The TCP family is a group of plant-specific transcription factors whose DNA binding properties have not been studied in detail. Here, we examine TCP4 by both biochemical and structural analyses to describe the DNA binding mechanisms of this family of proteins and predict a fold that the domain might adopt.
Abstract
The TCP transcription factors control multiple developmental traits in diverse plant species. Members of this family share an ∼60-residue-long TCP domain that binds to DNA. The TCP domain is predicted to form a basic helix-loop-helix (bHLH) structure but shares little sequence similarity with canonical bHLH domain. This classifies the TCP domain as a novel class of DNA binding domain specific to the plant kingdom. Little is known about how the TCP domain interacts with its target DNA. We report biochemical characterization and DNA binding properties of a TCP member in Arabidopsis thaliana, TCP4. We have shown that the 58-residue domain of TCP4 is essential and sufficient for binding to DNA and possesses DNA binding parameters comparable to canonical bHLH proteins. Using a yeast-based random mutagenesis screen and site-directed mutants, we identified the residues important for DNA binding and dimer formation. Mutants defective in binding and dimerization failed to rescue the phenotype of an Arabidopsis line lacking the endogenous TCP4 activity. By combining structure prediction, functional characterization of the mutants, and molecular modeling, we suggest a possible DNA binding mechanism for this class of transcription factors.
INTRODUCTION
The TCP class of genes is found only in plants and is represented by the first three identified members TEOSINTE BRANCHED1 in maize (Zea mays), CYCLOIDEA in Antirrhinum majus, and PCF-coding gene in rice (Oryza sativa; Cubas et al., 1999). Members belonging to this class are important regulators of plant growth and development and control multiple traits in diverse plant species, including flower and petal asymmetry (Luo et al., 1996, 1999), plant architecture (Doebley et al., 1997; Aguilar-Martínez et al., 2007), leaf morphogenesis (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007) and senescence (Schommer et al., 2008), embryo growth (Tatematsu et al., 2007), and circadian rhythm (Pruneda-Paz et al., 2009). At the cellular level, most TCP genes modulate cell proliferation in the axillary organs of plants (Nath et al., 2003; Li et al., 2005; Hervé et al., 2009; Martin-Trillo and Cubas, 2010) either by directly controlling the transcription of cyclin (Li et al., 2005) or by inducing differentiation of the cells (Nath et al., 2003; Efroni et al., 2008).
The TCP genes code for transcription factors that share an ∼60-residue homologous region called the TCP domain (Cubas et al., 1999), common to all the members. The domain has been found and analyzed in many proteins belonging to the plant kingdom (Reeves and Olmstead, 1997; Citerne et al., 2003; Navaud et al., 2007). Phylogenetic studies show that there are two major classes of the TCP domain, class I and II, and sequence analyses have derived a common consensus for both TCP classes (Cubas et al., 1999). The TCP domain mediates binding of the TCP proteins to GC-rich DNA sequence motifs in vitro (Kosugi and Ohashi, 2002). These motifs have been identified as cis-elements in many plant genes coding for proteins such as CYCLIN (Li et al., 2005), PCNA (Kosugi and Ohashi, 1997), RADIALIS (Costa et al., 2005), LIPOXIGENASE2 (Schommer et al., 2008), CIRCADIAN ASSOCIATED1 (Pruneda-Paz et al., 2009), etc. Thus, the TCP genes code for a group of transcription factors that directly modulate the transcriptional status of many plant genes.
Transcription factors of plant and animal origin are classified into five major families primarily based on the DNA binding domain they contain (Pabo and Sauer, 1992; Stegmaier et al., 2004; Qian et al., 2006). Members of the basic helix-loop-helix (bHLH) family of transcription factors are characterized by the presence of a DNA binding domain that contains a basic region that interacts with the target DNA, followed by a HLH region required for dimerization (Murre et al., 1994; Atchley and Fitch, 1997). The helix-turn-helix class of transcription factors has a DNA binding domain that has three helices separated by turns, where the third helix makes the most extensive contact with the major groove of the target DNA (Wintjens and Rooman, 1996). The Zn-finger class of transcription factors is characterized by the presence of a Zn-coordinating domain that is required for both DNA interaction and protein–protein dimerization (Struhl, 1989). The β-scaffold transcription factors form the only group that contacts the minor groove of the target DNA via a β-scaffold domain (Luscombe et al., 2000). Proteins that do not fall into any of the four classes described above are generally classified as “others” and contain different DNA binding domains, such as the AP2/EREBP-related domain, copper fist domain, pocket domain, or SAND domain (Stegmaier et al., 2004).
The TCP domain shares little sequence similarity with any of the transcription factor families described above; a standard blast search using the TCP domain as a query fails to yield any homologous proteins other than the TCP members. Secondary structure prediction revealed that the TCP domain forms a basic region followed by two helices separated by a loop (Kosugi and Ohashi, 1997; Cubas, et al., 1999), even though it shares little sequence similarity with the classical bHLH domain. Reports on plant bHLH proteins have excluded the TCP proteins from analysis (Buck and Atchley, 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003). TCP proteins bind to DNA elements that are different from those recognized by bHLH proteins (Murre et al., 1994; Kosugi and Ohashi, 2002). The TCP domain is thus a novel DNA binding domain specific to the plant kingdom, with unknown biochemical properties and DNA binding specificities. Structure of any TCP protein has not yet been determined.
In this study, we report biochemical and DNA binding properties of the TCP domain and identification of the residues involved in binding the target DNA.
RESULTS
Sequence Analysis of the TCP Domain
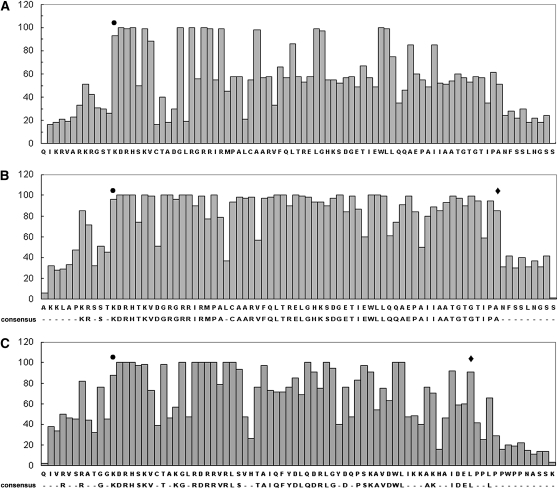
To delineate the TCP domain, we analyzed 206 TCP proteins from 21 species across the plant kingdom currently available in public databases. We aligned a region of 80 residues that includes the reported ∼60-residue-long TCP domain (Figure 1A). Phylogenetic analysis showed two distinct classes (Figure 1B; see Supplemental Figure 1 online; Cubas et al., 1999). To determine the start and the end of the TCP domain, we calculated the percentage of occurrence of amino acid residues in each position (here, we call it unique residue frequency [URF]). Figure 2A shows that high sequence conservation starts from an N-terminal Lys (marked with a circle), which is conserved in 93% of the members. The position preceding this Lys is poorly conserved with highest URF of 26% for Thr. Therefore, we considered this Lys as the start point of the TCP domain. The end of the domain was less obvious; the highest URF declined gradually at the C terminus, making the demarcation difficult. Hence, we decided to analyze the two classes separately. A total of 121 class I and 85 class II proteins (see Supplemental Tables 1 and 2 online) were aligned separately as described above. Analysis of URF values revealed clear end points in these two classes. The domain ended at an Ala (marked with a diamond in Figure 2B) with highest URF of 85% for class I and a Leu (marked with a diamond in Figure 2C) with highest URF of 91% for class II. The frequency of occurrence declined C-terminal to these positions. Comparable domain limits were obtained for both class I and class II proteins using frequency of similar amino acid residues, as opposed to unique residues. We thus concluded that the class I and II domains are 62 and 58 residues long, respectively. We then determined the consensus motifs of the two TCP classes based on the residues with ≥50% conservation (Figures 2B and 2C). Approximately 98 and 80% of residues in class I and II TCP domains, respectively, showed ≥50% URF values.
Figure 1.
Sequence Analysis of the TCP Domain.
(A) Sequence alignment of 80 residues, including the TCP domain from 57 TCP proteins representing all the clades. The class I domain has an extended sequence conservation at the C terminus (not shaded in this alignment). The class II domain has a three-residue insertion in the basic domain that is absent from class I.
(B) A neighbor-joining phylogram with midpoint rooting places these two domains in two major and distinct clades: class I (light gray) and class II (dark gray).
Figure 2.
Amino Acid Conservation in the TCP Domain.
The URF values (see Results) are plotted along the y axis, and the amino acid positions of the domain are shown along the x axis. Analysis is performed with 206 TCP domains belonging to both the classes (A), 121 class I domains (B), or 85 class II TCP domains (C). Predicted left and right borders of the domains are marked by circles and diamonds, respectively. The consensus sequences using ≥50% URF value are given below the x axis.
Secondary Structure Prediction Analysis
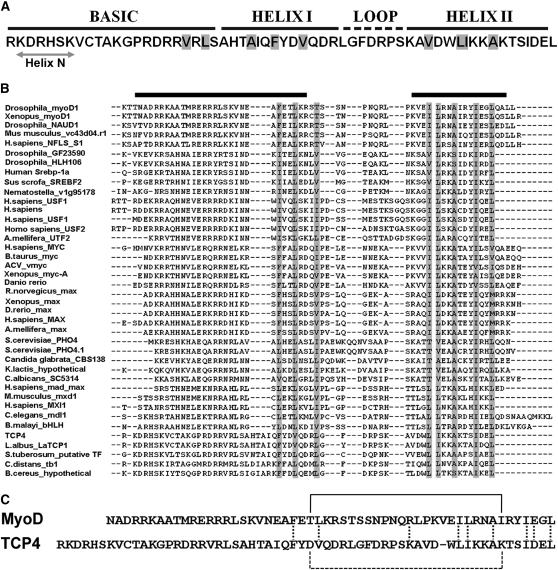
To analyze the predicted structural properties of the TCP domain, secondary elements, solvent accessibility, conformational disorder, and DNA binding properties were studied using various prediction methods. Secondary structure prediction analysis for the core DNA binding domain (Figure 2C) suggested a HLH motif at the C terminus (Cubas et al., 1999; Figure 3A; see Supplemental Figure 2A online) (Rost and Sander, 1993; Jones, 1999; Rost et al., 2004). Although a short stretch of helix (helix N in Figure 3A) was predicted at the N terminus with limited confidence, disorder prediction algorithms suggested the entire basic region to be disordered (Schlessinger and Rost, 2005; Galzitskaya et al., 2006; Ishida and Kinoshita, 2007; Schlessinger et al., 2007) (see Supplemental Figure 2B online). One prediction method based on helix transition theory suggested an extension of helix I up to Pro-58 in the basic region (Muñoz and Serrano, 1994). Most residues of the basic segment were predicted to be exposed (Rost and Sander, 1994), with the buried residues located mainly on the two helices (Figure 3A). Prediction programs, based on chemical and structural features of DNA binding residues in available protein-DNA complexes, identified involvement of the residues in the basic region of the TCP domain in DNA interaction (Ahmad and Sarai, 2005; Hwang et al., 2007; Ofran et al., 2007; Andrabi et al., 2009; see Supplemental Figures 2C and 2D online).
Figure 3.
Sequence Comparison between TCP and bHLH Domains.
(A) Sequence of the TCP4 domain and the predicted bHLH secondary structure are shown. A predicted short helix at the N terminus (helix N) is indicated. The residues predicted to be buried are shaded in gray.
(B) TCP4 sequence profile (see Results) was aligned with the profiles generated for the bHLH proteins of known three-dimensional structure. The residues that are fully conserved or substituted for similar amino acids are highlighted in gray. The helical regions for the MyoD structure are shown by black bars on the top of the alignment.
(C) Sequence alignment of MyoD and TCP4 DNA binding domains, highlighting the similarities in the C-terminal HLH region. The residue conservation in the HLH domains is shown by vertical dotted lines. van der Waals contact between Thr-33 and Ile-56 of MyoD (solid line) and a probable salt bridge interaction between corresponding residues Asp-75 and Lys-97 in TCP4 (broken line) are shown. No significant sequence similarity is observed in the N-terminal basic region.
To identify the probable fold that the TCP domain might adopt, threading and fold prediction programs were used. None of the four methods applied (McGuffin et al., 2000; Shi et al., 2001; Przybylski and Rost, 2004; Kelley and Sternberg, 2009; see Methods) predicted any known fold with high significance level (see Supplemental Figure 2E online).
To examine the conservation of residues, if any, between the TCP domain and the bHLH proteins, we performed sequence profile alignment, as opposed to sequence alignment (Pei et al., 2003). To generate profiles, sequences of bHLH proteins with known three-dimensional structures were used to search for homologs from the protein sequence database. A similar set of homologs was obtained using TCP4 sequences as the query. Homologs with ≥40% sequence identity were aligned initially to generate profiles, followed by an alignment of multiple profiles (Figure 3B). The alignment showed that the hydrophobic residues involved in dimerization and interhelical interface formation within a monomer of the bHLH proteins are conserved in the TCP domain (Ferré-D'Amaré et al., 1993; Ma et al., 1994). Sequence similarity was, however, less obvious at the N terminus, and the secondary structure predicted for the TCP domain did not match with that of bHLH proteins. Only short sequence motifs, such as RDRR and RLS, in the N terminus of TCP4 were found in the MyoD basic region (Ma et al., 1994). Based on this analysis, a manual sequence alignment between the TCP domain of TCP4, and the bHLH domain of MyoD, a representative of the bHLH family, was generated (Figure 3C).
DNA Binding Properties of the TCP Domain
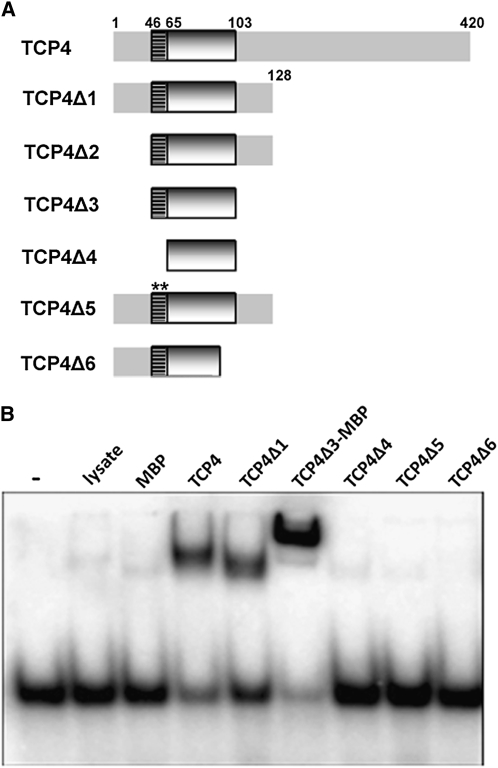
To compare the function of the TCP domain with other DNA binding domains, we characterized the biochemical properties of the TCP4 domain. The minimum DNA binding domain of TCP4 is predicted to be a region from 46 to 103 residues (Figures 2C and 3B). To show this experimentally, we performed electrophoretic mobility shift assays (EMSA) using the consensus target DNA of TCP4 (GTGGTCCC; Schommer et al., 2008) and bacterial lysates containing various truncated forms of the protein (Figure 4). Full-length TCP4 (420 residues), TCP4Δ1 (1 to 128 residues), TCP4Δ2 (45 to 128 residues), and TCP4Δ3 (45 to 103 residues) showed efficient binding with the target DNA, indicating that residues 45 to 103 contain all the information for DNA binding. When the basic region (residues 46 to 65) was deleted, the domain (TCP4Δ4) did not bind to DNA, suggesting that it is essential for DNA binding. To test whether the conserved Gly-Pro pair in the basic domain is required for DNA binding, we mutated both these residues to Leu. The G57L;P58L mutation (TCP4Δ5) completely abolished DNA binding, suggesting that an unstructured basic domain with an internal loop is essential for DNA binding. Residues 87 to 103 are predicted to form helix II that takes part in dimerization. When a part of this helix was deleted (TCP4Δ6), the truncated protein failed to bind DNA.
Figure 4.
Delineation of TCP4 DNA Binding Domain.
(A) Deletion versions of TCP4 are shown schematically. Numbers denote the amino acid positions on the full-length TCP4 protein. The basic region is shown as a hatched box, and the HLH region is depicted as a shaded box. The G57L;P58L double mutant is indicated by two asterisks.
(B) EMSA gel showing the retardation (top bands) of the free oligonucleotide (bottom bands) by TCP4 mutants (bacterial extract was used as the protein source). Oligo alone and bacterial lysate transformed with pRSET-C only are shown by (−) and (lysate), respectively. MBP, maltose binding protein.
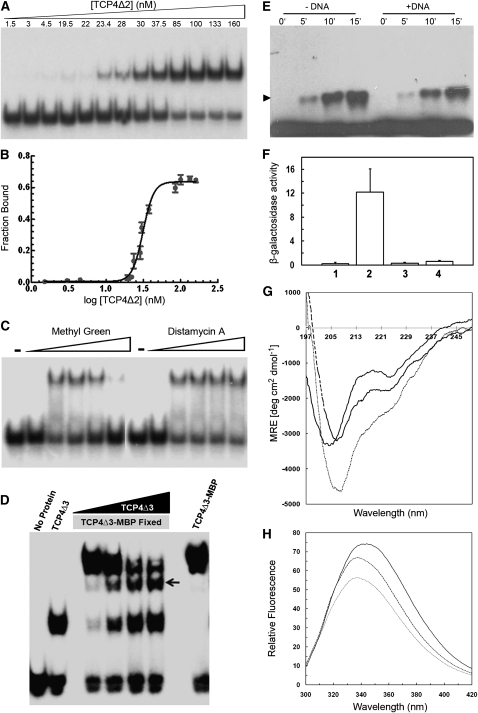
To study the strength of interaction between the TCP domain and the target DNA, we determined the equilibrium dissociation constant (Kd) of TCP4 with the consensus sequence. A fixed amount of radioactively labeled target DNA was incubated with increasing amounts of purified TCP4Δ2 protein, and the DNA-protein complex was analyzed by EMSA (Figure 5A). The fraction of DNA bound to the protein showed a sigmoidal dependence on protein concentration, indicating cooperative binding (Figure 5B). Fitting this data to the Hill equation (see Methods), we obtained the Kd value as 31.3 ± 2.2 nM and a Hill coefficient of 6.1.
Figure 5.
Biochemical Characterization of the TCP Domain.
(A) EMSA gel showing that a fixed amount of radioactively labeled target oligo incubated with increasing amount of purified TCPΔ2 retards progressively increasing amount of oligos. Numbers above the gel indicate concentration of dimer TCPΔ2.
(B) Fraction of bound oligo was quantified from (A) and plotted as a function of dimer protein concentration. Analysis of this data by Hill equation yielded an equilibrium dissociation constant (Kd) of 31.3 ± 2.2 nM.
(C) EMSA gel performed with purified TCP4Δ1 and target DNA probe in the presence of increasing amount of methyl green or distamycin A. Free oligo in the absence or the presence of 500 μM dye are shown as (−) and (+), respectively.
(D) Detection of homodimerization of the TCP4 DNA binding domain by EMSA. A band of intermediate mobility (indicated by an arrow) appeared when purified TCP4Δ3 (7 kD) and TCP4Δ3-MBP (50.2 kD) were incubated together with the 32P-labeled consensus oligos. The intermediate band increased in intensity when increasing amounts of TCP4Δ3 were incubated with a fixed amount of TCP4Δ3-MBP.
(E) Immunoblot of purified hexa-His-tagged TCP4Δ1 incubated with 0.01% of glutaraldehyde for increasing time points and probed with anti-(His)6 antibody. The monomeric TCP4 protein is seen here as a continuous band at the base of the denaturing polyacrylamide gel and the dimer band (arrowhead) of increasing intensity formed with increasing time of incubation. Addition of target oligo to the reaction mixture (+DNA) slightly inhibited dimer formation.
(F) The strength of protein–protein interaction, as performed by yeast two-hybrid assay, has been shown as β-galactosidase activity. Each experiment was repeated thrice, and the standard errors are shown as error bars along the y axis. The bait in all the assays is TCP4Δ1. Proteins used as prey are no protein (1), TCP4 (2), I93N (3), and I100T (4) mutants.
(G) CD spectra of 5 μM purified TCP4Δ2 with 0 μM (solid line), 1 μM (dashed line) and 5 μM (dotted line) target DNA. The spectra are corrected for the contribution of buffer (for solid line) or buffer plus oligo (for dashed and broken lines).
(H) Relative fluorescent intensity spectra of 5 μM purified, truncated TCP4 in the absence (solid line) or presence of 1 μM (dashed line) and 5 μM (dotted line) target oligo.
To determine whether TCP4 binds to major or minor groove of the DNA double helix, we performed EMSA in the presence of chemicals that discriminate between the major and the minor grooves. The major groove binding dye methyl green (Kim and Norden, 1993) displaced increasing amounts of purified TCP4Δ1 from the DNA-protein complex when used in increasing concentration (Figure 5C). By contrast, the minor groove binding dye Distamycin A (Chiang et al., 1994) failed to inhibit protein-DNA complex formation. This demonstrated that the TCP domain interacts with the major groove of DNA.
Most transcription factors bind to target DNA as dimers. To determine the dimerization status of TCP4 in solution, we performed size exclusion chromatography using purified TCP4Δ3-MBP fusion protein with a predicted molecular mass of 50.2 kD for the monomer (see Supplemental Figures 3A to 3C online). By comparing with the elution volumes of the standard protein markers, we determined the molecular mass of TCP4Δ3-MBP as ∼112 kD (see Supplemental Figure 3D online), as expected for a dimeric fusion protein. Approximately 15% of the protein eluted as monomer. To demonstrate direct dimer formation by DNA-bound TCP4, we performed EMSA experiments using radioactively labeled target DNA and two forms of TCP4 protein with different molecular masses. When used individually, purified TCP4Δ3 and TCP4Δ3-MBP retarded the mobility of the target DNA to different extents. Using both these proteins together in an EMSA experiment, we detected an additional retarded band corresponding to the dimeric band between these two forms of the protein (Figure 5D). Thus, we show that TCP4 protein can form dimers both in the absence and in the presence of the target DNA.
Dimerization of TCP4 is further supported by in vitro cross-linking experiments. Glutaraldehyde covalently links two monomers of a dimeric protein by linking two Lys residues through formation of a Schiff's base (Payne, 1973). When purified His-tagged TCP4Δ1 was incubated with 0.01% glutaraldehyde, a dimer band was detected in an immunoblot probed with anti-His-tag antibody (Figure 5E), indicating that TCP4 forms a dimer in solution. To test whether dimer formation increases upon DNA binding, we performed the cross-linking experiment in the presence of target DNA. The dimer band intensity did not increase in the presence of DNA, possibly because the free TCP4 protein alone can form dimer in solution (see Supplemental Figures 3C and 3D online).
To understand if TCP4 forms dimers in vivo, we performed a yeast two-hybrid assay. Since TCP4 alone, in fusion with GAL4 DNA binding domain, can strongly activate the reporter gene (see Supplemental Figure 4 online), we made a truncated version of TCP4 lacking the activation domain and fused it with the DNA binding domain of GAL4 to use as bait in the two-hybrid assay. The full-length TCP4 fused to the activation domain of GAL4 was used as prey. Activity of the reporter gene β-galactosidase was used as a read-out for the extent of dimerization in liquid assay (Figure 5F). Use of the wild-type TCP4 as prey resulted in significantly higher reporter activity compared with bait alone, demonstrating that TCP4 can dimerize in the absence of the target DNA. When either of the two conserved Ile residues in the C terminus was mutated, the mutant proteins (I93N and I100T) failed to form dimers with wild-type TCP4, suggesting that they play important roles in dimer formation.
Secondary structure analysis predicted that the basic region of TCP4 is largely unstructured (Figure 3A; see Supplemental Figure 2B online). To test if helicity is induced upon target DNA binding, we measured the α-helicity of purified TCP4Δ2 protein in the absence and the presence of the target DNA using circular dichroism (CD) spectroscopy. The CD spectrum of TCP4Δ2 showed two minima at 203 and 222 nm (Figure 5G), indicative of a mixture of α-helix (10%) and random coil (40%). The secondary structure increased upon addition of increasing amount of target DNA. In an equimolar mixture of oligonucleotide and protein, the θ208 and θ222 increased by 35 and 92% respectively, over the values shown by the protein alone. The corresponding increase in θ222 was ∼200% when 50% trifluoroethanol, a known inducer of α-helical structure, was added (see Supplemental Figure 5 online). Analysis of the spectra in Figure 5G showed a 3.5-fold increase in α-helix upon addition of DNA (from 10 to 34%), even though ∼30% of random coil was also predicted (Unneberg et al., 2001).
Induction of structural integrity of the domain was also reflected in tertiary structure as measured by the environment of Trp-91 on helix II. Changes in the Trp environment result in changes in fluorescence intensity and emission maximum and are often used as a measure for structural changes in the protein. The TCP4 domain alone has an emission λmax of 344 nm when excited at 280 nm, indicating that the lone Trp in the domain is partially exposed and is in a hydrophilic environment. Upon addition of equimolar oligonucleotide and protein, λmax shifted toward the blue end of the spectrum by 9 nm (to 335 nm), indicating that the Trp is more buried and sequestered from solvent in the protein-DNA complex (Figure 5H). There was, however, a small decrease in fluorescence intensity upon DNA binding, possibly due to quenching by an adjacent charged group (Sendak et al., 1996).
Identification of Residues in the TCP4 Domain Involved in DNA Binding
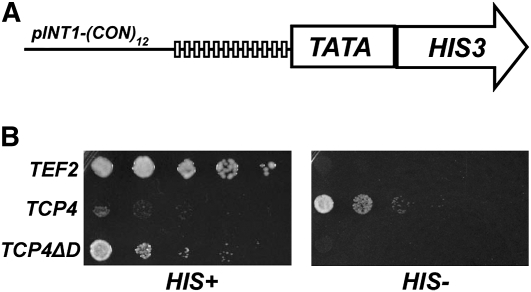
To identify the residues important for DNA binding in the TCP4 domain, we used a yeast-based functional screening of random point mutants. We generated a yeast strain by integrating 12 tandem copies of the TCP4 target sequence upstream of a minimal promoter and a HIS3 reporter gene (Figure 6A). This strain grew normally on nutrient medium supplemented with His. Growth, however, was completely inhibited in the absence of His supplemented with 25 mM 3-amino-1,2,4-triazole (3-AT), a chemical used to inhibit basal levels of HIS3 product (Figure 6B). We noticed that expression of full-length TCP4 protein under TEF2 promoter greatly reduced the growth of this strain on His-containing medium (Figure 6B). These colonies initially appeared smaller than the wild type, although the colony size eventually became comparable to the wild type as growth progressed. A truncated TCP4 lacking the DNA binding domain did not reduce yeast growth, indicating that growth retardation is dependent on the DNA binding ability of the protein. When grown in the absence of His, only the strain expressing full-length TCP4 grew.
Figure 6.
Expression of Full-Length TCP4 Protein Inhibits Yeast Growth.
(A) Schematic representation of the construct pINT1-(CON)12 with 12 copies of TCP4 consensus sequence (GTGGTCCC, vertical boxes) upstream of a minimal promoter (TATA) and a HIS3 reporter gene.
(B) Growth analysis of the yeast strain harboring either no transgene (TEF2), TCP4 gene under TEF2 promoter (TCP4), or TCP4 lacking the binding domain under TEF2 promoter (TCP4ΔD) performed in presence (HIS+) or absence (HIS−) of His supplement.
We used the differential growth of yeast colonies as a preliminary screening step for mutants of TCP4 with altered DNA binding ability. The domain region of TCP4 was subjected to random mutagenesis using error-prone PCR and introduced into the yeast strain under control of the TEF2 promoter. The bigger and fast-growing colonies, likely to harbor a mutated domain with reduced DNA binding ability, were isolated from a mixture of colonies having different sizes. In a second step of screening, these colonies were streaked onto nutrient medium lacking His and supplemented with 25 mM 3-AT. The bigger colonies in the first screen did not grow in this medium, whereas small colonies grew as big as the wild type, confirming that the bigger colonies of the first screen failed to bind to the TCP4 target sequence and thereby could not activate the HIS3 reporter gene. Plasmids from these colonies were isolated and transformed into Escherichia coli for sequencing.
One hundred and twenty-two colonies tested positive for both screens were sequenced, and the results are shown in Supplemental Table 3 online. Of 32 mis-sense point mutants obtained in the screen, 28 mapped within the TCP domain. Further analysis placed four mutants in the basic domain, 11 in helix I, six in the loop, and seven in helix II.
A Gross Structural Model of the TCP Fold Bound to Target DNA
The random mutagenesis yielded only four point mutants in the basic region, making it difficult to determine its role in DNA binding. Therefore, we made further changes in this region by site-directed mutagenesis. To help identify the DNA binding residues for mutagenesis, we first generated a gross structural model of the TCP4 DNA binding domain using the homodimeric structure of Myo-D in complex with its target DNA as template (Ma et al., 1994). Profile-based sequence alignment suggested sequence similarity at the HLH region between the TCP domain and the bHLH domain. Therefore, we first modeled this region of TCP4 using the HLH structure of MyoD as a template. Modeling the basic region of TCP4 was more challenging due to the lack of sequence similarity. Although this region in MyoD forms a helical structure in continuation with helix I, this possibility is limited in TCP4 due to the presence of a Gly-57–Pro-58 pair. To determine the compatibility of a continuous helix up to Pro-58, we modeled this region as a helix, continuous with helix I, using DNA-bound MyoD structure as a template. The nature of base-specific and backbone contacts found in the bHLH and other DNA binding protein complexes (Luscombe et al., 2001), the polarity of bases in the major groove of TCP4 target DNA, and the prediction of base-specific contacts for each residue in the TCP4 basic region (see Supplemental Figure 2D online) were considered to optimize the model in this region.
A short, conserved stretch of basic residues (KDRHSK) at the N-terminal end of the TCP4 basic domain is expected to form a helix (Figure 3A) and interact with the DNA bases (see Supplemental Figures 2C and 2D online). This region was modeled as a helix that is perhaps stabilized by a salt bridge between Asp-47 and Lys-51. It was then oriented with respect to the helix I (starting from Pro-58) and the target DNA to form the maximum number of favorable DNA backbone contacts that include a possible base specific interaction involving His-49 and an adenine base.
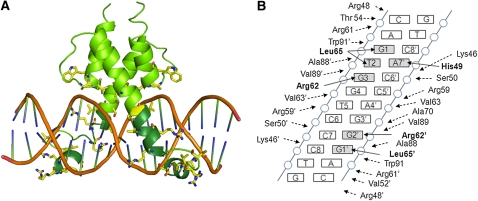
The base sequence of the DNA found in the template was computationally mutated to TCP4 target sequence (see Supplemental Figure 6 online) and was refined by energy minimization. For optimization of the interactions between the TCP domain and the target DNA, 10,000 steps of energy minimization were performed for this TCP4-DNA complex, and the final model is presented in Figure 7. The model predicted that the basic region forms two helical stretches separated by a loop: one at the N terminus (helix N) and the other at the C terminus in continuation with helix I. Besides, the basic region interacts with the major groove of the target DNA through several conserved residues. We used these predicted interactions (Figure 7B) as the basis for generating site-directed mutations and generated point mutants in nine positions that are likely to be important for DNA binding.
Figure 7.
Modeling the Three-Dimensional Structure of the TCP4-DNA Complex.
(A) Model of TCP4 domain in dimer form bound to the B-form target DNA sequence GTGGTCCC (in orange). The basic domain is shown in dark green, and residues that interact with the DNA backbone are shown in yellow.
(B) Summary of the amino acid–DNA interaction in the TCP4 domain. Residues that make direct contacts with DNA bases (gray rectangle) are shown in bold. A, adenine; T, thymidine; G, guanine.
DNA Binding Properties of the Mutants
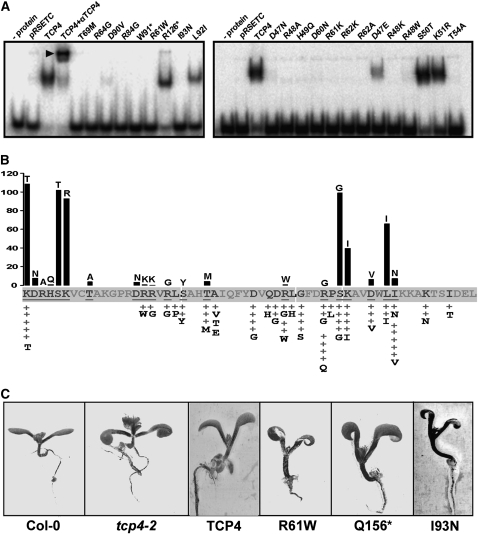
We tested the DNA binding abilities of the point mutants by in vitro and in vivo assays (Figures 8A and 8B). DNA binding ability of the random mutants was tested in vivo in the yeast system. The principle of this test was this: if a mutant is unable to bind to the TCP4 target sequence, it would fail to activate the HIS3 gene and, thereby, the colony growth would be inhibited at a minimum 3-AT concentration when grown in the absence of His. If the mutation did not affect DNA binding, then the colony growth would be affected at higher concentration of 3-AT. Thus, the maximum concentration of 3-AT required to inhibit yeast growth was used as a measure of strength of the mutation. A scale of 1 to 5 was assigned to the mutants, where + denotes a strong mutation with the least DNA binding capability, whereas +++++ denotes a weak mutation with DNA binding capability comparable to the wild-type domain. The results showed that the weakest mutations mapped to the loop region, whereas the strongest mutations occurred in the basic region and in the helix II (Figure 8B; see Supplemental Table 3 online).
Figure 8.
DNA Binding of TCP4 Mutants.
(A) EMSA gels showing the interaction of various mutants of TCP4 and the target oligo. The names of the mutants are given at the top of the lanes and radiolabeled oligo only is shown as (− protein). Bacterial cell extracts were used as protein source, and the lysate from bacterial culture that contained empty vector is shown as pRSETC. The supershift (arrowhead) of the wild-type TCP4-DNA complex was brought about by adding anti-(His)6 antibody to the reaction mixture. The asterisk indicates truncated protein due to premature stop codon in the gene.
(B) The band intensities in (A) were quantified and shown as a bar diagram in (B). The wild-type residue is shown at the base of the bar, whereas the residue it is mutated to is shown at the top. The intensity of the wild-type TCP4-DNA complex band was considered as 100%, and other values are expressed relative to this value. EMSA was performed for only those mutants where the underlined residues in (B) were mutated. The gray-colored residues in (B) indicate that no mutants in those positions were isolated. The activities of the random mutants as assessed in vivo (see Results and Supplemental Table 3 online) are expressed as + signs and the substituted residues are written below it.
(C) Arabidopsis seedlings from the wild type (Columbia-0 [Col-0]), TCP4 null mutant (tcp4-2), and various transgenic lines are shown for their epinasty phenotype of cotyledons. Col-0 has flat cotyledons that bend downwards when TCP4 is mutated (tcp4-2). This phenotype can be rescued by introducing a wild-type TCP4. However, a truncated TCP4 with the first 156 amino acids intact (Q156*) and two point mutants (R61W and I93N) fail to rescue epinasty.
To test the DNA binding abilities of the mutants in vitro, we performed EMSA experiments with the target DNA and the mutant proteins expressed in E. coli. All the mutant proteins used in EMSA were stable in E. coli as tested by immunoblot analysis using anti-His-tag antibody (see Supplemental Figure 7 online). Mutants in the basic region, except K46T, D47E, S50T, and K51R, failed to bind to DNA (Figures 8A and 8B). Thr-69 in helix1 is conserved in the entire TCP4 clade, and its mutation to Met abolished DNA binding. Three mutants in helix II were studied for their DNA binding properties. The 90th position is usually occupied by an acidic residue, and the D90V mutant failed to bind to DNA. The conservative mutation in L92I retained almost half of wild-type DNA binding ability. I93N lost binding ability completely, but a bulky hydrophobic residue was tolerated (I93V) at this position. The effect of the mutations on DNA binding was quantified from the intensity of the retarded bands of DNA-protein complex and shown in Figure 8B.
Functional Evaluation of DNA Binding Mutants in Planta
To find out whether the mutations affecting DNA binding in vitro also affect TCP4 function in planta, we expressed some of these mutant proteins in an Arabidopsis thaliana line that lacked a functional TCP4 protein (Figure 8C). Plants carrying the null allele of TCP4 show strong epinasty of the cotyledons (Schommer et al., 2008). This phenotype is caused by the lack of TCP4 protein alone, since it could be rescued by expressing the wild-type protein under its own promoter. A truncated TCP4 protein lacking the predicted activation domain but retaining the DNA binding domain failed to rescue the phenotype. When the R61W and I93N mutants were expressed under the endogenous promoter of TCP4 in the null mutant background, they failed to rescue the epinasty phenotype, indicating that these mutant proteins are nonfunctional in vivo.
DISCUSSION
Transcription factors common to plants and animals possibly originated in an ancient organism ancestral to both these kingdoms. Each eukaryotic lineage then invented its own set of factors specific to the lineage, either by innovation or by sequence divergence through mutations. The plant lineage shares several groups of transcription factors with their animal counterparts, including MYB, bHLH, HMG box, and Zn-coordinating proteins. On the other hand, several groups of transcription factors are unique to the plant kingdom and may perform plant-specific functions. This group includes the AP2/EREBP, NAC, WRKY, ARF, and TCP families (Riechmann et al., 2000). In Arabidopsis, TCP members constitute ∼1.6% of all transcription factors and play important roles by modulating transcription activities of target genes involved in diverse processes, such as floral symmetry, organ shape, plant architecture, circadian rhythm, and leaf senescence (Luo et al., 1996; Doebley et al., 1997; Nath et al., 2003; Palatnik et al., 2003; Aguilar-Martínez et al., 2007; Schommer et al., 2008; Pruneda-Paz et al., 2009; Martin-Trillo and Cubas, 2010).
All TCP members share a conserved domain of ∼60 residues in length, called the TCP domain, which is predicted to form a bHLH secondary structural motif (Cubas et al., 1999). The TCP domain, however, shares no obvious sequence similarity with the canonical bHLH domain found commonly in both plants and animals. Indeed, members of the TCP family have been classified in a group different from plant bHLH proteins (Buck and Atchley, 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003). Therefore, the TCP domain may represent a novel DNA binding structure. Although >200 TCP sequences have been reported in the database from various plant species, little is known about their DNA binding properties and structural similarities with other DNA binding domains. Here, we determined the biochemical properties of the TCP domain and its interaction with DNA and suggested a gross structural model for the TCP domain bound to its target DNA.
Features Unique to the TCP Domain
Sequence analysis suggests that the TCP domains of the member proteins share higher degree of similarity than other family of transcription factors with similar folds; >80% of residues in the TCP domains show ≥50% URF values (Figures 2B and 2C). The corresponding URF value for Arabidopsis bHLH proteins is only ∼30% (Atchley et al., 1999; Toledo-Ortiz et al., 2003). This is possibly reflected in the higher sequence conservation in the target DNA sequence of several TCP members for which the binding sites have been determined (Kosugi and Ohashi, 1997, 2002; Costa et al., 2005; Li et al., 2005; Schommer et al., 2008; Pruneda-Paz et al., 2009). A few residues are also conserved at both flanks of the core domain (Figures 2B and 2C), likely to be important in DNA binding specificities, as found in homeobox proteins (Joshi et al., 2007).
The basic residues implicated in DNA binding (see Supplemental Figure 2C online) are highly conserved in all the members of the TCP family. A small group of bHLH proteins in plants and animals lacks the basic residues and negatively regulates the DNA binding activity of other members by forming heterodimers (Benezra et al., 1990; Ellis et al., 1990; Toledo-Ortiz et al., 2003). The TCP family does not seem to include such negative regulators; all TCP members analyzed here have at least eight positively charged residues in the basic domain. By contrast, the founding member of the negative regulator class in bHLH, Id, has only one positive residue in the basic region as opposed to seven in MyoD (Benezra et al., 1990).
The Basic Region and TCP–DNA Interaction
The basic region of the TCP domain is required for binding to the major groove of DNA double helix (Figures 4, 5C, and 7A). This region is mostly unstructured (Figure 3A; see Supplemental Figure 2B online), and helix formation is partly induced upon DNA binding (Figure 5G). We noticed that even though the model predicts mostly helical structure for the DNA-bound TCP4 domain (Figure 7A), the CD (Figure 5G) shows only ∼34% helix in the DNA-bound domain. This may be due to the presence of the His-tag that constitutes ∼40% of the fusion protein. Induction of helicity upon target DNA binding is also observed in case of bHLH domain (Ferré-D'Amaré et al., 1994; Carroll, et al., 1997). Affinities of DNA–protein interaction are comparable in TCP and bHLH domains (Figures 5A and 5B; Künne et al., 1996; Winston et al., 1999; Shklover et al., 2007). The cooperative nature of the DNA–protein interaction observed for the TCP domain suggests the presence of monomers, incapable of binding to DNA, at low protein concentrations. A small fraction of monomer is indeed detected in the analytical gel filtration experiment (see Supplemental Figure 3C online). Cooperative DNA binding indicates a threshold-dependent, sharp, on–off switch for activation of target genes by TCP4 protein, as demonstrated in the case of Bicoid protein in Drosophila (Burz et al., 1998; Lebrecht et al., 2005). The TCP domain forms a stable homodimer (Figure 5D; see Supplemental Figure 3 online) mediated by the C-terminal HLH fold. Mutations in the residues predicted to be present in the dimer interface abolish DNA binding ability (Figures 8A and 8B), suggesting that only the dimer form of TCP4 can bind to DNA. Strong homodimerization is also seen in the closest homolog of TCP4 in rice, PCF5 (Kosugi and Ohashi, 2002; Os-TCP4 in Figure 1B). These data demonstrate that, despite the lack of overall sequence similarity, the TCP domain and the canonical bHLH domain share similar biochemical properties and modes of DNA interaction.
Profile alignment revealed that the TCP sequence shares a fair degree of sequence similarity with the bHLH domains especially in the C-terminal HLH region (Figure 3B). The residues involved in dimer formation and on the interface of the two helices within a protomer are conserved between these two domains. The TCP domain was modeled based on the sequence similarities found in this domain with MyoD of known structure (Figure 7A; Ma et al., 1994). Sequence similarity is less obvious in the basic region, which is predicted to make direct contacts with DNA bases (see Supplemental Figures 2C and 2D online; Ma et al., 1994). Presence of a Gly-Pro pair would prevent formation of a long helix in continuation with helix I (Figure 3A). Besides, the basic region contains a three-residue insertion preceding the Gly-Pro pair in the class II TCP domain (Thr-Ala-Lys in TCP4) that possibly forms a loop (Figure 7) required for DNA binding (Figure 4). A Gly-Pro pair is also found in the basic region of a bHLH subclass represented by E2F and is important for DNA binding (Jordan et al., 1994). However, E2F forms a winged-helix structure that includes β-sheets (Zheng et al., 1999), a secondary element that is not predicted for the TCP domain (Figures 3A and 7).
The TCP model predicts residues, such as His and Arg, to be involved in DNA interaction, similar to those found in the bHLH domains (Ferré-D'Amaré et al., 1993; Ma et al., 1994; Figure 3B). There are, however, several differences in DNA interaction between these two domains. All the DNA-contacting residues of the bHLH domain are located on extended helix I of the DNA-bound protein. By contrast, while two base-interacting residues in TCP4 (Arg-62 and Leu-65) are present on helix I, His-49 is located on the predicted helix N that is oriented at an angle with helix I (Figure 7). His-49 of TCP4 is, however, present in the sequence context (KDRHSKV) similar to that of several bHLH members, such as CBF1 (KDSHKEV; Ferré-D'Amaré et al., 1993). A Glu is conserved in all bHLH proteins (Glu-118 in MyoD; Ma et al., 1994) and contributes extensively in sequence-specific DNA binding. Although Asp-60 of TCP4 (RDRR) is present in a sequence context similar to MyoD (RERR), it is unlikely to take part in base contact with the target DNA (Figure 7) because a shorter side chain at this position is incompatible with DNA recognition in the MAX protein (Ferré-D'Amaré et al., 1993).
To summarize, the TCP domain and the bHLH domain share many biochemical properties that define their DNA binding activities. Critical residues in the C-terminal region of both the domains are conserved and are involved in dimer formation. The major differences in sequence and biochemical properties lie in the N-terminal basic domains. Although this region is of similar size and contains positively charged residues, the presence of a Gly-Pro pair restricts the extension of helix I in TCP4. Besides, a three-residue insertion preceding the Gly-Pro pair possibly forms a loop between proposed helix N and helix I. There are several differences in the residues involved in base contact of the target DNA in the bHLH and the TCP domains. An extended basic region with an ancillary DNA binding stretch in the Maf bZIP protein shows DNA interaction different from its other family members (Dlakić et al., 2001). Our results, however, suggest that the overall orientation of the TCP domain may be similar to that of the bHLH domain. The lengths of these two DNA binding regions are comparable and their target DNA elements are also of similar size. Besides, all the base-contacting residues reside on helices in these two proteins.
Evolution of the TCP Domain
The TCP domain is thought to have a distinct evolutionary origin from that of bHLH domain mainly because of the lack of clear sequence similarity. Besides, the core target DNA sequences recognized by these groups of proteins are different; most bHLH proteins bind to palindromic E-box sequence (CANNTG), whereas TCP proteins bind to the partially palindromic core sequence TGGNCC (Kosugi and Ohashi, 2002; Schommer et al., 2008). This study, however, suggests that they might have a common origin with comparable overall fold. Here, we predicted three TCP residues, His-49, Arg-62, and Leu-65, that directly contact the DNA bases. A common feature of the bHLH basic regions is that the base-interacting residues follow a (i, i+iv) distance relationship on a long helix. By contrast, the first base-interacting residue of TCP4 (His-49) is separated by at least 12 amino acids from the other two. Even though all three residues occupy predicted α-helical positions, the long intervening region makes a loop structure that separates helix N from helix I. It is possible that the TCP domain evolved from the canonical bHLH domain with a short insertion in the basic region that resulted in splitting a long helix into two. Such changes may have occurred in an ancestral plant preceding the emergence of land plants (Navaud et al., 2007), since the evolution of the TCP domain is thought to have coincided with the evolution of the phragmoplast, a plant cell–specific structure formed during late cytokinesis. Further amino acid changes in the TCP domain may have followed to accommodate binding to a DNA with a new core sequence. Molecular structure of the TCP-DNA complex would be able to test this possibility and shed more light into the structural and evolutionary similarities of these two protein families.
METHODS
Mutagenesis of the TCP4 Domain
For the yeast one-hybrid assay, a yeast strain containing TCP4 binding sequences upstream of the HIS3 reporter gene was generated. Two complementary oligonucleotides containing 12 copies of the TCP4 consensus sequence [(GTGGTCCC)12 and (GGGACCAC)12] were synthesized, converted into double-stranded DNA by annealing, and cloned into the NotI-SpeI site of the pINT1:HIS3NB vector (a gift from P.B.F. Ouwerkerk, Leiden University; Meijer et al., 1998). The SacI-NcoI fragment of the resulting construct, which contains the TCP4 binding sequences fused to a minimal promoter and driving the HIS3 reporter gene, was integrated into PDH locus of the yeast strain Y187 to generate the strain Y187-CON. Random mutants of the TCP domain were generated by PCR reaction using error-prone DNA polymerase as per the instruction manual (GeneMorph II random mutagenesis kit; Stratagene) and screened based on yeast one-hybrid assay. In short, error-prone PCR reaction was performed using the plasmid pTEF2:TCP4 as template and a TEF2 promoter-specific oligo (5′-CTTCGACTATGCTGGAGGCAGAGATG-3′) and a TCP4-specific oligo (5′-GCTATCGAATCTGAATCCATTGACG-3′) as primers. The PCR reaction was set in 50 μL volume containing 1× Mutazyme reaction buffer, 200 μM each deoxynucleotide triphosphate, 125 ng of each primer, 2.5 units of Mutazyme DNA polymerase, and 100 ng of template plasmid DNA (pTEF2:TCP4). The high amount of template DNA (100 ng) was used to obtain the low frequency (0 to 3) of mutagenesis. PCR reactions were performed in Bio-Rad thermal cycler programmed for 94°C for 1 min (94°C for 30 s, 57°C for 30 s, and 72°C for 1 min) for 30 cycles followed by 72°C for 10 min. The PCR product was quantified on 1.2% agarose gel by comparing the staining intensity with that of 1.1-kb gel standard. Y187-CON yeast strain containing pTEF2:TCP4 plasmid was transformed with 500 ng of PCR product to mobilize the mutagenized TCP domain into pTEF2:TCP4 by in vivo homologous recombination. Site-directed mutants were generated by the megaprimer inverse PCR method (Kirsch and Joly, 1998). PCR products were digested by DpnI restriction enzyme and transformed into Escherichia coli (DH5α). Mutant plasmids were screened by PCR and restriction digestion and then confirmed by sequencing.
One- and Two-Hybrid Assays
The full-length TCP4 open reading frame (for pTEF2:TCP4) or the TCP4 region lacking the binding domain (for pTEF2:TCP4ΔD) were PCR amplified using appropriate primers, and the products were cloned in BamHI-XhoI sites of pJV340 (alias pPS189; Pillai et al., 2001) downstream of pTEF2 promoter to generate pTEF2:TCP4 and pTEF2:TCP4ΔD constructs. Sequence integrity was confirmed with restriction digestion and sequencing.
For spot assays, yeast cells grown in synthetic dropout glucose medium for 1 d were adjusted to OD600 of 0.1 and diluted to various concentrations (OD600 of 0.1, 0.05, 0.01, 0.005, 0.001, and 0.0005). Aliquots (∼5 μL) from each dilution were spotted onto SD-glucose medium and incubated for 3 d at 30°C. A Bio-Rad gel documentation system was used to photograph yeast plates in white light.
One- and two-hybrid assays were performed in the yeast strain Y187 (Y187:ura3-52, trp-901, Ura3::GALUAS-Gal1TATA-LacZ ; Harper et al., 1993). Cells were transformed with the LiOAc/ssDNA/PEG method (Yeast Protocol Handbook; Clonetech; Gietz et al., 1992) and plated onto synthetic dropout medium lacking Leu and His for one-hybrid assays or Leu and Trp for two-hybrid assays. Selection medium for one-hybrid assay was supplemented with 0 to 100 mM 3-AT (Sigma-Aldrich) to investigate the binding strength of mutant proteins. β-Galactosidase activity was assayed by filter assay or liquid culture assay according to procedures described (Yeast Protocol Handbook) using X-Gal or o-nitrophenyl-β-D-galactopyranoside, respectively, as substrates. One unit of β-galactosidase activity was defined as hydrolysis of 1 μmol of o-nitrophenyl-β-D-galactopyranoside to o-nitrophenol and D-galactose per min per cell.
EMSA
EMSA was performed with crude lysates or purified proteins. Protein samples were run on SDS-PAGE and stained by Coomassie Brilliant Blue or transferred to polyvinylidene fluoride membrane (HybondP; GE Healthcare) for immunoblot analysis with horseradish peroxidase–conjugated anti-His antibody. DNA probe was made by end labeling double-stranded oligo (5′- ACATCTATGTGGTCCCCCTCATCT-3′) containing one TCP4 binding site with [γ-32P]ATP using the polynucleotide kinase. DNA-protein binding was performed in 10 μL reaction mixture containing 10 fmol probe, 1× binding buffer (20 mM HEPES-KOH at pH 7.8, 100 mM KCl, 1 mM EDTA, 0.1% BSA, 10 ng Herring sperm DNA, and 10% glycerol) and ∼1.5 μg bacterial lysate protein. The mixture was incubated for 30 min and loaded on an 8% native polyacrylamide gel. Electrophoresis was conducted at 4 V/cm for 45 min with 0.5× Tris-borate buffer at room temperature. Gel was autoradiographed using phosphor imaging.
For Kd determinations, protein titration was performed in the presence of constant amount of radiolabeled DNA (5 nM) using EMSA. The radioactivity for each band was quantified using Fuji Multi Gauge v2.3 software (FUJIFILM Medical Systems), and the fraction bound was plotted as a function of protein concentration. The data were fitted to the Hill equation (fraction bound = [protein]n/([protein]n + Kdn)), where n is the Hill coefficient and Kd is the apparent dissociation constant, using GraphPad Prism software version V (GraphPad Software).
Sequence Analysis and Bioinformatics
TCP4 protein sequences were obtained from public databases (http://datf.cbi.pbu.edu.cn; www.tigr.org), and sequence alignment was performed using the ClustalW program with default settings (Protein gap open penalty, 10; protein gap extension penalty, 0.2; protein matrix, Gonnet; protein ENDGAP, −1; protein GAPDIST, 4) (Thompson et al., 1994). Percentages of conservation of amino acid residues were obtained from Jalview option of ClustalW program. The alignment file was fed as input to Boxshade server 3.2 for better visualization. The phylogenetic tree was generated using the neighbor-joining method (Saitou and Nei, 1987) to produce a midpoint rooted tree and was conducted in MEGA4 (Tamura et al., 2007).
Protein Purification and Analytical Gel Filtration
His-tag TCP4Δ1, TCP4Δ2, and point mutants were expressed in E. coli (BL21 DE3) using pRSET C expression vector (Invitrogen). Bacterial culture at ∼0.6 OD600 was induced with 1 mM isopropyl-β-D-thiogalactoside for 3 h. Cells were harvested, suspended in buffer (50 mM NaH2P04, pH 8.0, 300 mM NaCl, 50 μg of lysozyme, and protease inhibitor cocktail [Roche]) and disrupted by sonication. Lysate was centrifuged and protein purified from the supernatant using Ni-NTA column as per instructions (Novagen). Eluted protein was dialyzed in 20 mM phosphate buffer, pH 8.0, and 150 mM NaCl. Protein concentration was determined by Bradford assay at A260. Purity was analyzed in a 15% SDS-PAGE. Analytical gel filtration was performed using a 25 mL Sephadex G200 FPLC column (Millipore) calibrated using the marker proteins aldolase, canalbumin, ovalbumin, and chymotrypsinogen A. A total of 150 μg of purified TCP4Δ3-MBP (200 μL) was injected and eluted with buffer (50 mM NaH2PO4, pH 8, and 150 mM NaCl). Elution was monitored by absorbance at 215 nm.
Glutaradehyde Cross-Linking Experiment
Cross-linking was performed by incubating ∼2.3 μM purified TCP4Δ1 in 0.01% glutaradehyde for required durations with or without equimolar amounts of 24-bp target DNA. Monomer and dimer populations were visualized by resolving the mixture on 12% SDS-PAGE followed by immunoblot analysis with anti-HIS tag antibody (Qiagen).
Spectroscopy
CD measurements were done on a Jasco J-810 spectropolarimeter. Spectra were recorded in a 1-mm path length quartz cuvette in 20 mM phosphate buffer, pH 8.0, and 100 mM NaCl with or without trifluoroethanol. Protein samples were incubated with double-stranded oligo at 25°C for 30 min before collecting the spectra. Samples were scanned at 2-nm steps at 25°C with 1-s averaging time. Each spectrum is an average of 10 scans. Fluorescence emission spectra were recorded on a Fluorolog-Tau-3-Fluorometer (Horiba-Jobin-Yvon) equipped with a 450-W Xenon short-arc lamp. TCP4Δ2 (5 μM) was incubated with varying amount of target DNA in buffer same as in CD at 21°C. Spectra were collected at 8 nm band-pass with excitation at 280 nm (2 nm band-pass).
Plant Expression Vectors and Arabidopsis Transformation
Genes coding for wild-type TCP4, TCP4Q156*, and point mutants (R61W and I93N) were cloned in the binary vector pCAMBIA2200 (http://www.cambia.org/daisy/cambia/585.html) under the control of 2.6-kb upstream region of TCP4 and introduced into Arabidopsis thaliana by Agrobacterium tumefaciens (GV3101)–mediated floral dip method (Weigel and Glazebrook, 2002). Kanamycin-resistant seedlings were selected and photographed using a Nikon Coolpix camera. Presence of transgenes was analyzed by PCR (primers specific to the pCAMBIA2200 NPT gene, 5′-GAACTCGTCAAGAAGGCGATAGAAG-3′ and 5′-CTATGACTGGGCACAACAGACAATC-3′). Transgenic plants were grown until the next generation to confirm the phenotype.
Structure Prediction and Molecular Modeling of the TCP Domain
Secondary structure, disorder, and other prediction analyses were performed using appropriate prediction methods (see Results). For profile analysis, the sequence of each of the members belonging to the bHLH family of proteins available in the Structural Classification of Proteins (http://scop.mrc-lmb.cam.ac.uk/scop/) was used as query to search the nonredundant database using the BLAST program. For each query, the five top hits with >50% sequence identity were chosen as representatives. Sequence profiles were generated for each member by aligning the sequences of the query with the close homologs, and conservation scores were computed (see Supplemental Figure 8 online) using the SCORECONS server (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/valdar/scorecons_server.pl).
DNA binding region of TCP4 was modeled using MyoD as the template. Using SYBYL tools (JPR Technologies), the base sequence of DNA found in the template was first mutated to TCP4 target sequence in silico. The conformations of substituted bases were then carefully adjusted using MANIP (Massire and Westhof, 1998) to align with the backbone and also to favor the base pair interactions. Two extra bases were added in the B-DNA conformation at each end to make the target DNA long enough to satisfy all protein-DNA contacts. Positions on these extra bases were adjusted manually to merge smoothly with the rest of the backbone. Structural information, such as hydrogen bonding pattern, solvent accessibility, and secondary structure topology, in the sequence alignment were assessed with JOY tool (Mizuguchi et al., 1998). The DNA model thus obtained was further refined by 100 steps of energy minimization using the Tripos 5.2 force field in SYBYL MAXIMIN tool (Clark et al., 1989). The HLH region of TCP4 was modeled based on the known structure of MyoD HLH region and on sequence conservation among the intra- and interhelical residues between these two regions (see Supplemental Figure 9 online). The basic stretch C-terminal to Gly-Pro pair was modeled as a helix in continuity with helix I. The other basic stretch at the N terminus was also modeled as α-helix (termed here as helix N) and placed at the major groove of the target DNA. Energy minimization of TCP4-DNA complex was performed in 10,000 steps using the AMBER tool (Assisted Model Building with Energy Refinement; Cheatham and Young, 2001; Ponder and Case, 2003). Protein structure and interactions between molecules (Figure 7; see Supplemental Figure 10 online) were visualized by PyMol (www.pymol.org) and Chimera (Pettersen et al., 2004). The model was evaluated using the Verify3D server (http://nihserver.mbi.ucla.edu/Verify_3D/). The residue-wise structural compatibility scores were compared with that of the corresponding residues in the MyoD structure (see Supplemental Figure 11 online).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TCP4 (AT3G15030), pINT1:HIS3NB (AY061966), and pCAMBIA2200 (AF234313). Accession numbers for the genes used in phylogenetic analyses can be found in Supplemental Tables 1 and 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Supplemental Figure 1. Evolutionary Relationship of TCP Genes.
Supplemental Supplemental Figure 2. Secondary Structure and Disorder Prediction of the TCP Domain.
Supplemental Figure 3. Purification and Analytical Gel Filtration of TCP Protein.
Supplemental Figure 4. β-Galactosidase Filter Assay in Yeast to Demonstrate Autoactivation of TCP4.
Supplemental Figure 5. CD Spectra of TCP4Δ2.
Supplemental Figure 6. The Consensus Sequences of the Target DNA of TCP4 and MyoD.
Supplemental Figure 7. Immunoblot Analysis of the Mutant TCP4 Proteins.
Supplemental Figure 8. Conservation Scores for the Profile-Based Alignment of TCP and bHLH Proteins.
Supplemental Figure 9. Comparison of the Predicted HLH Structure of TCP4 with MyoD HLH Domain.
Supplemental Figure 10. Energy Minimization of the Predicted TCP4 Domain.
Supplemental Figure 11. Verify 3D Structure Compatibility Score.
Supplemental Table 1. List of 121 Class I TCP Proteins Used for Sequence Analysis.
Supplemental Table 2. List of 85 Class II TCP Proteins Used for Sequence Analysis.
Supplemental Table 3. Summary of Random Mutagenesis Results.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Figure 1.
Supplemental Data Set 2. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 1.
Acknowledgments
We thank V. Nagaraja, B. Gopal, Arnab China, and T.S. Girish for help in biophysical experiments; Abdur Rahaman, Ashis Nandi, Mansi Gupta, and M. Subhashini for useful comments on the manuscript; Kavitha S. Rao for help in producing Supplemental Figure 4; P.B.F. Ouwerkerk of Leiden University for giving us the pINT1:HIS3NB vector; and A. Vaishnavi for help in cloning and sequencing of mutants. P.A. and M.D.G. were supported by fellowships from Council for Scientific and Industrial Research, Government of India; U.N. was supported by a grant from Department of Biotechnology, Government of India (BT/PR/5096/AGR/16/431/2004); and N.S. was supported by Department of Biotechnology, Government of India as a part of institution-wide support for structural biology.
References
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S., Sarai A. (2005). PSSM based prediction of DNA-binding sites in proteins. BMC Bioinformatics 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi M., Mizuguchi K., Sarai A., Ahmad S. (2009). Prediction of mono- and di-nucleotide-specific DNA-binding sites in proteins using neural networks. BMC Struct. Biol. 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley W.R., Fitch W.M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 94: 5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley W.R., Terhalle W., Dress A. (1999). Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48: 501–516 [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lockshort D., Turner D.L., Weintraub H. (1990). The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59 [DOI] [PubMed] [Google Scholar]
- Buck M.J., Atchley W.R. (2003). Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56: 742–750 [DOI] [PubMed] [Google Scholar]
- Burz D.S., Rivera-Pomar R., Jäckle H., Hanes S.D. (1998). Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17: 5998–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A.S., Gilbert D.E., Liu X., Cheung J.W., Michnowicz J.E., Wagner G., Ellenberger T.E., Blackwell T.K. (1997). SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 11: 2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T.E., Young M.A. (2001). Molecular dynamics simulation of nucleic acids: Successes, limitations and promise. Biopolymers 56: 232–256 [DOI] [PubMed] [Google Scholar]
- Chiang S.Y., Welch J., Rauscher F.J., III, Beerman T.A. (1994). Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA on DNA. Biochemistry 33: 7033–7040 [DOI] [PubMed] [Google Scholar]
- Citerne H., Luo D., Pennington R.T., Coen E., Cronk Q.C.B. (2003). A phylogenetic investigation of CYCLOIDEA-Like TCP genes in the leguminosae. Plant Physiol. 131: 1042–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M., Cramer R.D., Opdenbosch N.V. (1989). Validation of the general purpose Tripos 5.2 force field. J. Comput. Chem. 10: 982–1012 [Google Scholar]
- Costa M.M., Fox S., Hanna A.I., Baxter C., Coen E. (2005). Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–510116236768 [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Dlakić M., Grinberg A.V., Leonard D.A., Kerppola T.K. (2001). DNA sequence-dependent folding determines the divergence in binding specificities between Maf and other bZIP proteins. EMBO J. 20: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H.M., Spann D.R., Posakony J.W. (1990). extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell 61: 27–36 [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Pognonec P., Roeder R.G., Burley S.K. (1994). Structure and function of the b/HLH/Z domain of USF. EMBO J. 13: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Prendergast G.C., Ziff E.B., Burley S.K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363: 38–45 [DOI] [PubMed] [Google Scholar]
- Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. (2006). FoldUnfold: Web server for the prediction of disordered regions in protein chain. Bioinformatics 22: 2948–2949 [DOI] [PubMed] [Google Scholar]
- Gietz D., St. Jean A., Woods R.A., Schiestl R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M.C., Ramboer A., Lacout F., Tremousaygue D. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Gou Z., Kuznetsov I.B. (2007). DP-Bind: A web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23: 634–636 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K. (2007). PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35: W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Jordan K.L., Haas A.R., Logan T.J., Hall D.J. (1994). Detailed analysis of the basic domain of the E2F1 transcription factor indicates that it is unique among bHLH proteins. Oncogene 9: 1177–1185 [PubMed] [Google Scholar]
- Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S. (2007). Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim S.K., Norden B. (1993). Methyl green. A DNA majorgroove binding drug. FEBS Lett. 315: 61–64 [DOI] [PubMed] [Google Scholar]
- Kirsch R.D., Joly E. (1998). An improved PCR mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 26: 1848–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Künne A.G.E., Meierhans D., Allemann R.K. (1996). Basic helix-loop-helix protein MyoD displays modest DNA binding specificity. FEBS Lett. 391: 79–83 [DOI] [PubMed] [Google Scholar]
- Lebrecht D., Foehr M., Smith E., Lopes F.J.P., Vanario-Alonso C.E., Reinitz J., Burz D.S., Hanes S.D. (2005). Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl. Acad. Sci. USA 102: 13176–13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M. (2000). An overview of the structures of protein-DNA complexes. Genome Biol. 1: reviews001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N.M., Laskowski R.A., Thornton J.M. (2001). Amino acid–base interactions: A three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 29: 2860–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P.C., Rould M.A., Weintraub H., Pabo C.O. (1994). Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459 [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Massire C., Westhof E. (1998). MANIP: An interactive tool for modeling RNA. J. Mol. Graph. Model. 16: 197–205 [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Meijer A.H., Ouwerkerk P.B.F., Hoge J.H. (1998). Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14: 1407–1416 [DOI] [PubMed] [Google Scholar]
- Mizuguchi K., Deane C.M., Blundell T.L., Johnson M.S., Overington J.P. (1998). JOY: Protein sequence-structure representation and analysis. Bioinformatics 14: 617–623 [DOI] [PubMed] [Google Scholar]
- Muñoz V., Serrano L. (1994). Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1: 399–409 [DOI] [PubMed] [Google Scholar]
- Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Ouong M.W., Rivera R.R., Stuiver M.H. (1994). Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta 1218: 129–135 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Ofran Y., Mysore V., Rost B. (2007). Prediction of DNA-binding residues from sequence. Bioinformatics 23: i347–i353 [DOI] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Pabo C.O., Sauer R.T. (1992). Transcription factors: Structural families and principles of DNA recognition. Annu. Rev. Biochem. 61: 1053–1095 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Payne J.W. (1973). Polymerization of proteins with glutaraldehyde. Biochem. J. 135: 867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Sadreyev R., Grishin N.V. (2003). PCMA: Fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19: 427–428 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera – A visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pillai B., Sampath V., Sharma N., Sadhale P. (2001). Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. J. Biol. Chem. 276: 30641–30647 [DOI] [PubMed] [Google Scholar]
- Ponder J.W., Case D.A. (2003). Force fields for protein simulations. Adv. Protein Chem. 66: 27–85 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybylski D., Rost B. (2004). Improving fold recognition without folds. J. Mol. Biol. 341: 255–269 [DOI] [PubMed] [Google Scholar]
- Qian Z., Cai Y.-D., Li Y. (2006). Automatic transcription factor classifier based on functional domain composition. Biochem. Biophys. Res. Commun. 347: 141–144 [DOI] [PubMed] [Google Scholar]
- Reeves P., Olmstead R.G. (1997). Evolution of the TCP gene family in Asteridae: Cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol. Biol. Evol. 20: 1997–2009 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. (1993). Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. USA 90: 7558–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. (1994). Conservation and prediction of solvent accessibility in protein families. Proteins 20: 216–226 [DOI] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. (2004). The PredictProtein server. Nucleic Acids Res. 32: W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schlessinger A., Punta M., Rost B. (2007). Natively unstructured regions in proteins identified from contact predictions. Bioinformatics 23: 2376–2384 [DOI] [PubMed] [Google Scholar]
- Schlessinger A., Rost B. (2005). Protein flexibility and rigidity predicted from sequence. Proteins 61: 115–126 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: 1991–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendak R.A., Rothwarf D.M., Wedemeyer W.J., Houry W.A., Scheraga H.A. (1996). Kinetic and thermodynamic studies of the folding/unfolding of a tryptophan-containing mutant of ribonuclease A. Biochemistry 35: 12978–12992 [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell T.L., Mizuguchi K. (2001). FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310: 243–257 [DOI] [PubMed] [Google Scholar]
- Shklover J., Etzioni S., Weisman-Shomer P., Yafe A., Bengal E., Fry M. (2007). MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 35: 7087–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmaier P., Kel A.E., Wingender E. (2004). Systematic DNA-binding domain classification of transcription factors. Genome Inform. 15: 276–286 [PubMed] [Google Scholar]
- Struhl K. (1989). Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14: 137–140 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2007). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unneberg P., Merelo J.J., Chacón P., Morán F. (2001). SOMCD: Method for evaluating protein secondary structure from UV circular dichroism spectra. Proteins 42: 460–470 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Winston R.L., Millar D.P., Gottesfeld J.M., Kent S.B.H. (1999). Characterization of the DNA binding properties of the bHLH domain of Deadpan to single and tandem sites. Biochemistry 38: 5138–5146 [DOI] [PubMed] [Google Scholar]
- Wintjens R., Rooman M. (1996). Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262: 294–313 [DOI] [PubMed] [Google Scholar]
- Zheng N., Fraenkel E., Pabo C.O., Pavletich N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]