Single-electron reductions of host plant benzoquinones are catalyzed by a parasitic plant quinone oxidoreductase. This is one of the first steps in the haustorium development signal transduction pathway.
Abstract
Parasitic plants in the Orobanchaceae develop haustoria in response to contact with host roots or chemical haustoria-inducing factors. Experiments in this manuscript test the hypothesis that quinolic-inducing factors activate haustorium development via a signal mechanism initiated by redox cycling between quinone and hydroquinone states. Two cDNAs were previously isolated from roots of the parasitic plant Triphysaria versicolor that encode distinct quinone oxidoreductases. QR1 encodes a single-electron reducing NADPH quinone oxidoreductase similar to ζ-crystallin. The QR2 enzyme catalyzes two electron reductions typical of xenobiotic detoxification. QR1 and QR2 transcripts are upregulated in a primary response to chemical-inducing factors, but only QR1 was upregulated in response to host roots. RNA interference technology was used to reduce QR1 and QR2 transcripts in Triphysaria roots that were evaluated for their ability to form haustoria. There was a significant decrease in haustorium development in roots silenced for QR1 but not in roots silenced for QR2. The infrequent QR1 transgenic roots that did develop haustoria had levels of QR1 similar to those of nontransgenic roots. These experiments implicate QR1 as one of the earliest genes on the haustorium signal transduction pathway, encoding a quinone oxidoreductase necessary for the redox bioactivation of haustorial inducing factors.
INTRODUCTION
Plant-derived phenolic molecules play critical roles in coordinating symbioses between plant roots and associated organisms in the rhizosphere (Siqueira et al., 1991; Hirsch et al., 2002). From microbes to nematodes, pathogens, and symbionts, the behavior of rhizosphere communities are controlled to a large extent by small phenolic molecules presented by or released from plant roots (Bais et al., 2006; Faure et al., 2009). Phenolic molecules are also thought to coordinate interactions between roots of different plants (McCully, 2007, 1999). Certain phenolics, such as syringic acid and catechin, have been implicated as causative agents in allelopathy, the phenomenon whereby plants release phytotoxic compounds that inhibit the growth of nearby plants. While the effects of allelopathy have apparently been known for over 2000 years (Willis, 2007), its study has been hampered by the difficulty reproducing the pathological phenotype (Inderjit and Nilsen, 2003).
The most obvious phenotypes associated with root–root interactions are presented by parasitic species of Orobanchaceae (Kuijt, 1969). In the presence of host plants, these parasitic plants develop globular haustoria on their roots that attach to and penetrate host roots (Musselman, 1980; Riopel and Timko, 1995). Upon successful invasion of the host root, the haustorium matures into a parasite-specific organ that functions as the physiological bridge through which the parasite robs the host of water, minerals, and nutrients. In some obligate parasites, notably the agronomically devastating parasitic weeds Striga and Orobanche, the tip meristem of the primary radical develops into a terminal haustorium that needs to successfully invade a host before seedling development advances. Most Orobanchaceae are facultative parasites that can mature without parasitizing a host. In a field where neighboring host roots are available, facultative parasites develop haustoria at lateral positions on their roots. A single parasite root can have several haustoria attached to one or more host roots (Marvier, 1998). Facultative parasites are typically generalist feeders with a broad host range. For example, the parasitic plant Triphysaria versicolor parasitizes a wide range of monocots and dicots, including Arabidopsis thaliana, Medicago truncatula, maize (Zea mays), and rice (Oryza sativa; Thurman, 1966; Estabrook and Yoder, 1998; Jamison and Yoder, 2001).
The development of haustoria on Orobanchaceae roots is initiated by contact or close proximity to a host root. Previous investigations showed that water-soluble exudates of host roots induced haustoria when applied to parasite root tips in vitro (William, 1961; Atsatt et al., 1978; Riopel and Musselman, 1979). Haustorium ontogeny is similar between different Orobanchaceae, with the caveat that lateral haustoria do not terminally differentiate the tip meristem (Okonkwo and Nwoke, 1978; Baird and Riopel, 1984; Joel and Losner-Goshen, 1994). Upon contact of the parasite root with that of a host, there is an almost immediate cessation of parasite tip growth. This is soon followed by an isodiametric expansion of cortical cells within the parasite root that result in a noticeable bump at or near the tip meristem within 24 h. There is a concomitant elongation of epidermal cells into long, densely positioned haustorial hairs that are capable of adhering to host tissues (Baird and Riopel, 1985). Cortical swelling and haustorial hair proliferation are visual phenotypes of early haustorium development that occur prior to host contact. Once the parasite has firmly attached to the host, a penetration peg invades the host epidermis and cortex by a combination of physical and enzymatic processes until it reaches the host stele. Within a few days of host contact, a successful haustorium will have invaded the host and established a functional connection between host and parasite vascular systems.
The first chemical haustorium-inducing factors (HIFs) identified were the flavonoids xenognosin A and xenognosin B. These were isolated from a fractionation of gum tragacanth, a commercially available, water-soluble mixture of dried Astragalus sap (Steffens et al., 1982). The first and only HIF isolated to date from host roots is 2,6-dimethoxy-p-benzoquinone (DMBQ) (Chang and Lynn, 1986). Benzoquinones are produced in plants by biosynthesis on the shikimate acid pathway, by oxidative decarboxylation of phenolic acids, and by the enzymatic degradation of cell wall phenols by peroxidases and laccases (Caldwell and Steelink, 1969). Interestingly, DMBQ was identified from sorghum (Sorghum bicolor) roots only after they were physically abraded or coincubated with Striga cultures (Chang and Lynn, 1986). HPLC analyses showed that coincubation of root washes with Striga results in the generation of DMBQ through peroxidase-mediated oxidation of cell wall components (Lynn and Chang, 1990). Later experiments demonstrated that hydrogen peroxide generated at the Striga radical tip activates host plant peroxidases, which convert host cell wall phenols into haustorial-inducing benzoquinones (Kim et al., 1998; Keyes et al., 2007). The active extraction of HIFs from host roots provides a mechanism by which Striga ensures proximity to a host root prior to haustorial commitment.
The identification of natural HIF molecules has led to evaluation of other phenolic derivatives for their ability to induce haustoria (Riopel and Timko, 1995; Albrecht et al., 1999). Several active HIFs have been identified, including the simple phenolics syringic acid and vanillic acid; flavonoids, such as xenognosin A and peonidin; and p-benzoquinones, like DMBQ. Not all HIF molecules are equally active, and different concentrations or times of exposure are needed for optimal haustoria development. For example, haustorium initiation with syringic acid requires several more hours of exposure or severalfold higher concentrations than DMBQ. HPLC analyses showed that haustoria activity is dependent on syringic acid being enzymatically oxidized to DMBQ (Lynn and Chang, 1990).
An important insight into the mechanism of haustorium signaling resulted from the observation that different haustorium-inducing benzoquinones had similar first-half volt redox potentials (Smith et al., 1996). This led to the hypothesis of a redox model for HIF signaling in which semiquinone intermediates, formed during redox cycling between quinone and hydroquinone states, initiate haustorium development. This model was evaluated with the chemical spin trap cyclopropyl-p-benzoquinone. A single-electron reduction of the cyclopropyl ring of cyclopropyl-p-benzoquinone activates a reactive electrophilic center that irreversibly inhibits haustorium development in Striga in response to DMBQ (Zeng et al., 1996). The redox model hypothesizes that the first step in HIF recognition is the univalent reduction of benzoquinone to semiquinone.
Quinone redox changes are catalyzed by quinone oxidoreductases (EC 1.6.5), a subfamily of medium-chain dehydrogenase/reductases conserved in plants, primates, yeasts, and eubacteria (Persson et al., 2008). We previously isolated cDNAs from Triphysaria roots representing transcripts predicted to encode two classes of quinone oxidoreductases (Matvienko et al., 2001b). Transcripts for both QR1 and QR2 are rapidly upregulated in Triphysaria roots as a primary response to treatment with DMBQ and other quinones (Matvienko et al., 2001b). Based on sequence homologies, QR1 was classified as a member of the ζ-crystallin-like quinone oxidoreductases (EC 1.6.5.5) (Thorn et al., 1995; Edwards et al., 1996) and QR2 as a quinine-reducing flavoprotein (EC 1.6.5.2) (Sparla et al., 1996; Matvienko et al., 2001b). Later purifications of the QR2 enzyme showed that it catalyzes NAD(P)H-dependent quinone reduction with substrate and inhibitor specificity consistent with its placement into the Diaphorase family (Sparla et al., 1999; Wrobel et al., 2002).
This manuscript describes experiments examining the role of these two quinone oxidoreductases in haustorium development. RNA gel blot analyses were used to characterize the expression of QR1 and QR2 transcripts under haustorium-forming and nonforming conditions. Hairpin RNA interference (RNAi) technology was used to reduce transcript levels of these transcripts in Triphysaria roots that were subsequently assayed for their ability to form haustoria with different HIFs. Biochemical analyses of the QR1 enzyme were consistent with its placement into the ζ-crystallin family of quinone oxidoreductases. These results are consistent with the haustorium signaling system being redox activated by radicals produced by the QR1 reaction with HIFs.
RESULTS
QR1 Is Transcriptionally Regulated by Host Root Contact
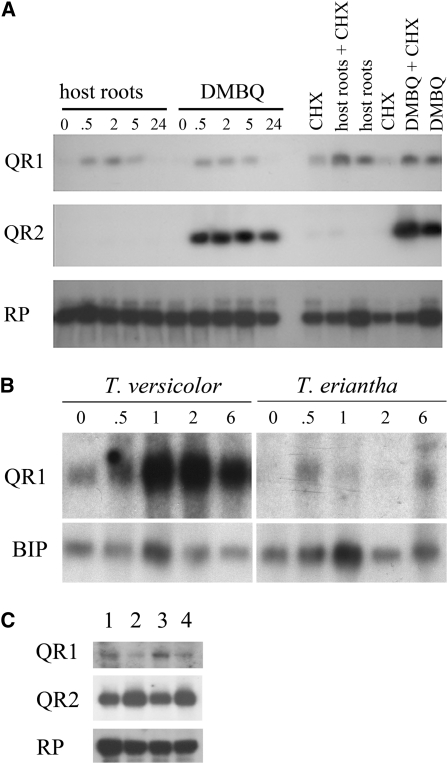
We had previously shown that QR1 and QR2 transcripts are rapidly induced in a primary response to exposure to certain quinones and naphthoquinones (Matvienko et al., 2001b). The regulation of these transcripts during contact with host roots was subsequently investigated by overlaying Triphysaria roots with those of aseptically grown Arabidopsis for different times before isolating RNA. As seen in Figure 1A, contact with Arabidopsis roots resulted in an upregulation of the QR1 message levels by 30 min. Steady state RNA levels reached a maximum approximately 5 h after induction, and by 24 h after induction, transcript levels had returned to preinduction levels (Figure 1A). QR1 transcript levels were similarly regulated by host root contact and DMBQ.
Figure 1.
Transcriptional Regulation of QR1 in Response to Host Root Contact.
(A) RNA was extracted from T. versicolor roots after exposure to either Arabidopsis (host) roots or DMBQ for 0 to 24 h. RNA gel blots were probed with QR1, QR2, or the ribosomal protein RP as a loading control. In the six lanes on the right side of the gel, the roots were first treated with cycloheximide (CHX) for 40 min prior to treatment with host roots or DMBQ.
(B) RNA was extracted from T. versicolor and T. eriantha roots after exposure to 10 μM DMBO for 0 to 6 h. The resultant RNA gel blot was probed with QR1 and the loading control BIP.
(C) RNA was isolated from Arabidopsis roots (1 and 2) or root tips (3 and 4) after treatment with water (lanes 1 and 3) or DMBQ (lanes 2 and 4) for 2 h. Resultant RNA gel blots were probed with QR1, QR2, or the loading control ribosomal protein RP.
By contrast, there was no apparent regulation of QR2 by host root contact, though QR2 was strongly induced by DMBQ. Transcriptional regulation of QR2 in response to DMBQ, and QR1 in response to DMBQ and host contact, occurred in the presence of the protein synthesis inhibitor cycloheximide, indicative of an early, primary response to the haustorium-inducing stimuli (Figure 1A) (Abel and Theologis, 1996).
The ability to develop haustoria in response to DMBQ shows natural variation among different Triphysaria species and collections (Jamison and Yoder, 2001). In one field collection of T. versicolor, >90% of the seedlings develop haustoria when treated with DMBQ, whereas in a collection of T. eriantha, <10% formed haustoria (Jamison and Yoder, 2001). To evaluate the expression of QR1 in haustoria forming and nonforming plants, we treated roots of T. versicolor and T. eriantha with 10 μM DMBQ, isolated RNA 0 to 6 h later, and probed the resultant blot with QR1 or the constitutive gene BIP. As shown in Figure 1B, there was significantly greater upregulation of QR1 message in T. versicolor roots than in T. eriantha. This experiment shows that QR1 is differentially regulated in natural variants that have differential haustorial responses to DMBQ.
The genomic sequence of QR1 was cloned and sequenced. The gene contains three introns and four exons. A BLAST search of sequenced plant genomes identified one homologous gene in Arabidopsis, three in poplar, and one in rice. These best hit genes had the same intron-exon structure as QR1, with some variation in intron size, though positioned at nearly identical locations within the homologs. The conservation of intron position, but not size, is typical of homologous genes across taxonomic boundaries (Fedorov et al., 2002).
To examine expression of the Arabidopsis homolog of QR1, we treated Arabidopsis roots with DMBQ or water and then probed the RNA gel blot with the T. versicolor QR1 cDNA. Interestingly, there was no apparent upregulation of the Arabidopsis homolog hybridizing to the QR1 probe. If anything, the steady state level of the Arabidopsis transcript was reduced after exposure to DMBQ (Figure 1C). This is consistent with our earlier studies indicating that QR1 is not transcriptionally regulated by DMBQ in nonparasitic plants (Matvienko et al., 2001b).
Hairpin-Mediated Silencing of QR1 Reduces Haustorium Development
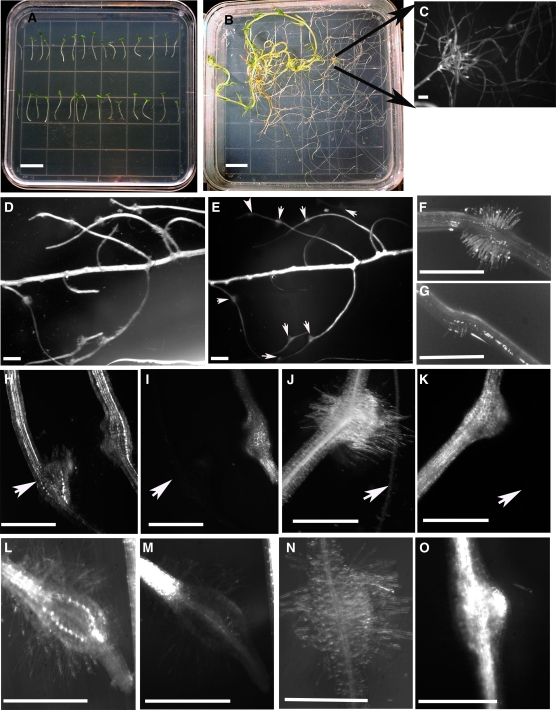
Hairpin RNAi vectors targeted against the QR1 and QR2 transcripts (pHpQR1 and pHpQR2, respectively) were transformed into Triphysaria roots via Agrobacterium rhizogenes. Transgenic roots were identified on the basis of yellow fluorescent protein (YFP) expression, and nontransgenic roots were removed by cutting. The percentage of Triphysaria with at least one YFP expressing root ranged from 20 to 40% depending on the transformation experiment (Table 1). YFP fluorescence was variable in different transgenic roots, ranging from very bright to minimal. Even weakly expressing roots could be identified because the autofluorescence of Triphysaria roots under the YFP filter set is very low (Figures 2E, 2I, and 2K). To increase the number of transgenic root tips for any given transformation event, YFP expressing roots were cut at the tips to stimulate root proliferation at the cut site (Figures 2B and 2C).
Table 1.
Efficiency and Phenotypes of Transgenic Roots
| Phenotype | pHG8-YFP | pHpQR1 | pHpQR2 | pHpQR2-QR1 | P Level |
| Transformation efficiencya | 28 ± 2.2 | 33 ± 5.3 | 30 ± 3.2 | 31 ± 4.3 | 0.37 |
| n = 1271 | n = 1144 | n = 1138 | n = 1051 | ||
| YFP-positive roots per plantb | 1.8 ± 0.4 | 1.6 ± 0.3 | 1.7 ± 0.2 | 1.6 ± 0.1 | 0.83 |
| n = 352 | n = 378 | n = 339 | n = 323 | ||
| Root length (cm)c | 3.5 ± 1.8 | 3.4 ± 1.7 | 3.2 ± 1.6 | 3 ± 1.5 | 0.18 |
| n = 49 | n = 50 | n = 49 | n = 51 | ||
| Lateral roots per YFP rootc | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.82 |
| n = 49 | n = 50 | n = 49 | n = 51 | ||
| Root growth no DMBQ (mm/24 h)c | 2.6 ± 0.4 | 2.7 ± 0.5 | 2.6 ± 0.4 | 2.6 ± 0.3 | 0.72 |
| n = 40 | n = 40 | n = 40 | n = 40 | ||
| Root growth with DMBQ (mm/24 h)c | 2.7 ± 0.5 | 2.6 ± 0.4 | 2.7 ± 0.5 | 2.6 ± 0.5 | 0.22 |
| n = 40 | n = 40 | n = 40 | n = 40 |
Values are mean ± sd with n = total number of plants or roots in all replicates. Means are the averages of three independent transformation experiments per construction.
Percentage of plants with at least one yellow root.
Twenty-one days after transformation.
Thirty-three days after transformation.
Figure 2.
Rhizogenes Transformation and Haustorium Phenotypes.
Photos in (A), (B), (D), (F), (G), (H), (J), (L), and (N) are bright-field images. Photos in (C), (E), (I), (K), (M), and (O) are epifluorescence images showing YFP expression. Bars = 1 cm for (A) and (B) and 1 mm for (C) to (G).
(A) Triphysaria seedlings on day 0 of transformation.
(B) A branched Triphysaria root 12 weeks after transformation. Six weeks prior to this photograph, a root tip was cut to stimulate new root development at or near the cut site. Root tips were cut a second time resulting in the highly branched transgenic root seen in (C).
(C) A magnification of a section of (B) showing a cluster of YFP expressing roots, all derived from a single transformation event.
(D) and (E) Transgenic roots formed multiple haustoria as marked by arrowheads.
(F) A completely developed haustorium on a pHG8-YFP root.
(G) A partially developed haustorium on a pHpQR1 root.
(H) and (I) A nontransgenic and pHG8-YFP transgenic root side by side, both with a haustorium. The arrow points to the nontransgenic root.
(J) and (K) Haustorium developing on a pHG8-YFP transgenic root formed in response to an Arabidopsis host root (arrowhead).
(L) and (M) A transgenic haustorium 24 h after initiation. There is little YFP expression in the newly formed haustorium.
(N) and (O) A transgenic haustorium 48 h after initiation. YFP expression within the haustorium is similar to other regions of the root.
[See online article for color version of this figure.]
Figures 2E, 2I, 2K, 2M, and 2O show YFP expressing roots with developed haustoria. Interestingly, YFP fluorescence was not observed in newly developed haustoria but only after some maturation time (Figures 2M and 2O). This might be indicative of a general reduction in protein synthesis during haustorium development or of a specific redirection of transcription away from the mas 2 promoter driving YFP. We also noted that there was little or no expression of YFP in haustorial hairs.
Transformation frequencies did not significantly differ between the different hairpin vectors. Similarly, the growth rates and the morphologies of YFP-expressing roots did not differ between different transformations (Table 1). This has not been the case for all of the hairpin constructions we have transformed into Triphysaria; hairpin constructions for some genes have resulted in significant morphological aberrations to the parasite root or could not be recovered in transgenics at all (data not shown).
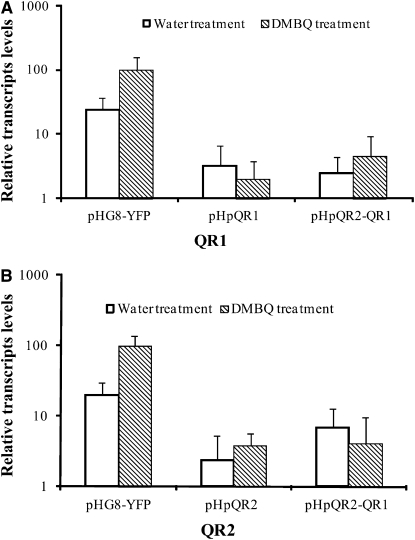
Transcript levels of QR1 and QR2 were quantified by real-time PCR in transgenic roots treated with DMBQ or water. The constitutive T. versicolor gene QAN8 was used as internal standard for each reaction. Figure 3A shows that the steady state transcript level of QR1 was reduced about 50-fold in pHpQR1 roots compared with roots transformed with the parent vector pHG8-YFP. Transcript abundance of QR2 was reduced in pHpQR2 roots about the same order of magnitude (Figure 3B). Triphysaria roots transgenic for the double hairpin pHpQR2-QR1 showed reduced levels of both targeted transcripts (Figures 3A and 3B). These experiments demonstrated that the hairpin constructions reduced steady state transcript levels of the targeted genes.
Figure 3.
Transcript Levels of QR1 and QR2 in Transgenic Roots.
Steady state transcript levels of QR1 and QR2 were determined by real-time PCR and normalized to the constitutively expressed gene QAN8 for each sample. Data are means ± sd of two technical replicates of three to four plants (n = 6 to 8). Expression levels in pHG8-YFP transgenic roots treated with DMBQ was set to 100%. Note the log scale y axis. (A) QR1 expression.
(B) QR2 expression.
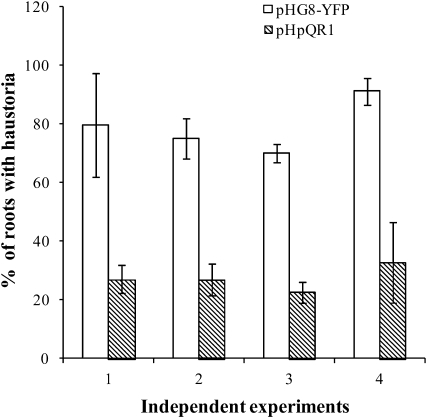
We assayed the competence of transgenic Triphysaria roots to form haustoria in response to Arabidopsis root contact. Haustorium development in roots transgenic for the empty vector pHG8-YFP or pHpQR2 was 80 to 90% (Table 2). This is approximately the same level of induction observed in nontransgenic seedlings. By contrast, transgenic roots bearing pHpQR1 initiated significantly fewer haustoria (∼30%). Figure 4 shows the variation in haustorium development across four independent transformation experiments. In each case, haustorium development was reduced in pHpQR1 transgenics by ∼70%. A similar result was obtained when Medicago exudates were used to stimulate haustorium development (Table 2). Transformants with the double hairpin pHpQR2-QR1 made fewer haustoria than nontransgenic roots, but the reduction was not as pronounced as that seen in the single hairpin transformants.
Table 2.
Haustorium Development in Transgenic Roots
| Percentage of Roots with Haustoriaa |
|||||
| Treatment | Seedlingsb | pHG8-YFP | pHpQR2 | pHpQR1 | pHpQR2-QR1 |
| Host rootsc | 87 ± 9 | 84 ± 11 * | 80 ± 10* | 28 ± 6** | 49 ± 4*** |
| n = 90 | n = 113 | n = 100 | n = 112 | n = 103 | |
| Host root exudatesd | 85 ± 3 | 81 ± 11 * | 87 ± 9* | 28 ± 12** | 40 ± 8*** |
| n = 214 | n = 118 | n = 145 | n = 98 | n = 131 | |
| DMBQe | 82 ± 5 | 87 ± 15* | 87 ± 9* | 28 ± 7** | 48 ± 9*** |
| n = 186 | n = 113 | n = 129 | n = 123 | n = 69 | |
| Syringic acidf | 82 ± 4 | 81 ± 7* | 81 ± 11* | 25 ± 4** | 50 ± 11*** |
| n = 239 | n = 145 | n = 126 | n = 124 | n = 130 | |
| 2-Methyl benzoquinoneg | 45 ± 4 | 45 ± 8* | 43 ± 5* | 12 ± 4** | 24 ± 7*** |
| n = 239 | n = 114 | n = 127 | n = 119 | n = 129 | |
| Peonidinh | 71 ± 10 | 64 ± 3* | 67 ± 9* | 27 ± 9** | 44 ± 6*** |
| n = 196 | n = 155 | n = 109 | n = 118 | n = 106 | |
| Water | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| n = 186 | n = 104 | n = 97 | n = 102 | n = 117 | |
Values are means ±sd of 3 to 11 plates with four to six independently transformed plants with 1 to 30 roots (n = total number of roots in all replicates). Analysis of variance was determined with the General Linear Model procedure and mean separation with the least significance difference method using Statistical Analysis Software 9.1 (SAS Institute). Pairwise comparisons of treatments within a row labeled with a different number of asterisks are significantly different at α = 0.05. Haustoria were counted 24 h after treatment except with host roots.
Seedling data were not included in statistical analysis because they had different ages.
Triphysaria roots were placed in contact with Arabidopsis roots for 5 d.
Medicago host exudates were obtained as described in Methods.
The 30 μM 2,6-DMBQ was from Pfalz and Bauer.
The 1000 μM syringic acid was from Sigma-Aldrich.
The100 μM 2-MBQ was from Sigma-Aldrich.
The30 μM peonidin chloride was from Indofine Chemical.
Figure 4.
Haustorium Development in Response to DMBQ Is Reduced in pHpQR1 Roots.
Transgenic roots of pHG8-YFP and pHpQR1 were exposed to 30 μM DMBQ and haustoria scored 24 h later. Each pair of bars represents an independent transformation experiment using pHpQR1 and the parent vector pHG8-YFP. Roots treated with water did not form haustoria. Data are means ± sd. Within each experiment, there were four to nine plates with four to seven plants each. The total number of pHG8-YFP roots examined per experiment ranged from 237 to 279, and the number of pHpQR1 roots examined per experiment ranged from 111 to 189.
We investigated the effectiveness of different classes of HIFs to trigger haustorium development in the RNAi roots (Table 2). Haustorium development is initiated in Triphysaria by treatment with syringic acid, 2-methylbenzoquinone, 2,6-dimethoxybenzoquinone, and peonidin, though at different optimal concentrations (Albrecht et al., 1999). When these chemical HIFs were applied to the transgenic roots, the same pattern was observed as with DMBQ; roots transgenic for pHpQR1 or pHpQR2-QR1 made fewer haustoria than did the roots transgenic for the parent vector pHG8-YFP or pHpQR2 (Table 2). Haustorium development was reduced in pHpQR1 transgenic roots to about a third of that obtained with controls (untransformed seedlings or those transformed with the empty parent vector). In all cases, transgenic roots containing the double construction made significantly more haustoria than pHpQR1, but fewer than pHG8-YFP.
Approximately 30% of the YFP roots obtained from the pHpQR1 transformations developed haustoria. In many cases, these haustoria were less well developed than seen in control roots. Often, only a few haustorial hairs were observed without noticeable swelling (Figures 2F and 2G). These partially formed haustoria appear similar to haustoria obtained with suboptimal HIF concentrations or shortened incubation times (Smith et al., 1990).
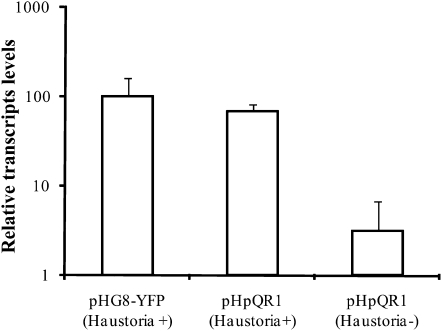
Transcript abundance was compared in pHpQR1 roots that made haustoria versus pHpQR1 roots that did not. Because the amount of RNA obtained from a single root was insufficient for quantification, four to six haustorium forming roots were pooled for RNA isolation. Figure 5 shows that the QR1 expression in pHpQR1 transgenics that formed haustoria was near the level of the transformants with the empty vector (pHG8-YFP). By contrast, pHpQR1 transgenics that did not develop haustoria in response to DMBQ expressed QR1 at an ∼20-fold lower level.
Figure 5.
Quantification of QR1 Expression in pHpQR1 Roots with Haustoria.
Steady state mRNA levels of QR1 relative to QAN8 were determined by real-time PCR in pHpQR1 transgenic roots that either made (+) or did not make (−) haustoria in response to DMBQ. Data are means ± sd of two technical replicates of three to four pHG8-YFP plants and three to four4 pHpQR1 plants without haustoria. Data for pHpQR1 plants without haustoria were obtained from four pools of four to six plants in each pool (n = 6 to 8). Expression level in DMBQ treated pHG8-YFP was set to 100%. Note the log scale y axis.
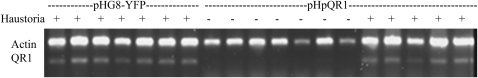
We assayed additional pools of haustorium forming and nonforming pHpQR1 roots by limited cycle PCR. In each case, if a Triphysaria root formed a haustorium, the QR1 transcript was observed (Figure 6). By contrast, all of the 13 pHpQR1 transgenics that did not make haustoria had QR1 levels below that detected by the assay (seven of these shown in Figure 6). These experiments show a direct correlation between abundance of QR1 transcripts and haustorium initiation.
Figure 6.
QR1 Expression in Haustoria-Forming and Nonforming Roots.
Limited cycling PCR (30 cycles) was used to identify pHpQR1 transcripts in transgenic roots that either did (+) or did not (−) make haustoria. The amplified QR1 product is labeled; Actin was used as an endogenous RT-PCR standard. cDNA generated from pools of four to six roots formed haustoria was used. Representative experimental replicates are shown.
QR1 Encodes an NADPH-Dependent Quinone Oxidoreductase
The QR1 protein was expressed in Escherichia coli and purified by a combination of ion exchange and affinity chromatography. Oxidation of NADPH was monitored as the change in absorbance at 550 in the presence of enzyme and electron accepting quinones (Table 3). The most active substrate was 9,10-phenanthrenequinone (PAQ). The reaction was NADPH specific, and no oxidation of NADH was observed when NADH was used. The second most active substrate was 1,2-naphthoquinone followed by 5-hydroxy-1,4-naphthoquinone (juglone) and 2,6-DMBQ. QR1 was incapable of reducing menadione, which distinguishes it from the diaphorase or QR2 class of enzymes. Also, QR1 did not reduce diamide, distinguishing it from the Arabidopsis ζ-crystallin protein P1-ZCr (Mano et al., 2000). QR1 also shows no activity on 2,3-dimethoxy-5-methyl-1,4-bezoquinone or 1,4-napthoquinone (Table 3).
Table 3.
Substrate Specificity of QR1 Enzyme
| Catalytic Rates (Kcat (s−1)]a |
|||
| QR1 |
P1-ZCr, AT-AER |
ZCr |
|
| Substrate | T. versicolor | (AT5G16970) | Cavia porcellus |
| PAQ | 23.3 | 98 | 19 |
| 1,2-Napthoquinone | 2.44b | 54c | 39 |
| 5-Hydroxy-1,4-napthoquinone (juglone) | 0.31 | 1.9 | 4.9 |
| DMBQ | 0.09 | ND | ND |
| 2-Methylnaphthalene-1,4-dione (menadione) | 0 | 0 | 0 |
| 2,3-Dimethoxy-5-methyl-1,4-bezoquinone | 0 | ND | ND |
| 1,4-Benzoquinone | |||
| 1,4-Napthoquinone | 0 | 3c | 1d |
| Diamide | 0b | 95 | 0 |
Kcat (s−1) values determined from the maximum velocities with 200 μM NADPH and 100 μM acceptor.
Activity determined with MTT tetrazolium dye.
Kcat (s−1) values determined from the maximum velocities with 50 μM NADPH and 25 μM acceptor.
Kcat (s−1) values determined from the maximum velocities with 50 μM NADPH and 250 μM acceptor.
NADPH oxidation rates were measured as change in absorbance at 610. Rate = average of three rate measurements (μmol 0.5 mL−1 min−1). Kcat (s−1) = rate × 10−6/(7.5 × 10−12)/60.
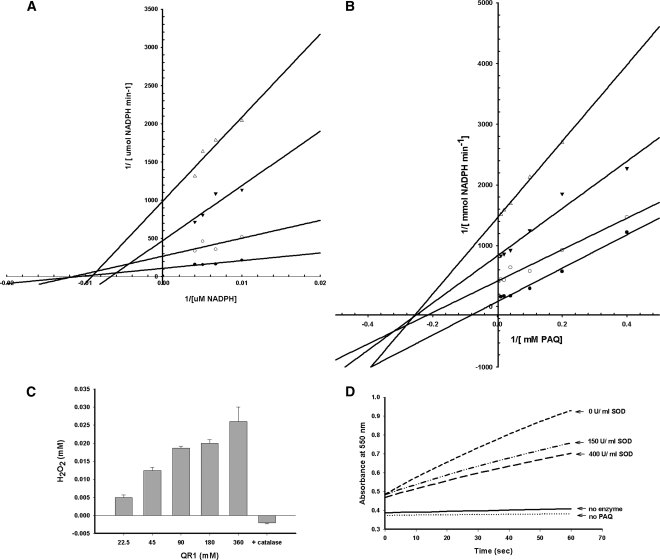
Cytochrome c is a mitochondrial protein that transfers single electrons between cytochrome oxidase complexes (Fridovich, 1970). The reduction of cytochrome c requires a superoxide anion (O2•−) and hence is inhibited by superoxide dismutase (McCord and Fridovich, 1969; Iyanagi and Yamazaki, 1970). As seen in Figure 7D, there was no change in absorbance at 550 nm in the absence of enzyme or in the absence of quinone (no PAQ). However, in the presence of QR1 enzyme and PAQ, cytochrome c was linearly reduced at a rate of 13.75 μM min−1. Cytochrome c reduction was inhibited ∼50% by the addition of superoxide dismutase, evidence of a superoxide anion being generated during the QR1 catalyzed reduction of PAQ. Cytochrome c was not similarly reduced with the QR2 enzyme (data not shown).
Figure 7.
Enzymatic Activity and Substrate Specificity of QR1.
(A) and (B) Inhibition of activity by dicumarol as measured by varying concentrations of PAQ (A) or NADPH (B). The four lines in each graph indicate different concentrations of dicumarol. Closed circles correspond to 0 μM dicumarol, open circles correspond to 10 μM dicumarol, closed triangles correspond to 25 μM dicumarol, and open triangles correspond to 100 μM dicumarol. Each point represents the mean of triplicate trials.
(C) H2O2 generated from different concentration of QR1 in the standard PAQ reduction assay. Addition of 0.1 mg/mL of catalase abolished the hydrogen peroxide signal.
(D) Reduction of cytochrome c by QR1. Activity was followed by recording the cytochrome c reduction at 550 nm. Lines show absorbance changes over time without enzyme, without PAQ, the complete assay system without superoxide dismutase, and the complete assay system with superoxide dismutase concentrations 150 and 400 units/mL.
Figures 7A and 7B show the quinone oxidoreductase inhibitor dicumarol inhibited QR1 activity by 90%. The kinetics of dicumarol inhibition suggests a mixed, noncompetitive inhibition with respect to both NADPH and PAQ, similar to the Arabidopsis P1-ZCr (Mano et al., 2000). Figure 7C shows that hydrogen peroxide is generated during the QR1 catalyzed reduction of PAQ. Inclusion of catalase removes H2O2 from the reaction.
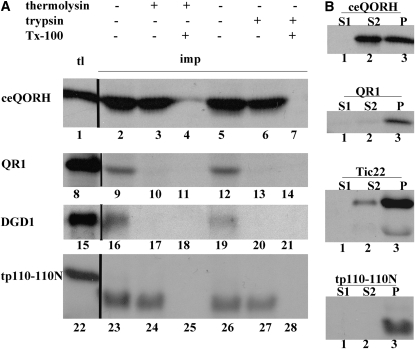
In BLAST searches, the Arabidopsis protein with the highest percentage of homology to QR1 encodes the protein ceQORH at locus AT4G13010. The ceQORH protein binds to chloroplasts via a noncanonical binding site that does not require an N-terminal cleavable targeting sequence for localization (Miras et al., 2002, 2007). To test whether this is also the case for QR1, we performed in vitro import assay using pea seedling chloroplasts. Chloroplasts were used for these studies to mirror those used in previous studies (Inoue et al., 2006). Proteins were radiolabeled, incubated with pea (Pisum sativum) seedling chloroplasts under import conditions, and then treated with either thermolysin, a protease that cannot penetrate the outer membrane (Cline et al., 1984), or trypsin, a protease that can reach the space between the outer and inner membranes of the chloroplast envelope (Jackson et al., 1998a). The fates of the imported proteins were determined by SDS-PAGE and fluorography. Four control protein precursors were included in these reactions; the outer envelope protein DGD1 (Froehlich et al., 2001), the inner envelope protein tp110-110N (Jackson et al., 1998b), and two proteins peripherally associated with the chloroplast inner envelope, ceQORH (Miras et al., 2002) and Tic22 (Kouranov et al., 1998a).
As shown in Figure 8A, a slight but significant amount of QR1 was recovered in the chloroplast without any apparent change in its mobility on SDS-PAGE. The imported QR1 was almost completely digested by both thermolysin (lane 10) and trypsin (lane 13), as was the outer envelope protein DGD1 (Froehlich et al., 2001). By contrast, the Arabidopsis protein ceQORH was resistant to both proteases, as was the inner envelope protein derived from tp110-110N. These results confirm the previous report that ceQORH is targeted to the chloroplast inner envelope (Miras et al., 2002, 2007) and suggests that ceQORH faces the stroma side of the membrane.
Figure 8.
In Vitro Import into Chloroplasts.
Radiolabeled proteins were incubated with intact chloroplasts for 20 min under the import condition at which time the chloroplasts were reisolated and fractions containing imported proteins subjected to SDS-PAGE, followed by fluorography. Black lines separate grouping of images from different portions of the same gel. The “tl” lane shows 10% of the translation product subjected to the assay; “imp” lanes represent the imported protein.
(A) Chloroplasts with imported protein were treated with thermolysin or trypsin as indicated by the + and − symbols. Triton X-100 (Tx-100) was also included in some reactions.
(B) Chloroplasts were hypotonically lysed and fractionated by centrifugation into a supernatant that contains soluble proteins (S1) and a pellet. The pellet was resuspended in Na2CO3 and fractionated by centrifugation into a supernatant containing peripheral membrane proteins (S2) and a pellet (P).
We tested the nature of the association of the imported proteins with chloroplasts by alkaline treatment (Figure 8B). Two proteins peripherally associated to the chloroplast inner envelope, ceQORH and Tic22, were released by the Na2CO3 wash (lanes 2 and 8), whereas the integral membrane protein derived from tp110-110N was recovered in the pellet fraction (lane 12). QR1 was found almost exclusively in the pellet fraction (lane 6). Collectively, these results suggest that QR1 may be targeted integrally into the outer membrane of the chloroplast envelope, although we cannot completely rule out the possibility that the association of QR1 with isolated chloroplasts may be nonspecific. Nonetheless, the behavior of QR1 in the import assay is clearly different from that of its Arabidopsis homolog.
DISCUSSION
Many of the biological activities ascribed to phenols and quinones, whether as medical pharmacological agents or as rhizosphere community mediators, are redox associated (O'Brien, 1991; Appel, 1993). For example, the reduction of quinonic antitumor agents is required for their bioactivation (Testa, 1995). Similarly juglone, the phytotoxic naphthoquinone ascribed to black walnut allelopathy, is synthesized as the nontoxic hydrojuglone. Hydrojuglone is then activated to the toxic juglone upon release from the plant and exposure to oxygen (Lee and Campbell, 1969). Haustorium initiation in parasitic plants is thought to be induced by redox cycling of the exogenous haustorium-inducing factor (Smith et al., 1996). For this reason, we investigated the function of quinone oxidoreductase genes in parasite roots.
Quinones can be reduced by either one or two electron transfer mechanisms (Figure 9). Single electron transfers are catalyzed by oxidoreductases typically involved in electron transport and metabolism (Testa, 1995). These reactions generate highly reactive free radical semiquinones that in the presence of oxygen are rapidly oxidized to the quinone, with the concomitant formation of the superoxide anion radical O2•− (Figure 9). Semiquinones and superoxide radicals are cytotoxic, as are the hydroxyl radicals formed subsequently by the Fenton reaction (Burkitt, 2003). By contrast, two electron quinone reductions catalyzed by enzymes like DT-diaphorase do not generate radical semiquinones, but form relatively stable hydroquinones that, depending on the functional groups present, can be further stabilized by conjugation (Cadenas et al., 1992; Testa, 1995). Two electron transfers protect the cell against reactive oxygen intermediates generated by univalent reducing enzymes (Lind et al., 1990; Ross et al., 2004).
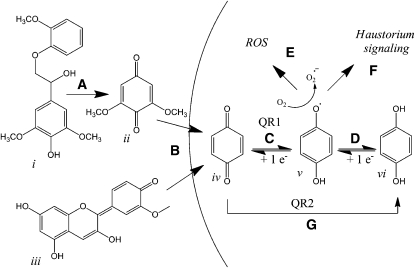
Figure 9.
Schematic Model of QR1 and QR2 Action.
(A) The lignin component (i) is oxidatively decarboxylated to DMBQ (ii) through the actions of fungal peroxidases, host peroxidases, or mild abrasion (Caldwell and Steelink, 1969; Keyes et al., 2007).
(B) DMBQ (ii) and peonidin (iii) enter the parasite cells, possibly by nonselective diffusion (Shann and Blum, 1987). Peonidin is drawn in one of several possible conformations (Brouillard and Cheminat, 1982).
(C) The p-benzoquinone motive (iv) is reduced by a single electron to a free radical semiquinone (v) by the action of QR1.
(D) Further nonenzymatic reduction of the semiquinone leads to an equilibrium with the relatively stable hydroquinone (vi) (Testa, 1995).
(E) In the presence of oxygen, the semiquinone promotes the generation of superoxide anion-radicals (O2•−), which are further modified to highly toxic hydroxyl radicals.
(F) The haustorium signaling pathway is initiated either by the radical semiquinone or by the ROS species generated in air.
(G) QR2 encodes a two electron detoxifying reduction that avoids the radical semiquinone.
Sequence homologies placed QR1 into the ζ-crystalline-like quinone oxidoreductase family (Edwards et al., 1996). The archetypal protein in this family is ζ-crystallin, a highly abundant structural protein present in the lens of guinea pigs that when mutated can be responsible for cataracts (Huang et al., 1987, 1990; Rao et al., 1992). The metabolic activity of guinea pig ζ-crystallin catalyzes quinone reductions via a single-electron reducing mechanism. The ζ-crystallin reaction produces a radical semiquinone that generates reactive oxygen species (ROS) under aerobic conditions (Rao et al., 1992; Porté et al., 2009). In plants, the best characterized ζ-crystallin is P1-ZCr (also known as AT-AER arkenal reductase or NADPH:2-alkenal α,β-hydrogenase) encoded by the Arabidopsis AT5G16990 locus. P1-ZCr was originally isolated from a cDNA expression library on the basis of its ability to confer resistance to the oxidizing drug diamide in Saccharomyces cerevisiae (Babiychuk et al., 1995). The enzyme was later characterized as an NADPH-dependent reductase active on quinones, azodicabonyls, and 2-alkenals (Mano et al., 2000, 2002). Transgenic tobacco (Nicotiana tabacum) plants with elevated AER levels exhibited significantly less damage than control plants when exposed to 4-hydroxy-(2E)-nonenal or methyl viologen, leading the authors to propose the enzyme detoxifies reactive aldehydes produced by lipid peroxidases (Mano et al., 2005). The biochemical activities of QR1 are similar to animal and plant ζ-crystallins; QR1 is an NADPH-specific quinone oxidoreductase that catalyzes the reduction of various quinone substrates, but not menadione. The reaction generates hydrogen peroxide, and, in the presence of PAQ, QR1 catalyzes the reduction of cytochrome c. These properties are consistent with QR1 encoding a single-electron reducing quinone reductase.
By contrast, QR2 is related to a family of flavoproteins that catalyze the two-electron reduction of short-chain acceptors, including benzoquinone, juglone, and menadione (Matvienko et al., 2001b; Wrobel et al., 2002). This class of quinone reductases is best typified by the mammalian quinone reductase NQOR or DT-diaphorase (Ross et al., 2004). These enzymes reduce quinones by a two-electron transfer mechanism that does not generate the semiquinone or associated ROS (Testa, 1995). Hence, NQOR is a detoxification enzyme that is involved in cellular defense against electrophilic quinones (Ross et al., 2004). Typical for this class of enzymes, the heterologously expressed QR2 protein uses either NADH or NADPH as electron donor and acts on a much broader range of quinones than TvQR1 (Wrobel et al., 2002). Broad substrate specificity is typical for xenobiotic metabolizing enzymes and results from plastic active sites that can accommodate a range of structures (Testa, 1995; Faig et al., 2001).
Both QR1 and QR2 are transcriptionally upregulated in a primary response to exposure to benzoquinones and naphthoquinones (Matvienko et al., 2001b). Transcription of xenobiotic metabolizing enzymes is often regulated by their substrates and has been observed in plants, animals, and fungi (Prestera and Talalay, 1995; Testa, 1995; Akileswaran et al., 1999; Laskowski et al., 2002; Greenshields et al., 2005). DT-diaphorase (menadione reductase activity) is induced in mammalian cells treated with a wide range of electrophilic chemical carcinogens (Cadenas et al., 1992). Xenobiotic responsive elements) upstream of the rat quinone reductase gene have been identified that are responsible for its transcriptional activation by exogenous toxins (Favreau and Pickett, 1991).
Transcriptional regulation of QR1 is distinct from that of QR2 because QR1 is induced by contact with Arabidopsis roots, but QR2 is not. One explanation for QR2's responsiveness to DMBQ, but not host roots, might be that DMBQ is not the active inducer on the Arabidopsis root surface. Host peroxidases activated by Striga ROS are necessary to release sufficient DMBQ from sorghum roots to induce haustoria, and the concentration of DMBQ at the Arabidopsis root surface may not be sufficient to induce the QR2 promoter (Kim et al., 1998). QR1 is induced with both DMBQ and direct host root contact. Lateral haustorium development in Triphysaria and Striga is enhanced by tactile stimuli in addition to chemical HIFs (Baird and Riopel, 1984; Wolf and Timko, 1991). Overlaying Arabidopsis roots onto those of Triphysaria provides both types of stimuli. It is possible that both QR1 and QR2 promoters contain DMBQ-responsive elements, but QR1 has additional transcriptional enhancers responsive to tactile stimuli. The homologous QR1 transcript in Arabidopsis is not induced by either DMBQ or host root contact, and its promoter is predicted to lack both types of enhancers. In vertebrate evolution, the ζ-crystallin quinone oxidoreductase protein has been independently recruited as a taxon-specific crystallin at least twice, once in hystricomorpha rodents and once in the Camelidae. High expression of ζ-crystallin in lens results from the addition of a lens specific enhancer into the ζ-crystallin promoter (Lee et al., 1994; Fernald, 2006). In these animals, the metabolic ζ-crystallin enzyme was recruited as a lens structural protein through a promoter mutation. A similar event may have occurred in the promoter of QR1 to change its normal expression pattern to one inducible by host root contact.
RNA silencing was used to investigate the functions of QR1 and QR2 in parasitic plant roots. Haustorium development was significantly reduced in transgenic roots silenced for QR1, but not QR2 transcripts. Transgenic roots for pHpQR1 had about a 10-fold reduction in QR1 transcripts compared with nontransgenic roots, and these roots developed haustoria about 3 to 4 times less frequently than roots transgenic for the empty donor vector (pHG8-YFP) or pHpQR2. The low percentage of transgenic pHpQR1 plants that did form haustoria were found to have elevated levels of QR1 transcripts relative to pHpQR1 transgenics that did not develop haustoria. The correlation between steady state QR1 transcript levels and haustorium development was found to extend to natural variants isolated from the field. T. versicolor plants that effectively form haustoria in response to DMBQ also regulate QR1 in response to DMBQ. By contrast, T. eriantha plants that do not effectively form haustoria with DMBQ treatment also do not upregulate QR1. The correlation between the high abundance of QR1 transcripts and haustorium development therefore was seen in both transgenic and natural populations.
While the double hairpin construction pHpQR2-QR1 reduced steady state levels of both QR1 and QR2 transcripts, the frequency of haustorium development was consistently higher than that obtained with the single target hairpin constructions. It might be that the single hairpin constructions are more efficiently transcribed and processed into RNAi than the dual hairpin, though no differences were seen in the quantitative PCR analyses. Alternatively, reduction in QR2 transcript levels might counter the effects of silencing QR1. As shown in Figure 9, QR1 and QR2 both act on HIFs but to different effects; QR1 activates ROS activity, while QR2 functions to limit ROS. By reducing QR2 activity, more benzoquinone substrate remains available for the semiquinone-generating activity of QR1. The predicted increase in haustorial inducing activity in silenced QR2 was not observed because control, nontransgenic plants are already at optimal haustoria-forming levels. However, when the haustoria formation rate is reduced by silencing QR1, the increase in haustoria formation with QR2 silenced may result from the increased pool of quinone substrate. These results are consistent with the model shown in Figure 9 where QR1 generates a radical semiquinone that is an initial step in the haustorium development signal transduction pathway, while QR2 acts to remove the signal through an existing detoxification system (Smith et al., 1996; Zeng et al., 1996).
It is not clear if the semiquinone directly activates a redox-mediated signal transduction pathway or if activation of signal transduction requires a ROS. Redox signal transduction pathways are common in both plant and animal systems (Ahmad et al., 2008; Jones, 2008). The generation of ROS may also be directly involved in morphological changes associated with development of the parasitic organ. ROS are involved in both tip growth and diffuse cell growth (Carol and Dolan, 2006). ROS have been detected in rapidly growing cells of maize roots and cucumber (Cucumis sativus) and Arabidopsis seedlings using electron paramagnetic resonance (Liszkay et al., 2004; Renew et al., 2005). In growing primary roots of maize, wall-loosening reaction driven by reactive oxygen intermediates can lead up to 50% length increase per hour (Liszkay et al., 2004). In this process, plasma membrane NADPH oxidases produce superoxide radicals that are then converted into polysaccharide-cleaving hydroxyl radicals (OH) by cell wall peroxidase activity (Liszkay et al., 2004). The rapid cortical cell expansion in early haustorium development may be driven by a similar ROS mechanism. In root hair development, ROS accumulation is specifically at the hair dome, but not at the other parts of the wall of the trichoblast (Foreman et al., 2003; Carol et al., 2005). ROS continue to accumulate at the tip of the growing root until the hair stops growing. ROS controls root hair development by activating plasma membrane calcium ion channels, which leads to the production of calcium gradient needed for tip growth (Foreman et al., 2003). Rapid ROS accumulation catalyzed by QR1 may also be associated with the growth of long densely positioned haustoria hairs. Arguing against the involvement of ROS in the direct modification of the haustorial cells is the observation that Striga radicals treated with DMBQ had reduced levels of ROS (Keyes et al., 2007). This suggests that ROS levels are controlled during haustorium development, at least in Striga, to a greater degree than might be predicted if they were directly used as developmental catalysts.
METHODS
Plants
Triphysaria is a genus of broad host range, facultative parasites that grow as annuals in coastal and grassland stands along the Pacific Coast from Baja to British Columbia (Hickman, 1993). Seeds of the outcrossing species Triphysaria versicolor andTriphysaria eriantha were collected from an open pollinated population growing in a pasture land south of Napa California. Arabidopsis thaliana Columbia seeds were obtained from the ABRC. Seeds from Medicago truncatula Jemalong A17 were obtained from Douglas R. Cook (University of California, Davis, CA).
Plasmid Constructions
The mas2' promoter (Velten et al., 1984) was PCR amplified from pFGC5941 (Gendler et al., 2008) and inserted into the NcoI site at the start of the YFP reporter gene in pBIN-YFP (Subramanian et al., 2006). The mas-YFP fusion was then cloned into the SacI site of pHellsgate8 to allow visual screening of transgenic roots (Helliwell et al., 2002). This vector, called pHG8-YFP, was the parent of subsequent hairpin constructions.
The hairpin RNAi vectors used to silence QR1 and QR2 gene transcripts were constructed as follows. A 300-nucleotide region toward the 3′ end of the QR1 open reading frame and a 436-nucleotide region toward the 5′ end of QR2 open reading frame were amplified by PCR using primers modified with attB recombinase sites. The PCR products were gel purified using a Qiagen gel extraction kit and recombined into the Gateway donor vector pDONR211 following the manufacturer's protocols (Invitrogen). To make a hairpin targeted to both genes simultaneously, the 300-nucleotide QR1 and 436-nucleotide QR2 fragments were denatured and annealed with a single primer containing a 20-mer sequence homologous to the QR1 fragment and a 20-mer sequence homologous to the QR2 sequence. The chimeric fragment was PCR amplified using an outer set of QR1 and QR2 primers that amplified across both fragments and the junction primer. The various primers used in these constructions are given in Supplemental Table 1 online. They were designed using Primer 3 software (Rozen and Skaletsky, 2000) and synthesized by Integrated DNA Technologies.
The donor vector constructions were confirmed by sequencing, and a second Gateway recombination reaction was performed with each amplified target into the pHG8-YFP parent vector, yielding three hairpin vectors: pHpQR1, pHpQR2, and pHpQR2-QR1. The vectors were confirmed by restriction digestions and transformed into Agrobacterium rhizogenes MSU440 by electroporation (Sonti et al., 1995).
Triphysaria Root Transformation
The A. rhizogenes–mediated transformation of Triphysaria roots was performed as previously described with some modifications (Tomilov et al., 2007). Prior to transformation, the A. rhizogenes was grown in MGL plates (Walkerpeach and Velten, 1994) containing 400 μM acetosyringone. After dipping the excised ends of Triphysaria seedlings into the A. rhizogenes culture, inoculated plantlets were placed on sugar-free, 0.25× Hoagland medium containing 400 μM acetosyringone. The plates were placed vertically in a 16°C growth room for 5 d and then transferred into a 25°C growth room with a 16-h light period at 150 μE light intensity. About 3 weeks later, transgenic roots were identified by visualization of YFP fluorescence with a Zeiss Stemi SV11 dissecting microscope equipped with a YFP filter set with excitation HQ500/20, dichroic beam splitter Q515LP, and emission HQ535/30. Images were captured with a mounted CCD camera (Sensys air-cooled; Photometrics) using SimplePCI.1 software (Compix).
Non-YFP roots were removed with a scalpel, and seedlings with transgenic roots were transferred onto new plates containing 0.25× Hoagland medium with 7.5 g/L sucrose and 300 mg/L of the antibiotic timentin (SmithKline Beecham Pharmaceuticals). To increase the quantity of transgenic root material, selected transgenic roots were cut 2 to 3 mm behind the tip, resulting in several new roots emerging from the cut surface that could be assayed 2 to 6 weeks later. Transformation efficiencies were calculated as the number of plants with at least one yellow root. Root growth rates were determined by marking the tips of the roots and measuring their length 24 h later with a light microscope containing a reticule.
Haustorium Assays
Haustorium development in transgenic roots was monitored following contact with host roots, host root exudates, or chemical HIFs as previously described (Jamison and Yoder, 2001). Host root contact was realized by overlaying YFP-expressing transgenic roots of Triphysaria seedlings growing on the surface of an agar plate with those of Arabidopsis for 5 d. We determined the number of YFP roots that made haustoria and expressed that as a fraction of the total number of YFP roots in contact with a host root.
Host exudates were made by growing M. truncatula plants in Magenta boxes containing 20 mL of 0.5% agar for 10 to 14 d. After removing the plants, the agar was centrifuged at 20,000 rpm for 30 min and the supernatant filter sterilized. Chemical HIFs were dissolved in water or methanol and then diluted in water. For both the exudates and HIF treatments, 2 mL were added to each plate of Triphysaria seedlings, which were held horizontal for 2 h before returning to their vertical orientation in the 25°C culture room. Haustoria were identified 24 h later as localized haustorial hair proliferation and tip swelling.
Transcriptional Analyses
Triphysaria or Arabidopsis roots were harvested from agar plates before and after treatment with HIFs and frozen in liquid nitrogen. The tissue was ground in liquid nitrogen and RNA isolated using the TRIzol reagent (Invitrogen). RNA gel blot hybridizations were performed as previously described (Matvienko et al., 2001a).
For quantitative RT-PCR, RNA was treated with DNase 1 prior to further purification with RNeasy Mini Spin columns (Qiagen). One microgram of RNA for each sample was converted to cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). The reverse transcription reaction was diluted 20-fold, and 2 μL was PCR amplified in the presence of SYBR green and assayed in real time with a LightCycler480 (Roche). Only reactions with a single melting peak were considered in the analysis.
Transcript levels were measured in duplicate for each of three to four biological replicates. Target gene expression was expressed relative to the constitutively expressed gene QAN8. Expression calculations used the standard curve method taking into account the efficiencies of the PCR reactions, calculated by log-linear regression LightCycler480 analysis software (Roche).
Expression of QR1 Protein in Escherichia coli
The coding region of QR1 was cloned into the E. coli protein expression vector pBAD-Topo (Invitrogen). The construct was transformed into E. coli Top 10 and grown to mid log phase in Luria-Bertani broth at 23°C with gentle shaking. The QR1 transcript was induced by adding l-arabinose to a final concentration of 0.02% (w/v). Cells were collected by centrifugation 4 h later, resuspended in extraction buffer (50 mM sodium phosphate, pH 7.2, 10 mg/mL lysozyme, 10 μg/mL DNase, and 5 μg/mL RNase) and lysed by freeze-thawing. After a brief ultrasonic treatment of the cell extract to fragment the bacterial DNA, the suspension was centrifuged and the supernatant loaded onto a Bio-Rex 70 cation exchange resin equilibrated with 50 mM sodium phosphate buffer, pH 7.2 (Bio-Rad). The protein was eluted with two bed volumes of 1 M NaCl, concentrated by ultrafiltration through a 10,000 molecular weight cutoff membrane (Millipore) and resuspended in binding buffer (50 mM sodium phosphate, pH 7.2, 0.5 M NaCl, 20 mM β-mercaptoethanol, and 10 mM imidazole). The suspension was loaded onto a nickel-NTA agarose column, washed with binding buffer, and eluted with 100 mM imidazole. The protein was again concentrated by ultrafiltration and the buffer exchanged for 10 mM sodium phosphate containing 10% glycerol. The enzyme was stored at −20°C. Protein concentrations were determined by the Bradford Protein Assay (Bio-Rad) and analyzed by SDS-PAGE under reducing conditions on 4 to 12% acrylamide gels.
Enzymatic Assays
The standard quinone reductase assay was conducted aerobically at room temperature in 0.5-mL reaction volumes containing 100 mM PIPES buffer, pH 6.5, 200 μM NADPH, and 100 μM quinone substrates. The enzyme had maximal activity at pH 6.5, and activity was reduced 50% at pH 6.0 or 7.0. The reactions were initiated by addition of enzyme and NADPH oxidation assayed as the change in absorbance at 340 nm with a Beckman DU 600 spectrophotometer in kinetic mode at room temperature. Reduction rates were corrected for the noncatalytic oxidation of NADPH in the presence of certain electron acceptors. Some quinones had a high background absorbance at 340 nm, and in these cases, enzymatic reduction was monitored by coupled reduction of the tetrazolium dye 3-(4,5-dimethylthiazo-2-yl)-2,5-diphyltetrazolium bromide (MTT), which was added to the standard mix at 0.3 mg mL−1 (Prochaska and Santamaria, 1988). Reduction of MTT was monitored at 610 nm using absorption coefficient of 11.3 mM−1 cm−1.
Cytochrome c reduction assays were performed in 0.5 mL final volume of 50 mM Tris, pH 7.6, 200 μM NADPH, 25 μM PAQ, and 40 μM cytochrome c. Reactions were started by the addition of enzyme and the reduction of cytochrome c monitored at 550 nm using an absorption coefficient of 29 mM−1 cm−1.
For inhibition studies, the QR1 enzyme was preincubated with 10 μM dicumarol (3,3′-methylenebis-(4-hydroxycoumarin)) for 5 min, at which time the reaction was initiation by addition of NADPH and PAQ.
Hydrogen peroxide was assayed using the Amplex-Red Hydrogen Peroxide Assay Kit (Molecular Probes). In selected reactions, catalase was added to a final concentration of 0.1 mg/mL.
Chloroplast Import Assays
Radiolabeled proteins were prepared from cDNAs encoding QR1, ceQORH (stock number U10519 from the ABRC), and three control proteins from pea (Pisum sativum), DGD1 (Froehlich et al., 2001), tic22 (Kouranov et al., 1998b), and tp110-110N (Lübeck et al., 1997), using the TNT T7 Coupled Reticulocyte Lysate System (Promega) and l-[35S] Met (Perkin-Elmer Life Sciences). Labeled proteins were incubated with chloroplasts isolated from 12-d-old pea seedlings in import buffer (50 mM HEPES-KOH and 330 mM sorbitol, pH 8.0) containing 3 mM ATP at room temperature for 20 min (Inoue et al., 2006). After import, intact chloroplasts were reisolated by centrifugation through 40% Percoll, washed with the import buffer, fractionated by SDS-PAGE, and observed by fluorography.
For protease treatments, reisolated chloroplasts were resuspended in import buffer and incubated on ice for 30 min with trypsin (0.1 mg = 1410 BAEE [N-benzoyl-l-arginine ethyl ester] units/mg chlorophyll equivalent chloroplasts; Sigma-Aldrich) or thermolysin (0.1 mg mg−1 chlorophyll equivalent chloroplasts in import buffer containing 2 mM CaCl2). The trypsin reaction was quenched with trypsin inhibitor (1 mg/mg trypsin; Sigma-Aldrich), 0.1 mg of which had a capacity to inactivate 2100 BAEE units of trypsin. To select the reactions to control for protease treatments, 1% Triton X-100 was added.
For Na2CO3 treatment, chloroplasts containing imported proteins were suspended in hypotonic lysis buffer (10 mM HEPES and 10 mM MgCl2, pH 8.0) and separated into supernatant and pellet fractions by centrifugation at 16,000g at 4°C for 20 min. The pellet fractions were further resuspended in 0.1 mM Na2CO3 on ice for 5 min and then separated again into supernatant and pellet fractions by centrifugation at 16,000g at 4°C for 20 min.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: T. versicolor QR1 sequence, AF304461; T. versicolor QR2 sequence, AF304462; T. versicolor RB sequence, DR169571.1; T. versicolor BIP sequence, DR169582.1; T. versicolor QAN8 sequence, DR170086.1; and Arabidopsis ceQORH protein, NP_193037.
Author Contributions
P.C.G.B. performed gene silencing experiments, T.F. performed the biochemical analyses with QR1, A.T., N.B.T., and D.J.-M. performed the hybridization analyses, Q.N. performed the QR1 gene structure analysis, K.I. performed chloroplast import assays, J.I.Y. designed and supervised the research, and J.I.Y. and P.C.G.B. wrote the manuscript with editing from the other authors.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Sequences of Primers Used.
Supplementary Material
Acknowledgments
This project was funded by National Science Foundation Grant 0236545. P.C.G.B. was supported in part by a UC Davis Plant Sciences Fellowships and Fulbright Fellowship. We thank Gino Cortopassi for technical assistance and David Lynn for insightful discussions.
References
- Abel S., Theologis A. (1996). Early genes and auxin action. Plant Physiol. 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P., Sarwat M., Sharma S. (2008). Reactive oxygen species, antioxidants and signaling in plants. J. Plant Biol. 51: 167–173 [Google Scholar]
- Akileswaran L., Brock B.J., Cereghino J.L., Gold M.H. (1999). 1,4-Benzoquinone reductase from Phanerochaete chrysosporium: cDNA cloning and regulation of expression. Appl. Environ. Microbiol. 65: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht H., Yoder J.I., Phillips D.A. (1999). Flavonoids promote haustoria formation in the root parasite Triphysaria. Plant Physiol. 119: 585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel H.M. (1993). Phenolics in ecological interactions - The importance of oxidation. J. Chem. Ecol. 19: 1521–1552 [DOI] [PubMed] [Google Scholar]
- Atsatt P.R., Hearn T.F., Nelson R.L., Heineman R.T. (1978). Chemical induction and repression of haustoria in Orthocarpus purpurascens (Scophulariaceae). Ann. Bot. (Lond.) 42: 1177–1184 [Google Scholar]
- Babiychuk E., Kushnir S., Bellesboix E., Van Montagu M., Inze D. (1995). Arabidopsis thaliana NADPH oxidoreductase homologs confer tolerance of yeasts toward the thiol-oxidizing drug diamide. J. Biol. Chem. 270: 26224–26231 [DOI] [PubMed] [Google Scholar]
- Baird W.V., Riopel J.L. (1984). Experimental studies of haustorium initiation and early development in Agalinis purpurea (L.) Raf. (Scrophulariaceae). Am. J. Bot. 71: 803–814 [Google Scholar]
- Baird W.V., Riopel J.L. (1985). Surface characteristics of root haustorial hairs of parasitic Scrophulariaceae. Bot. Gaz. 146: 63–69 [Google Scholar]
- Bais H.P., Weir T.L., Perry L.G., Gilroy S., Vivanco J.M. (2006). The role of root exudates in rhizosphere interations with plants and other organisms. Annu. Rev. Plant Biol. 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Brouillard R., Cheminat A. (1982). Structural transformations of anthocyanins in water. Anthocyanins as Food Colors, Markakis P., (New York: Academic Press; ), pp. 1–38 [Google Scholar]
- Burkitt M.J. (2003). Chemical, biological and medical controversies surrounding the Fenton reaction. Prog. React. Kinet. Mech. 28: 75–103 [Google Scholar]
- Cadenas E., Hochstein P., Ernster L. (1992). Pro-and antioxidant functions of quinones and quinone reductases in mammalian cells. Adv. Enzymol. Relat. Areas Mol. Biol. 65: 97–146 [DOI] [PubMed] [Google Scholar]
- Caldwell E.S., Steelink C. (1969). Phenoxy radical intermediate in the enzymatic degradation of lignin model compounds. Biochim. Biophys. Acta 184: 420–431 [DOI] [PubMed] [Google Scholar]
- Carol R.J., Dolan L. (2006). The role of reactive oxygen species in cell growth: Lessons from root hairs. J. Exp. Bot. 57: 1829–1834 [DOI] [PubMed] [Google Scholar]
- Carol R.J., Takeda S., Linstead P., Durrant M.C., Kakesova H., Derbyshire P., Drea S., Zarsky V., Dolan L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Chang M., Lynn D.G. (1986). The haustorium and the chemistry of host recognition in parasitic angiosperms. J. Chem. Ecol. 12: 561–579 [DOI] [PubMed] [Google Scholar]
- Cline K., Werner-Washburne M., Andrews J., Keegstra K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.J., Barton J.D., Rossjohn J., Thorn J.M., Taylor G.L., Ollis D.L. (1996). Structural and sequence comparisons of quinone oxidoreductase, zeta-crystallin, and glucose and alcohol dehydrogenases. Arch. Biochem. Biophys. 328: 173–183 [DOI] [PubMed] [Google Scholar]
- Estabrook E.M., Yoder J.I. (1998). Plant-plant communications: Rhizosphere signaling between parasitic angiosperms and their hosts. Plant Physiol. 116: 1–7 [Google Scholar]
- Faig M., Bianchet M.A., Winski S., Hargreaves R., Moody C.J., Hudnott A.R., Ross D., Amzel L.M. (2001). Structure-based development of anticancer drugs: Complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones. Structure 9: 659–667 [DOI] [PubMed] [Google Scholar]
- Faure D., Vereecke D., Leveau J.H.J. (2009). Molecular communication in the rhizosphere. Plant Soil 321: 279–303 [Google Scholar]
- Favreau L.V., Pickett C.B. (1991). Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J. Biol. Chem. 266: 4556–4561 [PubMed] [Google Scholar]
- Fedorov A., Merican A.F., Gilbert W. (2002). Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl. Acad. Sci. USA 99: 16128–16133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald R.D. (2006). Casting a genetic light on the evolution of eyes. Science 313: 1914–1918 [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Fridovich I. (1970). Quantitative aspects of the production of superoxide anion radical by milk xanthine oxidase. J. Biol. Chem. 245: 4053–4057 [PubMed] [Google Scholar]
- Froehlich J., Benning C., Dormann P. (2001). The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J. Biol. Chem. 276: 31806–31812 [DOI] [PubMed] [Google Scholar]
- Gendler K., Paulsen T., Napoli C. (2008). ChromDB: The Chromatin Database. Nucleic Acids Res. 36: 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenshields D.L., Liu G.S., Selvaraj G., Wei Y.D. (2005). Differential regulation of wheat quinone reductases in response to powdery mildew infection. Planta 222: 867–875 [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Wesley S.V., Wielopolska A.J., Waterhouse P.M. (2002). High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Hickman J.C. (1993). The Jepson Manual: Higher Plants of California. (Berkeley, CA: University of California Press; ). [Google Scholar]
- Hirsch A.M., Bauer W.D., Bird D.M., Cullimore J., Tyler B., Yoder J.I. (2002). Molecular signals and receptors- Controlling rhizosphere interactions between plants and other organisms-by chance or intent? Ecol. 84: 858–868 [Google Scholar]
- Huang Q.-L., Du X.-Y., Stone S.H., Amsbaugh D.F., Datiles M., Hu T.-S., Zigler J.S. (1990). Association of hereditary cataracts in strain 13/N guinea-pigs with mutation of the gene for [zeta]-crystallin. Exp. Eye Res. 50: 317–325 [DOI] [PubMed] [Google Scholar]
- Huang Q.L., Russell P., Stone S.H., Zigler J.S. (1987). Zeta-crystallin, a novel lens protein from the guinea pig. Curr. Eye Res. 6: 725–732 [DOI] [PubMed] [Google Scholar]
- Inderjit, Nilsen E.T. (2003). Bioassays and field studies for allelopathy in terrestrial plants: Progress and problems. Crit. Rev. Plant Sci. 22: 221–238 [Google Scholar]
- Inoue K., Furbee K., Uratsu S., Kato M., Dandekar A., Ikoma Y. (2006). Catalytic activities and chloroplast import of carotenogenic enzymes from citrus. Physiol. Plant. 127: 561–570 [Google Scholar]
- Iyanagi T., Yamazaki I. (1970). One-electron transfer reactions in biochemical systems. 5. Difference in mechanism of quinone reduction by NADH dehydrogenase and NAD(P)H dehydrogenase (DT-diaphorase). Biochim. Biophys. Acta 216: 282–294 [DOI] [PubMed] [Google Scholar]
- Jackson D., Froehlich J., Keegstra K. (1998a). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273: 16583–16588 [DOI] [PubMed] [Google Scholar]
- Jackson D.T., Froehlich J.E., Keegstra K. (1998b). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273: 16583–16588 [DOI] [PubMed] [Google Scholar]
- Jamison D.S., Yoder J.I. (2001). Heritable variation in quinone-induced haustorium development in the parasitic plant Triphysaria. Plant Physiol. 125: 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D.M., Losner-Goshen D. (1994). The attachment organ of the parasitic angiosperms Orobanche cumana and O. aegyptiaca and its development. Can. J. Bot. 72: 564–574 [Google Scholar]
- Jones D.P. (2008). Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 295: C849–C868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes W.J., Palmer A.G., Erbil W.K., Taylor J.V., Apkarian R.P., Weeks E.R., Lynn D.G. (2007). Sernagenesis and the parasitic angiosperm Striga asiaticae. Plant J. 51: 707–716 [DOI] [PubMed] [Google Scholar]
- Kim D., Kocz R., Boone L., Keyes W.J., Lynn D.G. (1998). On becoming a parasite: Evaluating the role of wall oxidases in parasitic plant development. Chem. Biol. 5: 103–117 [DOI] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D. (1998b). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Chen X.J., Fuks B., Schnell D.J. (1998a). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt J. (1969). The Biology of Parasitic Flowering Plants. (Berkeley, CA: University of California Press; ). [Google Scholar]
- Laskowski M.J., Dreher K.E., Gehring M.A., Abel S., Gensler A.L., Sussex I.M. (2002). FQR1. A novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol. 128: 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.C., Gonzalez P., Wistow G. (1994). ζ-Crystallin: A lens-specific promoter and the gene recruitment of an enzyme as a crystallin. J. Mol. Biol. 236: 669–678 [DOI] [PubMed] [Google Scholar]
- Lee K.C., Campbell R.W. (1969). Nature and occurence of juglone in Juglans nigra L. Hortic. Sci. 4: 297–298 [Google Scholar]
- Lind C., Cadenas E., Hochstein P., Ernster L. (1990). DT-diaphorase: Purification, properties, and function. Methods Enzymol. 186: 287–301 [DOI] [PubMed] [Google Scholar]
- Liszkay A., van der Zalm E., Schopfer P. (2004). Production of reactive oxygen intermediates (O-2(center dot-), H2O2, and (OH)-O-center dot) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 136: 3114–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübeck J., Heins L., Soll J. (1997). A nuclear-coded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol. 137: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn D.G., Chang M. (1990). Phenolic signals in cohabitation: Implications for plant development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 497–526 [Google Scholar]
- Mano J., Babiychuk E., Belles-Boix E., Hiratake J., Kimura A., Inzea D., Kushnir S., Asada K. (2000). A novel NADPH:diamide oxidoreductase activity in Arabidopsis thaliana P1 zeta-crystallin. Eur. J. Biochem. 267: 3661–3671 [DOI] [PubMed] [Google Scholar]
- Mano J., Belles-Boix E., Babiychuk E., Inzé D., Torii Y., Hiraoka E., Takimoto K., Slooten L., Asada K., Kushnir S. (2005). Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol. 139: 1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano J., Belles-Boix E., Babiychuk E., Van Montague M., Inze D., Kushnir S., Asada K., Slooten L. (2002). Arabidopsis P1 zeta-crystallin forms an O-2-dependent electron sink in tobacco leaves. Plant Cell Physiol. 43: S70 [Google Scholar]
- Marvier M.A. (1998). A mixed diet improves performance and herbivore resistance of a parasitic plant. Ecol. 79: 1272–1280 [Google Scholar]
- Matvienko M., Torres M.J., Yoder J.I. (2001a). Transcriptional responses in the hemiparasitic plant Triphysaria versicolor to host plant signals. Plant Physiol. 127: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M., Wojtowicz A., Wrobel R., Jamison D., Goldwasser Y., Yoder J.I. (2001b). Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 25: 375–387 [DOI] [PubMed] [Google Scholar]
- McCord J.M., Fridovich I. (1969). Superoxide dismutase. J. Biol. Chem. 244: 6049–6055 [PubMed] [Google Scholar]
- McCully M. (2007). Rhizosphere allelopathy. Allelopathy J. 19: 75–84 [Google Scholar]
- Miras S., Salvi D., Ferro M., Grunwald D., Garin J., Joyard J., Rolland N. (2002). Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 277: 47770–47778 [DOI] [PubMed] [Google Scholar]
- Miras S., Salvi D., Piette L., Seigneurin-Berny D., Grunwald D., Reinbothe C., Joyard J., Reinbothe S., Rolland N. (2007). Toc159-and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J. Biol. Chem. 282: 29482–29492 [DOI] [PubMed] [Google Scholar]
- Musselman L.J. (1980). The biology of Striga, Orobanche, and other root parasitic weeds. Annu. Rev. Phytopathol. 18: 463–489 [Google Scholar]
- O'Brien P. (1991). Molecular mechanisms of quinone toxicity. Chem. Biol. Interact. 80: 1–41 [DOI] [PubMed] [Google Scholar]
- Okonkwo S.N.C., Nwoke F.I.O. (1978). Initiation, development and stucture of the primary haustorium in Striga gesnerioides (Scrophulariaceae). Ann. Bot. (Lond.) 42: 455–463 [Google Scholar]
- Persson B., Hedlund J., Jornvall H. (2008). The MDR superfamily. Cell. Mol. Life Sci. 65: 3879–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porté S., Crosas E., Yakovtseva E., Biosca J.A., Farrés J., Fernández M.R., Parés X. (2009). MDR quinone oxidoreductases: The human and yeast [zeta]-crystallins. Chem. Biol. Interact. 178: 288–294 [DOI] [PubMed] [Google Scholar]
- Prestera T., Talalay P. (1995). Electrophile and antioxidant regulation of enzymes that detoxify carcinogens. Proc. Natl. Acad. Sci. USA 92: 8965–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochaska H.J., Santamaria A.B. (1988). Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: A screening assay for anticarcinogenic enzyme inducers. Anal. Biochem. 169: 328–336 [DOI] [PubMed] [Google Scholar]
- Rao P.V., Krishna C.M., Zigler J.S. (1992). Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens - A novel NADPH-quinone oxidoreductase. J. Biol. Chem. 267: 96–102 [PubMed] [Google Scholar]
- Renew S., Heyno E., Schopfer P., Liszkay A. (2005). Sensitive detection and localization of hydroxyl radical production in cucumber roots and Arabidopsis seedlings by spin trapping electron paramagnetic resonance spectroscopy. Plant J. 44: 342–347 [DOI] [PubMed] [Google Scholar]
- Riopel J., Musselman L. (1979). Experimental initiation of haustoria in Agalinis purpurea. Am. J. Bot. 66: 570–575 [Google Scholar]
- Riopel J.L., Timko M.P. (1995). Haustorial initiation and differentiation. Parasitic Plants, Press M.C., Graves J.D., (London: Chapman and Hall; ), pp. 39–79 [Google Scholar]
- Ross D., Siegel D., Helmut S., Lester P. (2004). NAD(P)H:Quinone Oxidoreductase 1 (NQO1, DT-Diaphorase), functions and pharmacogenetics. Methods in Enzymology, Sies H., Packer L., (London: Academic Press; ), pp. 115–144 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols, Krawetz S., Misener S., (Totowa, NJ: Humana Press; ), pp. 365–386 [DOI] [PubMed] [Google Scholar]
- Shann J.R., Blum U. (1987). The uptake of ferulic and p-hydroxybenzoic acids by Cucumis sativus. Phytochem. 26: 2959–2964 [Google Scholar]
- Siqueira J.O., Safir G.R., Nair M.G. (1991). Stimulation of vesicular-arbuscular mycorrhiza formation and growth of white clover by flavanoid compounds. New Phytol. 118: 87–93 [Google Scholar]
- Smith C.E., Dudley M.W., Lynn D.G. (1990). Vegetative/parasitic transition: control and plasticity in Striga development. Plant Physiol. 93: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.E., Ruttledge T., Zeng Z., O'Malley R.C., Lynn D.G. (1996). A mechanism for inducing plant development- the genesis of a specific inhibitor. Proc. Natl. Acad. Sci. USA 93: 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonti R.V., Chiurazzi M., Wong D., Davies C.S., Harlow G.R., Mount D.W., Signer E.R. (1995). Arabidopsis mutant deficient in T-DNA Integration. Proc. Natl. Acad. Sci. USA 92: 11786–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F., Tedeschi G., Pupillo P., Trost P. (1999). Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett. 463: 382–386 [DOI] [PubMed] [Google Scholar]
- Sparla F., Tedeschi G., Trost P. (1996). NAD(P)H-(quinone-acceptor) oxidoreductase of tobacco leaves is a flavin mononucleotide-containing flavoenzyme. Plant Physiol. 112: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens J.C., Lynn D.G., Kamat V.S., Riopel J.L. (1982). Molecular specificity of haustorial induction in Agalinis purpurea (L.) Raf. (Scrophulariaceae). Ann. Bot. (Lond.) 50: 1–7 [Google Scholar]
- Subramanian C., Woo J., Cai X., Xu X.D., Servick S., Johnson C.H., Nebenfuhr A., von Arnim A.G. (2006). A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J. 48: 138–152 [DOI] [PubMed] [Google Scholar]
- Testa B. (1995). The Metabolism of Drugs and Other Xenobiotics: Biochemistry of Redox Reactions. (New York: Academic Press; ). [Google Scholar]
- Thorn J.M., Barton J.D., Dixon N.E., Ollis D.L., Edwards K.J. (1995). Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 249: 785–799 [DOI] [PubMed] [Google Scholar]
- Thurman L.D. (1966). Genecological Studies in Orthocarpus Subgenus Triphysaria. (Berkeley, CA: University of California Press; ). [Google Scholar]
- Tomilov A.A., Tomilova N.B., Yoder J.I. (2007). Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225: 1059–1071 [DOI] [PubMed] [Google Scholar]
- Velten J., Velten L., Hain R., Schell J. (1984). Isolation of a dual plant promoter fragment from the Ti plasmid of Agbrobacterium tumefaciens. EMBO J. 3: 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkerpeach C.R., Velten J. (1994). Agrobacterium-mediated gene transfer to plant cells: Cointegrate and binary vector systems. Plant Molecular Biology Manual, Gelvin S., Schilperoort R., (Dordrecht, The Netherlands: Kluwer; ), pp. 1–19 [Google Scholar]
- William C.N. (1961). Growth and morphogenesis of Striga seedlings. Nature 189: 378–381 [Google Scholar]
- Willis R.J. (2007). The History of Allelopathy. (Dordrecht, The Netherlands: Springer; ). [Google Scholar]
- Wolf S.J., Timko M.P. (1991). In vitro root culture - A novel approach to study the obligate parasite Striga-asiatica (L) Kuntze. Plant Sci. 73: 233–242 [Google Scholar]
- Wrobel R.L., Matvienko M., Yoder J.I. (2002). Heterologous expression and biochemical characterization of an NAD(P)H:quinone oxidoreductase from the hemiparasitic plant Triphysaria versicolor. Plant Physiol. Biochem. 40: 265–272 [Google Scholar]
- Zeng Z.X., Cartwright C.H., Lynn D.G. (1996). Cyclopropyl-p-benzoquinone - A specific organogenesis inhibitor in plants. J. Am. Chem. Soc. 118: 1233–1234 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.