This work shows how a specific class of small RNAs respond to auxin and quantitatively regulate root branching, an important adaptive trait in plants. These small RNAs and their target transcription factors form a self-regulatory gene network through multiple feedback loops. This ensures a quantitative control of lateral root development and modulation of auxin effects.
Abstract
Plants adapt to different environmental conditions by constantly forming new organs in response to morphogenetic signals. Lateral roots branch from the main root in response to local auxin maxima. How a local auxin maximum translates into a robust pattern of gene activation ensuring the proper growth of the newly formed lateral root is largely unknown. Here, we demonstrate that miR390, TAS3-derived trans-acting short-interfering RNAs (tasiRNAs), and AUXIN RESPONSE FACTORS (ARFs) form an auxin-responsive regulatory network controlling lateral root growth. Spatial expression analysis using reporter gene fusions, tasi/miRNA sensors, and mutant analysis showed that miR390 is specifically expressed at the sites of lateral root initiation where it triggers the biogenesis of tasiRNAs. These tasiRNAs inhibit ARF2, ARF3, and ARF4, thus releasing repression of lateral root growth. In addition, ARF2, ARF3, and ARF4 affect auxin-induced miR390 accumulation. Positive and negative feedback regulation of miR390 by ARF2, ARF3, and ARF4 thus ensures the proper definition of the miR390 expression pattern. This regulatory network maintains ARF expression in a concentration range optimal for specifying the timing of lateral root growth, a function similar to its activity during leaf development. These results also show how small regulatory RNAs integrate with auxin signaling to quantitatively regulate organ growth during development.
INTRODUCTION
The initiation of lateral roots plays a crucial role in plant development, since it determines the architecture of the root system and, thus, stability as well as nutrient and water uptake potential for the entire organism. Lateral root development is a typical example of a canalized developmental process (i.e., buffered against perturbation; Siegal and Bergman 2002), yet roots strongly adapt to the local environment to maximize acquisition of water and nutrients from the soil. In recent years, it has become clear that lateral roots initiate from a small number of pericycle cells (initiation) that differentiate into a primordia and grow out of the primary root (emergence) (Hardtke, 2006; De Smet et al., 2006; Parizot et al., 2008; Petricka and Benfey, 2008). Auxin is a morphogenetic trigger for lateral root formation (Benková et al., 2009), and its local maximum acts as an instructive signal for initiation of these organs (Dubrovsky et al., 2008). Many of auxin's actions are mediated by transcription factors of the auxin response factor (ARF) family, and several ARFs play critical roles in lateral root development (Okushima et al., 2005b; Wilmoth et al., 2005).
Small RNAs, such as microRNAs (miRNAs) and trans-acting short-interfering RNAs (tasiRNAs), control many aspects of development in eukaryotes. As negative regulators of gene expression, they can act as developmental switches to shut down gene expression programs. Alternatively, small RNAs can fine-tune gene expression to quantitatively adapt developmental processes to endogenous or environmental fluctuations and therefore act as canalization factors (Li et al., 2009). Several reports have implicated miRNAs in the modulation of auxin action during lateral root development supporting this model (Guo et al., 2005; Mallory et al., 2005; Gifford et al., 2008; Yoon et al., 2010).
tasiRNAs belong to a plant-specific class of endogenous small RNAs, whose biogenesis requires an initial miRNA-mediated cleavage of their precursors. The cleavage product is then converted to double-stranded RNA through RNA-DEPENDENT RNA POLYMERASE6 (RDR6) activity and sequential DICER-LIKE4 (DCL4)-mediated cleavage events (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Adenot et al., 2006). Of the four tasiRNAs precursors identified (TAS1-4) in Arabidopsis thaliana, cleavage of TAS3 is unique since it requires the specific action of the miR390/ARGONAUTE7 (AGO7) complex for tasiRNA production (Montgomery et al., 2008). miR390 and TAS3 tasiRNAs define a pathway that regulates leaf patterning and developmental timing by repressing the ARF family members ARF2, ARF3, and ARF4 (Figure 1A) (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). We and others have previously reported that TAS3, AGO7, and miR390 are expressed in root tissues (Hirsch et al., 2006; Montgomery et al., 2008); however, the function of this pathway in root development is unclear.
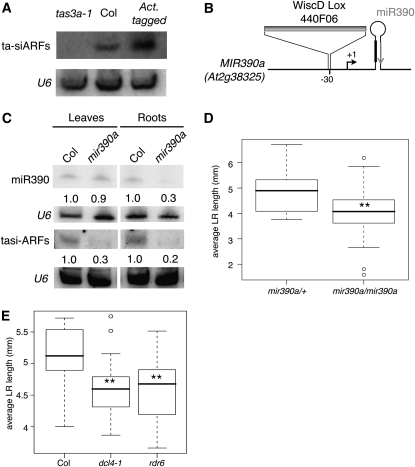
Figure 1.
Altered Levels of TAS3a Modify Root Architecture.
(A) Schematic representation of the TAS3 pathway. miR390-loaded AGO7 cleaves the TAS3 precursor RNA. The cleavage product is converted into a double-stranded RNA by RDR6 and SGS3 and then diced into tasiARFs by DCL4 and DRB4. tasiARFs inhibit ARF2, ARF3, and ARF4 mRNA expression.
(B) Root architecture of 10-d-old seedlings of the wild type (Columbia [Col]), an overexpression line (35S:TAS3a), the GABI 626B09 activation tagging line (Act. tagged), and the GABI 621G08 mutant (tas3a-1; Adenot et al., 2006). Bars = 10 mm.
(C) Measurement of the average lateral root (LR) length in the different genotypes. Distribution of the population (n > 22) is represented by box plots. Differences with the wild type are indicated (***, P < 0.001; *, P < 0.05; Student's t test).
(D) Numbers of lateral root primordia at specific developmental stages in 8-d-old wild-type, tas3a-1, and activation-tagged TAS3a roots (expressed as stages 1 to 7, according to [Malamy and Benfey, 1997]; mean ± se, n = 12 for each group of seedlings).
Here, we show that mutations affecting the abundance of TAS3-derived tasiRNAs lead to quantitative changes in the rate of lateral root growth. miR390 is induced during lateral root initiation and triggers the local production of tasiRNAs. In the lateral root primordium, the tasiARFs reduce the activity of ARF2, ARF3, and ARF4, thereby promoting lateral root growth. In addition, ARF2, ARF3, and ARF4 are required for proper miR390 expression through different feedback mechanisms. Thus, auxin, miR390, TAS3, and their ARFs targets define a regulatory network quantitatively controlling lateral root growth. This complex network acts to fine-tune local auxin responses and thus provides robustness and flexibility to lateral root growth.
RESULTS
TAS3a Controls Lateral Root Growth
To determine the role of TAS3a (At3g17185) during root development, we first analyzed the effects of increased levels of TAS3a on root architecture. We identified an activation-tagged line in the GABI-Kat collection (Rosso et al., 2003) in which TAS3a transcript levels were elevated >100-fold compared with wild-type plants (see Supplemental Figure 1 online). In these plants, the average length of lateral roots increased by 1.5-fold (Figures 1B and 1C), whereas primary root length and lateral root density did not differ from the wild type (see Supplemental Figures 2A and 2B online). To confirm that these effects were caused by TAS3a overexpression, we analyzed the root architecture of transgenic plants in which TAS3a is expressed from the 35S promoter (35S:TAS3a). As in the activation-tagged line, TAS3a transcripts levels were increased 100-fold and 35S:TAS3a plants had longer lateral roots than wild-type controls (Figures 1B and 1C; see Supplemental Figure 1 online), while primary root length or lateral root density were unchanged (see Supplemental Figures 2A and 2B online). We then analyzed the root architecture of the tas3a-1 mutant (Adenot et al., 2006), which has only 40% of wild-type TAS3a transcript levels (see Supplemental Figure 1 online). In contrast with the elongated lateral roots in 35S:TAS3a, tas3a-1 mutant plants showed shorter lateral roots than wild-type controls, demonstrating that TAS3a transcript levels quantitatively correlate with lateral root length (Figures 1B and 1C). To gain further insight into the developmental basis for the lateral root defect of TAS3a mutants, we quantified the distribution of stages of lateral root primordia in wild-type and mutant roots (Figure 1D). Plants overexpressing TAS3a had twice as many stage 5-7 lateral root primordia than the wild type, whereas in tas3a-1 mutants, the number of stage 1-2 primordia was increased by 50% (Figure 1D). The total number of emerged and nonemerged (stage 1-7) primordia did not differ across the different lines tested (see Supplemental Figures 2C and 2D online), suggesting that TAS3a regulates the rate of primordia progression through the developmental stages, rather than the initiation process. To further analyze this, we quantified the effect of TAS3a levels on cell elongation and cell proliferation, two postemergence processes that could contribute to the overall change in lateral root length. The size of both emerged lateral root meristems and cortical cells was reduced in tas3a-1 mutants but unchanged in plants overexpressing TAS3a compared with controls (see Supplemental Figures 2E and 2F online). This result indicated that TAS3a is required but not limiting in the control of cell proliferation and cell expansion postemergence. Thus, the differences in lateral root length induced by modified TAS3a levels reflect changes in rates of developmental progression during lateral root formation and emergence. This suggested that TAS3a acts as a positive regulator of lateral root growth.
The Abundance of TAS3a-Derived Small RNAs Correlates with Lateral Root Length
The biogenesis of the biologically active TAS3-derived tasiRNAs (hereafter called tasiARFs) is dependent on miR390-mediated cleavage of TAS3a (Montgomery et al., 2008). Thus, we used RNA gel blotting to directly quantify tasiARFs and found increased amounts in the activation-tagged allele and 35S:TAS3a roots compared with the wild type, whereas tasiARFs were undetectable in tas3a-1 mutant plants (Figure 2A; Adenot et al., 2006). The positive correlation between TAS3a levels, tasiARF abundance, and the growth rate of lateral roots suggested that the effect of TAS3a on root architecture is mediated by the tasiARFs. To further test this hypothesis, we analyzed the phenotype of plants in which tasiARFs production from TAS3a was compromised. To this end, we first characterized the root architecture of mutants impaired in the production of miR390. miR390 and tasiARF levels were strongly reduced in roots but not in leaves of mir390a mutants (Figures 2B and 2C; see Supplemental Figure 3 online), indicating that the MIR390a locus contributed the majority of miR390 levels in roots, whereas miR390 is produced by both the MIR390a and MIR390b loci in leaves. Lateral roots were shorter in mir390a mutants compared with heterozygous mir390a/+ plants (Figure 2D). We then analyzed plants with mutations in DCL4 and RDR6, two enzymes critical for tasiARFs, which act downstream of miR390-mediated cleavage (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Gasciolli et al., 2005; Xie et al., 2005). Both mutants had shorter lateral roots than the wild type (Figure 2E). Taken together, these results confirmed that tasiARF abundance is instrumental in controlling lateral root length. Interestingly, the phenotypes of the tas3a, activation-tagged, and 35S:TAS3a lines were limited to lateral roots (see Supplemental Figure 2A online), suggesting a specific function of tasiARFs in lateral root development.
Figure 2.
Modulation of tasiARF Abundance Correlates with Lateral Root Length.
(A) RNA gel blot analysis of 15μg of leaves RNA of tas3a-1 mutant (GABI 621G08), wild-type, and activation-tagged (GABI 629B09) lines. The blot was hybridized with probes complementary to TAS3 tasiRNA (Gasciolli et al., 2005), and U6 snRNA served as a loading control.
(B) Schematic representation of the MIR390a locus. Position of the transcription initiation start identified by 5′ RACE is indicated by the arrow, the mature miR390 is indicated on the stem loop by a gray arrow, and the miR390* is indicated by a thick line. Position of the WiscDs insertion 30 bp upstream of the +1 is indicated.
(C) RNA gel blot analysis of 15 μg of leaves or root RNA from 10-d-old wild-type (Col) or mir390a plants hybridized with miR390 or tasiARFs. U6 snRNA served as a loading control, and numbers are the ratios of miR390 to U6 signal. This experiment was done twice with similar results.
(D) and (E) Measurement of the average lateral root (LR) length in the indicated genotypes.
(D) The average lateral root length is reduced in homozygous mir390a plants compared with heterozygous (mir390a/+) plants.
(E) The average lateral root length is reduced in dcl4-1 and rdr6 mutants compared with wild-type controls. Distribution of the populations (n > 18) is represented by box plots. **, P < 0.01; Student's t test.
TAS3a-Derived tasiARFs Are Produced and Active during Lateral Root Development
To elucidate the spatio-temporal basis of tasiARFs action during lateral root development, we determined the expression patterns of TAS3a and miR390. We first examined the expression pattern of a pTAS3a:GUS (for β-glucuronidase) reporter fusion construct. GUS expression was detected throughout the root in the parenchyma cells of the differentiated central cylinder (Figures 3A and 3A'), but it was absent from lateral root primordia. By contrast, a pMIR390a:GUS-GFP (for green fluorescent protein) reporter fusion construct showed GUS expression only in the proximal primary root meristem and the basal cells of the lateral root primordia (Figure 3B). Transverse sections of emerging primordia indicated that the MIR390a promoter is active in the mesenchymal cells of the central cylinder, the pericycle, and the flanks of the developing primordia (Figure 3B'). To test if the pMIR390a:GUS-GFP reporter faithfully reflected the spatio-temporal pattern of miR390 activity, we used a miR390-GFP sensor that is degraded in cells where miR390 is present (see Methods and Figure 3E). We found GFP to be specifically excluded from lateral root primordia of plants expressing the miR390 sensor (Figure 3F). By contrast, GFP was readily detectable in lateral root primordia of plants expressing a mutated form of the sensor, which was immune to miR390 action (Figure 3G). These results confirmed that the absence of GFP in cells of the primordia is caused by miR390 and demonstrated that miR390 is produced and active in lateral root primordia. Next, we tested the activity of tasiARFs in these cells using a sensor construct with tasiARF-sensitive GUS expression (Chitwood et al., 2009; Schwab et al., 2009). Comparison of GUS expression patterns in plants expressing sensitive (Figures 3D and 3D') and insensitive (Figures 3C and 3C') tasiARF sensors revealed that tasiARFs are active in lateral root primordia.
Figure 3.
The Localized Expression of miR390 Governs Local tasiARF Production in Incipient Lateral Roots.
(A) to (D’) Expression of reporters for TAS3a (A), MIR390a (B), an ARF3-based tasiARF sensor, and its control ([C] and [D]; Fahlgren et al., 2006) was observed in lateral root primordia either on intact 10-d-old lateral root primordia ([A] to [D]) or on transverse sections ([A’] to [D’]). The dashed lines in (A) to (D) indicate the position of transverse sections shown in (A’) to (D’). Pericycle (p), xylem (x), and phloem (ph) poles are indicated. Arrowheads in (A’) indicate pTAS3a:GUS expression in the phloem poles and parenchyma cells of the central cylinder. The arrowhead in (C’) indicates tasiARF activity in the center of the primordium. Bars = 75 μm.
(E) Schematic representation of the miR390 sensor constructs. In the wild type (WT sensor), which is sensitive to miR390 action, a wild-type miR390 binding site from TAS3a (gray line) is cloned downstream of GFP, whereas in the mutated version (Mut. sensor), the miR390 binding site contains five mismatches.
(F) and (G) Expression of the wild-type and mutated miR390 sensor in stage 4 lateral root primordia of 10-d-old plants. The dashed lines indicate the contour of the primordia on the confocal section and the transmitted light images (insets). Bars = 30 μm.
Taken together, our results showed that the expression of miR390 and TAS3a overlap at the base of initiating lateral root primordia, which leads to a spatially restricted production of tasiARFs. This suggested that miR390 activity closely defines the expression pattern of tasiARFs.
Developmental Signals Controlling miR390 Expression
Having identified miR390 as a key regulator of tasiARFs production and, thus, lateral root development, we wanted to gain further insight into the developmental signals controlling miR390 expression. To this end, we analyzed pMIR390a:GUS-GFP expression patterns during lateral root development using confocal microscopy. GFP activity was detected in all dividing pericycle cells of stage 1 and 2 primordia (Figures 4A and 4B), while at stage 3, pMIR390a:GUS-GFP expression defined a cup-shaped domain at the base of the primordia that extended into the central cylinder (Figures 4C and 4D). Since a local accumulation of auxin is an early marker for lateral root initiation (Dubrovsky et al., 2008), we used the reporter pDR5rev:erRFP as a proxy for auxin accumulation (Gallavotti et al., 2008). We observed that MIR390a expression and DR5 reporter activity overlapped only in stage 1-2 primordia and then segregated (Figures 4E to 4H). This result indicated that the local auxin maximum is unlikely to be the primary signal affecting miR390 accumulation during lateral root formation. In addition, pMIR390a:GUS-GFP expression was detected in the parenchyma cells of one xylem pole before any pericycle division (Figures 4I and 4J), in cells where auxin did not accumulate (Figure 4E). Thus, the onset of miR390 expression preceded the first steps of lateral root initiation, marked by auxin accumulation in the pericycle cells and their subsequent asymmetric division. Once the lateral root is initiated, MIR390a is expressed at the base and flanks of the primordium.
Figure 4.
miR390 Expression during Early Stages of Lateral Root Formation.
(A) to (D) Confocal observation of pMIR390a:GUS-GFP reporter during early stages of lateral root development (Malamy and Benfey, 1997). GFP signal is in green, nuclei are stained by DAPI (blue), and the position of the xylem is marked by a dashed line. The inset in (D) is a view of a primordium from the top.
(E) to (H) Confocal observation of pMIR390a:GUS-GFP and pDR5rev:erRFP reporters during early stages of lateral root development. GFP signal is in green, red fluorescent protein (RFP) is in red, and yellow indicates area of overlapping signals. The closed arrowhead in (E) indicates expression of the GFP reporter in the dividing pericycle cells, whereas the open arrowhead is expression in the xylem mesenchymal cells.
(I) and (J) Observation of pMIR390a:GUS-GFP reporter before pericyle division. p, pericycle; e, endodermis; cx, cortex; ep, epidermis.
(I) Confocal section showing expression of the GFP reporter in the xylem mesenchymal cells (open arrowhead).
(J) Transverse section showing expression of the GUS reporter in the xylem mesenchymal cells (open arrowhead).
Bars = 30 μm.
To determine the connection between lateral root development and endogenous miR390 expression, we monitored its accumulation in plants where development of the lateral roots was synchronously induced by treatment with an inhibitor of polar auxin transport (1-N-naphthylphthalamic acid [NPA]) followed by an auxin (indole-3-acetic acid [IAA]) treatment. RNA gel blot analysis indicated that miR390 expression gradually increased 6 h after auxin treatment (Figure 5A) corresponding to the onset of lateral root initiation (Vanneste et al., 2005). After 24 h of auxin treatment (corresponding to stage 2-3), expression of miR390 was increased up to fourfold when compared with starting point of the time series (Figure 5A), whereas in the same conditions, levels of miR156 and miR160 were not or only marginally affected (Figure 5A). miR390 accumulation was suppressed in roots cotreated for 24 h with cycloheximide, an inhibitor of protein biosynthesis, indicating that MIR390a is not a primary auxin response gene (Figure 5B). We then determined which of the two MIR390 loci (Montgomery et al., 2008) responded to induction of lateral root formation. miR390 did not accumulate in auxin-treated roots of mir390a mutants, confirming that in roots, miR390 mostly originates from the MIR390a and not from the MIR390b locus (Figure 5C). Furthermore, RT-PCR analysis showed an increase in the accumulation of the MIR390a precursor, suggesting that miR390 accumulation during lateral root initiation could be controlled at the transcriptional level (Figure 5D). This hypothesis was also consistent with the increased activity of the pMIR390a:GUS-GFP reporter we observed in synchronously induced lateral roots (Figures 5E and 5F).
Figure 5.
miR390 Expression Responds to Auxin during Lateral Root Induction.
(A) RNA gel blot analysis of 15 μg of root RNA from 10-d-old wild-type plants during a time course of 10 μM auxin (IAA) treatment after 24 h of 10 μM NPA pretreatment. The blot was successively probed with DNA complementary to miR390, miR156, and miR160. U6 snRNA served as a loading control.
(B) RNA gel blot analysis of 15 μg of root RNA from 10-d-old wild-type plants. Plants were pretreated with 10 μM NPA and then for another 24 h with either DMSO (–), 10 μM cycloheximide (CHX), 10 μM IAA, or both (CHX+IAA). U6 snRNA served as a loading control.
(C) RNA gel blot analysis of 15 μg of root RNA from 10-d-old wild-type or mir390a plants treated (+) or untreated (−) for 24 h with 10 μM IAA after 24 h of 10 μM NPA pretreatment. In (A) to (C), numbers are the ratios of miR390 to U6 signal. These experiments were done twice with similar results.
(D) RT-PCR analysis of root RNA from 10-d-old wild-type plants treated for 24 h with 10 μM IAA (+) or untreated (−) after NPA pretreatment. Primers amplify specifically the MIR390a precursor and ACTIN2 (loading control) from cDNA (RT+) but not from genomic DNA (RT−).
(E) and (F) Confocal analysis of pMIR390a:GUS-GFP expression in 10-d-old wild-type plants treated (+IAA) or untreated (Control) for 10 h with 10 μM IAA after NPA pretreatment. GFP is in green, and cell walls are stained by propidium iodide in red. Bars = 50 μm.
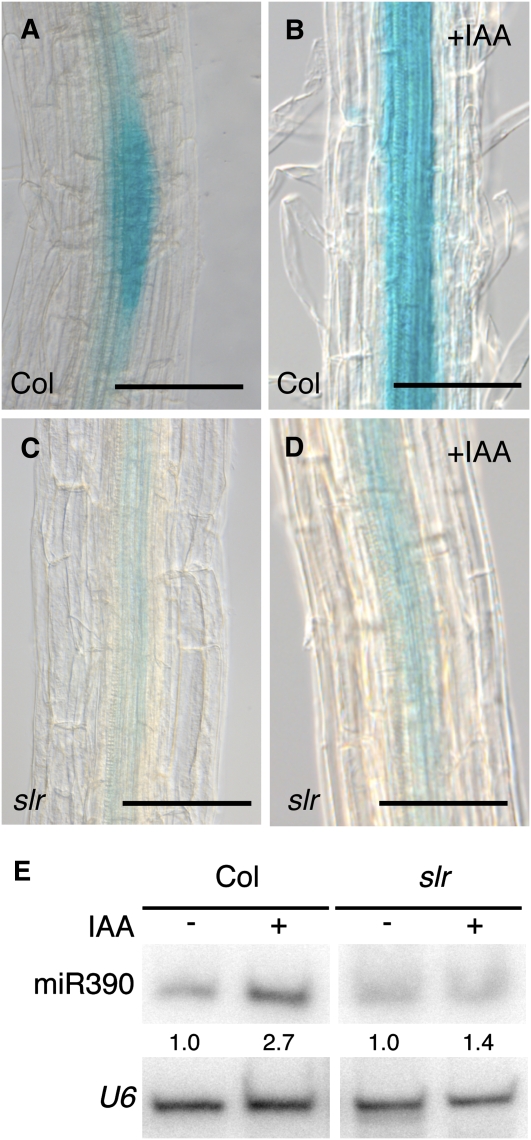
Taken together, these results suggested that miR390 expression responded to the morphogenetic effects of auxin during lateral root formation. To test this functionally, we examined the expression of the pMIR390a:GUS-GFP reporter in the solitary root (slr) mutant. In this mutant, auxin perception, but not its accumulation, is impaired in the pericycle. Consequently, the slr mutant does not form lateral roots (Fukaki et al., 2002). pMIR390a:GUS-GFP expression was severely reduced in the slr background, and only a faint staining in the parenchyma cells of the xylem remained, supporting the idea that most of pMIR390a:GUS-GFP expression is dependent on lateral root initiation (Figure 6A versus 6C). When treated with auxin, only a modest increase in GUS staining was observed in the slr background compared with the wild type (Figure 6D versus 6B), indicating that a developmental signal different from auxin but produced by the developing lateral root primordium controls miR390 induction. This result was further confirmed by comparing the abundance of endogenous miR390 in wild-type and slr plants upon synchronous induction of lateral root formation by NPA/auxin treatment. miR390 accumulated to lower levels in slr mutant plants than in the wild type (1.4- versus 2.7-fold; Figure 6E). Together, these results showed that miR390 expression is set in the xylem mesenchymal cells before the auxin peak and the first pericycle cell division occurs, marking lateral root primordia initiation. Then, a signal produced by the developing lateral root primordium in response to the auxin peak restricts miR390 expression at the base and flanks of the newly formed primordium.
Figure 6.
miR390 Expression Depends on Signals from the Developing Lateral Root Primordium.
(A) to (D) Visualization of pMIR390a:GUS-GFP activity in 10-d-old wild-type or slr mutant plants treated with 10 μM IAA ([B] and [D]) or untreated ([A] and [C]) after 24 h of NPA pretreatment. GUS assay development times were equal for (A) to (D).
(E) RNA gel blot analysis of 15 μg of root RNA from 10-d-old wild-type or slr plants treated with 10 μM IAA for 24 h (+) or untreated (−) after NPA pretreatment. U6 snRNA served as a loading control, and numbers are the ratios of miR390 to U6 signal. This experiment was done twice with similar results.
tasiARF Targets Control Lateral Root Development and miR390 Accumulation
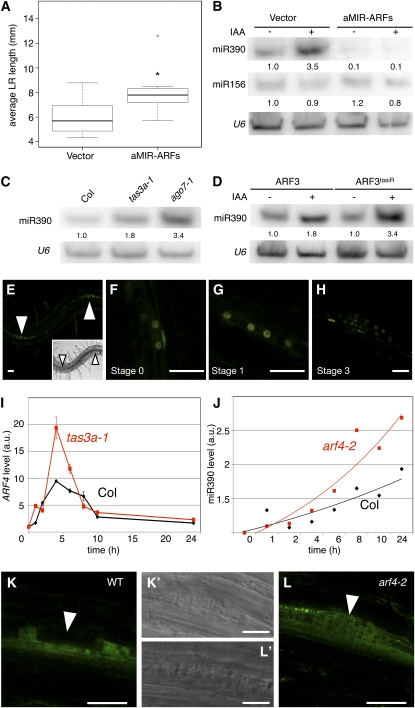
Our results showed that lateral root formation stimulates miR390 expression triggering the biogenesis of tasiARFs, which in turn promoted the growth of the newly formed primordia. To elucidate the role of tasiARFs during root development, we investigated the contribution of their targets ARF2, ARF3, and ARF4 (Peragine et al., 2004; Allen et al., 2005; Williams et al., 2005) to the control of lateral root growth. We first expressed an artificial miRNA (aMIR-ARF), which simultaneously knocks down these three ARFs (Alvarez et al., 2006) (see Supplemental Figure 4 online). Plants expressing the 35S:aMIR-ARF construct had longer lateral roots than plants transformed with a control vector (Figure 7A). This phenotype was similar to 35S:TAS3a plants, confirming that the tasiARF targets contribute to the control of lateral root growth. We then tested the contribution of each individual target ARF using arf2, arf3, or arf4 single mutants. Albeit more modest than the effects of the 35S:aMIR-ARFs, each mutant showed longer lateral roots than the wild-type plants (see Supplemental Figure 5 online), indicating that the combined action of the three tasiARFs targets regulates lateral root growth.
Figure 7.
ARF2/ARF3/ARF4 Control Lateral Root Growth, and ARF4 Is Required for miR390 Expression.
(A) Measurement of average lateral root length in 10-d-old seedlings expressing either the 35S:aMIR-ARFs (aMIR-ARFs) or the empty vector (Vector). Distribution of the population (n > 9) is represented by box plots. aMIR-ARFs plants have longer lateral roots than the vector controls (*, P < 0.05; Student's t test).
(B) RNA gel blot analysis of 15 μg of root RNA from 10-d-old plants expressing either 35S:aMIR-ARFs or the empty vector. The plants were treated (+) with 10 μM IAA or untreated (−) after 24 h of 10μM NPA pretreatment.
(C) RNA gel blot analysis of 15 μg of roots RNA from plants of the indicated genotype.
(D) RNA gel blot analysis of 15 μg of root RNAs from 10-d-old plants expressing either ARF3:ARF3:GUS or its tasiARF-resistant version. The plants were treated as in (B). In (B) to (D), U6 snRNA served as a loading control, and numbers are the ratios of probe to U6 signal. These experiments were done twice with similar results.
(E) to (H) Confocal observation of the pARF4:nls-3xGFP reporter construct during early stages of lateral root development. Arrowheads in (E) indicate lateral root primordia, and the inset shows transmitted light image of the same field. Bars = 20 μm.
(I) Quantitative RT-PCR analysis of ARF4 transcripts in the wild type (black) and tas3a-1 mutants (red) during a time course of 10 μM auxin (IAA) treatment after 24 h of 10 μM NPA pretreatment. Values, expressed in arbitrary units (a.u.), are averages of two replicates, and error bars represent se.
(J) miR390 abundance in wild-type and arf4-2 plants during a time course of 10 μM auxin (IAA) treatment after 24 h of 10 μM NPA pretreatment. Quantification of the miR390 signal was performed as in Figure 5A.
(K) and (L) Confocal observation of the pMIR390a:GUS-GFP reporter in a stage 3 primordium expressed in wild-type (K) or arf4-2 mutant backgrounds (L). The arrowheads in (K) and (L) indicate the central zone of the primordium, whereas transmitted light picture of the same regions are shown in (K’) and (L’). Bars = 30 μm.
Control of miRNA expression by their target is a recurring motif in animal gene circuits, and several examples have been recently reported in plants (Tsang et al., 2007; Gutierrez et al., 2009; Wu et al., 2009). We thus investigated whether miR390 accumulation depends on the ARFs during lateral root formation. To this end, we triggered the synchronous induction of lateral roots by NPA/auxin treatment in 35S:aMIR-ARFs plants. In response to auxin, miR390 accumulated to lower levels in 35S:aMIR-ARFs plants than in vector-transformed control plants (Figure 7B). Reciprocally, increased miR390 accumulation was observed in tas3a-1 and ago7-1 mutant plants in which all three tasiARFs targets overaccumulate (Figure 7C), strongly suggesting that the tasiARF-regulated ARFs are required for miR390 accumulation. We also quantified the abundance of miR390 in auxin-treated roots of plants expressing a wild-type or tasiARF-resistant form of ARF3. Plants expressing the tasiARF-resistant form of ARF3 accumulated more miR390 than plants expressing the wild-type form of ARF3 (Figure 7D). Taken together, these results indicate that MIR390a and at least ARF3 are connected by a positive feedback loop.
ARF4 Confines the miR390 Expression Pattern
A prominent feature of the MIR390a expression pattern is its progressive exclusion from the center of the developing primordium at stage 3 (Figure 4C). Hence, we investigated whether ARFs also may be involved in the spatial restriction of MIR390a expression and its consequence in the control of lateral root growth.
Because ARF4 is regulated during lateral root initiation (Vanneste et al., 2005), we examined the expression pattern of a pARF4:nls3xGFP reporter in lateral roots. GFP was detected in very young lateral root primordia (Figure 7C) and was already detectable in pericycle cells that had not yet divided (stage 0) but already had the typical round nuclei (Figure 7D). pARF4:nls3xGFP expression persisted until stage 3 (Figures 7E and 7F), indicating that ARF4 expression overlaps spatially and temporally with MIR390a expression during lateral root initiation.
To further study the interplay between TAS3 and ARF4, we monitored endogenous ARF4 levels by quantitative RT-PCR in wild-type and tas3a-1 plants. In plants for which lateral root development was synchronously induced by NPA/auxin treatment, ARF4 expression increased within 1 h of auxin treatment, peaked after 6 h, and dropped to basal levels after 24 h (Figure 7I). In tas3a-1 plants, ARF4 levels were 1.7- to 2-fold higher at 4 to 6 h after treatment compared with wild-type plants (Figure 7I), indicating that tasiARFs inhibit ARF4 accumulation at the early stages of lateral root formation. We then investigated whether miR390 accumulation depends on ARF4 during lateral root formation. We triggered the synchronous induction of lateral roots by NPA/auxin treatment and monitored miR390 accumulation in the wild type and the arf4-2 mutant. In response to auxin, miR390 accumulated to slightly higher levels in arf4-2 plants than in the wild-type control (Figure 7J). RT-PCR analysis of MIR390a precursor levels in arf4-2 and a second allele (arf4-7) further confirmed that ARF4 is a negative regulator of MIR390a expression (see Supplemental Figure 6 online). We then looked at expression of the pMIR390a:GUS-GFP reporter in the lateral root primordium of the arf4-2 mutant. By stage 3, cells located at the center of the primordium express only faint levels of the pMIR390a:GUS-GFP reporter compared with the flanks (Figures 4C, 7K, and 7K', arrowhead). In the arf4-2 mutant, the cells at the center and at the flanks of the primordium expressed comparable levels of the reporter (Figures 7L and 7L', arrowhead). This result demonstrates that ARF4 contributes to the restriction of MIR390a expression to the base and the flanking cells of the primordium, whereas together with ARF2 and/or ARF3, they define a homeostatic regulatory loop controlling miR390 expression (Figure 8).
Figure 8.
A Model for the Role of the miR390/tasiARF/ARF Module during Lateral Root Growth.
The diagram illustrates the spatial expression patterns of TAS3a, miR390, and ARF4 in a lateral root primordium. Hatched regions indicate the territories of overlapping gene expression. The cell layers of the primary root are indicated (x, xylem; p, pericycle; e, endodermis; cx, cortex; ep, epidermis). TAS3a accumulates in the vasculature, miR390 in the xylem, and the pericycle and the primordium in the base and flanks. The positive feedback of ARFs on miR390 supports a homeostatic model in which miR390 and ARF abundance are tightly regulated, whereas the mutual repression of miR390 and ARF4 helps to reinforce the miR390 expression pattern by removing it from the center of the primordium. Dashed arrows indicate indirect relationships.
DISCUSSION
In this work, we investigated how the tasiRNA pathway controls lateral root growth and development. Our results suggest a model in which miR390 expression is activated in the mesenchymal cells of the xylem prior to lateral root initiation (Figure 8). miR390 then allows the production of tasiARFs that repress their targets ARF3 and ARF4 in the new primordium. Positive and negative feedback by ARF2, ARF3, and ARF4 ensure the proper expression of miR390 and regulate lateral root growth. Our results uncover a regulatory network involved in auxin signaling during lateral root formation and reveal a potentially widespread feature of regulatory small RNAs to quantitatively control organ growth.
Regulation of Development Timing during Lateral Root Growth
Our results indicate that TAS3a is a potent regulator in the timing of lateral root growth prior to emergence. Our analysis of lateral root cell elongation and proliferation, two postemergence processes, revealed that tas3a-1 mutants have smaller meristems and cells than the wild type (see Supplemental Figure 2 online). However, because plants overexpressing TAS3a do not have larger cells or meristems, the effects of TAS3a loss of function could be secondary consequences of an altered developmental timing at earlier stages.
Because the TAS3 pathway also affects the timing of leaf development, our results point to a convergence of its role in roots and leaves. In leaves, tasiARFs posttranscriptionally regulate the abundance of ARF3 and ARF4, which are transcription factors that promote the expression of adult traits and consequently control the entry into the adult phase (Fahlgren et al., 2006; Hunter et al., 2006). Mutations that impair tasiARFs production accelerate this transition, and adult leaves are produced earlier. In roots, mutations that impair tasiARFs production cause an overaccumulation of young lateral root primordia (stages 1 to 4), whereas plants with elevated tasiARFs levels exhibit an increase of later stages (5 to 7).
During lateral root formation, the activation of the newly formed meristem is a crucial transition, which occurs around stage 4 (Laskowski et al., 1995). Thus, one could speculate that ARF2, ARF3, and ARF4 contribute to the repression of meristem activation and that the miR390/TAS3/tasiARFs pathway maintains these ARFs in an activity range that allows proper growth of the newly formed meristem. Consistent with this hypothesis, we observed that the reduction of tasiARF abundance resulted in higher levels of ARFs and a delayed activation of the meristem, whereas an increase in tasiARF abundance or ARF inactivation resulted in precocious meristem activation. Interestingly, our results show that tasiARFs control ARF4 abundance rather than its timing of accumulation, indicating that other regulatory mechanisms independent of miR390 and TAS3a likely affect the temporal pattern of ARF4 expression.
miR390 Expression during Lateral Root Development
MIR390a displays a dynamic expression pattern during lateral root formation. Initially expressed in the mesenchymal cells of the central cylinder, MIR390a expression extends into the pericycle cells concomitant to the first asymmetric cell divisions, where it colocalizes with an auxin maximum. From stage 3 onward, MIR390a is expressed at the base and flanks of the developing primordium (Figures 3 and 4). We show that ARF4 is required to suppress miR390 expression from the center of the primordium and hence contributes to the definition of its expression pattern. However, the absence of any canonical auxin response elements in the MIR390a promoter and the suppression of induction upon auxin/cycloheximide cotreatment suggest that the effects of auxin/ARF4 are probably indirect.
Although miRNAs are thought to act largely cell autonomously (Parizotto et al., 2004; Alvarez et al., 2006; Schwab et al., 2006; Tretter et al., 2008), the trafficking of some miRNAs over short distances and in specific developmental contexts remains a possibility. Our results show that miR390 acts in the whole lateral root primordium (Figure 3), a domain slightly broader than the one defined by the MIR390a reporter (limited to the flanks and base of the primordium; Figures 3 and 4), suggesting that miR390 might act non-cell-autonomously across a few cells, in agreement with observations made in the maize (Zea mays) and Arabidopsis shoot apex (Chitwood et al., 2009; Nogueira et al., 2009). The mechanisms regulating the range of miR390 activity are not known but could include regulation of its biogenesis, stability, or movement through modulation of intercellular permeability. Furthermore, the tasiARF sensor revealed tasiARF activity in lateral root primordia (Figures 2A and 2B) several cell layers away from the cells where TAS3a and MIR390a are coexpressed (in the central cylinder), consistent with tasiARFs intercellular mobility. Non-cell-autonomous activity of tasiRNAs has been postulated based on their requirement for DCL4, which produces siRNAs that associate with mobile silencing (Dunoyer et al., 2005; Bouché et al., 2006; Deleris et al., 2006). This model is supported by both the non-cell-autonomous silencing mediated by an artificial transgene-based system, which produces siRNAs from a tasiRNA-like precursor (Tretter et al., 2008), and by the observation that the tasiARFs act at distance from their site of production in the shoot apical meristem (Chitwood et al., 2009; Schwab et al., 2009).
Quantitative Regulation of Development by Small RNAs
35S:TAS3a and 35S:aMIR-ARFs plants have longer lateral roots than wild-type controls (Figure 7A), whereas the length of lateral roots in arf2, arf3, and arf4 single mutants is only marginally affected (see Supplemental Figure 5 online). This suggests that the role of ARF4 during lateral root growth could be restricted to fine-tuning the regulatory system. In the absence of ARF4, the functions of ARF2 and ARF3 are still sufficient to maintain an almost normal development of lateral roots through a homeostatic regulatory loop. Thus, the simultaneous inactivation of multiple targets may be critical for the full activity of the miR390/TAS3 module and a more general requirement for regulation of developmental processes by miRNAs (Voinnet, 2009).
Our results establish that the tasiARFs targets contribute differently to miR390 expression. First, simultaneous impairment of ARF2, ARF3, and ARF4 function with an artificial miRNA reduces the expression of miR390, whereas increasing the abundance of all three ARFs or ARF3 alone results in higher miR390 accumulation. This positive feedback of ARF2 and/or ARF3 on miR390 has the potential to ensure tight control of the miR390-TAS3-ARFs module activity. This homeostatic model for miR390/ARF function during lateral root formation maintains the activity of ARFs within an optimal range. Second, ARF4 has a specific function in the spatial restriction of miR390 expression via a negative feedback mechanism in the central primordial cells. The coexistence of both mechanisms for overlapping targets illustrates the importance of a finely tuned activity of the miR390/TAS3/tasiARF module. Our results differ from a recent report describing the auxin induction of miR390 in roots and its role in lateral root development (Yoon et al., 2010). The main discrepancy concerns the respective contribution of the MIR390a and MIR390b loci to miR390 accumulation and response to auxin. The 5′ rapid amplification of cDNA ends (RACE) mapping and mir390a mutant analysis revealed that MIR390a is the major contributor of miR390 accumulation in the root (Figure 2C), in agreement with our sensor data (Figure 3F). By contrast, in situ hybridization data presented by Yoon et al. could not determine the locus of origin of miR390 because MIR390a and MIR390b encode the same mature miRNA. In addition, the pMIR390a:GUS and pMIR390b:GUS reporter used by Yoon et al. encompassed parts of the miR390 precursor, whereas ours corresponded to the actual nontranscribed genomic region, which could account for discrepancies between our data. Moreover, we showed that MIR390a is auxin inducible during the early stages of lateral root initiation (stage 0 to 2/3). On the contrary, Yoon et al. describe the effects of auxin on older lateral roots. This is a major difference between the two studies. We have not studied the effect of auxin on older primordia; therefore, we cannot exclude that MIR390b is induced at later stages. However, our results using the mir390a mutant firmly establish that upon the first stages of auxin-induced lateral root initiation, only the MIR390a locus is active (Figures 2C and 5C).
To conclude, we show how miR390-TAS3 tasiRNA-ARF2/3/4 integrate with auxin signaling to regulate lateral root growth, in addition to miR167-ARF8 modulation of lateral root meristem activation in response to nitrogen availability (Gifford et al., 2008). Negative and positive feedback loops of miRNA/target regulons have been described for the posttranscriptional regulation of miRNA homeostasis in plants (Xie et al., 2003; Vaucheret et al., 2004, 2006; Rajagopalan et al., 2006) and for the transcriptional regulation of gene networks in animal development (Tsang et al., 2007). Two recent reports describe additional miRNA/target regulons in plant development. First, expression of miR156 and miR172 depends on their targets, the transcription factors of the SPL and AP2 families, and is crucial for the vegetative phase transition in Arabidopsis (Wu et al., 2009). Second, formation of adventitious roots involves a complex regulatory network including cross-regulation of miR160/167 homeostasis by direct and indirect targeting of ARF transcription factors (Gutierrez et al., 2009). Here, we show that the reciprocal feedback between miRNA and their targets can be extended to the tasiRNA pathway in which an miRNA controls abundance of its target using intermediary and potentially mobile siRNAs. Transcription factors and miRNAs are the major trans-acting regulators that determine the dynamic equilibrium of transcriptional networks at each developmental stage (Hobert, 2008). Our results underline the importance of reciprocal miRNA/transcription factor regulatory feedback loops in the control of plant organ growth in response to a specific morphogenetic trigger.
METHODS
Plant Material
All lines used in this study are in the Arabidopsis thaliana Col ecotype background. The tas3a-1 (GABI_621G08), rdr6 (sgs2-1), dcl4-1, tasiARF sensors (pARF3:ARF3:GUS and pARF3:ARF3tasiR-GUS), slr, arf2-6, arf3-2, and arf4-2 (salk_070506) have been previously described (Fukaki et al., 2002; Gasciolli et al., 2005; Okushima et al., 2005a, 2005b; Pekker et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Chitwood et al., 2009; Schwab et al., 2009). The arf4-7 allele was identified in the SALK collection (salk_028804C) (Alonso et al., 2003). Sequencing of T-DNA junctions confirmed that the T-DNA was inserted in the 5′ untranslated region of ARF4 (At5g60450) 376 bp upstream of the ATG. The activation-tagged TAS3a allele (GABI_626B09) was identified in the GABI-KAT collection (Rosso et al., 2003). Sequencing of T-DNA junctions revealed that the T-DNA was inserted 102 bp upstream of TAS3a (At3g17185). The mir390a line (WiscDsLox440F06; mir390a-2) was identified in the Wisconsin DsLox collection (Woody et al., 2007). Sequencing of T-DNA junctions revealed that the insertion is located 30 bp upstream of the MIR390a (At2g38325) transcriptional start site.
Growth Conditions
Soil-grown plants were propagated in a greenhouse (23°C). For in vitro conditions, plants were grown on 0.5× Murashige and Skoog (MS)/0.8% agar (MS agar) plates in controlled-environment chambers under the following conditions: 150 μmol photon·m−2·s−1 luminance, 16 h light, and 23°C temperature. For synchronous induction of lateral root development, plants were germinated on nylon sheets (SEFAR NITEX 03-100/44) on 0.5× MS agar for 8 d, transferred to 0.5× MS agar + 10μM NPA for 24 h, and shifted to 0.5× MS agar + 10 μM IAA for the indicated time.
Phenotypic Analysis
For quantification of root morphology, plates were scanned after 10 d of growth and examined under a binocular microscope to determine the number of emerged lateral root primordia. Measurements of primary root length and lateral root length were made on the scanned picture using Image-J (http://rsb.info.nih.gov/ij/). Measurements of cell and meristem size were performed as described (Cazalé et al., 2009). We used R (www.r-project.org) for statistical analysis and graphing of the data.
Construction of Reporter and Sensor Transgenes
For the 35S:TAS3a construct, TAS3a (At3g17185) was amplified (primers N-0081/82) from genomic DNA to generate a Gateway (Invitrogen) entry clone in pDONR221, which was then recombined in a homemade Gateway-compatible version of pCHF3 (Jarvis et al., 1998). For pTAS3a:GUS reporters, we amplified with primers N-0087/88 550bp of regulatory sequence able to rescue the phenotype of tas3a-1 mutants (Adenot et al., 2006) to generate an entry clone that was then recombined in the pMDC163 vector (Curtis and Grossniklaus, 2003). pMIR390a:GUS-GFP and pMIR390b:GUS-GFP reporters were built by amplifying 2.6 and 0.5 Kbp, respectively, of genomic DNA upstream of the transcription initiation start of At2g38325 (MIR390a) and At5g58465 (MIR390b) with primers N-0154/155 and N-0156/157, generating entry clones (pENTR-D; Invitrogen), which were then recombined in pKGWFS7 (Karimi et al., 2007). The tasiARFs sensors were described by Fahlgren et al. (2006). For the miR390 sensor constructs, a 200-bp fragment of TAS3a containing either the wild-type 3′ miR390 binding site (wild-type sensor) or a mutated 3′ site that impairs proper miR390 recognition (mutated sensor) was amplified by PCR (using primers N-2016/2017 and 2018) and placed downstream of GFP under the control of the 35S promoter using gateway-based cloning. The mutated version was obtained using a 3′ primer that introduces five point mutations between positions 1 and 11 of the miR390 binding site. For the DR5rev:erRFP reporter, DR5rev:erRFP was amplified by PCR (primers N-2166/2167) and cloned to generate an entry clone, then recombined in pHGWL7 (Karimi et al., 2007). The 35S:aMIR-ARF construct was described previously (Alvarez et al., 2006). For the pARF4:nls-3xGFP constructs, 2.0 kb upstream of the ARF4 start codon was amplified and conventionally cloned into a pGREENII-based vector containing the nuclear localized 3xGFP sequence and a NOS transcriptional terminator (primers ARF4PFWD and ARF4PREV). This construct was introduced into Agrobacterium tumefaciens strain GV3101 harboring pGREENII helper plasmid pSOUP. Sequences of all primers used can be found in Supplemental Table 1 online, and all constructs were checked by sequencing. Vectors besides pARF4:nls-3xGFP were introduced in Agrobacterium (ASE strain), and all constructs were transformed into plants by floral dipping (Weigel and Glazebrook, 2002).
GUS and Confocal Analysis
GUS activity was assayed at 37°C for 6 to 18 h using 2 mM ferri/ferrocyanide as described (Weigel and Glazebrook, 2002). Transverse sections were obtained after GUS staining using the protocol described by De Smet et al. (2004), mounted in Eukitt (EMS), and photographed on a DMI-6000 microscope (Leica Microsystems). For confocal imaging, roots were mounted in 5% glycerol and directly imaged on a TCS-SP2 upright microscope (Leica Microsystems) with 488-nm/543-nm excitation, 488/543 beamsplitter filter, and 515 ± 15 nm (green channel) and 610 ± 25 nm (red channel) detection windows. Transmitted light was also collected. All images were acquired with similar gain adjustments. Counterstaining of cell walls was achieved by 5 min of incubation in 100 μg·mL−1 propidium iodide. For 4',6-diamidino-2-phenylindole (DAPI) staining of the nuclei, plants were fixed for 45 min in 4% paraformaldehyde in MTSB (Müller et al., 1998), washed in 2 mM glycine, and mounted in Vectashield (Vector Laboratories) containing 1.5 μg/mL DAPI. Plants were imaged for GFP signal as described above and finally imaged for DAPI with a 364-nm UV laser (no beamsplitter filter set, detection window of 415 to 550 nm).
RNA Extraction, RNA Blot Assays, Quantitative RT-PCR, and RACE Analysis
Total RNA was extracted as described (Mallory et al., 2001). For RNA gel blot analysis, 15 μg of RNA were separated by denaturing (7 M Urea) 15% polyacrylamide gel electrophoresis, blotted to a nylon membrane (Hybond NX; GE Healthcare), and cross-linked as described (Pall et al., 2007). miRNA probes were prepared by end labeling antisense oligonucleotides with 32P using T4 polynucleotide kinase (Fermentas). RNA gel blots were hybridized (Mallory et al., 2001) with the miRNA probe together with U6 probe, stripped, and reprobed successively with different miRNAs. Nonsaturated signals were quantified on a Molecular Dynamics Storm 840.
For quantitative RT-PCR analysis, 4 μg of total RNA was treated with RNase-free DNase (Fermentas) and reverse transcribed (Superscript II; Invitrogen). cDNA was diluted four times and used for amplification. A parallel reaction without reverse transcriptase was systematically performed and used as a control for DNA contamination. Quantitative PCR was performed in capillaries on a Roche LightCycler thermocycler using the manufacturer's instructions. Two reference genes (AT1G13320 and AT4G26410; empirically identified for their stable expression across a wide range of conditions [Czechowski et al., 2005]) were used to normalize our signal. Efficiency of each primer pair was determined beforehand. For nonquantitative PCR, Taq polymerase (Fermentas) was used; amplification was stopped after 25 cycles and analyzed on agarose gels. All primers used are described in Supplemental Table 1 online.
The 5′ RACE was performed using a FirstChoice RLM-RACE Kit (Ambion) following the manufacturer's instructions. Twelve cloned RACE fragments were sequenced to map the MIR390a and MIR390b transcription start. All primers used are described in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TAS3a (AT3G17185), MIR390a (AT2G38325), MIR390b (AT5G48465), ARF2 (AT5G62000), ARF3 (AT2G33860), ARF4 (AT5G60450), ACTIN2 (AT3G18780), and quantitative PCR references (AT1G13320 and AT4G26410; Czechowski et al., 2005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Level of Expression of TAS3a (At3g17185) in Loss- and Gain-of-Function Alleles.
Supplemental Figure 2. Phenotypic Characterization of the Root System of Plants Deregulated for the TAS3 tasiRNA Pathway.
Supplemental Figure 3. MIR390b Expression Pattern.
Supplemental Figure 4. ARF Expression in aMIR-ARFs and Wild-Type Plants.
Supplemental Figure 5. Phenotypic Characterization of ARF Mutant Root Phenotypes.
Supplemental Figure 6. ARF4 and MIR390a Expression in arf4 Knockdown Mutants.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank B. Ben Amor for initial experiments, T. Beeckman for the slr mutant, J. Carrington for the tasiARF sensors, and A. Gallavotti for the DR5rev:erRFP construct. We thank A. Leibfried, J. Lohmann, and G. Cristofari for their critical reading of the manuscript. This work was supported by an ANR-GENOPLANT grant (RIBOROOT-ANR06 GPLA 011) and has benefited from the facilities of the Imagif Cell Biology Unit of the Gif campus (www.imagif.cnrs.fr), which is supported by the Conseil Général de l'Essonne.
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Ivanchenko M.G., Friml J., Shishkova S., Dubrovsky J.G. (2009). A morphogenetic trigger: Is there an emerging concept in plant developmental biology? Trends Plant Sci. 14: 189–193 [DOI] [PubMed] [Google Scholar]
- Bouché N., Lauressergues D., Gasciolli V., Vaucheret H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé A.C., Clément M., Chiarenza S., Roncato M.A., Pochon N., Creff A., Marin E., Leonhardt N., Noël L.D. (2009). Altered expression of cytosolic/nuclear HSC70–1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 60: 2653–2664 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- De Smet I., Chaerle P., Vanneste S., De Rycke R., Inzé D., Beeckman T. (2004). An easy and versatile embedding method for transverse sections. J. Microsc. 213: 76–80 [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benkova E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.S., Xie Q., Fei J.F., Chua N.H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J.D., Pacurar D.I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S. (2006). Root development–branching into novel spheres. Curr. Opin. Plant Biol. 9: 66–71 [DOI] [PubMed] [Google Scholar]
- Hirsch J., Lefort V., Vankersschaver M., Boualem A., Lucas A., Thermes C., d'Aubenton-Carafa Y., Crespi M. (2006). Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 140: 1192–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2008). Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Karimi M., Bleys A., Vanderhaeghen R., Hilson P. (2007). Building blocks for plant gene assembly. Plant Physiol. 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M.J., Williams M.E., Nusbaum H.C., Sussex I.M. (1995). Formation of lateral root meristems is a two-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Li X., Cassidy J.J., Reinke C.A., Fischboeck S., Carthew R.W. (2009). A microRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Bartel D.P., Bartel B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Ely L., Smith T.H., Marathe R., Anandalakshmi R., Fagard M., Vaucheret H., Pruss G., Bowman L., Vance V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T., Chitwood D.H., Madi S., Ohtsu K., Schnable P.S., Scanlon M.J., Timmermans M.C. (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Mitina I., Quach H.L., Theologis A. (2005a). AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 43: 29–46 [DOI] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005b). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G.S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A. (2007). Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizot B., et al. (2008). Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol. 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Benfey P.N. (2008). Root layers: Complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schwab R., Maizel A., Ruiz-Ferrer V., Garcia D., Bayer M., Crespi M., Voinnet O., Martienssen R.A. (2009). Endogenous TasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS One 4: e5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal M.L., Bergman A. (2002). Waddington's canalization revisited: developmental stability and evolution. Proc. Natl. Acad. Sci. USA 99: 10528–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter E.M., Alvarez J.P., Eshed Y., Bowman J.L. (2008). Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 147: 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. (2007). MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A.C., Bartel D.P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. 2002. Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Williams L., Carles C.C., Osmont K.S., Fletcher J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102: 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth J.C., Wang S., Tiwari S.B., Joshi A.D., Hagen G., Guilfoyle T.J., Alonso J.M., Ecker J.R., Reed J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Kasschau K.D., Carrington J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Yoon E.K., Yang J.H., Lim J., Kim S.H., Kim S.K., Lee W.S. (2010). Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 38: 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.