Viral movement proteins (MPs) enable viral pathogens to pass through cell walls by increasing the size exclusion limit (SEL) of plasmodesmata (PD). Cucumber mosaic virus (CMV) MP was found to bind and sever actin filament (F-actin) in vitro, and such severing was required for CMV MP-induced increase in PD SEL.
Abstract
Plant viral movement proteins (MPs) enable viruses to pass through cell walls by increasing the size exclusion limit (SEL) of plasmodesmata (PD). Here, we report that the ability of Cucumber mosaic virus (CMV) MP to increase the SEL of the PD could be inhibited by treatment with the actin filament (F-actin)–stabilizing agent phalloidin but not by treatment with the F-actin–destabilizing agent latrunculin A. In vitro studies showed that CMV MP bound globular and F-actin, inhibited actin polymerization, severed F-actin, and participated in plus end capping of F-actin. Analyses of two CMV MP mutants, one with and one without F-actin severing activities, demonstrated that the F-actin severing ability was required to increase the PD SEL. Furthermore, the Tobacco mosaic virus MP also exhibited F-actin severing activity, and its ability to increase the PD SEL was inhibited by treatment with phalloidin. Our data provide evidence to support the hypothesis that F-actin severing is required for MP-induced increase in the SEL of PD. This may have broad implications in the study of the mechanisms of actin dynamics that regulate cell-to-cell transport of viral and endogenous proteins.
INTRODUCTION
Plasmodesmata (PD) are channels within plant cell walls that enable communication between adjacent cells. It is well established that PD allow trafficking of macromolecules. Such cell-to-cell movement of macromolecules can be specific or nonspecific and is regulated during plant development and differentiation (for reviews, see Haywood et al., 2002; Roberts and Oparka, 2003; Cilia and Jackson, 2004; Ruiz-Medrano et al., 2004; Lough and Lucas, 2006). Therefore, the regulation of PD is an important mechanism of developmental regulation in plants (Zambryski and Crawford, 2000; Kim et al., 2002, 2005; Zambryski, 2004; Kobayashi et al., 2007).
Viral movement proteins (MPs) are crucial for the spread of viruses from cell to cell. MPs enable viral pathogens to pass through cell walls by increasing the size exclusion limit (SEL) of PD (for review, see Lucas, 2006). Previous studies have demonstrated that the introduction of viral MPs by microinjection or expression of green fluorescent protein (GFP)–tagged MPs, including the Cucumber mosaic virus (CMV) MP, results in an increase in the SEL of the PD in various types of plant cells (Fujiwara et al., 1993; Vaquero et al., 1994; Ding et al., 1995; Itaya et al., 1997). Therefore, MPs are useful tools for studying the mechanisms of plasmodesmal regulation (Lazarowitz and Beachy, 1999). Models have been proposed to describe how viral MPs increase the PD SEL to facilitate viral RNA (vRNA) trafficking (Heinlein et al., 1995, 1998; Carrington et al., 1996; Lazarowitz and Beachy, 1999; Boyko et al., 2000a, 2000b; Tzfira et al., 2000; Zambryski and Crawford, 2000; Aaziz et al., 2001; Boevink and Oparka, 2005). These models suggest that cytoskeletal components are involved in the transport of MPs to PD sites, as well as in the transfer of the viral genome from cell to cell through the PD.
It has been shown that microtubules can interact with the MP of Tobacco mosaic virus (TMV) and, together with the endoplasmic reticulum (ER), play a role in viral movement (Heinlein et al., 1995, 1998; Más and Beachy, 1999, 2000; Boyko et al., 2000a, 2000b, 2002; Sambade et al., 2008). Tobamovirus MPs possess a conserved sequence motif, which shares similarity with a region in tubulins that mediates lateral contact between microtubule protofilaments (Boyko et al., 2000b). Therefore, MPs may mimic tubulin assembly surfaces to propel vRNA transport through a dynamic process that is driven by microtubule polymerization (Boyko et al., 2000b). In addition, it has been reported that some host proteins, such as movement protein binding 2C and microtubule end binding protein 1a, bind both microtubules and TMV MP (Kragler et al., 2003; Curin et al., 2007; Brandner et al., 2008) and play significant roles in virus infectivity (Brandner et al., 2008; Ruggenthaler et al., 2009).
Studies of TMV have indicated that microtubules may be involved in MP degradation but not in cell-to-cell trafficking (Reichel and Beachy, 1998; Más and Beachy, 1999; Gillespie et al., 2002). However, there is also evidence that opposes the hypothesis that microtubules target the MP for degradation (Ashby et al., 2006). In addition, overexpression of calreticulin, an ER-based protein that binds TMV MP, interferes with plasmodesmal targeting of TMV MP and results in the redistribution of this MP to the microtubular network. Cell-to-cell spread of TMV is significantly compromised when calreticulin is overexpressed (Chen et al., 2005). Therefore, microtubules may function to remove excess MP from the ER. Microtubule-disrupting agents have been widely used in studies ruling out the involvement of microtubules in viral cell-to-cell movement (Gillespie et al., 2002; Kawakami et al., 2004). However, a recent report argues that tubulin-GFP is not a suitable marker for microtubule disruption by these inhibitors (Seemanpillai et al., 2006).
Alternatively, the actin cytoskeleton may be involved in viral movement. Biochemical assays have demonstrated that TMV MP binds actin directly in vitro (McLean et al., 1995). The 126-kD protein, a constituent of TMV virus replication complexes (VRCs), traffics along microfilaments (Liu et al., 2005). The Cauliflower mosaic virus protein P6 forms highly motile cytoplasmic inclusion bodies that can also move along microfilaments (Harries et al., 2009a). In addition, actin filaments may be important for the targeting of viral proteins to PD sites and therefore affect virus movement. The targeting of TMV MP to PD requires the actin/ER network. Disruption of actin filaments by specific inhibitors reduces PD localization of TMV MP (Wright et al., 2007). Both the actin cytoskeleton and myosin VIII are required for the targeting of the viral heat shock protein 70 homolog (Hsp70h) to PD sites (Prokhnevsky et al., 2005; Avisar et al., 2008). Moreover, intracellular movement and spread to adjacent cells of TMV VRCs can be blocked by inhibitors of filamentous actin and myosin but not by microtubule inhibitors (Kawakami et al., 2004). TMV movement can be reduced substantially if the actin cytoskeleton is blocked either by specific inhibitors or actin gene silencing (Liu et al., 2005). In addition, the microinjection results regarding the opening of PD upon the presence of F-actin depolymerizing factors published by Ding et al. (1996) suggest that F-actin depolymerization induces PD opening.
However, there are many contradictions with respect to the role of the actin cytoskeleton in intra- and intercellular viral movement. In contrast with Hsp70h, none of the actin cytoskeleton-specific inhibitors interfere with the PD targeting of TMV MP (Prokhnevsky et al., 2005). Transient expression of the actin binding domain 2 (ABD2) of Arabidopsis thaliana fimbrin 1 fused to GFP (Sheahan et al., 2004) to inhibit myosin-based motility has shown that the intracellular transport of TMV is independent of the actin cytoskeleton but occurs via the ER. Furthermore, the actin cytoskeleton may regulate ER-mediated transport via myosin. TMV cell-to-cell movement can continue in the absence of an intact actin cytoskeleton (Hofmann et al., 2009). In addition, plant viruses may have differing requirements for actin and myosin for their intercellular movement (Harries et al., 2009b).
Therefore, the mechanism regulating the way in which the cytoskeleton functions in cell-to-cell viral trafficking remains controversial. In this study, we investigated the role of the actin cytoskeleton in CMV MP-induced changes in PD SEL. In addition, we asked whether MP binding remodels actin filaments. Our results show that the CMV MP-induced increase in PD SEL was inhibited by phalloidin, an actin-stabilizing agent. However, the increase in PD SEL was restored when actin filaments were depolymerized with the agent latrunculin A prior to injection of CMV MP and phalloidin, suggesting that actin depolymerization may be involved in CMV MP-mediated regulation of the PD. In vitro analysis revealed that recombinant CMV MP bound both G- and F-actin. CMV MP also inhibited actin polymerization, severed filaments, and capped the plus end of the filaments. When expressed in cells, CMV MP localized to the PD, rendering it incapable of severing cytoplasmic F-actin. By contrast, CMV MP mutants without a PD localization pattern increased the PD SEL effectively when F-actin severing was unaffected. However, if the mutation affected severing activity, the SEL remained unaltered, suggesting that F-actin severing is necessary for CMV MP-mediated regulation of the PD SEL. Furthermore, the TMV MP also exhibited F-actin severing activity, and its capacity to increase PD SEL was inhibited by treatment with phalloidin. Our study suggests that F-actin severing by viral MPs may play an important role in increasing the SEL of the PD.
RESULTS
Requirement of the Actin Cytoskeleton in the CMV MP-Induced Increase in Plasmodesmal SEL
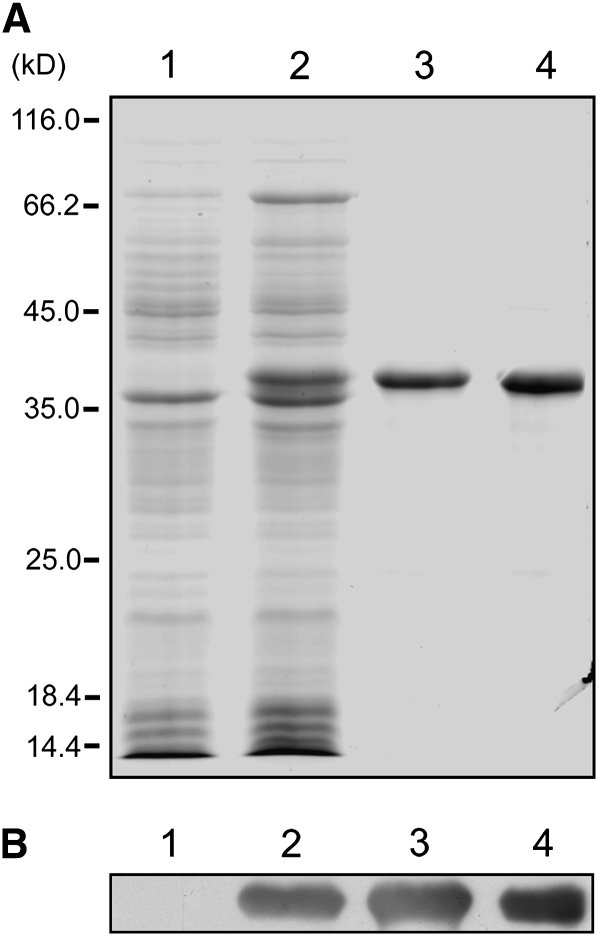
Recombinant CMV MP was required for investigating the interaction between CMV MP and actin in vitro. Therefore, the CMV MP gene was cloned, and the expressed protein was purified as described in Methods. SDS-PAGE showed that CMV MP was produced and purified from Escherichia coli (Figure 1A). An anti-CMV MP antibody recognized a specific band in extracts from bacteria induced to express recombinant CMV MP, in the inclusion body and in the purified fractions, but not in extracts from non-transformed bacteria (Figure 1B).
Figure 1.
Purification of Recombinant CMV MP.
(A) SDS-PAGE analysis of CMV MP recombinant protein purification. Lane 1, total extract from nontransformed bacterial cells (15 μg); lane 2, total extract from bacterial cells induced with IPTG to express CMV MP (20 μg); lane 3, total protein from the inclusion body (1 μg); lane 4, purified CMV MP recombinant protein (1.5 μg).
(B) Immunoblot analysis of protein extracts from nontransformed bacteria (lane 1), IPTG-induced bacteria (lane 2), the inclusion body (lane 3), and purified protein (lane 4) using an anti-CMV MP antibody.
Previous reports indicated that CMV MP potentiates cell-to-cell movement of fluorescein-conjugated dextran (Ding et al., 1995). To confirm this function of CMV MP, the activity of our purified recombinant CMV MP was tested by coinjecting the protein and fluorescent probes of various molecular masses into mesophyll cells from tobacco (Nicotiana benthamiana). The SEL of the PD was then assessed. Lucifer Yellow CH (LYCH), which has a molecular mass of 457 D, diffused freely from the injected cell into neighboring cells, whereas the higher molecular mass fluorescein isothiocyanate (FITC)-dextran was usually confined to the injected cell (Figures 2A and 2B, Table 1). However, the proportion of cell-to-cell movement of FITC-dextran increased substantially when cells were coinjected with recombinant CMV MP (Figure 2C, Table 1). Coinjection of CMV MP with 4-kD FITC-dextran increased cell-to-cell movement from 23 to 71% (Table 1). In these experiments, the cell-to-cell movement of the 4-kD FITC-dextran alone was rather high compared with the results reported by Wolf et al. (1989) who used Nicotiana tabacum cv Xanthi as the microinjection material. This may reflect an overall higher SEL of N. benthamiana PD. Coinjection of CMV MP and 10-kD FITC-dextran increased cell-to-cell movement from 6 to 41% (Table 1). However, no cell-to-cell movement of the 20-kD FITC dextran was observed when cells were coinjected with CMV MP (Table 1). M5, a mutant of CMV MP that has an Ala residue substituted for Tyr-144 and Asp-145 of the wild-type protein and a defect in cell-to-cell movement, was used as a negative control (Ding et al., 1995; Canto and Palukaitis, 2005). Microinjection of M5 did not appear to affect cell-to-cell movement of the 10-kD FITC-dextran (Figure 2D, Table 1). These results demonstrate that recombinant CMV MP increases PD SEL, which is consistent with the results of Ding et al. (1995); thus, the recombinant protein can be used in further experiments.
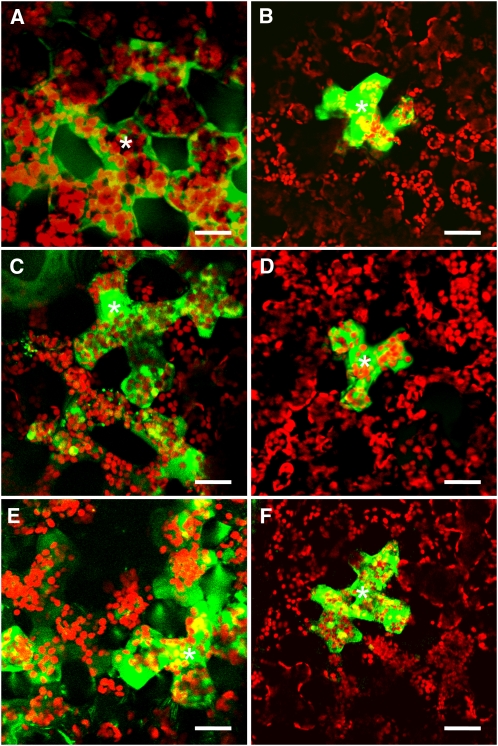
Figure 2.
The Actin Cytoskeleton Is Involved in the CMV MP-Induced Increase in Plasmodesmal SEL.
Microinjection into mesophyll cells of (A) LYCH, which is 457 D, and (B) 10-kD FITC-dextran alone or in addition to (C) 10 μM CMV MP, (D) 10 μM M5, (E) 2 μM cytochalasin D (CD), or (F) 2 μM phalloidin and 10 μM CMV MP. Asterisks indicate the cells injected. Bars = 20 μm.
Table 1.
Effect of CD, Lat A, or Phalloidin on the CMV MP-Induced Increase in PD SEL between Tobacco Mesophyll Cells
| Injection | Total Cells Observed | Cells Permitting Movement |
| LYCH (457 D) | 17 | 16 (94%) |
| 4-kD FITC-dextran | 13 | 3 (23%) |
| 4-kD FITC-dextran + MP | 14 | 10 (71%) |
| 10-kD FITC-dextran | 16 | 1 (6%) |
| 10-kD FITC-dextran + MP | 17 | 7 (41%) |
| 20-kD FITC-dextran + MP | 10 | 0 (0%) |
| 10-kD FITC-dextran + M5 | 15 | 2 (13%) |
| 10-kD FITC-dextran + M8 | 15 | 8 (53%) |
| Cytochalasin D + 10-kD FITC-dextran | 10 | 6 (60%) |
| 10-kD FITC-dextran (cells were preincubated with 0.1 μM Lat A for 30 min) | 10 | 8 (80%) |
| Phalloidin + 10-kD FITC-dextran + MP | 14 | 2 (14%) |
| Phalloidin + LYCH (457 D) | 11 | 10 (91%) |
| Phalloidin + 10-kD FITC-dextran + MP (cells were preincubated with 0.1 μM Lat A for 30 min) | 13 | 9 (69%) |
Fluorescence was observed in one to three cells beyond the injected cell in all cases, except in experiments with Lat A, where fluorescence was observed in two to five cells.
To test whether the actin cytoskeleton was involved in the CMV MP-induced increase in PD SEL, various inhibitors of actin polymerization or depolymerization were used in the microinjection experiments. Two inhibitors of actin polymerization, cytochalasin D (CD) and latrunculin A (Lat A), and one inhibitor of actin depolymerization, phalloidin, were used.
Consistent with Ding et al. (1996), coinjection with CD increased the 10-kD FITC-dextran movement into neighboring cells from 6 to 60% (Figure 2E, Table 1). Likewise, incubation with Lat A for 30 min prior to injection increased cell-to-cell movement of 10-kD FITC-dextran from 6 to 80% (Table 1). However, when actin polymerization was stabilized with phalloidin, cell-to-cell movement of 10-kD FITC-dextran was inhibited, even in the presence of CMV MP (Figure 2F, Table 1). Cell-to-cell movement decreased from 41 to 14% under these conditions, whereas LYCH moved freely between cells and remained at a similar level in the presence of phalloidin (Table 1). However, when tobacco leaves were pretreated with Lat A for 30 min prior to microinjection with CMV MP and phalloidin, cell-to-cell movement of 10-kD FITC-dextran increased to 69% (Table 1). Together, these results demonstrate that the stabilization of F-actin inhibited the CMV MP-induced increase in PD SEL, indicating that actin depolymerization was required for this activity.
Interestingly, similar results were obtained in experiments with TMV MP. Cell-to-cell movement of 10-kD FITC-dextran increased from 6 to 60% in the presence of TMV MP. This value dropped to 10% when cells were coinjected with TMV MP and phalloidin. Pretreatment of tobacco leaves with Lat A also restored cell-to-cell movement of 10-kD FITC-dextran to 80% when cells were coinjected with TMV MP and phalloidin (see Supplemental Figure 1 and Supplemental Table 1 online). These observations suggest that TMV MP may have similar mechanisms to increase PD SEL as do CMV MP.
CMV MP Binds G- and F-Actin in Vitro
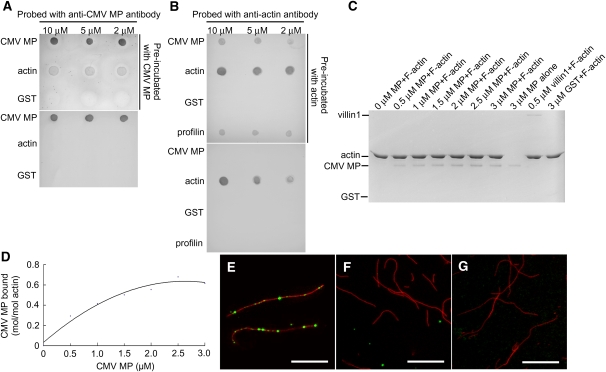
To determine whether CMV MP binds and disrupts actin filaments directly, we analyzed the ability of recombinant CMV MP to bind G- and F-actin. Immunoblotting was used to analyze the ability of recombinant CMV MP to bind G-actin. Actin was purified from rabbit skeletal muscle. Various concentrations of CMV MP and G-actin were spotted onto a nitrocellulose membrane, which was subsequently incubated with CMV MP for 2 h at room temperature, followed by probing with an anti-CMV MP antibody. The anti-CMV MP antibody detected CMV MP and also recognized G-actin spotting after incubation with CMV MP, whereas only CMV MP, but not G-actin spotting, was recognized in the absence of CMV MP incubation (Figure 3A). Conversely, when the membrane was incubated with G-actin for 2 h at room temperature prior to probing with an anti-actin antibody, G-actin and CMV MP spotting were detected. Only the actin spots were recognized in the absence of G-actin incubation (Figure 3B). These results demonstrate that recombinant CMV MP binds G-actin in vitro. Neither antibody recognized glutathione S-transferase (GST) (Figures 3A and 3B). The actin binding protein profilin was used as a positive control (Figure 3B).
Figure 3.
Recombinant CMV MP Binds G- and F-Actin in Vitro.
(A) Aliquots of 2, 5, and 10 μM of CMV MP, G-actin, and GST were spotted onto nitrocellulose membrane, which was preincubated with or without 4 μM CMV MP, and the membrane was then probed with anti-CMV MP antibody.
(B) Aliquots of CMV MP, G-actin, GST, and profilin were spotted onto nitrocellulose and then preincubated with or without 4 μM G-actin prior to probing with anti-actin antibody.
(C) Cosedimentation experiments with preformed F-actin (formed from 1 μM G-actin) incubated with various concentrations of recombinant CMV MP. Villin1 was used as a positive control, and GST was the negative control.
(D) Densitometry of the results shown in (C). Binding to F-actin was saturated at a stoichiometry of 0.68 mol CMV MP per mol of G-actin.
(E) to (G) In vitro immunofluorescence labeling experiments confirmed that CMV MP localized to F-actin. CMV MP was incubated with Alexa488-phalloidin stabilized F-actin (shown in red) and then stained with anti-CMV MP antibody followed by TRITC-conjugated goat anti-rabbit IgG (shown in green).
(E) Merged image of F-actin and CMV MP.
(F) Merged image of F-actin and denatured CMV MP.
(G) Merged image of F-actin and TRITC-conjugated goat anti-rabbit IgG alone. Bars = 10 μm in (E) to (G).
Cosedimentation experiments were performed to assess the ability of recombinant CMV MP to bind F-actin. Preformed F-actin (formed from 1 μM G-actin) was incubated with various concentrations of recombinant CMV MP. After centrifugation, supernatants and pellets were harvested and analyzed using SDS-PAGE. The results show that CMV MP cosedimented with F-actin in the pellet in a concentration-dependent manner and reached saturation in the presence of excess CMV MP (2.5 μM) (Figures 3C and 3D). Arabidopsis villin1 (Huang et al., 2005) was used as a positive control. No cosedimentation was observed with GST, which was used as a negative control (Figure 3C).
To further test whether CMV MP can bind F-actin in vitro, immunofluorescence labeling experiments were performed. Alexa488-phalloidin–stabilized F-actin was incubated with recombinant CMV MP and then stained with an anti-CMV MP antibody followed by a secondary fluorescent-labeled antibody. Samples were visualized on a microscope equipped with a CCD camera. These studies revealed that recombinant CMV MP formed dot-like structures along F-actin (Figure 3E). These structures were not detected when CMV MP was denatured by boiling (Figure 3F) or when stained with secondary antibody alone (Figure 3G). Collectively, our results demonstrate that CMV MP can bind G- and F-actin in vitro.
CMV MP Inhibits Actin Polymerization and Severs Actin Filaments in Vitro
The observation that CMV MP binds to G- and F-actin suggests that CMV MP may interact with and exert its effects on actin filaments directly. Therefore, we analyzed the effect of CMV MP on actin polymerization. Actin was purified from rabbit skeletal muscle and labeled with pyrene. Polymerization of pyrene-labeled actin was monitored over time in the presence of various concentrations of CMV MP. The polymerization rate and quantity of F-actin at the equilibrium stage decreased in the presence of CMV MP in a concentration-dependent manner (Figure 4A), indicating that CMV MP had an inhibitory effect on actin polymerization.
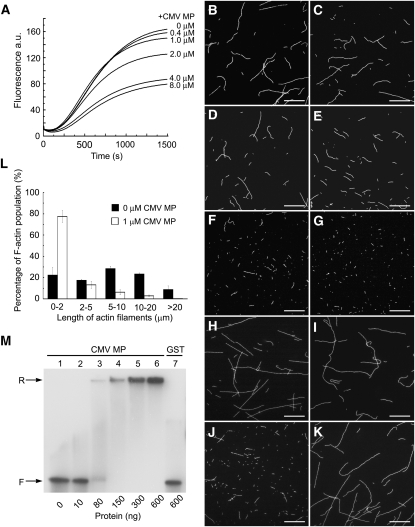
Figure 4.
Recombinant CMV MP Inhibits Actin Polymerization and Fragments Actin Filaments.
(A) Actin polymerization was performed with a 3.6 μM actin solution containing 5% pyrene-labeled actin in the presence of various concentrations of recombinant CMV MP (0, 0.4, 1, 2, 4, and 8 μM). The time course of actin polymerization was recorded by measuring pyrene fluorescence. a.u., arbitrary units. (B) to (I) Preformed F-actin (formed from 1 μM G-actin) was incubated with 0, 0.1, 0.2, 0.5, 1, and 2 μM recombinant CMV MP (B) to (G), 1 μM GST (H), or 1 μM Alexa488-phalloidin prior to the addition of 2 μM recombinant CMV MP (I). Actin filaments were then visualized by the addition of 1 μM Alexa488-phalloidin and observed with a CCD camera.
(J) and (K) One hundred nanograms of CMV RNA3 were mixed with (J) or without (K) 2 μg recombinant CMV MP, and then the mixture was added to preformed F-actin (formed from 1 μM G-actin). Bars = 10 μm in (B) to (K).
(L) The length of F-actin was measured after incubation with or without 1 μM recombinant CMV MP. Values presented are the mean ± se (n = 3).
(M) Gel mobility shift assay for RNA binding by recombinant CMV MP. About 2 ng 32P-labeled CMV RNA3 transcript was incubated with increasing amounts of recombinant CMV MP (lanes 1 to 6) or GST (lane 7) in 20 μL of binding buffer. After incubation on ice for 30 min, the mixtures were electrophoresed in 1% (v/v) nondenaturing agarose gel. The amounts of proteins added are indicated. The electrophoretic positions of free and retarded probes are marked. F, free probe; R, retarded probe. Each experiment was repeated at least three times, yielding similar results.
Furthermore, actin filaments were visualized by Alexa488-phalloidin staining after incubation with various concentrations of CMV MP. These studies demonstrate that actin filaments were fragmented in the presence of CMV MP and that the amount of fragmentation increased with the concentration of CMV MP (Figures 4B to 4G). Fragmentation of F-actin was not observed when CMV MP was replaced with GST (Figure 4H). Incubation of preformed F-actin with Alexa488-phalloidin before the addition of CMV MP inhibited fragmentation of F-actin by CMV MP (Figure 4I). In addition, the average length of the F-actin filaments in the presence of CMV MP was only 22% that of filaments not exposed to CMV MP (Figure 4L).
Previous reports have indicated that CMV MP may bind to viral RNA, forming an MP-RNA complex to transport the virus genome from cell to cell (Ding et al., 1995; Li and Palukaitis, 1996; Nagano et al., 2001; Nurkiyanova et al., 2001). To confirm that our recombinant CMV MP was able to bind the vRNA, a gel mobility shift assay was performed. 32P-labeled CMV RNA3 was incubated with increasing amounts of purified CMV MP, and the mixtures were analyzed by electrophoresis on a nondenaturing agarose gel. As shown in Figure 4M, most of the labeled CMV RNA3 migrated as free probe when low amounts of CMV MP were added. However, the labeled probes were retarded and barely entered the gel at higher molar ratios of CMV MP:RNA. By contrast, the labeled CMV RNA3 migrated as free probe when GST was added, which was used as a negative control. The absence of any intermediate retarded bands indicated that CMV MP bound in a cooperative manner to CMV RNA3, which is consistent with the results reported by Li and Palukaitis (1996). The retarded probes barely entered the gel, and this may be due to a highly cooperative binding between MP and the labeled probe (Nurkiyanova et al., 2001; Isogai and Yoshikawa, 2005). A competition binding assay with unlabeled CMV RNA3 competitor further confirmed that the purified CMV MP was able to bind vRNA in vitro (see Supplemental Figure 2A online). To assay whether CMV MP can still fragment F-actin in the presence of its vRNA, we performed F-actin fragmenting experiments in the presence of CMV MP and CMV RNA3. The results showed that actin filaments were fragmented in the presence of CMV MP and CMV RNA3 (Figure 4J). No obvious fragmentation of F-actin was observed when only the CMV RNA3 was added without CMV MP (Figure 4K).
Interestingly, similar results were obtained with TMV MP. The gel mobility shift assay and the competition binding assay showed that the purified TMV MP was functional to bind vRNA in vitro, which is consistent with Citovsky et al. (1990) (see Supplemental Figures 3H and 2B online). Fragmentation of F-actin by TMV MP was also observed, and the fragmentation was not inhibited in the presence of TMV RNA or CMV RNA3, indicating that TMV MP can also fragment F-actin. The results of F-actin fragmentation by TMV MP in the absence or presence of CMV RNA3 are presented in Supplemental Figures 3A to 3G online.
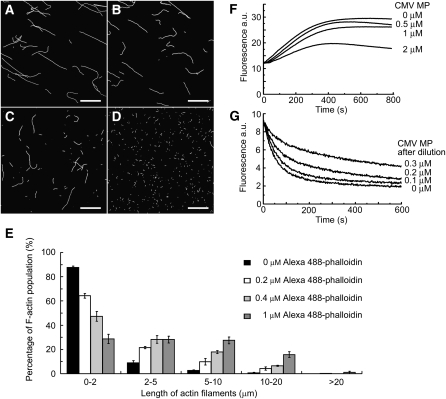
This reduction in F-actin length could be a result of F-actin depolymerization or F-actin severance by CMV MP. To determine the cause of reduced F-actin length, F-actin was preformed and incubated with saturating (the molar concentration of phalloidin is equal to that of actin) and nonsaturating (the molar concentration of phalloidin is lower than that of actin) concentrations of Alexa488-phalloidin prior to incubation with CMV MP. When preformed F-actin (formed from 1 μM G-actin) was incubated with a saturating concentration of phalloidin (1 μM), the fragmenting effect of CMV MP was completely inhibited (Figure 5A). However, when the phalloidin concentration was reduced to 0.4 μM, CMV MP was able to fragment the preformed F-actin (Figures 5B and 5E). The length of F-actin fragments decreased as the concentration of Alexa488-phalloidin decreased to 0.2 and 0 μM (Figures 5C to 5E). No significant changes in F-actin length were observed in the presence of various concentrations of Alexa488-phalloidin when CMV MP was not added. The results suggest that CMV MP has a severing, rather than a depolymerizing, effect on actin filaments.
Figure 5.
CMV MP Severs F-Actin and Caps the Plus End of F-Actin.
(A) to (D) Preformed F-actin (formed from 1 μM G-actin) was incubated with various concentrations of Alexa488-phalloidin followed by 1 μM recombinant CMV MP. After incubation, Alexa488-phalloidin was added to a final concentration of 1 μM to visualize the actin filaments, which were observed with a CCD camera. Results of preincubation with 1 μM (A), 0.4 μM (B), 0.2 μM (C), and 0 μM (D) of phalloidin. Bars = 10 μm.
(E) The F-actin lengths at the various concentrations of Alexa488-phalloidin were assessed. Each experiment was repeated at least three times. Values presented are the mean ± se (n = 3).
(F) Analysis of CMV MP capping activity through measurement of pyrene fluorescence using a spectrofluorometer. Various concentrations of recombinant CMV MP (0, 0.5, 1, and 2 μM) were incubated with preformed F-actin (formed from 1 μM G-actin) for 5 min to form seeds. An aliquot of F-actin seeds was added to a cocktail containing 1 μM G-actin (5% pyrene-labeled) and 4 μM profilin.
(G) Effects of F-actin dilution in the presence of CMV MP. Various concentrations of CMV MP (0, 2.5, 5, and 7.5 μM) were preincubated with preformed F-actin (formed by 5 μM G-actin) for 5 min before the solution was diluted 25-fold with G-buffer.
Next, we asked whether CMV MP is involved in actin filament capping. Various concentrations of CMV MP were incubated with F-actin to form seeds for polymerization. The seed was then added to a cocktail of 5% pyrene-labeled G-actin and profilin, and actin polymerization was monitored by measuring pyrene fluorescence. Under these conditions, the elongation rate depended on the availability of the barbed ends of the F-actin seeds because G-actin was saturated with profilin to prevent polymerization from the pointed ends. The polymerization rate and quantity of F-actin at equilibrium decreased in the presence of CMV MP in a concentration-dependent manner (Figure 5F), indicating that CMV MP had a capping effect on the barbed ends of F-actin. To rule out the possibility that the reduction in fluorescence could be due to the fragmentation of the F-actin, preformed F-actin (formed from 2 μM G-actin containing 5% pyrene-labeled G-actin) was incubated with various concentrations of CMV MP for 5 min, and then pyrene fluorescence was measured. The results show that no significant decrease in pyrene fluorescence occurred in the presence of various concentrations of CMV MP compared with the control (see Supplemental Figure 4 online). Therefore, the observed fluorescence decrease in the elongation experiment was due to a capping effect of CMV MP on F-actin and not to F-actin fragmentation.
To test this capping activity further, dilution experiments were performed. In these experiments, F-actin (formed from 5 μM G-actin containing 5% pyrene-labeled G-actin) was incubated with various concentrations of CMV MP for 5 min and the mixture was then diluted 25-fold. Time courses of pyrene fluorescence were measured immediately. The results revealed that the depolymerization rate of F-actin decreased in the presence of CMV MP in a concentration-dependent manner (Figure 5G), further confirming that CMV MP had a capping effect by binding and blocking depolymerization from the barbed ends of actin filaments.
Lack of CMV MP-Induced Cytoplasmic F-Actin Severing Due to the Localization of CMV MP in the PD
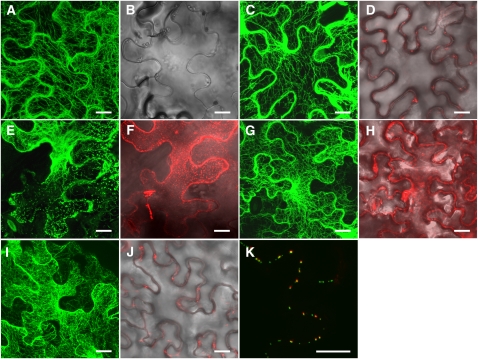
To investigate whether CMV MP severs F-actin in cells, agroinfiltration experiments were conducted. To visualize the actin filaments, a plasmid (GFP-fABD2) encoding a GFP fusion protein that binds actin was introduced into tobacco cells via agroinfiltration. Fine actin networks were visualized in cells expressing GFP-fABD2 (Figures 6A and 6B). When both GFP-fABD2 and CMV MP-mCherry were expressed in cells, no obvious change in the actin cytoskeleton was observed (Figures 6C and 6D). Approximately 76% of cells expressing both GFP-fABD2 and CMV MP-mCherry displayed normal actin filaments compared with ∼75% of cells that expressed GFP-fABD2 alone. Therefore, no evidence of CMV MP fragmentation of cytoplasmic actin filaments was observed. However, in vitro analysis demonstrated that CMV MP-mCherry alone retained the ability to sever actin filaments (see Supplemental Figure 5 online). Analysis of the subcellular localization of CMV MP-mCherry showed that this fusion protein was present along the cell wall (Figure 6D). When TMV MP-GFP was also expressed in cells, CMV MP colocalized with TMV MP, indicating that CMV MP localized to PD sites in vivo (Figure 6K). CMV MP is localized to PD, as has been shown previously by immunolabeling and GFP fusion (Oparka et al., 1996; Blackman et al., 1998) and here with mCherry fusion. It is possible that the PD localization of CMV MP prevents it from having access to and severing cytoplasmic F-actin.
Figure 6.
CMV MP Does Not Sever Cytoplasmic F-Actin Due to Its Localization in the Cell.
(A) and (B) Visualization of F-actin in tobacco leaf cells expressing GFP-fABD2.
(C) and (D) Effect of GFP-fABD2 and CMV MP-mCherry coexpression on the cytoplasmic F-actin network (C) and localization along the cell wall (D).
(E) and (F) Results of coexpression of GFP-fABD2 and M8-mCherry on the cytoplasmic F-actin network and cytoplasmic distribution.
(G) and (H) Coexpression of GFP-fABD2 and mCherry.
(I) and (J) Coexpression of GFP-fABD2 and M5-mCherry.
(K) Merged image of CMV MP-mCherry and TMV MP-GFP. Projections of confocal stacks are shown in each panel.
Bars = 20 μm.
To test this hypothesis, we used another mutant of CMV MP, M8, in which Asp-20 and Asp-21 are substituted with Ala residues. A previous report showed that M8 is distributed uniformly throughout the cytoplasm and nucleus due to an inability to localize to PD sites (Canto and Palukaitis, 2005). After coagroinfiltration with GFP-fABD2 and M8-mCherry, short fragments of cytoplasmic F-actin and actin patches were dispersed in cells (Figures 6E and 6F). Such disruption of actin filaments by expression of M8 did not occur frequently, and actin filaments were disrupted in ∼25% of cells expressing both GFP-fABD2 and M8-mCherry. However, when GFP-fABD2 and mCherry were coagroinfiltrated into the tobacco leaf cells, disruption of the actin network was not observed (Figures 6G and 6H). In addition, actin networks remained normal in cells expressing GFP-fABD2 and M5-mCherry (Figures 6I and 6J). These results indicate that the observed lack of cytoplasmic F-actin severing could be due to the localization of CMV MP to PD sites.
Necessity for F-Actin Severing Activity in the CMV MP-Induced Increase in the PD SEL
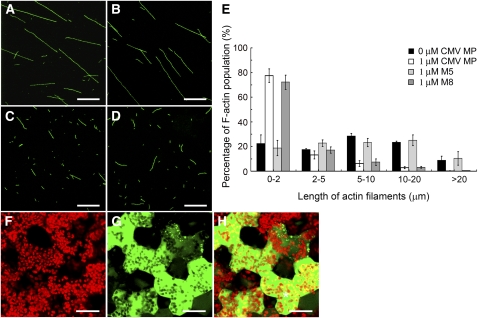
The CMV MP mutants M5 and M8 were used to further investigate the requirement of F-actin severing activity in the CMV MP-induced increase in the PD SEL. Cosedimentation experiments showed that both M5 and M8 bound F-actin directly in vitro (see Supplemental Figure 6 online). Fluorescence experiments were performed to test the F-actin severing activity of the two mutant proteins. Preformed F-actin (formed from 1 μM G-actin) was incubated with M5, M8, and CMV MP, followed by incubation with Alexa488-phalloidin to stabilize and stain the actin filaments. The length of F-actin in each treatment was visualized and measured. The data demonstrate that the average length of the actin filaments in the presence of M5 did not significantly differ from the average length in the absence of CMV MP (Figures 7A, 7B, and 7E). However, the actin filaments were predominantly fragmented in the presence of M8 and CMV MP (Figures 7C to 7E). These results illustrate that, while M5 does not possess F-actin severing activity, M8 retains this function.
Figure 7.
F-actin Severing Is Necessary for the CMV MP-Induced Increase in PD SEL.
(A) to (D) Preformed F-actin (formed from 1 μM G-actin) was incubated without CMV MP (A) or with 1 μM M5 (B), 1 μM M8 (C), or 1 μM CMV MP (D). Bars = 10 μm.
(E) Measurement of the actin filament length for each treatment. Each experiment was repeated at least three times. Values presented are the mean ± se (n = 3).
(F) to (H) Effect of M8 microinjection with 10-kD FITC-dextran on PD SEL. Chloroplast (F), 10-kD FITC-dextran (G), and merged (H) image of (F) and (G). Asterisk in (H) indicates the injected cell. Bars = 20 μm.
Microinjection of M5 with 10-kD FITC-dextran showed that this mutant did not increase the PD SEL (Figure 2D, Table 1). By contrast, M8 induced an increase in the PD SEL, as observed when this CMV MP mutant was coinjected with 10-kD FITC-dextran (Figures 7F to 7H, Table 1). Cell-to-cell movement of 10-kD FITC-dextran occurred in 53% of cells coinjected with M8, which was comparable to movement of 10-kD FITC-dextran in the presence of CMV MP (Table 1). These results establish that F-actin severing activity is necessary for CMV MP to induce an increase in the PD SEL. Based upon the localization of the CMV MP to PD, it is possible that this is where the severing of F-actin occurs.
DISCUSSION
CMV MP Increases PD SEL by Disrupting F-Actin
Although increasing the PD SEL is necessary for viral spread from cell to cell, this is not the only necessary step for viral trafficking. In fact, the actin cytoskeleton may participate in multiple steps during this event. The integrity of F-actin is needed for viral movement. Treatment with actin or myosin inhibitors has been reported to block the intracellular movement of TMV VRCs, preventing viral spread to adjacent cells (Kawakami et al., 2004). The 126-kD protein, a constituent of the TMV VRCs, traffics along microfilaments (Liu et al., 2005). Together with the ER, actin participates in targeting the TMV MP to PD sites (Wright et al., 2007). The Cauliflower mosaic virus protein P6 inclusion bodies, which associate with the actin/ER network, also traffic along microfilaments (Harries et al., 2009a). Although there are reports suggesting that the actin cytoskeleton is not involved in the targeting of TMV MP to PD sites, it is required for plasmodesmal localization of the viral Hsp70 homolog (Prokhnevsky et al., 2005; Avisar et al., 2008).
However, disruption of the actin cytoskeleton may be required for increasing the SEL of PD. Microinjection of actin-disrupting drugs or the actin binding protein profilin, which has been shown to depolymerize actin filaments rapidly after being injected into cells (Staiger et al., 1994), induces an increase in PD SEL (Ding et al., 1996). This study demonstrated that the cell-to-cell movement of 10-kD FITC-dextran induced by CMV MP was inhibited when actin filaments were stabilized by phalloidin. However, the capacity for SEL increase was restored when tobacco leaves were pretreated with Lat A prior to microinjection with CMV MP and phalloidin. In addition, CMV MP inhibited actin polymerization and severed actin filaments in vitro. M5, a CMV MP mutant lacking F-actin severing activity, also lost its ability to increase the PD SEL. Although this study and that of Ding et al. (1996) demonstrates that the SEL, as measured with molecular mass probes, is elevated upon disruption of the actin cytoskeleton, it cannot be concluded that viral movement between cells is enhanced because intracellular movement of the virus may still be blocked. Nevertheless, our data indicate that depolymerization of F-actin is necessary for the CMV MP-induced increase in PD SEL. Furthermore, our investigation of TMV MP also shows that this protein has an F-actin severing activity, and inhibition of F-actin depolymerization also blocks the increase of PD SEL induced by TMV MP. These observations reinforce our hypothesis that the depolymerization of F-actin is necessary for the MP-induced increase in PD SEL, at least for CMV MP and TMV MP.
Therefore, both the integrity and depolymerization of F-actin are required for viral movement. Our data provide a possible explanation to those seemingly contradictory observations. Analysis of cells that express both GFP-fABD2 and CMV MP-mCherry indicated that CMV MP does not sever cytoplasmic F-actin but localizes to PD sites, a finding consistent with Itaya et al. (1998). Studies of the localization of viral MPs in cells indicate that these proteins target the PD (Oparka et al., 1996, 1997; Blackman et al., 1998; Itaya et al., 1998). Therefore, the targeting of MPs to the PD is important for allowing CMV MP to be sequestered from cytoplasmic F-actin and to disrupt F-actin specifically at these sites.
Investigation of the CMV MP mutant M8, which does not localize to PD, showed that this protein possesses cytoplasmic F-actin severing activity in cells. On the other hand, microinjection of M8 increases the SEL of PD. This is an apparent contradiction due to the mislocalization of M8. We assume that the M8 protein diffuses to the PD site when it is microinjected into the cell at a relatively high concentration. Because the F-actin near the orifice of the PD may regulate PD SEL, M8 that has diffused to the PD site may break down F-actin there.
However, the total number of cells exhibiting cytoplasmic F-actin severing was relatively small. Therefore, other mechanisms may be involved in protecting cytoplasmic F-actin from CMV MP severing. For example, viral MPs may be confined to the ER (Heinlein et al., 1998; Reichel and Beachy, 1998; Lazarowitz and Beachy, 1999) and separated from the cytoplasmic actin cytoskeleton. Based on these observations, we hypothesize that the integrity of F-actin within the cytoplasm is necessary for intracellular movement of VRCs, whereas disruption of the actin cytoskeleton at PD sites may be involved in increasing the SEL of the PD.
To further test our model, many critical issues need to be addressed in future studies. Immunolabeling implicates the presence of actin and myosin in PD (White et al., 1994; Blackman and Overall, 1998; Radford and White, 1998; Volkmann et al., 2003). Additional evidence, for instance, direct biochemical isolation of these proteins from PD, is necessary to further establish this. It is also possible that F-actin is present near the orifice of PD and that the dynamic changes of such actin play a role in regulating PD SEL. Regardless of whether F-actin is present in or near PD, a more critical mechanistic question is whether MPs interact with it locally to regulate PD transport. Many studies have shown that the actin cytoskeleton has a close association with the ER system in plant cells, and it is of interest to determine whether MPs interact with actin at the ER and thereby affect the intracellular movement of VRCs (Goosen-de Roo et al., 1983; Quader et al., 1987; Lichtscheidl et al., 1990; Uetake and Peterson, 2000). Answers to these questions will be critical for unraveling the detailed mechanism by which MPs elevate PD SEL.
Actin Cytoskeleton Remodeling for Regulation of PD SEL
Many studies have shown that non-cell-autonomous proteins can move between cells in various tissues and at different developmental stages in plants (Perbal et al., 1996; Brand et al., 2000; Reiser et al., 2000; Haywood et al., 2002; Lough and Lucas, 2006). However, little is known about how PD SEL is regulated during the movement of macromolecules. Our study suggests that the regulation of actin structure is involved in the movement of macromolecules between plant cells. Although there is no direct evidence to show that the actin cytoskeleton is involved in the trafficking of non-cell-autonomous macromolecules through the PD, it is possible that plant cells use a mechanism similar to that of viruses to regulate the PD in non-cell-autonomous protein trafficking. Our study shows that CMV MP has several characteristics in common with actin binding proteins (ABPs) from both plants and animals. For example, like actin depolymerizing factor/cofilin and villin, CMV MP binds both G- and F-actin in vitro (Bretscher and Weber, 1980; Ressad et al., 1998). CMV MP also has the ability to sever actin filaments, similar to some of the ABPs, including members of the gelsolin/villin protein family, actin depolymerizing factor/cofilin, and formin (Harris et al., 2004; Huang et al., 2004; Andrianantoandro and Pollard, 2006; Xiang et al., 2007). In addition, CMV MP plays a role in capping the plus end of F-actin, which has been reported for the capping proteins CapZ, gelsolin, and formin (Yu et al., 1990; Huang et al., 2003, 2004; Kovar et al., 2003; Michelot et al., 2005). Therefore, MPs may mimic certain ABPs to remodel the actin cytoskeleton and increase the SEL of the PD.
Considering that MPs might adopt an endogenous mechanism to increase PD SEL, the use of ABPs to remodel the actin cytoskeleton at the PD site may be a general mechanism for the trafficking of macromolecules through this channel. This implies that ABPs participate in the regulation of the PD and allow the trafficking of macromolecules between plant cells. Thus, it will be of significant interest in the future to investigate whether the actin cytoskeleton and ABPs together participate in the trafficking of non-cell-autonomous macromolecules between plant cells.
METHODS
Plant Materials
Wild-type Nicotiana benthamiana plants were grown, as described previously by Martins et al. (1998). Six- to eight-week-old plants were used for the agroinfiltration and microinjection experiments.
Plasmid Construction
For Escherichia coli expression, the CMV MP (strain Fny) coding sequence was amplified by PCR and cloned into the BamHI and SacI sites of vector pET30a to generate His-tagged recombinant CMV MP. The CMV MP mutant M5, which has Ala substituted for Tyr-144 and Asp-145 of the wild-type protein (Ding et al., 1995), was generated with the TaKaRa MutanBEST site-directed mutagenesis kit (TaKaRa Bio), using a pMD18-T-CMV MP simple vector (TaKaRa) as a template. Another mutant, M8, which has Asp-20 and Asp-21 substituted with Ala (Canto and Palukaitis, 2005), was also generated, as described for M5. To create the CMV MP-mCherry fusion protein, the BamHI/SalI PCR fragment was cloned into pET30a. There is a 4× Gly linker between CMV MP and mCherry cDNAs in the fusion construct.
All plasmids used for agroinfiltration experiments were constructed in the pGD vector (Goodin et al., 2002). CMV MP-mCherry, M8-mCherry, M5-mCherry, and mCherry were cloned into the BglII and SalI sites of pGD, respectively. The XhoI and SalI cDNA PCR fragment of TMV MP-GFP was cloned into the pGD vector. All cDNA sequences were verified by sequence analysis.
Preparation of Recombinant Proteins and Anti-CMV MP Antibodies
A pET30a plasmid containing the CMV MP cDNA was transformed into E. coli strain BL21, and expression of the CMV MP recombinant protein was induced by the addition of isopropylthio-β-galactoside (IPTG) prior to bacterial cell harvest. Inclusion bodies containing the recombinant CMV MP were isolated, as described by Ding et al. (1995). For further purification, inclusion bodies were dissolved on ice in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3) containing 8 M urea. Cold PBS was then added gradually to decrease the concentration of urea to 4 M. After centrifugation at 20,000g for 30 min at 4°C, the purified inclusion bodies were mixed with Ni-NTA beads (Qiagen), which had been pre-equilibrated for 2 h at 4°C with PBS containing 4 M urea. The recombinant protein was renatured by decreasing the urea concentration to 0 M with PBS at 4°C and washing with buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0). Renatured recombinant CMV MP was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). The collected protein was dialyzed immediately against G-buffer without ATP (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.01% NaN3, and 0.5 mM DTT) for 3 h during which the buffer was changed three times. The recombinant CMV MP was stored at –80°C for future use.
M5, M8, and CMV MP-mCherry recombinant proteins were induced, as described above, and purified on Ni-NTA beads (Qiagen) under native conditions according to the manufacturer's instructions.
Protein samples were analyzed using SDS-PAGE with 12% acrylamide gels and visualized by staining with Coomassie Brilliant Blue R 250 (Sigma-Aldrich). The purified CMV MP recombinant protein was centrifuged at 20,000g for 30 min at 4°C before use.
Purified recombinant CMV MP was used to elicit polyclonal antiserum production in rabbits. The CMV MP polyclonal antibody was purified using a Protein A Sepharose CL-4B column (GE Heathcare) and a CMV MP-conjugated CNBr-Sepharose 4B column (GE Heathcare). To test whether the antibody recognized CMV MP specifically, bacterial protein extracts were assessed by immunoblotting. The blots were probed with purified anti-CMV MP antibody (1:1000) in TBST (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 0.05% Tween 20).
Microinjection
The fluorescent probes LYCH, as well as 4-, 10-, and 20-kD FITC-conjugated dextran (Sigma-Aldrich), were used to measure the PD SEL. The concentration of the fluorescent probes in the microinjection needles was 1 mM in 5 mM KHCO3, pH 8.0. For the coinjection experiments, recombinant CMV MP, M5, and M8 were used at a concentration of 10 μM with the various fluorescent probes.
Microinjection was performed using an Olympus BX50-W1 microscope with a ×40 objective (LMPlanF1; Olympus). The injection system was controlled by a Narishige ONM-1 manipulator. The needles were prepared in the lab. The microinjection samples were prepared using 3 × 4-cm2 fragments cut from the sixth leaf of tobacco plants. A portion of the lower epidermis was peeled off, and the mesophyll cells were injected. The injected samples were observed using a Zeiss LSM 510 META confocal microscope with a ×20 objective (Plan-NEOFLUAR, numerical aperture [NA] of 0.5; Zeiss). On average, three cells were injected per leaf, and more than three leaves from different plants were used for each treatment.
CD, Lat A, and phalloidin (Sigma-Aldrich) were used to disrupt or stabilize F-actin. Samples were prepared as described above. CD (2 μM) and phalloidin (2 μM) were coinjected with 10-kD FITC-dextran. For experiments with Lat A, samples were incubated with 0.1 μM Lat A for 30 min prior to microinjection.
Actin Binding Assays
Actin was purified from rabbit skeletal muscle acetone powder (prepared as described in Spudich and Watt, 1971) by Sephacryl S-300 chromatography (as described in Pollard, 1984) in G-buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT, and 0.01% NaN3). Pyrene-labeled actin was labeled on Cys-374 with pyrene iodoacetamide (Molecular Probes) as described by Huang et al. (2003).
The ability of CMV MP to bind to G-actin was tested by immunoblotting. Aliquots (2 μL) of 2, 5, and 10 μM CMV MP were spotted onto nitrocellulose membranes and allowed to dry. Identical aliquots of G-actin and GST were also spotted. The membrane was then incubated with 3% BSA in TBST buffer overnight at 4°C. Three milliliters of 4 μM recombinant CMV MP was then added to the membrane, which was then incubated for 2 h at room temperature. The membrane was washed with TBST and then incubated for another 2 h at room temperature with anti-CMV MP antibody (1:1000) in a 3-mL volume. The unbound primary antibody was washed using TBST, and then 3 mL of alkaline phosphatase–conjugated goat anti-rabbit IgG (1:10,000) (Sigma-Aldrich) in TBST was applied to the membrane for 2 h at room temperature. Following additional washes in TBST, the bound antibody was visualized by BCIP/NBT color development substrate (Promega) according to the manufacturer's instructions. Incubation without recombinant CMV MP was used as a control. Nitrocellulose membrane spotted with CMV MP, G-actin, GST, and profilin was also incubated with 3 mL 4 μM rabbit actin, which was then detected using an anti-actin antibody (MP Biomedicals). An identical membrane that had not been incubated with G-actin was used as a control.
Cosedimentation experiments were performed to test the ability of CMV MP, M5, and M8 to bind to F-actin. All proteins were centrifuged at 100,000g for 1 h at 4°C before use. Actin was polymerized at 25°C for 30 min in KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole-HCl, pH 7.0). Preformed F-actin (formed by 1 μM G-actin) was incubated with various concentrations of CMV MP (0, 0.5, 1, 1.5, 2, 2.5, and 3 μM), M5 (0, 0.5, 1, 2, 4, 5, and 6 μM), and M8 (0, 0.5, 1, 2, 4, 5, and 6 μM) for 30 min at 4°C in a 100-μL reaction volume, respectively. After incubation, the samples were centrifuged at 100,000g for 20 min at 4°C. Aliquots of the supernatant and pellet were analyzed using SDS-PAGE with 12% acrylamide gels. The amount of CMV MP, M5, and M8 bound to F-actin was determined by densitometry using a Quantity One-4.5.0 scanner (Bio-Rad).
In vitro immunofluorescence labeling experiments were performed to test CMV MP binding to F-actin. Preformed F-actin was stabilized by Alexa488-phalloidin and then incubated with recombinant CMV MP at a molar ratio of 50:1 for 20 min at room temperature in KMEI buffer. Anti-CMV MP antibody (1:100) was added and incubated for 1 h at room temperature, followed by a 15-min room temperature incubation with TRITC-conjugated goat anti-rabbit IgG (1:200). A 1-μL aliquot was transferred onto slides pretreated with poly-l-Lys. CMV MP, which was boiled for 2 min to denature the protein, and staining with secondary antibody alone were included as negative controls. The samples were visualized on an Olympus BX51 microscope equipped with a CoolSNAP HQ CCD camera (Photometrics). Images were acquired using MetaMorph 6.0 (Universal Imaging).
Actin Polymerization Assays
The time course of actin polymerization was monitored according to Huang et al. (2003). Briefly, 3.6 μM G-actin containing 5% pyrenyl-actin in G buffer was polymerized in KMEI buffer with various amounts of CMV MP (0, 0.4, 1, 2, 4, and 8 μM). Pyrene fluorescence was measured with an F-4500 fluorescence spectrofluorometer (Hitachi). The experiments were repeated at least three times under the same conditions.
Gel Mobility Shift Assays
32P-labeled full-length RNA3 of Fny CMV was generated with T7 RNA polymerase (Promega) by in vitro transcription using [α-32P]UTP (10 μCi/μL) and purified with a RNAclean kit (Tiangen). The labeled transcript was heated at 95°C for 30 s and then immediately soaked on ice for 10 min before adding protein. About 2 ng 32P-labeled CMV RNA3 transcript was incubated with purified CMV MP (0, 10, 80, 150, 300, and 600 ng) and GST (600 ng) or TMV MP (0, 5, 20, 80, 200, and 400 ng) and GST (400 ng) in 20 μL of binding buffer (50 mM Tris-HCl, pH 7.0, and 50 mM NaCl). The mixtures were incubated on ice for 30 min and then electrophoresed in a 1% (v/v) nondenaturing agarose gel in 0.5× TBE at 150 V for 1 h on ice. The gels were dried and autoradiographed. Competition binding assays were performed as described by Ham et al. (2009). Various amounts of unlabeled CMV RNA3 (0, 10, and 100 ng) were mixed with CMV MP (200 ng) or TMV MP (300 ng), for 15 min, followed by the addition of 2 ng labeled CMV RNA3 and a further incubation for 15 min.
Observation of F-Actin Fragmentation Induced by CMV MP or Its Derivatives
Preformed F-actin (formed by 1 μM G-actin) was incubated with CMV MP at various concentrations (0, 0.1, 0.2, 0.5, 1, and 2 μM) in the presence of 0.2 mM ATP at 4°C for 30 min, followed by the addition of 1 μM Alexa488-phalloidin for an additional 10 min at 4°C (Molecular Probes). Samples were then diluted 10-fold with KMEI, and 1 μL was placed on a slide coated with poly-l-Lys. Samples were observed using an Olympus BX51 microscope equipped with a CoolSNAP HQ CCD camera (Photometrics) with a ×100 oil objective (1.3 NA). GST was used as a control. As another control, 1 μM Alexa488-phalloidin was added to preformed F-actin (formed by 1 μM G-actin) before incubation with 2 μM CMV MP.
To test whether CMV MP or TMV MP could still fragment F-actin in the presence of vRNA, the CMV RNA3 (100 ng) was mixed with CMV MP (2 μg) or TMV MP (2 μg) in 20 μL of binding buffer (50 mM Tris-Cl, pH 7.0, and 50 mM NaCl). After incubation on ice for 30 min, the mixtures were added to preformed F-actin (formed from 1 μM G-actin) as described above.
To establish whether CMV MP severs actin filaments, nonsaturating and saturating concentrations of Alexa488-phalloidin were tested according to the method described by Harris and Higgs (2006). Preformed F-actin (formed from 1 μM G-actin) was incubated with 0, 0.2, 0.4, or 1 μM Alexa488-phalloidin for 15 min at 4°C before the addition of CMV MP. Then, 1 μM recombinant CMV MP was added, and the samples were incubated for another 30 min at 4°C. Before visualization, Alexa488-phalloidin was added to all samples to a final concentration of 1 μM to detect actin filaments. The samples were observed as described above. The length of the actin filaments in the images was measured using Metamorph 6.0 (Universal Imaging). The lengths of actin filaments from five images were collected and calculated for each treatment. Each treatment was repeated at least three times. The data were processed using Excel (Microsoft).
To further test the F-actin severing activity of M5 and M8, fluorescence experiments were performed as described above. Preformed F-actin (formed from 1 μM G-actin) was incubated without CMV MP or with M5 (1 μM), M8 (1 μM), or CMV MP (1 μM), followed by incubation with 1 μM Alexa488-phalloidin to stabilize and stain the actin filaments, as described above. The length of F-actin in each treatment was measured using Metamorph 6.0 (Universal Imaging). Each treatment was repeated at least three times. The data were processed using Excel (Microsoft).
To analyze the F-actin severing activity of this fusion protein in vitro, preformed F-actin (formed by 1 μM G-actin) was incubated with CMV MP-mCherry at various concentrations (0, 0.5, 1, and 2 μM) as described above.
Capping and Dilution Assays
The capping activity of CMV MP was analyzed as described by Huang et al. (2003). The actin polymerization seeds for the capping assay were prepared by incubation of CMV MP (0, 0.5, 1, or 2 μM) with preformed F-actin (formed from 1 μM G-actin containing 5% pyrene-labeled G-actin) for 5 min at room temperature. A 20-μL aliquot of F-actin seeds was added to 1 μM G-actin (5% pyrene-labeled actin) and 4 μM profilin that had been premixed in KMEI buffer. Measurement of pyrene fluorescence was used to analyze the time course of actin polymerization. The experiments were repeated at least three times under the same conditions.
To test the capping activity of CMV MP further, dilution experiments were performed. Preformed F-actin (formed from 5 μM G-actin that contained 5% pyrene-labeled G-actin) was incubated with various concentrations (0, 2.5, 5, and 7.5 μM) of CMV MP in KMEI buffer for 5 min and then diluted 25-fold with G-buffer. The time courses were recorded by measuring pyrene fluorescence immediately, as described above. The experiments were repeated at least three times under the same conditions.
Agroinfiltration
pCAMBIA1300-GFP-fABD2, pGD-CMV MP-mCherry, pGD-M8-mCherry, pGD-M5-mCherry, pGD-TMV MP-GFP, and pGD-mCherry were expressed in N. benthamiana leaves by agroinfiltration according to Goodin et al. (2002). Agrobacterium tumefaciens (GV3101) was transformed with these constructs and grown to saturation in Luria-Bertani medium containing antibiotics for selection. The cultures were centrifuged and the bacterial cells were resuspended in infiltration solution (10 mM MgCl2, 10 mM MES, pH 5.6, and 100 μM acetosyringone) to an OD600 of 1 and then kept at room temperature for 2 h. Before coinfiltration, the suspensions were mixed equally (1:1) and then coinfiltrated into the abaxial sides of young leaves using a 2-mL syringe without a needle. The leaves were visualized on a Zeiss LSM510 confocal microscope with a ×40 oil objective (1.3 NA; Zeiss) 36 to 70 h after infiltration. For each construct, ∼100 cells were observed in three independent experiments.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: D10538 REGION, 120 to 959 (CMV MP) and AB369276 REGION, 4903 to 5709 (TMV MP).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Actin Cytoskeleton Is Involved in the TMV MP-Induced Increase in PD SEL.
Supplemental Figure 2. Competition Binding Assays with Recombinant CMV MP and TMV MP.
Supplemental Figure 3. Recombinant TMV MP Fragments Actin Filaments.
Supplemental Figure 4. CMV MP-Induced Fragmentation of F-Actin Does Not Result in the Reduction of Pyrene Fluorescence.
Supplemental Figure 5. Recombinant CMV MP-mCherry Severs F-Actin in Vitro.
Supplemental Figure 6. Both M5 and M8 Bind F-Actin in Vitro.
Supplemental Table 1. Effect of Lat A or Phalloidin on the TMV MP-Induced Increase in PD SEL between Tobacco Mesophyll Cells.
Acknowledgments
We thank Biao Ding at Ohio State University for critical reading and comments on the manuscript. N. benthamiana plants, TMV MP cDNA, CMV RNA3, TMV RNA, and the pGD vector were generously provided by Jialin Yu and Dawei Li of China Agricultural University. Profilin and villin1 were kindly provided by Shanjin Huang from the Institute of Botany, Chinese Academy of Sciences. We also thank Haiyun Ren of Beijing Normal University for providing the pCAMBIA1300-GFP-fABD2 vector. This research was supported by grants from the National Basic Research Program of China (2006CB100101), the 111 Project (B06003), and the National Natural Science Foundation of China (30830058 and 30721062) to M.Y.
References
- Aaziz R., Dinant S., Epel B.L. (2001). Plasmodesmata and plant cytoskeleton. Trends Plant Sci. 6: 326–330 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Ashby J., Boutant E., Seemanpillai M., Sambade A., Ritzenthaler C., Heinlein M. (2006). Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J. Virol. 80: 8329–8344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A.I., Dolja V.V. (2008). Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J. Virol. 82: 2836–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman L.M., Boevink P., Santa Cruz S., Palukaitis P., Oparka K.J. (1998). The movement protein of Cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell 10: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman L.M., Overall R.L. (1998). Immunolocalisation of the cytoskeleton to plasmodesmata of Chara corallina. Plant J. 14: 733–741 [Google Scholar]
- Boevink P., Oparka K.J. (2005). Virus-host interactions during movement process. Plant Physiol. 138: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V., Ashby J.A., Suslova E., Ferralli J., Sterthaus O., Deom C.M., Heinlein M. (2002). Intramolecular complementing mutations in Tobacco mosaic virus movement protein confirm a role for microtubule association in viral RNA transport. J. Virol. 76: 3974–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V., Ferralli J., Ashby J., Schellenbaum P., Heinlein M. (2000b). Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Boyko V., Ferralli J., Heinlein M. (2000a). Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 22: 315–325 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Brandner K., Sambade A., Boutant E., Didier P., Mely Y., Ritzenthaler C., Heinlein M. (2008). Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol. 147: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A., Weber K. (1980). Villin is a major protein of the microvillus cytoskeleton which binds both G and F actin in a calcium-dependent manner. Cell 20: 839–847 [DOI] [PubMed] [Google Scholar]
- Canto T., Palukaitis P. (2005). Subcellular distribution of mutant movement proteins of Cucumber mosaic virus fused to green fluorescent proteins. J. Gen. Virol. 86: 1223–1228 [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Kasschau K.D., Mahajan S.K., Schaad M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8: 1669–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Tian G.W., Gafni Y., Citovsky V. (2005). Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 138: 1866–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia M.L., Jackson D. (2004). Plasmodesmata form and function. Curr. Opin. Cell Biol. 16: 500–506 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Knorr D., Schuster G., Zambryski P. (1990). The P30 movement protein of Tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Curin M., Ojangu E.L., Trutnyeva K., Ilau B., Truve E., Waigmann E. (2007). MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of Tobacco mosaic virus movement protein in plants. Plant Physiol. 143: 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Kwon M.O., Warnberg L. (1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10: 157–164 [Google Scholar]
- Ding B., Li Q., Nguyen L., Palukaitis P., Lucas W.J. (1995). Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology 207: 345–353 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Giesmann-Cookmeyer D., Ding B., Lommel S.A., Lucas W.J. (1993). Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the Red clover necrotic mosaic virus movement protein. Plant Cell 5: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie T., Boevink P., Haupt S., Roberts A.G., Toth R., Valentine T., Chapman S., Oparka K.J. (2002). Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of Tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Goosen-de Roo L., Burggraaf P.D., Libbenga K.R. (1983). Microfilament bundles associated with tubular endoplasmic reticulum in fusiform cells in the active cambial zone of Fraxinus excelsior L. Protoplasma 116: 204–208 [Google Scholar]
- Ham B.K., Brandom J.L., Xoconostle-Cázares B., Ringgold V., Lough T.J., Lucas W.J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.S., Higgs H.N. (2006). Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 406: 190–214 [DOI] [PubMed] [Google Scholar]
- Harris E.S., Li F., Higgs H.N. (2004). The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279: 20076–20087 [DOI] [PubMed] [Google Scholar]
- Harries P.A., Palanichelvam K., Yu W., Schoelz J.E., Nelson R.S. (2009a). The Cauliflower mosaic virus protein p6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 149: 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P.A., Park J.W., Sasaki N., Ballard K.D., Maule A.J., Nelson R.S. (2009b). Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc. Natl. Acad. Sci. USA 106: 17594–17599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V., Kragler F., Lucas W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14: S303–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M., Epel B.L., Padgett H.S., Beachy R.N. (1995). Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M., Padgett H.S., Gens J.S., Pickard B.G., Casper S.J., Epel B.L., Beachy R.N. (1998). Changing patterns of localization of the Tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C., Niehl A., Sambade A., Steinmetz A., Heinlein M. (2009). Inhibition of Tobacco mosaic virus movement by expression of an actin-binding protein. Plant Physiol. 149: 1810–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Chaudhry F., Franklin-Tong V.E., Staiger C.J. (2004). A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 279: 23364–23375 [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai M., Yoshikawa N. (2005). Mapping the RNA-binding domain on the Apple chlorotic leaf spot virus movement protein. J. Gen. Virol. 86: 225–229 [DOI] [PubMed] [Google Scholar]
- Itaya A., Hickman H., Bao Y., Nelson R., Ding B. (1997). Cell-to-cell trafficking of Cucumber mosaic virus movement protein: Green fluorescent protein fusion produced by biolistic gene bombardment in tobacco. Plant J. 12: 1223–1230 [Google Scholar]
- Itaya A., Woo Y., Masuta C., Bao Y., Nelson R.S., Ding B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S., Watanabe Y., Beachy R.N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 101: 6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Cho E., Crawford K., Hempel F.D., Zambryski P.C. (2005). Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 2227–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Hempel F.D., Sha K., Pfluger J., Zambryski P.C. (2002). Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129: 1261–1272 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Otegui M., Krishnakumar S., Mindrinos M., Zambryski P. (2007). Increased Size Exclusion Limit2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by Profilin. J. Cell Biol. 161: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F., Curin M., Trutnyeva K., Gansch A., Waigmann E. (2003). MPB2C, a microtubule-associated plant protein binds to and interferes with cell to-cell transport of Tobacco mosaic virus movement protein. Plant Physiol. 132: 1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S.G., Beachy R.N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Palukaitis P. (1996). Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 216: 71–79 [DOI] [PubMed] [Google Scholar]
- Lichtscheidl I.K., Lancelle S.A., Hepler P.K. (1990). Actin-endoplasmic reticulum complexes in Drosera. Their structural relationship with the plasmalemma, nucleus, and organelles in cells prepared by high pressure freezing. Protoplasma 155: 116–126 [Google Scholar]
- Liu J.Z., Blancaflor E.B., Nelson R.S. (2005). The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 138: 1853–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough T.J., Lucas W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57: 203–232 [DOI] [PubMed] [Google Scholar]
- Lucas W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- Martins C.R.F., Johnson J.A., Lawrence D.M., Choi T.-J., Pisi A., Tobin S., Lapidus L.D., Wagner J.D.O., Ruzin S., McDonald K., Jackson A.O. (1998). Sonchus yellow net rhabdovirus nuclear viroplasms contain polymerase-associated proteins. J. Virol. 72: 5669–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Beachy R.N. (1999). Replication of Tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intercellular distribution of viral RNA. J. Cell Biol. 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Beachy R.N. (2000). Role of microtubules in the intracellular distribution of Tobacco mosaic virus movement protein. Proc. Natl. Acad. Sci. USA 97: 12345–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean B.G., Zupan J., Zambryski P.C. (1995). Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H., Mise K., Furusawa I., Okuno T. (2001). Conversion in the requirement of coat protein in cell-to-cell movement mediated by the Cucumber mosaic virus movement protein. J. Virol. 75: 8045–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurkiyanova K.M., Ryabov E.V., Kalinina N.O., Fan Y.C., Andreev I., Fitzgerald A.G., Palukaitis P., Taliansky M. (2001). Umbravirus-encoded movement protein induces tubule formation on the surface of protoplasts and binds RNA incompletely and non-cooperatively. J. Gen. Virol. 82: 2579–2588 [DOI] [PubMed] [Google Scholar]
- Oparka K.J., Boevink P., Santa Cruz S. (1996). Studying the movement of plant viruses using green fluorescent protein. Trends Plant Sci. 1: 412–418 [Google Scholar]
- Oparka K.J., Prior D.A.M., Santa Cruz S., Padgett H.S., Beachy R.N. (1997). Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of Tobacco mosaic virus (TMV). Plant J. 12: 781–789 [DOI] [PubMed] [Google Scholar]
- Perbal M.C., Haughn G., Saedler H., Schwarz-Sommer Z. (1996). Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122: 3433–3441 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky A.I., Peremyslov V.V., Dolja V.V. (2005). Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 79: 14421–14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quader H., Hofmann A., Schnepf E. (1987). Shape and movement of the endoplasmic reticulum in onion bulb epidermis cells: Possible involvement of actin. Eur. J. Cell Biol. 44: 17–26 [Google Scholar]
- Radford J.E., White R.G. (1998). Localization of a myosin-like protein to plasmodesmata. Plant J. 14: 743–750 [DOI] [PubMed] [Google Scholar]
- Reichel C., Beachy R.N. (1998). Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 95: 11169–11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L., Sánchez-Baracaldo P., Hake S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42: 151–166 [PubMed] [Google Scholar]
- Ressad F., Didry D., Xia G.X., Hong Y., Chua N.-H., Pantaloni D., Carlier M.-F. (1998). Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 273: 20894–20902 [DOI] [PubMed] [Google Scholar]
- Roberts A.G., Oparka K.J. (2003). Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 26: 103–124 [Google Scholar]
- Ruggenthaler P., Fichtenbauer D., Krasensky J., Jonak C., Waigmann E. (2009). Microtubule-associated protein AtMPB2C plays a role in organization of cortical microtubules, stomata patterning, and tobamovirus infectivity. Plant Physiol. 149: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cazares B., Kragler F. (2004). The plasmodesmatal transport pathway for homoeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7: 641–650 [DOI] [PubMed] [Google Scholar]
- Sambade A., Brandner K., Hofmann C., Seemanpillai M., Mutterer J., Heinlein M. (2008). Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9: 2073–2088 [DOI] [PubMed] [Google Scholar]
- Seemanpillai M., Elamawi R., Ritzenthaler C., Heinlein M. (2006). Challenging the role of microtubules in Tobacco mosaic virus movement by drug treatments is disputable. J. Virol. 80: 5807–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction. 1. Biochemical studies of interaction of tropomyosin–troponin complex with actin and proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger C.J., Yuan M., Valenta R., Shaw P.J., Warn R.M., Lloyd C.W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4: 215–219 [DOI] [PubMed] [Google Scholar]
- Tzfira T., Rhee Y., Chen M.-H., Kunik T., Citovsky V. (2000). Nucleic acid transport in plant-microbe interactions: The molecules that walk through the walls. Annu. Rev. Microbiol. 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Uetake Y., Peterson R.L. (2000). Spatial associations between actin filaments, endoplasmic reticula, mitochondria and fungal hyphae in symbiotic cells of orchid protocorms. Mycoscience 41: 481–489 [Google Scholar]
- Vaquero C., Turner A.P., Demangeat G., Sanz A., Serra M.T., Roberts K., Garcia-Luque I. (1994). The 3a movement protein from Cucumber mosaic virus increases the gating capacity of plasmodesmata in transgenic tobacco plants. J. Gen. Virol. 75: 3193–3197 [DOI] [PubMed] [Google Scholar]
- Volkmann D., Mori T., Tirlapur U.K., König K., Fujiwara T., Kendrick-Jones J., Baluška F. (2003). Unconventional myosins of the plant-specific class VIII: endocytosis, cytokinesis, plasmodesmata/pit-fields, and cell-to-cell coupling. Cell Biol. Int. 27: 289–291 [DOI] [PubMed] [Google Scholar]
- White R.G., Badelt K., Overall R.L., Vesk M. (1994). Actin associated with plasmodesmata. Protoplasma 180: 169–184 [Google Scholar]
- Wolf S., Deom C.M., Beachy R.N., Lucas W.J. (1989). Movement protein of Tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Wood N.T., Robert A.G., Chapman S., Boevink P., MacKenzie K.M., Oparka K.J. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic 8: 21–31 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). Actin binding protein 29 from lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Johnston P.A., Südhof T.C., Yin H.L. (1990). gCap39, a calcium ion- and polyphosphoinositide-regulated actin capping protein. Science 250: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Zambryski P., Crawford K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16: 393–421 [DOI] [PubMed] [Google Scholar]
- Zambryski P. (2004). Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J. Cell Biol. 164: 165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]