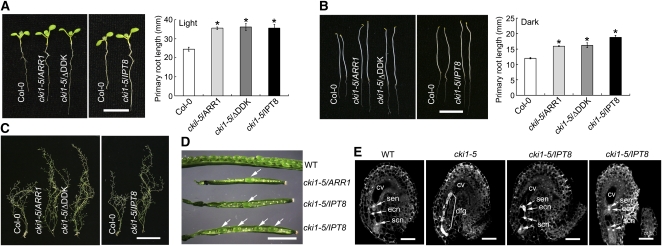
Because CKI1 acts upstream of AHP genes to regulate cytokinin signaling as well as plant growth and development, we then asked if the activation of a downstream component of the phosphorelay was able to rescue the cki1 mutant phenotype. To test this possibility, a CKI1:ARR1 construct was made and initially transformed into CKI1/cki1-8 heterozygous plants. In later experiments, we found that floral dipping with Agrobacterium tumefaciens cultures, that were either transformed with a binary vector or not transformed, was able to cause partial recovery of homozygous cki1-8 mutant plants. We found that Agrobacterium cultures could significantly induce CKI1 expression in cki1-8 plants for unknown reasons in an allele-specific manner that did not occur in wild-type and cki1-5 flowers (see Supplemental Figure 16 online). Presumably, the partial rescue effect on the cki1-8 mutant was likely caused by the Agrobacterium-induced expression of CKI1. Therefore, we used the null cki1-5 mutant (Pischke et al., 2002) in all of the related experiments described below.
In CKI1/cki1-5 plants transformed with an empty vector (pER8), no homozygous cki1-5 plants were recovered from the tested population (159 independent transgenic lines; Table 2). We analyzed 100 independent transgenic lines (T1) of CKI1/cki1-5 plants transformed with a CKI1:ARR1 transgene that were resistant to hygromycin (conferred by the binary vector pER8). Seven of these transgenic lines exhibited enlarged plant size and remarkably reduced fertility, a phenotype similar to that of cki1-8 (Table 2). Genotyping confirmed that these seven lines were indeed homozygous for the T-DNA insertion at the cki1-5 locus, indicating that the CKI1:ARR1 transgene is able to partially rescue the cki1-5 mutant phenotype. When an ARR1ΔDDK transgene, which was shown to be a constitutively activated form (Sakai et al., 2001), was used in the experiment, a similar rescue frequency (8.96%) was observed (Table 2).
Table 2.
Rescue of the cki1-5 Mutant Phenotype by ARR1 and IPT8 Transgenes
| Genotypea | Constructb | Testedc | cki1-5/−d | Percentage |
| cki1-5/+ | – | >500 | 0 | 0.00 |
| cki1-5/+ | Empty vector | 159 | 0 | 0.00 |
| cki1-5/+ | ARR1 | 100 | 7 | 7.00 |
| cki1-5/+ | ARR1ΔDDK | 201 | 18 | 8.96 |
| cki1-5/+ | ARR1D94E | 47 | 4 | 8.51 |
| cki1-5/+ | ARR1D94N | 233 | 15 | 6.44 |
| cki1-5/+ | IPT8 | 58 | 2 | 3.45 |
Heterozygous cki1-5/+ plants were transformed with different constructs.
All transgenes were placed under the control of the CKI1 promoter and were cloned into the binary vector pER8 (Zuo et al., 2000) that carried a hygromycin selectable marker gene. “Empty vector” refers to the use of pER8 in the transformation. “–” Denotes self-pollinated nontransgenic plants.
“Tested” refers to the tested T1 transgenic lines that were resistant to hygromycin and carried a cki1-5 mutation. For nontransgenic plants, the number refers to the analyzed progeny obtained from self-pollinated cki1-5/+ plants that carried a cki1-5 mutation.
“cki1-5/−” refers to homozygous lines (transgenics) or families (self-pollinated) that were obtained from the tested populations by PCR genotyping and phenotypic analysis.
We also transformed CKI1/cki1-5 plants with two additional mutant ARR1 transgenes, in which the conserved Asp-94 (D94) residue in the receiver domain was substituted with a Glu (D94E) or an Asn (D94N). The Asp-94 residue is presumed to receive a phosphoryl group from AHP proteins upon the activation of the cytokinin pathway. Therefore, ARR1D94E, similar to ARR1ΔDDK, may act as a constitutively active mutant by mimicking phosphorylation, whereas ARR1D94N is presumably an inactive form due to its inability to be phosphorylated at Asp-94. We have recovered homozygous cki1-5 T1 transgenic plants transformed with CKI1:ARR1D94E or CKI1:ARR1D94N at a similar efficiency as obtained in the CKI1:ARR1-transformed CKI1/cki1-5 plants (Table 2), suggesting that both transgenes were able to rescue the cki1-5 mutant phenotype. In a previous study, a mutant transgene ARR2D80N, analogous to ARR1D94N in this study, also showed a phenotype similar to wild-type ARR2, and the ARR2D80N mutation did not affect its ability to enhance ARR6-LUC expression in the absence or the presence of cytokinin (Hwang and Sheen, 2001). It has been proposed that ectopic overexpression of ARR2D80N may bypass the negative regulation of the cytokinin pathway and thus activate the cytokinin pathway without an input signal (Hwang and Sheen, 2001). An alternative explanation could be that nonphosphorylated ARR1 and ARR2, when ectopically overexpressed, may have the basal transcriptional competence to allow them to bind to and subsequently activate their target genes. Nevertheless, these results suggest that the ARR1 transgene, but not an empty vector, can partially rescue the cki1-5 mutant phenotype.
Because of the low frequency of seed setting (∼10 to 40 seeds per plant; see below), we amplified these transgenic lines (at least two independent lines for each construct) for an additional two to three generations (T2 through T4) and obtained thousands of seeds for the analyses described below. All of these rescued cki1-5 plants displayed a phenotype similar to cki1-8 plants, with characteristics of an enlarged size of seedlings/plants, longer primary roots, delayed termination of flower production, and abortive female gametogenesis, which resulted in setting seeds at low frequency (Figures 10A to 10D). In cki1-5 plants rescued with the ARR1 transgene, development of shoot procambial cells appeared to be normal (see Supplemental Figure 6 online).
Figure 10.
ARR1 and IPT8 Transgenes Partially Rescue the cki1-5 Mutant Phenotype.
(A) Left: Seven-day-old seedlings of the wild type (Col-0) and cki1-5 carrying CKI1:ARR1 (cki1-5/ARR1), CKI1:ARR1/ΔDDK (cki1-5/ΔDDK), or CKI1:IPT8 (cki1-5/IPT8) transgenes, respectively. T2 (CKI1:IPT8) or T3 (other transgenics) seedlings grown on GM under continuous white light are shown. Right: Primary root length of wild-type (Col-0) and cki1-5 transgenic plants. Data presented are mean values of three independent experiments (n > 15 in each experiment) with standard deviations. Asterisk indicates statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
(B) Left: Five-day-old seedlings germinated and grown on GM in the dark. See (A) for other details. Right: Primary root length of wild-type (Col-0) and cki1-5 transgenic plants. Data presented are mean values of three independent experiments (n > 15 in each experiment) with standard deviations. Asterisk indicates statistically significant differences compared with wild-type plants (Student's t test, P < 0.01).
(C) Eight-week-old soil-grown plants of the wild type (Col-0) and cki1-5 carrying different transgenes, as shown in the panel.
(D) Seed development in siliques of wild-type and cki1-5/− plants carrying a CKI1:ARR1 (cki1-5/ARR1) or CKI1:IPT8 (cki1-5/IPT8) transgene. Arrows denote seeds in the cki1-5/ARR1 and cki1-5/IPT8 siliques.
(E) CLSM of ovules derived from wild-type, cki1-5/+ (cki1-5), and cki1-5/− plants transformed with the CKI1:IPT8 transgene (cki1-5/IPT8) flowers as indicated on the top of the images. Approximate 50% of ovules in cki1-5/+ flowers show various defects as previously observed (Pischke et al., 2002). Among 222 examined ovules in cki1-5/IPT8 flowers, 13 show relatively normal development as shown in the figure. cv, central cell vacuole; ecn, egg cell nucleus; dfg, degenerated female gametophyte; scn, synergid cell nucleus; sen, secondary nucleus.
Bars = 10 mm in (A) and (B), 10 cm in (C), 2 mm in (D), and 25 μm in (E).
No CKI1 expression was detected in the flowers of the rescued cki1-5 plants (transformed with ARR1 or ARR1ΔDDK) (see Supplemental Figure 17A online), thus ruling out the possibility that the rescued phenotype was caused by residual expression of CKI1. Instead, expression of the ARR1 or ARR1ΔDDK transgenes was detected in flowers of the rescued cki1-5 plants, using primer pairs specific for these transgenes by qRT-PCR analysis (see Supplemental Figures 17B and 17C online). Taken together, these results indicate that ARR1 is able to partially complement the cki1-5 mutant phenotype.
Ectopic Expression of IPT8 in the CKI1-Expressing Domain Partially Rescues the cki1-5 Mutant Phenotype
Although the cytokinin receptors are dispensable in female gametogenesis, we asked if CKI1 activity could be functionally substituted by receptor-activated signaling. If this is the case, an increased cytokinin level in the CKI1-expressing domain, which presumably activates cytokinin receptor-mediated signaling, should be able to rescue the cki1 phenotype. To test this possibility, we constructed a CKI1:IPT8 transgene and transformed it into CKI1/cki1-5 heterozygous plants. IPT8 encodes an isopentenyl transferase that catalyzes the first rate-limiting reaction of the cytokinin biosynthesis pathway, and overexpression of IPT8 causes an elevated cytokinin level in planta (Sun et al., 2003). Among the 58 tested independent transgenic lines, we recovered two lines that are homozygous for the T-DNA insertion at the cki1-5 locus (Table 2).
Because of the relatively low rescue frequency of the cki1-5 phenotype by IPT8, we then analyzed progeny obtained from self-pollinated transgenic T1 lines that were heterozygous for the T-DNA insertion at the CKI1 locus (cki1-5/+). If the IPT8 transgene is able to partially rescue the cki1-5 phenotype, homozygous cki1-5/− plants should be recovered from self-pollinated T2 cki1-5/+ plants. Among 10 T1 lines transformed by an empty vector, no cki1-5/− homozygous plants were recovered by analyzing 580 T2 plants. In 11 T1 lines transformed with the CKI1:IPT8 transgene, cki1-5/− homozygous plants were recovered from seven lines, and the rescue frequency ranged from 1.25 to 11.48% (see Supplemental Table 1 online). Taken together, these observations indicate that the CKI1:IPT8 transgene can partially rescue the cki1-5 female gametophytic defects.
T2 and T3 progenies derived from these IPT8-rescued plants showed a phenotype similar to that of cki1-8 and cki1-5 plants rescued by an ARR1 or an ARR1ΔDDK transgene (Figures 10A to 10C). CLSM revealed the presence of normal or relatively normal ovules in flowers of the IPT8-rescued cki1-5 plants (Figure 10E). Moreover, rarely-set seeds were observed in the siliques of the IPT8-rescued cki1-5 plants (Figure 10D).
In flowers of the IPT8-rescued transgenic plants, expression of the IPT8 transgene, but not of CKI1, was detected as analyzed by qRT-PCR (see Supplemental Figures 17A and 17D online). These results indicate that expression of IPT8 in the CKI1-expressing domain partially rescues the cki1-5 mutant phenotype, presumably by locally activating receptor-mediated cytokinin signaling.
DISCUSSION
The identification of CKI1 implied the possible involvement of a TCS-based mechanism in cytokinin signaling (Kakimoto, 1996). Subsequent studies revealed the pivotal role of the TCS-mediated phosphorelay in cytokinin signaling (Hwang et al., 2002; Heyl and Schmülling, 2003; Kakimoto, 2003; Ferreira and Kieber, 2005; Müller and Sheen, 2007). Despite these progresses, several key questions on the function of CKI1 remain unanswered. CKI1 shares considerable homology with the cytokinin receptors, and overexpression of CKI1 is able to evoke typical cytokinin responses that are indistinguishable from those in cytokinin-treated wild-type plants (Kakimoto, 1996; Hwang and Sheen, 2001; Yamada et al., 2001; Zheng et al., 2006). However, CKI1 does not appear to be a cytokinin receptor, owing to its incapability to bind to cytokinins and its cytokinin-independent manner of action. Therefore, it has long been questioned whether CKI1 is directly involved in cytokinin signaling during plant growth and development (Haberer and Kieber, 2002; Hwang et al., 2002; Kakimoto, 2003; Hwang and Sakakibara, 2006; Klumpler et al., 2009). Whereas CKI1 plays an essential role in female gametogenesis, it is an open question if this function of CKI1 in female gametophyte development is dependent on the TCS-mediated signaling or on an independent pathway (Pischke et al., 2002; Hejátko et al., 2003). Lastly, again because of female gametophytic lethality, the function of CKI1 beyond the gametophytic stage remains completely unknown.
In this study, we show that overexpression of CKI1 induces the cytokinin response in a CRE1/WOL-independent manner and is able to partially rescue the wol mutant phenotype. These results suggest that CKI1 may function downstream of CRE1/WOL or independently of the cytokinin receptors. However, biochemical analyses indicate that both CRE1/WOL and CKI1 are able to phosphorylate AHP1, 2, 3, and 5 (Mähönen et al., 2006a) and that CKI1 can dephosphorylate AHP1 and AHP2 (Nakamura et al., 1999), suggesting that these two histidine kinases act directly on downstream targets and do not need an intermediate component. In addition, the wol mutant and ahk2,3,4 triple mutants do not show phenotypic similarity with the cki1-8 mutant in vegetative growth and female gametogenesis, thus disfavoring a hypothesis that cytokinin receptor genes and CKI1 act in a linear pathway. More importantly, whereas ahk2,3,4 triple mutants have no response to cytokinin (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), the cki1-8 mutant shows a normal response to cytokinin. Lastly, genetic analysis reveals that AHP genes are essential for CKI1 activity. Together, these results suggest that the cytokinin receptors and CKI1 may act in parallel branches and that AHPs act as a converging point of TCS-mediated signaling that is independently activated by the cytokinin receptors and CKI1.
The TCS has also been implicated in certain physiological activities other than cytokinin signaling. For example, AHK1, another homolog of the cytokinin receptors, has been shown to be involved in both the stress response and seed maturation (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008). Interestingly, several type-A ARR genes are coexpressed with AHK1, and different combinations of multiple arr mutants show altered responses to osmotic stress (Wohlbach et al., 2008). These observations suggest that AHK1 may also adopt TCS to activate downstream responses, although it remains unknown if this response is dependent on AHPs and ARRs (Tran et al., 2010). In contrast with AHK1, however, CKI1 induces all examined cytokinin responses, and these responses are strictly dependent on the AHP loci. Moreover, the cki1-5 and ahp1,2,3,4,5 mutants show similar defects in female gametophyte development, and these defects in cki1-5 can be partially rescued by the activation of cytokinin signaling. Therefore, although both CKI1 and AHK1 use TCS to transmit specific signals, these two structurally related histidine kinases execute distinctive physiological activities, involving the cytokinin response and the stress response, respectively. Taken together, these observations indicate that CKI1 is capable of specifically activating the cytokinin signaling pathway.
Several lines of evidence presented in this study demonstrate that CKI1-regulated development of female gametophytes is dependent on the activation of the TCS-based signaling pathway. First, whereas CKI1-activated signaling is dependent on AHP genes, the ahp1,2,3,4,5 mutants show defective female gametophyte development, similar to that observed in cki1 mutants. Moreover, cki1 mutants and the ahp1,2,3,4,5 mutants also show a similar phenotype in flower development and male gametogenesis. These results suggest that CKI1 acts upstream of AHP and that CKI1-AHP–dependent signaling is essential for female gametophyte development. Second, ectopic expression of ARR1 in the CKI1-expressing domain partially rescues the cki1-5 mutant phenotype, indicating that cell- or tissue-specific activation of TCS-mediated signaling is able to bypass the essential requirement of the activity of CKI1 and AHP genes during female gametophyte development. Third, given that TCS-based signaling is required for female gametogenesis and that this requirement is functionally fulfilled by CKI1 in ovules, the activation of receptor-mediated cytokinin signaling should complement, at least partly, the cki1 mutant phenotype. Indeed, ectopic expression of IPT8 in the CKI1-expressing domain partially rescues the cki1-5 mutant phenotype, providing an additional line of evidence to support the essential role of the TCS-activated signaling in female gametophyte development. Notably, whereas the cki1 mutant shows impaired female gametogenesis, the receptor mutations do not appear to affect gametophyte development. The use of CKI1, instead of the conventional cytokinin-activated receptors, during female gametophyte development may represent a default signaling shortcut, which facilitates specific and rapid activation of the pathway in a small group of cells in a relatively short developmental window. Collectively, these results suggest that CKI1 positively regulates the TCS-based signaling via AHP and type-B ARR genes, and this genetic framework is essential for female gametophyte development.
Compared with the severe developmental abnormalities observed in the receptor mutants (ahk2,3,4 and wol) and the ahp1,2,3,4,5 mutant, cki1-8 shows a moderate phenotype during vegetative growth. However, in contrast with the small-sized ahk2,3,4 and ahp1,2,3,4,5 mutants, cki1-8 plants grow larger than wild-type plants. It is worth noting that, whereas the ahk2,3 double mutants have reduced growth of shoots and leaves (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), cre1/ahk4 single and ahk2,3 double mutants have longer primary roots and more lateral roots than wild-type plants (Riefler et al., 2006). Compared with these receptor mutants, cki1-8 shows a similar phenotype in root growth but an opposite phenotype in shoot and leaf growth, suggesting that the cytokinin receptors and CKI1 play overlapping yet distinctive roles during vegetative growth and development. Consistent with these developmental differences, the cytokinin receptors positively regulate both cell division and cell expansion (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), whereas CKI1 appears to negatively regulate cell growth in leaves, with little, if any, effects on cell division during vegetative growth. Notably, the ahp1,2-1,3,4,5 mutants show a similar phenotype as ahk2,3,4 and wol during vegetative growth but similar to that of cki1 mutants during female gametophyte development. By contrast, cki1-8 and ahp1,2-1,3,4,5 show similar reproductive phenotypes, but exhibit different phenotypes during vegetative growth. Based on these observations, we propose that, whereas CKI1-AHP-dependent signaling is essential for megagametogenesis, cytokinin receptor-AHP–mediated signaling mainly regulates vegetative growth and possibly other developmental programs as well.
Finally, because both cytokinin receptors and CKI1 are able to activate cytokinin signaling via AHPs, it is likely that these two types of HKs function redundantly, at least partially, in certain developmental stages. Notably, the ahk2,3,4 triple mutants are neither gametophyte nor embryo lethal, as one would expect for essential roles of cytokinin signaling in plant growth and development (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). In this regard, the presence of a cytokinin receptor-independent signaling mechanism mediated by the CKI1-AHP scheme may explain concerns on the viability of ahk2,3,4 triple mutants. Presumably, the cytokinin receptor activity is, at least partly, substituted by CKI1 during reproductive development. In agreement with this notion, the ahp1,2-2,3,4,5 mutant, which lacks AHP2 expression, but has residual AHP4 expression, has severe defects in female gametophyte development similar to those observed in cki1-8, and the rarely recovered ahp1,2-2,3,4,5 seedlings die during early developmental stages. In summary, the results presented here indicate that the cytokinin receptors and CKI1 function independently and complementarily to regulate the TCS-based signaling pathway upon perceiving distinctive signals during different developmental stages. In particular, the CKI1-AHP–dependent signaling pathway is crucial for female gametophyte development and vegetative growth.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana wild types Wassilewskija (Ws) and Columbia-0 (Col-0) were used in this study. The cki1-7 and cki1-8 mutants (Ws) have been previously described (Zheng et al., 2006). The cki1-5 (Col-0; CS6360) and ahp2-2 (Col-0; SALK_019024) mutants were obtained from the ABRC. The wol, ahk2,3,4, and ahp1,2,3,4,5 (all in Col-0) mutant seeds were kindly provided by Ykä Helariutta, Thomas Schmülling, and Joseph Kieber, respectively. Seeds were imbibed at 4°C for 3 d prior to germination. Unless indicated otherwise, plants were grown under continuous white light (100 μmoles m−2 s−1) at 22°C in soil or on GM (1× Murashige and Skoog salts, 1% sucrose, 1× B5 vitamin, 0.05% MES-KOH, and 0.3% Phytagel).
Agrobacterium tumefaciens–mediated transformation of Arabidopsis plants was performed by the floral dip method (Bechtold and Pelletier, 1998). The agrobacterial strain GV3101 was used in all transformation experiments.
Genetic Analyses and Genotyping
Reciprocal crosses and genetic analysis were performed as described (Pischke et al., 2002). Genotyping of cki1-5, cki1-7, cki1-8, and ahp1,2,3,4,5 mutants was performed by PCR with three primers as previously described (Hutchison et al., 2006; Zheng et al., 2006). Genotyping of wol was performed by digesting a PCR fragment spanning the mutated region with SpeI. This SpeI site was introduced by the PCR primer in the wild-type genome but not in the wol sequence (a derived cleaved amplified polymorphism sequence marker; see Supplemental Table 2 online for sequences of all primers used in this study).
Assays of Cytokinin Responses
Root elongation and shoot formation assays were performed as previously described (Sun et al., 2003; Zheng et al., 2006). Analysis of root vasculature was performed by basic Fuchsin Red staining according to Mähönen et al. (2000). After the staining, the sample was analyzed under a confocal microscope. Dark-induced leaf senescence was analyzed as described previously (Dong et al., 2007; Feng et al., 2007).
Microscopy Analyses
CLSM of ovule development was performed according to Christensen et al. (1997). Scanning electron microcopy was performed as previously described (Feng et al., 2007). Analysis of leaf development, including the analysis of cell size and the calculation of cell number per leaf, was performed essentially according to Nishimura et al. (2004).
Analysis of vascular development of inflorescence stems was performed essentially according to Hejátko et al. (2009). Semithin sections were made from the base of the inflorescence stem when the first silique appeared, following the same methods as described (Hejátko et al., 2009).
In Situ RNA Hybridization
In situ RNA hybridization was performed as previously described (Chen et al., 2007). To prepare probes, a 299-bp cDNA fragment amplified by PCR with primers P3 and P4 (see Supplemental Table 2 online for primer sequences) was subcloned in a pBluescript II SK− vector in two different orientations. The resulting constructs were used as templates for PCR amplification of linearized DNA fragments, using primer pairs M13F/P3 (for sense probe; with M13F anchored on the vector) and M13F/P4 (for antisense probe), respectively. The PCR products were gel purified and then used for in vitro transcription of sense or antisense probes with T7 RNA polymerases in the presence of digoxigenin-11-dUTP.
RT-PCR–DNA Gel Blot Analysis of CKI1 Expression
Poly(A) RNA was prepared from fresh or frozen plant materials using the Oligotex mRNA mini kit (Qiagen) according to the manufacturer's instructions. One microgram of poly(A) RNA was used as a template for the RT reaction using oligo(dT) as a primer. The resulting reactions were then subjected to PCR for ∼20 to 25 cycles (cycling condition: 94°C, 30 s; 56°C, 30 s; 72°C, 25 s), using appropriate primer pairs. Aliquots of the PCR products were separated on a 2% agarose gel, stained with ethidium bromide, photographed using a UV light box, and then subjected to DNA gel blot analysis. The probe was double labeled by radioactive 32P-dATP and 32P-dCTP using the Megaprime DNA labeling system kit (Amersham) or labeled by the DIG DNA labeling and detection kit (Roche) according to the manufacturer's instructions. Both methods yielded similar results.
Molecular Manipulations
All molecular manipulations were performed according to standard methods (Sambrook and Russell, 2001). Primer sequences are presented in Supplemental Table 2 online. To make CKI1:CKI1, a CKI1 cDNA clone was placed under the control of the putative CKI1 promoter (containing sequences ∼1.5 kb upstream from the translation start codon), and the resulting fragment was cloned into the Sse8337I and XbaI sites of a binary vector pX7 (Zuo et al., 2001). An ∼6.0-kb CKI1 genomic fragment, which contains the putative promoter sequence (∼2.0 kb upstream of the translation start codon) and the coding sequences, was PCR amplified, in-framed fused to a GFP cDNA fragment using a SmaI site, and then cloned between the XhoI and SpeI sites of pER8 (Zuo et al., 2000) to yield CKI1:CKI1-GFP. To generate constructs in which the CKI1 promoter controlled ARR1, ARR1ΔDDK (Sakai et al., 2001), ARR1D94E, ARR1D94N, and IPT8 transgenes, the CKI1 promoter and 5′-untranslated region sequences (the same sequences as for the CKI1:CKI1-GFP construct) were PCR amplified and then ligated to PCR-amplified cDNA (ARR1 and its mutated versions) or genomic sequences (IPT8; Sun et al., 2003) of these tested genes using a BamHI restriction site embedded in the PCR primers. For genotyping the ARR1D94E and ARR1D94N mutant genes, a HindIII site 15 bp downstream of the Asp-94 codon was eliminated during PCR, and this change (AAGCTT to AAaCTT) did not alter the encoded amino acid residue (both AAG and AAA encode Lys).
Analysis of gene expression by qRT-PCR was performed as previously described (Dong et al., 2007; Feng et al., 2007). Briefly, the first-strand cDNA was synthesized using ∼1 to 2 μg of total RNA primed by oligo(dT). SuperScript II (Invitrogen) was used for the RT reaction according to the manufacturer's instructions. After incubated with RNase H for 30 min at 37°C, 1 μL of the diluted RT reaction (usually ∼5- to 10-fold) was then used as a template of qPCR. Real-time qPCR was performed using ExTaq DNA polymerase (Takara Biotechnology) and the fluorescent intercalating dye SYBR-Green (iQ SYBR Green Super mix; Bio-Rad) according to the manufacturer's instructions. The reaction was run in a PTC200 DNA Engine gradient thermal cycler (MJ Research). ACTIN7 was used as an internal control in the qRT-PCR analysis. Relative expression of the target genes was analyzed with the delta-delta Ct method.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CKI1 (At2g47430), AHK2 (At5g35750), AHK3 (At1g27320), CRE1/WOL/AHK4 (At2g01830), AHP1 (At3g21510), AHP2 (At3g29350), AHP3 (At5g39340), AHP4 (At3g16360), AHP5 (At1g03430), ARR1 (At3g16857), ARR6 (At5g62920), ARR7 (At1g19050), IPT8 (At3g19160), WUS (At2g17950), and ACT7 (At5g09810).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The CKI1-OXi Gain-of-Function Mutant Phenotype.
Supplemental Figure 2. Phenotype of Transgenic Plants upon Inducible Overexpression of a CKI1-GFP Transgene.
Supplemental Figure 3. Female Gametophyte Development in cki1-8 Ovules.
Supplemental Figure 4. CKI1 Expression in Wild-Type Seedlings Analyzed by RT-PCR–DNA Gel Blot Analysis.
Supplemental Figure 5. Expression of CKI1 in Wild-Type and cki1-8 Ovules Analyzed by in Situ RNA Hybridization.
Supplemental Figure 6. Vascular Development of Inflorescence Stems in cki1-8 and cki1-5/ARR1.
Supplemental Figure 7. The cki1-8 Phenotype under the Nutrition-Restricted Condition.
Supplemental Figure 8. Reproductive Development Phenotype of the cki1-8 Mutant.
Supplemental Figure 9. Floral Development in cki1-8 Plants Induced with Estradiol.
Supplemental Figure 10. The Cytokinin Response in cki1-8.
Supplemental Figure 11. Root Xylem Development and Shoot Regeneration Phenotype in wol and ahp1,2-1,3,4,5 Mutants.
Supplemental Figure 12. Female Gametophyte Development in wol and ahk2,3,4 Mutants.
Supplemental Figure 13. Female Gametophyte Development in ahp1,2-1,3,4,5.
Supplemental Figure 14. Reproductive Development in the ahp1,2,3,4,5 Mutants.
Supplemental Figure 15. AHP4 Expression in ahp1,2-1,3,4,5 Seedlings.
Supplemental Figure 16. Agrobacterium-Induced CKI1 Expression in cki1-8 Flowers.
Supplemental Figure 17. Expression of CKI1 and the ARR1 and IPT8 Transgenes in Transgenic Plants.
Supplemental Table 1. Rescue of the cki1-5 Mutant Phenotype by CKI1:IPT8 Transgene in T2 Transgenic Plants.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Ykä Helariutta, Joseph Kieber, Tatsuo Kakimoto, Thomas Schmülling, and the ABRC for mutant seeds. We thank Tatsuo Kakimoto for sharing unpublished data. This work was supported by grants from the Ministry of Science and Technology of China (2007CB948203 and 2009CB941502) and the National Natural Science Foundation of China (90817107).
References
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Li H.-J., Shi D.-Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.-C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.A., King E.J., Jordan J.R., Drews G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64 [Google Scholar]
- Davies P.J. (1995). Plant Hormones: Physiology, Biochemistry, and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Press; ). [Google Scholar]
- Dong H., Deng Y., Mu J., Lu Q., Wang Y., Xu Y., Chu C., Chong K., Lu C., Zuo J. (2007). The Arabidopsis Spontaneous Cell Death1 gene, encoding a ζ-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 17: 458–470 [DOI] [PubMed] [Google Scholar]
- Feng H., Chen Q., Feng J., Zhang J., Yang X., Zuo J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A–2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira F.J., Kieber J.J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8: 518–525 [DOI] [PubMed] [Google Scholar]
- Glover B.J., Torney K., Wilkins C.G., Hanke D.E. (2008). CYTOKININ INDEPENDENT-1 regulates levels of different forms of cytokinin in Arabidopsis and mediates response to nutrient stress. J. Plant Physiol. 165: 251–261 [DOI] [PubMed] [Google Scholar]
- Haberer G., Kieber J.J. (2002). Cytokinins. New insights into a classic phytohormone. Plant Physiol. 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J., Pernisová M., Eneva T., Palme K., Brzobohatý B. (2003). The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol. Genet. Genomics 269: 443–453 [DOI] [PubMed] [Google Scholar]
- Hejátko J., Ryu H., Kim G.-T., Dobesová R., Choi S., Choi S.M., Soucek P., Horák J., Pekárová B., Palme K., Brzobohaty B., Hwang I. (2009). The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21: 2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel L.L., Nelson M.A., Richmond T.A., Bleecker A.B. (1994). The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol. 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Schmülling T. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Higuchi M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M.W., Maxwell B.B., Perdue T.D., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Chen H.-C., Sheen J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Sakakibara H. (2006). Cytokinin biosynthesis and perception. Physiol. Plant. 126: 528–538 [Google Scholar]
- Hwang I., Sheen J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Klumpler T., Pekárová B., Marek J., Borkovcová P., Janda L., Hejátko J. (2009). Cloning, purification, crystallization and preliminary X-ray analysis of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65: 478–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. (2006b). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.P., Higuchi M., Törmäkangas K., Miyawaki K., Pischke M.S., Sussman M.R., Helariutta Y., Kakimoto T. (2006a). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 16: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jurgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mok D.W., Mok M.C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2007). Advances in cytokinin signaling. Science 318: 68–69 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Kakimoto T., Imamura A., Suzuki T., Ueguchi C., Mizuno T. (1999). Biochemical characterization of a putative cytokinin-responsive His-kinase, CKI1, from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 63: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke M.S., Jones L.G., Otsuga D., Fernandez D.E., Drews G.N., Sussman M.R. (2002). An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 99: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani J.A., Hutchison C.E., Schaller G.E., Kieber J.J. (February 3, 2010). The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2010.04165.x [DOI] [PubMed]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Scheres B., Di Laurenzio L., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Sun J., Niu Q.-W., Tarkowski P., Zheng B., Tarkowska D., Sandberg G., Chua N.-H., Zuo J. (2003). The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol. 131: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Ishikawa K., Yamashino T., Mizuno T. (2002). An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: Overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol. 43: 123–129 [DOI] [PubMed] [Google Scholar]
- Tran L.-S.P., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav. 5: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S.P., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., Yamaguchi-Shinozaki K. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Miyata S., Yamaguchi-Shinozaki K., Shinozaki K. (2000). Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 478: 227–232 [DOI] [PubMed] [Google Scholar]
- Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., Shinozaki K. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach D.J., Quirino B.F., Sussman M.R. (2008). Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20: 1101–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Zheng B., Deng Y., Mu J., Ji Z., Xiang T., Niu Q.-W., Chua N.-H., Zuo J. (2006). Cytokinin affects circadian-clock oscillation in a phytochrome B- and Arabidopsis Response Regulator4-dependent manner. Physiol. Plant. 127: 277–292 [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Moller S.G., Chua N.H. (2001). Chemical-regulated, site-specific DNA excision in transgenic plants. Nat. Biotechnol. 19: 157–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.