Using large-scale quantitative analysis, this work reveals that grain shape and size are independent traits in both modern and primitive wheat and are under the control of distinct genetic components. Moreover, the phenotypic diversity in grain morphology found in modern commercial wheat is the result of a recent and severe bottleneck.
Abstract
Grain morphology in wheat (Triticum aestivum) has been selected and manipulated even in very early agrarian societies and remains a major breeding target. We undertook a large-scale quantitative analysis to determine the genetic basis of the phenotypic diversity in wheat grain morphology. A high-throughput method was used to capture grain size and shape variation in multiple mapping populations, elite varieties, and a broad collection of ancestral wheat species. This analysis reveals that grain size and shape are largely independent traits in both primitive wheat and in modern varieties. This phenotypic structure was retained across the mapping populations studied, suggesting that these traits are under the control of a limited number of discrete genetic components. We identified the underlying genes as quantitative trait loci that are distinct for grain size and shape and are largely shared between the different mapping populations. Moreover, our results show a significant reduction of phenotypic variation in grain shape in the modern germplasm pool compared with the ancestral wheat species, probably as a result of a relatively recent bottleneck. Therefore, this study provides the genetic underpinnings of an emerging phenotypic model where wheat domestication has transformed a long thin primitive grain to a wider and shorter modern grain.
INTRODUCTION
Wheat epitomizes the effectiveness of artificial selection and breeding in shaping a crop to suit human social and historical circumstances as well as economical incentives. The domestication of wild einkorn and emmer wheat around 10,000 years ago marked the transition from a hunter-gatherer society to an agrarian one with considerable effects on the evolution of human civilization. Moreover, the emergence of hexaploid, common or bread wheat, followed by further selection and extensive breeding, led to a crop species of significant financial and nutritional importance since it provides one-fifth of the calories consumed by humans today (Dubcovsky and Dvorak, 2007).
One of the main components of the domestication syndrome in cereals (i.e., the set of characters that distinguishes the domesticated species from its wild ancestors) is an increase in grain size (Fuller, 2007; Brown et al., 2009). Archaeobotanical evidence from around the Fertile Crescent region indicates that the transition from the diploid wild einkorn (Triticum monococcum ssp aegilopoides; AmAm) and tetraploid emmer wheat (Triticum turgidum ssp dicoccoides; BBAA) to the domesticated forms (T. monococcum ssp monococcum and T. turgidum ssp dicoccum, respectively) was associated with a trend toward larger grains (Feldman, 2001; Fuller, 2007). This phenomenon is thought to have occurred relatively quickly and preceded the transition to nonshattering/free-threshing (two of the most important components of the domestication syndrome) wheat forms (Fuller, 2007). Mainly because of its effect on yield, increasing grain size continues to be a major selection and breeding target in modern tetraploid (T. turgidum ssp durum) and hexaploid wheat (Triticum aestivum ssp aestivum; BBAADD).
Grain shape does not appear to have been a major component of the wheat domestication syndrome, in contrast with other cereal species, such as rice (Oryza sativa), where the domestication process involved strong selection both for grain size and shape (Kovach et al., 2007), but has been a relatively recent breeding target dictated by the market and industry requirements. Indeed, grain shape (and size), density, and uniformity are important attributes for determining the market value of wheat grain since they influence the milling performance (i.e., flour quality and yield). Theoretical models predict that milling yield could be increased by optimizing grain shape and size with large and spherical grains being the optimum grain morphology (Evers et al., 1990). Several other quality criteria used by the industry are influenced by grain morphology. Specific weight (kilograms of mass per liter bulk grain) is used extensively to grade wheat before milling, and it is thought to be related to the grain shape or size since these parameters determine the way the individual grain packs. Grain size was also found to be associated with various characteristics of flour, such as protein content and hydrolytic enzymes activity, which in turn determine baking quality and end-use suitability (Millar et al., 1997; Evers, 2000).
Though genetically and developmentally important, the phenotypic and genetic variation of wheat grain morphology is surprisingly understudied mainly due to the difficulty in quantifying this trait. Previous studies used a limited number of metrics that were analyzed discretely largely in single mapping populations (Giura and Saulescu, 1996; Campbell et al., 1999; Dholakia et al., 2003; Breseghello and Sorrels, 2007; Sun et al., 2009). This approach identifies only pairwise associations between traits and single-trait genetic effects; therefore, it is limited in providing a comprehensive model for the phenotypic and genetic structure of the quantitative traits. One approach is to integrate the different metrics into a low dimensional framework (i.e., a few variables that capture most of the trait variation) and subsequently use this to identify the genetic basis of the phenotypic relationship between grain size and shape. Another problem is that the use of single biparental mapping populations reveals only part of the genetic architecture of the traits and restrains identification of background-specific alleles. The inference power of quantitative analysis to determine the genetic architecture of a trait could be enhanced by analyzing, in the same experiment, multiple populations that represent a wider sample of genetic variation present (Holland, 2007).
Therefore, to gain deeper insights into the genetic basis of grain size and shape variation, several different populations of recombinant doubled haploids (DH) that capture a broad spectrum of the phenotypic variation present in the elite winter wheat germplasm pool were exploited. Furthermore, grain material from accessions of primitive wheat species and modern elite varieties were measured to determine the phenotypic structure of the traits and assess the extent of variation retained in domesticated wheat. We show that grain size is largely independent of grain shape both in the DH populations and in the primitive wheat species and that there is a significant reduction of phenotypic variation in grain shape in the breeding germplasm pool probably as a result of relatively recent bottleneck. This phenotypic structure is attributed to a distinct genetic architecture where common genetic components are involved in the control of those traits in different wheat varieties.
RESULTS
Variation and Heritability of Grain Size and Shape in DH Populations
Six morphometric parameters, 1000-grain weight (TGW), grain area, width (W), length (L), L/W ratio, and factor form density (FFD), which efficiently and reproducibly capture grain size and shape variation, were measured in a collection of six DH mapping populations (Table 1, Figure 1). All the measurements were performed using a digital grain analyzer assisted by an automatic image analysis suite that allowed high-throughput data collection from a large number of grains and lines. There were no significant differences between the parental lines for most of the traits (see Supplemental Table 1 online) with the exception of Beaver and Soissons that differ for area and L/W, Avalon and Cadenza that differ for length and L/W, and Savannah and Rialto that differ for TGW and FFD (see Supplemental Tables 1A, 1C, and 1E online). However, extensive transgressive segregation exists in the DH populations, with lines showing higher and lower phenotypic values from the parents for all traits (see Supplemental Figures 1 and 2 online). This indicates the polygenic inheritance of the traits with both parents contributing increasing and decreasing trait alleles. The genotypic and environmental effects both within and between different years were calculated for all populations and traits. Significant differences among DH lines (for each individual population) were found for all six traits (P < 0.001). Broad sense heritability was moderate to high for all the traits, ranging between 0.51 and 0.95 with grain length and L/W showing the highest heritability across all populations (see Supplemental Table 1 online).
Table 1.
DH Mapping Populations Studied
| Population | Abbreviation | No Lines | Environmentsa |
| Avalon × Cadenza | A × C | 202 DH | CF07, CF08 |
| Beaver × Soissons | B × S | 65 DH | CF07, CF08 |
| Shamrock × Shango | S × S | 76 DH | CF06, CF07 |
| Spark × Rialto | Sp × R | 112 DH | CF07 |
| Savannah × Rialto | Sa × R | 98 DH | CF08 |
| Malacca × Charger | M × C | 100 DH | CF07 |
CF, Church Farm, Norwich, UK.
Numerical suffixes show the years of which each experiment was carried out.
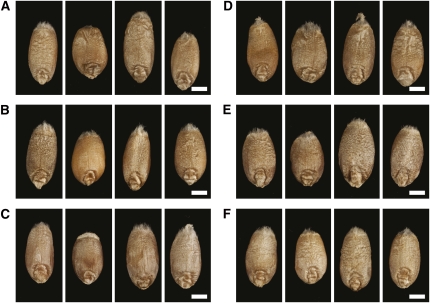
Figure 1.
Phenotypic Variation in Grain Size and Shape in Six DH Mapping Populations.
AxC (A), BxS (B), SxS (C), SaxR (D), SpxR (E), and MxC (F). Within each panel, the grains at the extremities correspond to the parental lines following the order (i.e., left or right) of the cross, while the two middle grains correspond to extreme DH lines. Bars = 2 mm.
[See online article for color version of this figure.]
Phenotypic Structure of Grain Size and Shape Variation
Simple linear correlation coefficients (Spearman's rho) were calculated between the morphological traits studied (see Supplemental Table 2 online). TGW is highly positively correlated with grain area, width, and FFD in all populations and years (r ≥ 0.75, P < 0.001) and moderately correlated with grain length (r ≥ 0.23, P < 0.001). The only exception is BxS, where TGW is not significantly correlated with grain length. Interestingly, the L/W ratio shows no significant or a very weak correlation (see Supplemental Table 2 online) with either of the two main grain size variables (TGW and grain area), suggesting that the relative proportions of the main growth axes of the grain, which largely describe grain shape, is independent of grain size.
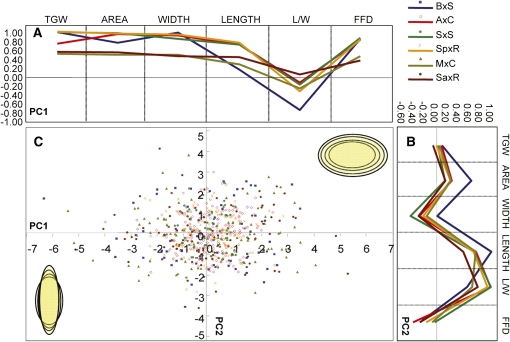
A principal component analysis (PCA) was performed to identify the major sources of variation in the morphometric data sets of each DH population (Figure 2) and on the population-wide data set (see Supplemental Table 3A online). PCA does that by identifying orthogonal directions, namely principal components (PCs), along which the trait variance is maximal (Jolliffe, 2002). The substantive importance of a given variable for a given factor can be gauged by the relative weight of the component loadings (Field, 2005). In this study, only variables with loading values of >0.4 were consider important and therefore used for interpretation following the criteria proposed by Stevens (2009) that take into account both the sample size and the percentage of shared variance between the variable and the component (Stevens, 2009). Two significant PCs, PC1 and PC2, were extracted for each DH population that capture 88.7 to 90.9% of the variation apparent in these populations (see Supplemental Table 3 online). Both PCs showed analogous organization in all six populations (Figure 2), with PC1 (55.6 to 67.1%) and PC2 (23.8 to 30.1%) capturing primarily variation in grain size and grain shape, respectively. Furthermore, PCA on a population-wide data set also identified two PCs, comparable to the ones identified for the individual DH populations, each of which explained 68.7 and 23.3% of the variation, respectively (see Supplemental Table 3A online). Therefore, PC1 describes grain size differences, where a proportional increase along both the longitudinal (length) and proximodistal (width) axes positively associates with an increase in grain area and subsequently grain weight (Figure 2C). On the other hand, PC2 captures primarily grain shape differences with L/W ratio and grain length being the main explanatory factors (Figure 2C).
Figure 2.
A Morphometric Model for Variation in Grain Morphology in Wheat Mapping Populations.
(A) and (B) Variation in grain size is captured by PC1 with both grain length and width having large effects, whereas PC2 describes variation in grain shape largely through changes in grain length. Component loading (i.e., correlations between the variables and factor) for PC1 (A) and PC2 (B) for each population are color coded.
(C) Score distribution for PC1 and PC2. Schematic representation of variation in grain size and shape captured by PC1 (x axis) and PC2 (y axis), respectively.
Genetic Architecture Is Consistent with the Phenotypic Structure for Grain Size and Shape Variation
The phenotypic model for the grain size and shape parameters (Figure 2) suggests that these two traits are probably under the control of distinct genetic components. To address this question, we identified the genetic basis underlying all six morphometric traits studied. Quantitative trait loci (QTL) analysis was performed on six DH populations for either two consecutive years (AxC, SxS, and BxS) or for 1 year only (SpxR, MxC, and SaxR). Consistent with the extensive transgressive segregation apparent in the morphometric data (see Supplemental Figures 1 and 2 online), numerous QTL with dispersed effects between the parents were identified (see Supplemental Figure 3 and Supplemental Tables 4 to 6 online). Specifically, 54 QTL were identified in AxC, 18 QTL in BxS, 10 QTL in SpxR, 10 QTL in MxC, 12 QTL in SaxR, and 13 QTL in SxS. The LOD scores and variation explained by each of these QTL range between 3.0 and 18.1, and 6.6 to 50.2%, respectively. In the AxC and BxS populations, where the broad sense heritability is very high for all the traits, most of the QTL are common between years for any given population (see Supplemental Figures 3A and 3C online). The strong positive correlations between the grain size variables (i.e., TGW, area, width, and FFD) and between the grain shape variables (i.e., L/W and length) can be attributed to cosegregating QTL with the same allelic effect. Indeed, QTL for the grain size variables cosegregated consistently in all populations and years. The same holds true for the QTL for grain length and L/W (see Supplemental Text 1 online).
These findings are consistent with the phenotypic architecture of the morphometric traits studied, where grain size is largely independent of grain shape in the individual populations as well as in the population-wide data set. To further substantiate this, QTL analysis was performed on the principal components (i.e., PC1 and PC2) extracted from each DH population (Figure 2). Similar approaches have been used before for the study of organ morphology in other species (Langlade et al., 2005; Feng et al., 2009).
A total of 25 QTL for PC1 and PC2 were identified in the six DH mapping populations with LOD scores ranging between 2.9 and 10.6, and the amount of variation explained was between 9.2 and 36.4% (see Supplemental Table 7 online). The majority of the QTL identified are located on five chromosomes, 1A, 3A, 4B, 5A, and 6A (two colocated QTL or more). Three QTL for PC2 were detected in the BxS, SpxR, and SaxR on chromosome 1A, each of which explained 29.2, 11.3, and 18.5% of the variation in grain shape, respectively (Figure 3; see Supplemental Figure 4 online). Meta-analysis identified two QTL at close proximity to each other, MQTL1 between markers psp3027 and wPt5374 and MQTL2 between GluA1 and s12/m25.6 on the consensus map for 1A (Figure 3, Table 2). Significant effects both for PC1 and PC2 were identified in AxC, BxS, and SpxR on chromosome 3A. Specifically, two QTL for PC2 were detected in AxC and SpxR populations around markers barc19 and wmc264, respectively, while one QTL for PC1 was identified in BxS around marker gwm2 (Figure 3). Meta-analysis revealed one meta-QTL that spanned the interval s635ACAG-barc19 (Figure 3, Table 2). Two QTL for PC1 were detected in BxS and AxC populations on chromosome 4B, around markers s14/m15.6 and wmc349, respectively (Figure 3). One meta-QTL was identified between the markers gwm149 and wmc47 on the wheat consensus map (Figure 3, Table 2).
Figure 3.
Genetic Structure Underlying Phenotypic Variation.
Common genetic components for grain size and shape in the DH mapping populations. Schematic representation of colocated QTL identified for PC1 and PC2 on five main chromosomes (i.e., 1A, 3A, 4B, 5A, and 6A) and their corresponding meta-QTL. QTL for each DH population are color coded. Meta-QTL are depicted in red. The length of each QTL denotes 2-LOD confidence intervals on the wheat consensus map (Somers et al., 2004).
Table 2.
Meta-QTL Identified across All Populations and Traits
| Traitsa |
PCb |
||
| Chromosome | Mapping Intervalc | Chromosome | Mapping Interval |
| 1A | barc263-wPt7030 | 1A | psp3027-wPt5374 |
| 1A | wPt-8347-s12/m25.6 | 1A | GluA1-s12/m25.6 |
| 2A | wmc819-gwm47 | 3A | s635ACAG-barc19 |
| 2D | cfd233-wmc41 | 3A | gwm149-wmc47 |
| 2D | gwm349-wmc167 | 5A | wPt4131-gwm293 |
| 3A | wPt1688-barc45 | 5A | wmc492-gwm666 |
| 3A | wPt9562-wPt9215 | 5A | cfa2185-wmc727 |
| 3A | barc19-cfa2262 | 6A | psp3029-wmc179 |
| 4B | s14/m15.6-wmc349 | 6A | wPt5549-wPt5480 |
| 5A | gwm443-cfa2104 | ||
| 5A | wmc492-wmc475 | ||
| 5A | wPt0654-wPt3069 | ||
| 5A | wmc110-wmc727 | ||
| 6A | wmc6807-psp3071 | ||
| 6A | cos07Tb-wPt5480 | ||
Meta-QTL identified for the individual morphometric traits. Only meta-QTL that correspond to more than two different populations are shown. A detailed description of the QTL for the individual traits and their corresponding meta-QTL can be found in Supplemental Text 1 online.
Meta-QTL identified for the individual PCs. Only meta-QTL that correspond to more than two different populations are shown.
Meta-QTL mapping intervals on the consensus map (Somers et al., 2004).
QTL both for PC1 and PC2 were identified in AxC, SxS, SpxR, and SaxR populations on chromosome 5A (Figure 3). Meta-analysis identified three meta-QTL in the intervals wPt4131-gwm293, wmc492-gwm666, and cfa2185-wmc727 (Figure 3, Table 2). Effects for PC1 were identified in AxC, MxC, and SaxR in the middle of chromosome 6A, whereas a QTL for PC2 was detected in AxC on the long arm of 6A (Figure 3). Two meta-QTL were identified: MQTL8 corresponds to the PC1-specific QTL between psp3029 and wmc179, and MQTL9 corresponds to PC2 QTL between wPt5549 and wPt5480 (Figure 3, Table 2).
Changes in Grain Size and Shape through Domestication and Breeding
Wheat domestication led predominantly to an increase in grain size, a phenomenon that has been further intensified by continuous breeding of hexaploid wheat. However, it is still unclear how these processes impacted on grain shape and how much of the grain size/shape variation apparent in the wild wheat species still remains in the modern wheat varieties and breeding germplasm.
To address these questions, ancestral wheat species originating from the broader Fertile Crescent region were analyzed for variation in grain size and shape (Figure 4). The collection that was analyzed includes all the known species of the genus Triticum based on the most commonly accepted taxonomic and phylogenetic classification for wheat (Feldman, 2001; Golovnina et al., 2007). Moreover, to assess the variation within each species, a comprehensive sampling and analysis of different accessions of each species was performed, each of which corresponded to a distinct geographical locale or country of origin (http://data.jic.bbsrc.ac.uk/cgi-bin/germplasm/triticeae/).
Figure 4.
Phenotypic Variation in Grain Size and Shape in Ancestral Wheat Species.
Representative grains are shown for 22 species and subspecies of the genus Triticum. Species are organized according to the ploidy level from diploids to hexaploids. The genome of each species is given in parentheses. T. sinskajae is a mutant free-threshing form of T. monococcum (Goncharov et al., 2007), T. militinae represents a free-threshing mutant form of T. timopheevii (Feldman, 2001), and T. vavilovii (BBAuAuDD) and T. zhukovskyi (BBAAAmAm) are hexaploid hulled wheats. Bar = 3 mm.
[See online article for color version of this figure.]
Extensive variation exists for grain area (mean ± sd = 22.8 mm2 ± 3.6, range = 18.4 mm2), length (mean ± sd = 8.5 ± 1.1 mm, range = 5.6 mm), and L/W (mean ± sd = 2.6 ± 0.6, range = 2.64; see Supplemental Figure 5 and Supplemental Table 8 online), whereas grain width is the least variable trait (mean ± sd = 3.3 ± 0.4 mm, range = 1.94 mm). Similar to the results from the DH populations, grain size appeared largely independent from grain shape in the primitive wheat species. Indeed, grain area showed no significant correlation with L/W (r = 0.13, P = 0.335), whereas length and width were significantly correlated with both area (r = 0.67, r = 0.45, P < 0.001, respectively) and L/W (r = 0.67, r = −0.75, P < 0.001, respectively).
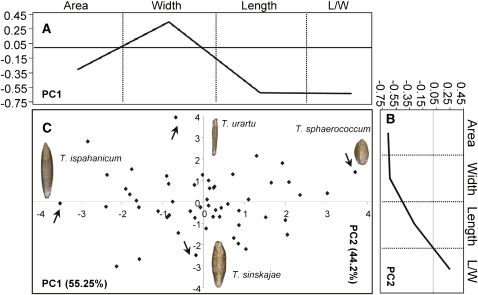
PCA identified two significant PCs that collectively explained 99.5% of the total variation of the traits in primitive wheat (Figure 5). For PC1 (55.25%), the main explanatory factors were L/W and grain length, whereas for PC2 (44.2%), grain area and width were the main explanatory factors (Figures 5A and 5B). Therefore, PC1 captured variation in grain shape primarily through changes in grain length, and PC2 captured variation in grain size through changes in width.
Figure 5.
A Morphometric Model for Variation in Grain Morphology in Ancestral Wheat Species.
Variation in grain shape and size is captured by PC1 and PC2, respectively.
(A) and (B) Component loading for PC1 (A) and PC2 (B), as in Figure 2.
(C) Score distribution for PC1 and PC2. Grains of the species that correspond to extreme size or shape phenotypes (arrows) are shown. Variation explained by each principal component is shown in parentheses.
[See online article for color version of this figure.]
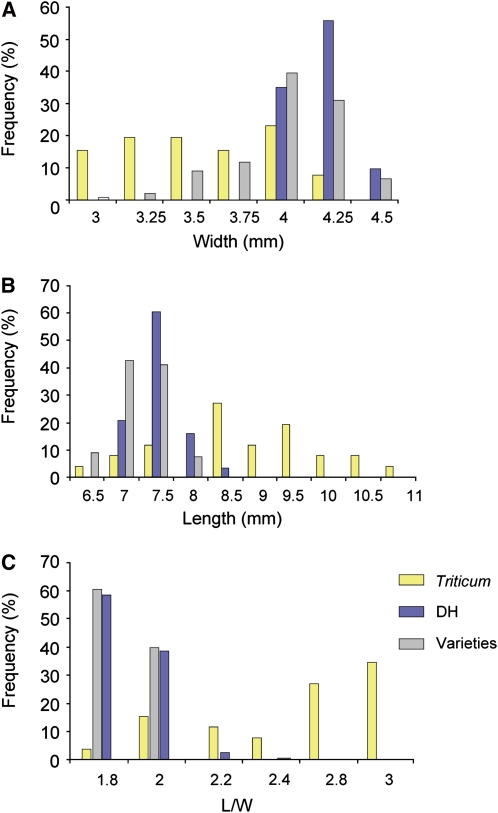
Triticum sphaerococcum and Triticum ispahanicum correspond to the two extreme grain shape phenotypes along PC1, with the latter having approximately twofold longer (L/W = 3.4) but almost as wide grains as T. sphaerococcum (L/W = 1.5) (Figure 5C). On the other hand, Triticum urartu and Triticum sinskajae represent the two extreme phenotypes in terms of grain size along PC2 (Figure 5C), with T. sinskajae having approximately twice the grain size (∼25 mm2 versus 14.6 mm2) primarily due to increased grain width (∼4 mm versus 2.2 mm). Thus, it is evident that broad variation both in grain size and shape exists in the primitive wheat species in contrast with the DH populations and modern varieties where grain shape variation is considerably reduced. Indeed, ∼70% of the variation captured by PC1 in the DH population data sets is attributed primarily to grain size differences with only ∼24% attributed to variation in grain shape (Figure 2) as opposed to ∼44 and ∼55% in the wild species, respectively. Much of the grain shape variation present in the primitive wheat species has been lost in the modern breeding germplasm probably due to selection for more uniform grain shape in the elite varieties. Figures 6A to 6C illustrate how grain dimensions (i.e., length and width) and their relative proportions (i.e., L/W) have changed during domestication and breeding. Specifically, grain width appeared markedly increased in the DH populations and in the collection of elite varieties examined (mean = 4.08 mm; see Supplemental Table 9 online), whereas grain length is decreased (mean = 7.3 mm) compared with the wild species (mean = 3.3 and 8.52 mm, respectively) (see Supplemental Table 8 online). These changes in grain axial dimensions resulted in grain shape modifications that led from a predominant long and thin primitive grain (meanL/W = 2.63) to a much shorter and wider grain in the DH populations and elite varieties (meanL/W = 1.78). Moreover, there is a much wider spectrum of grain shape phenotypes in the wild species (rangeL/W = 2.64, minimum = 1.54, maximum = 4.2) compared with the DH populations and elite varieties (rangeL/W ∼0.4, minimum ∼1.6, maximum ∼2.0).
Figure 6.
Changes in Grain Axial Dimensions in Ancestral and Modern Wheat.
Frequency distributions for grain width (A), length (B), and L/W (C) in the DH mapping populations, Triticum species, and wheat varieties.
[See online article for color version of this figure.]
DISCUSSION
We used high-throughput morphometric analysis to quantify grain size and shape variation and determine its underlying genetic basis in an extensive collection that included DH mapping populations, ancestral wheat species, and commercial varieties.
Quantitative analyses of the morphometric data revealed that grain size and shape are largely independent traits. This is unlikely to be the result of artificial selection during breeding since size and shape are also independent variables in primitive wheat. At the developmental level, this phenomenon may reflect differential modulation in growth (or growth arrest) along the main axes of the grain at different developmental stages. In tomato (Solanum lycopersicum), for example, loci have been identified that affect fruit shape but not size and vice versa (Frary et al., 2000; Tanksley, 2004). Specifically, the fw2.2 gene was shown to be a major determinant for fruit size but not shape variation (Frary et al., 2000), whereas the ovate (Liu et al., 2002) and sun (van der Knaap and Tanksley, 2001) loci alter fruit shape with little effect on size due to asymmetric growth in the longitudinal axis of the carpels and young fruit, respectively. The notion that certain developmental constraints during fruit or grain growth could lead to morphological changes is further corroborated by recent studies on grain size/shape genes in rice (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Takano-Kai et al., 2009). The GS3 locus was found to have major effects on grain length and weight and smaller effects on grain width (Fan et al., 2006), and the longer grains can be attributed to relaxed constraints during grain elongation (Takano-Kai et al., 2009). The GW2 gene was shown to alter grain width and weight and to lesser extend grain length owing to changes in the width of the spikelet hull (Song et al., 2007). Similarly, the SW5 gene has been reported to effect grain width by modulating the size of the outer glume (Shomura et al., 2008).
Recent advances in comparative sequence analysis between wheat and rice genomes have confirmed extensive synteny between the two species (Sorrells et al., 2003; Quraishi et al., 2009). This enables us to assess the positional correspondence between QTL identified in wheat and known QTL or loci that affect grain morphology in rice. Several QTL for grain weight and length (Thomson et al., 2003; Li et al., 2004) were reported on rice chromosome 3 and correspond to QTL for related traits identified in this study. Specifically, rice QTL for grain weight correspond to TGW and grain width QTL on the syntenic regions of wheat chromosomes 5A and 6A. The rice gw3.1 QTL (Thomson et al., 2003) and its underlying loci GS3 (Fan et al., 2006; Takano-Kai et al., 2009) correspond to the very strong wheat QTL for grain size on chromosome 4B. The GS3 effect on grain size is attributed primarily to alterations in grain length and less so in width (Fan et al., 2006). However, the grain weight QTL on wheat chromosome 4B cosegregates consistently only with grain width QTL, suggesting a different mechanism. It was recently reported (Li et al., 2010) that the orthologous gene of GS3 in maize (Zea mays) affects kernel weight but also through a different mechanism to that described in rice. The GW2 locus on rice chromosome 2 does not correspond to any of the identified wheat QTL. However, several grain weight, width, and length rice QTL correspond to grain size (TGW and grain width) wheat QTL on chromosome 6A. Such orthologous relationships between different species should remain speculative until positive confirmation through gene-specific comparative analysis is provided.
The observed phenotypic structure might reflect an evolutionary process in which grain size and shape have been selected and fixed independently over the wheat domestication. Our analysis clearly demonstrates that PC1 and PC2 are under the control of distinctly different genetic components in all of the mapping populations studied. Even when the various morphometric traits were considered separately, the QTL for either grain size or grain shape parameters were frequently colocated in the different mapping populations, indicating that common genetic components underlie the observed variation in grain morphology. Interestingly, there are a number of differences in the QTL distribution across the populations, even between populations that share a common parent, showing that identifying background-specific effects by analyzing multiple populations is crucial in determining the genetic architecture. This becomes more apparent when we consider the results from previous studies, most of which analyzed single populations.
In this study, QTL have been identified in most of the homeologous groups, but those on chromosomes 1A, 3A, 4B, 5A, and 6A have the largest and most consistent (across different populations) effects on grain size and/or shape.
Strong QTL for grain length and grain shape (PC2) were identified on chromosome 1A in three populations, BxS, SpxR, and SaxR. Effects for grain length on 1A have been previously reported. Grain length was negatively affected by chromosome 1A in a disomic population derived from a cross between long and short grain parental lines (Giura and Saulescu, 1996). A QTL for grain length was identified in a recombinant inbred line population between two Chinese winter elite varieties at an equivalent chromosomal position on 1A as the QTL in BxS, SpxR, and SaxR reported in this study (Sun et al., 2009).
Homoeologous group 3, especially chromosome 3A, has a strong effect both on grain size and shape across the DH populations studied here, yet QTL for related traits (grain length and area) have been reported only in one previously studied population on chromosome 3B (Campbell et al., 1999). The QTL for grain size and shape on chromosome 3A identified in this analysis are of particular interest since their mapping intervals coincide with the approximate position of the sphaerococcum locus (Salina et al., 2000). The sphaerococcoid mutation reduces grain length, thus significantly altering grain shape and size. The three genes S1, S2, and S3 of the sphaerococcoid mutation were mapped on chromosomes 3D, 3B, and 3A, respectively (Salina et al., 2000). The S3 locus was shown to be located between the centromeric markers gwm2 and gwm720 on 3A (Salina et al., 2000). This position corresponds to the mapping interval (highest LOD scores) of QTL for grain length and L/W in AxC and grain width in BxS. QTL for grain length in SaxR and grain width and L/W in MxC were adjacent to the S3 locus mapping interval mentioned above.
Strong grain size QTL have been identified on chromosomes 4B and 4D in the BxS and AxC populations. However, effects of chromosome 4B on grain length and width have been previously identified only in one population (Giura and Saulescu, 1996). The AxC and BxS populations segregate for the Rht-B1 and Rht-D1 homoeoalleles (frequently referred as dwarfing genes), respectively, that were shown to affect grain weight, among other traits (Flintham et al., 1997). Therefore, the proximity of these grain size QTL to Rht-B1 and Rht-D1 might indicate further pleiotropic effects of this loci on other grain size parameters. Chromosome 5A was found to have large effects on both grain size and shape-related parameters in four of the six populations studied. None of the previous studies identified QTL for related traits on this chromosome, but significant associations between loci on 5A and grain length were reported before (Breseghello and Sorrells, 2006). In this study, strong QTL for grain size-related traits have been identified on chromosome 6A in three DH populations (AxC, SaxR, and MxC) and in two previously described studies (Giura and Saulescu, 1996; Sun et al., 2009). Specifically, QTL for grain width (Sun et al., 2009) have been detected before at an approximately equivalent interval as the grain width QTL in the AxC and SaxR populations. Chromosome 6A was also found to have a positive effect on FFD (Giura and Saulescu, 1996) consistent with the effects detected in the AxC and SaxR populations.
Therefore, comparison of the QTL described here with those identified in other studies showed that allelic variation for some, for example, effects on 1A and 6A, appear to occur frequently in diverse germplasm. However, the majority of them appear unique to this study, most notably effects on 3A, 4B, and 5A.
Morphometric analysis shows that wheat grain evolved from a long and thin primitive grain to a much wider and shorter modern grain. Moreover, variation of grain shape appeared significantly reduced not only in the modern elite varieties but also in the mapping populations, suggesting that the natural genetic diversity present in the ancestral wheat species has been reduced during the development of the modern elite cultivars. One possibility is that diversity in grain shape was reduced as a result of the polyploidy speciation of T. aestivum. However, hexaploid wheat retained a relatively large percentage of the nucleotide diversity of A and B genomes found in the tetraploid ancestors, suggesting that ploidy differences imposed a weak barrier to gene flow, at least from the tretraploid ancestor, during the emergence of the hexaploid wheat (Dvorak et al., 2006; Haudry et al., 2007). Overrepresentation of grain size and shape QTL in the A and B genomes further supports this notion. The data presented here (see Supplemental Table 8 online) indicate that the phenotypic variation for the main grain shape parameters, grain length and L/W, has actually increased in the hexaploid species (T. aestivum ssp; rangeL = 3.1 mm; rangeL/W = 0.91) compared with that of the tetraploid ones (T. turgidum ssp; rangeL = 2.0 mm; rangeL/W = 0.6). The other possible explanations are that the reduction in grain shape variation apparent in the modern germplasm and elite varieties might have appeared either at very early stages of the evolution of common wheat (ssp aestivum) or at later stages after the emergence of the subspecies.
T. aestivum ssp spelta and ssp macha represent hexaploid hulled wheats (Feldman, 2001) and were very similar to the tetraploid species in terms of grain shape (see Supplemental Table 8 online). It is believed that T. aestivum ssp spelta, at least the Asiatic type, gave rise to nonhulled or free-threshing common wheat (ssp aestivum) (Feldman, 2001). Therefore, the reduced variation in grain shape observed in common wheat may have been the result of a bottleneck that occurred during the transition from the hulled to free-threshing form. The lowest trait value in the hexaploid species is provided by T. aestivum ssp sphaeroccocum, which is thought to have emerged from T. aestivum ssp aestivum as a result of mutation at the S gene and imposed a significant alteration in grain shape (Salina et al., 2000). Thus, part of the grain shape diversity found in hexaploid wheat (T. aestivum ssp) has risen relatively recently and most likely after the emergence of common wheat (ssp aestivum). This then suggests that strong mutations, such as the sphaeroccocum, were selected against at the early stages of wheat breeding possibly because of undesirable pleiotropic effects on other traits. Further selection at later stages for certain grain morphology and greater uniformity among the different varieties could have limited the genetic variation in the gene pool even more and subsequently resulted in the predominant grain shape found in the modern elite varieties.
The present genetic and phenotypic structure supports an emerging model for grain size and shape variation, where grain size has progressively increased through alterations both in grain width and length, followed at later stages by modifications in grain shape largely through changes in grain length. Elucidating the genetic basis of variation in grain size and shape in wheat is instrumental to the effort to improve yield potential and processing performance, especially in the current climate where food security is at the epicenter of crop research worldwide.
METHODS
Genetic Resources and Field Trials
The DH populations and genetics maps used in this study were generated as was described previously (Snape et al., 2007; Griffiths et al., 2009) and summarized in Table 1. Specifically, they are as follows: Avalon × Cadenza (AxC), Beaver × Soissons (BxS), Shamrock × Shango (SxS), Spark × Rialto (SpxR), Malacca × Charger (MxC), and Savannah × Rialto (SaxR). The populations were grown in randomized, replicated field trials (three replicates) at Church Farm, Norwich, UK, over two consecutive years: 2007 and 2008 (AxC and BxS), and 2006 and 2007 (SxS). The SpxR, MxC, and SaxR populations were grown only in 2007 and 2008, respectively. The lines were grown in large-yield plots (1 × 5 m2) following standard agronomic practices. Grain material of primitive wheat accessions was provided by the John Innes Centre Triticeae Collection. The accession numbers of the species studied are given in Supplemental Table 8 online, and further information can be found at http://data.jic.bbsrc.ac.uk/cgi-bin/germplasm/triticeae/.
Morphometric Analysis
Morphometric measurements were performed on 200 to 250 grains/line using the MARVIN grain analyzer (GTA; Sensorik). Specifically, TGW, grain width (W), length (L), and grain area were measured. The ratio L/W and the FFD were calculated. FFD describes the differences in grain density and the deviation of a shape from a cylindrical form and is given by: grain weight/(grain length * grain width) (Giura and Saulescu, 1996).
Statistical Analysis
Descriptive statistics and normality tests on the quantitative data were performed using GenStat v 11. Analysis of variance was performed to estimate the relative genetic contribution to trait variation for each population and year. Broad sense heritability was calculated by h2 = 1 − M2/M1, where M1 and M2 are the mean square values for genotype and genotype × environment (for the two years trials), respectively (Knap et al., 1985). Mean values of the three replicates for each year were used to calculate the correlation coefficients (Pearson's correlation and Spearman's rho) and for the QTL mapping. PCA was performed on each population (mean values for each year) and on a population-wide (P-W) data set using SPPS 12.0. For the extraction of PCs, the correlation matrix method was used. Only the factors with an eigenvalue ≥1 according to Kaiser's criterion were retained (Field, 2005).
QTL and Meta-QTL Analysis
The MapQTL 5.0 software (Van Ooijen, 2004) was used for the analysis of the quantitative data. Single-interval mapping was initially used to identify QTL followed by an automatic cofactor selection process (Van Ooijen, 2004). The resulted set of cofactors was then used in composite-interval mapping. A genome-wide threshold LOD value for significant QTL was set at 3.1 (P < 0.05) by performing 10,000 permutations of the original data.
Meta-QTL analysis was performed using Biomercator software v. 2.1. The published consensus map (Somers et al., 2004) was used as a reference map upon which the genetic linkage maps of the six populations were subsequently projected. QTL and 2-LOD confidence intervals were projected together with the genetic linkage maps. Meta-analysis was conducted initially for each population and chromosome followed by a population-wide analysis for meta-QTL across all the traits and years. The number of meta-QTL present was determined as the model that minimized the Akaike criterion (Arcade et al., 2004).
Author Contributions
V.C.G., J.W.S., J.H.D., and S.G. conceived and designed the experiments. V.C.G. and A.N. performed the experiments. V.C.G. analyzed the data. J.S., L.F., L.S., and S.O. provided technical support. V.C.G., J.W.S., and J.H.D. wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Frequency Distributions of the Morphometric Traits for AxC, BxS, and SxS.
Supplemental Figure 2. Frequency Distributions of the Morphometric Traits for Spark × Rialto, Savannah × Rialto, and Malacca × Charger.
Supplemental Figure 3. QTL Identified for the Morphometric Parameters in the Six Mapping Populations.
Supplemental Figure 4. Common Genetic Components for Grain Size and Shape in the DH Mapping Populations.
Supplemental Figure 5. Morphometric Traits for Triticum Species.
Supplemental Table 1. Trait Variation and Heritability in the Parental Lines and DH Mapping Populations.
Supplemental Table 2. Spearman Rank Correlation Coefficients between Morphometric Traits.
Supplemental Table 3. Principal Component Analysis on the Mapping Populations.
Supplemental Table 4. QTL for the Morphometric Traits in the AxC Population.
Supplemental Table 5. QTL for the Morphometric Traits in the SxS and BxS Populations.
Supplemental Table 6. QTL for the Morphometric Traits in the SpxR, MxC, and SaxR Populations.
Supplemental Table 7. QTL for the Principal Components in the AxC, SxS, BxS, SpxR, MxC, and SaxR Populations.
Supplemental Table 8. Morphometric Data on Triticum Species.
Supplemental Table 9. Morphometric Data on 61 Commercial Varieties.
Supplemental Text 1. Details of the QTL Locations and Effects on Each Homoeologous Group.
Supplementary Material
Acknowledgments
We thank Peter Shaw for helpful discussion and comments on the manuscript and Luzie Wingen for advice on QTL meta-analysis. We also thank Andrew Davis for photography. This work was supported by funding from the Biotechnology and Biological Sciences Research Council through the Crop Science Initiative (Grant BB/E00721x/1).
References
- Arcade A., Labourdette A., Falque M., Mangin B., Chardon F., Charcosset A., Joets J. (2004). BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20: 2324–2326 [DOI] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M.E. (2006). Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello F., Sorrels M.E. (2007). QTL analysis of kernel size and shape in two hexaploid wheat mapping populations. Field Crops Res. 101: 172–179 [Google Scholar]
- Brown T.A., Jones M.K., Powell W., Allaby R.G. (2009). The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol. Evol. 24: 103–109 [DOI] [PubMed] [Google Scholar]
- Campbell K.G., Bergman C.J., Gualberto D.G., Anderson J.A., Giroux M.J., Hareland G., Fulcher R.G., Sorrells M.E., Finney P.L. (1999). Quantitative trait loci associated with kernel traits in a soft x hard wheat cross. Crop Sci. 39: 1184–1195 [Google Scholar]
- Dholakia B.B., Ammiraju J.S.S., Singh H., Lagu M.D., Röder M.S., Rao V.S., Dhaliwal H.S., Ranjekar P.K., Gupta V.S., Weber W.E. (2003). Molecular marker analysis of kernel size and shape in bread wheat. Plant Breed. 122: 392–395 [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Akhunov E.D., Akhunov A.R., Deal K.R., Luo M.C. (2006). Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23: 1386–1396 [DOI] [PubMed] [Google Scholar]
- Evers A.D. (2000). Grain size and morphology: Implications for quality. Wheat Structure, Biochemistry and Functionality, Schofield D., (London: Royal Society of Chemistry; ), pp. 19–24 [Google Scholar]
- Evers A.D., Cox R.I., Shaheedullah M.Z., Withey R.P. (1990). Predicting milling extraction rate by image analysis of wheat grains. Asp. Appl. Biol. 25: 417–426 [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Feldman M. (2001). Origin of cultivated wheat. The World Wheat Book: A History of Wheat Breeding, Bonjean A.P., (Andover, UK: Intercept; ), pp. 3–56 [Google Scholar]
- Feng X., Wilson Y., Bowers J., Kennaway R., Bangham A., Hannah A., Coen E., Hudson A. (2009). Evolution of allometry in antirrhinum. Plant Cell 21: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A. (2005). Exploratory factors analysis. Discovering Statistics Using SPSS, Wright D.B., (London: SAGE Publications; ), pp. 619–680 [Google Scholar]
- Flintham J.E., Borner A., Worland A.J., Gale M.D. (1997). Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128: 11–25 [Google Scholar]
- Frary A., Nesbitt T.C., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Tanksley S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fuller D.Q. (2007). Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. (Lond.) 100: 903–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giura A., Saulescu N.N. (1996). Chromosomal location of genes controlling grain size in a large grained selection of wheat (Triticum aestivum L.). Euphytica 89: 77–80 [Google Scholar]
- Golovnina K.A., Glushkov S.A., Blinov A.G., Mayorov V.I., Adkison L.R., Goncharov N.P. (2007). Molecular phylogeny of the genus Triticum L. Plant Syst. Evol. 264: 195–216 [Google Scholar]
- Goncharov N.P., Kondratenko E., Bannikova S.V., Konovalov A.A., Golovnina K.A. (2007). Comparative genetic analysis of diploid naked wheat Triticum sinskajae and the progenitor T. monococcum accession. Russ. J. Genet. 43: 1248–1256 [PubMed] [Google Scholar]
- Griffiths S., et al. (2009). Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theor. Appl. Genet. 119: 383–395 [DOI] [PubMed] [Google Scholar]
- Haudry A., Cenci A., Ravel C., Bataillon T., Brunel D., Poncet C., Hochu I., Poirier S., Santoni S., Glemin S., David J. (2007). Grinding up wheat: A massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24: 1506–1517 [DOI] [PubMed] [Google Scholar]
- Holland J.B. (2007). Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10: 156–161 [DOI] [PubMed] [Google Scholar]
- Jolliffe I.T. (2002). Principal Component Analysis. (New York: Springer; ). [Google Scholar]
- Knap S.J., Stroup W.W., Ross W.M. (1985). Exact confidence intervals for heritability on a progeny mean basis. Crop Sci. 25: 192–194 [Google Scholar]
- Kovach M.J., Sweeney M.T., McCouch S.R. (2007). New insights into the history of rice domestication. Trends Genet. 23: 578–587 [DOI] [PubMed] [Google Scholar]
- Langlade N.B., Feng X., Dransfield T., Copsey L., Hanna A.I., Thebaud C., Bangham A., Hudson A., Coen E. (2005). Evolution through genetically controlled allometry space. Proc. Natl. Acad. Sci. USA 102: 10221–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Thomson M., McCouch S.R. (2004). Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168: 2187–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yang X., Bai G., Warburton M.L., Mahuku G., Gore M., Dai J., Li J., Yan J. (2010). Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 120: 753–763 [DOI] [PubMed] [Google Scholar]
- Liu J., Van Eck J., Cong B., Tanksley S.D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 99: 13302–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar S.J., Whitworth M.B., Evers A.D. (1997). Image analysis: The prediction and assessment of wheat quality and milling properties. Proceedings of the International Wheat Quality Conference, Steele J.L., Chung K.O., (Manhattan, KS: Grain Industry Alliance; ), pp. 141–151 [Google Scholar]
- Quraishi U.M., Abrouk M., Bolot S., Pont C., Throude M., Guilhot N., Confolent C., Bortolini F., Praud S., Murigneux A., Charmet G., Salse J. (2009). Genomics in cereals: From genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct. Integr. Genomics 9: 473–484 [DOI] [PubMed] [Google Scholar]
- Salina E., Börner A., Leonova I., Korzun V., Laikova L., Maystrenko O., Röder M.S. (2000). Microsatellite mapping of the induced sphaerococcoid mutation genes in Triticum aestivum. Theor. Appl. Genet. 100: 686–689 [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Snape J., Foulkes M., Simmonds J., Leverington M., Fish L., Wang Y., Ciavarrella M. (2007). Dissecting gene × environmental effects on wheat yields via QTL and physiological analysis. Euphytica 154: 401–408 [Google Scholar]
- Somers D.J., Isaac P., Edwards K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Sorrells M.E., et al. (2003). Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J.P. (2009). Exploratory and confirmatory factor analysis. Applied Multivariate Statistics for the Social Sciences (New York: Routledge, Taylor & Francis Group; ), pp. 325–381 [Google Scholar]
- Sun X.-Y., Wu K., Zhao Y., Kong F.-M., Han G.-Z., Jiang H.-M., Huang X.-J., Li R.-J., Wang H.-G., Li S.-S. (2009). QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 165: 615–624 [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., Padhukasahasram B., Bustamante C., Yoshimura A., Doi K., McCouch S. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S.D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16: S181–S189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M.J., Tai T.H., McClung A.M., Lai X.H., Hinga M.E., Lobos K.B., Xu Y., Martinez C.P., McCouch S.R. (2003). Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]
- van der Knaap E., Tanksley S.D. (2001). Identification and characterization of a novel locus controlling early fruit development in tomato. Theor. Appl. Genet. 103: 353–358 [Google Scholar]
- Van Ooijen J.W. (2004). MapQTL: Software for the Mapping of Quantitative Trait Loci in Experimental Populations. (Wageningen, The Netherlands: Kyazma; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.