Cellulases are ubiquitous enzymes that play an essential role during plant cell growth and development. This study demonstrates that the activity of a dinoflagellate cell wall–bound cellulase is required for cell cycle progression and that its expression is downregulated in response to specific inhibitors in the G2/M phase.
Abstract
Cellulose synthesis, but not its degradation, is generally thought to be required for plant cell growth. In this work, we cloned a dinoflagellate cellulase gene, dCel1, whose activities increased significantly in G2/M phase, in agreement with the significant drop of cellulose content reported previously. Cellulase inhibitors not only caused a delay in cell cycle progression at both the G1 and G2/M phases in the dinoflagellate Crypthecodinium cohnii, but also induced a higher level of dCel1p expression. Immunostaining results revealed that dCel1p was mainly localized at the cell wall. Accordingly, the possible role of cellulase activity in cell cycle progression was tested by treating synchronized cells with exogenous dCelp and purified antibody, in experiments analogous to overexpression and knockdown analyses, respectively. Cell cycle advancement was observed in cells treated with exogenous dCel1p, whereas the addition of purified antibody resulted in a cell cycle delay. Furthermore, delaying the G2/M phase independently with antimicrotubule inhibitors caused an abrupt and reversible drop in cellulase protein level. Our results provide a conceptual framework for the coordination of cell wall degradation and reconstruction with cell cycle progression in organisms with cell walls. Since cellulase activity has a direct bearing on the cell size, the coupling between cellulase expression and cell cycle progression can also be considered as a feedback mechanism that regulates cell size.
INTRODUCTION
Cellular growth, but not degradation of cellular constituents, is generally considered to be coupled to cell cycle progression, resulting in the homeostasis of cell size. In walled organisms, cell walls represent the boundary of the cell and define the cell size. To cope with the increase in cell size during cell growth, the cell wall must grow with the cell it encloses and endure all the changes the cell undergoes. Cell walls are not static, and changes in their architecture, which are defined by synthesis, degradation, and reorganization of individual cell wall components, play a key role in the regulation of many physiological and developmental processes.
Dissolution of cell wall polysaccharides during growth and morphogenesis requires concerted activities of different cell wall enzymes, such as cellulase (endo-β-1,4-glucanases) (Hoson, 1993; Cosgrove, 1999; Carpita and McCann, 2000) and expansins (Carpita and Gibeaut, 1993; McQueen-Mason and Cosgrove, 1995; Cosgrove, 1998) in plants and chitinase and glucanases in yeasts (Cabib et al., 2001; Adams, 2004). Glucans, including cellulose in plants and β-glucans in fungi, are the major components of most cell walls and are responsible for the rigidity of the cell wall structure. Although most of the cell wall glucans do not undergo remarkable turnover during vegetative growth (Cosgrove, 1998; Baladron et al., 2002), it has been proposed that site-directed hydrolysis of the cell wall glucans, mediated by β-glucanases, probably plays a central role in several morphogenetic processes in the cell cycle (Kuranda and Robbins, 1991; Cid et al., 1995; Smits et al., 2001; Baladron et al., 2002).
Based on the primary mode of attack on cellulose molecules, cellulases have been divided into exo- and endo-forms. Exoglucanases (cellobiohydrolases) hydrolyze cellulose by removing the cellobiose units from the nonreducing ends (Barras and Stone, 1969). Endoglucanases (carboxymethyl cellulases [CMCase]) randomly cleave or hydrolyze internal β-1,4-glucosidic bonds in cellulose and thereby decrease the polymer length. Plant endoglucanases constitute a large, ubiquitous family of enzymes (del Campillo, 1999; Molhoj et al., 2002) and are expressed heterogeneously in Arabidopsis thaliana and poplar (Populus alba) plants (Ohmiya et al., 2003; Park et al., 2003). Endoglucanases play an important role in many plant developmental processes, including abscission (Tucker et al., 1988; Kemmerer and Tucker, 1994; del Campillo and Bennett, 1996), fruit ripening (Fischer and Bennett, 1991), and adventitious root initiation (Kemmerer and Tucker, 1994). The positive correlation between the expression of some plant endoglucanases (e.g., KOR in Arabiodopsis) and rapid cell elongation suggests a role for endoglucanases in cell expansion (Nicol et al., 1998; Ohmiya et al., 2003). Recent studies have also highlighted the involvement of endoglucanases during cellulose synthesis (Lane et al., 2001; Molhoj et al., 2002; Peng et al., 2002; Robert et al., 2005) and cytokinesis in plants (Zuo et al., 2000; Lane et al., 2001).
Cell division requires the coordination of nuclear division with numerous cellular and morphogenesis processes. For example, cellular events such as DNA synthesis and spindle formation are monitored by the cell cycle machinery through different checkpoints (Hartwell and Weinert, 1989; Elledge, 1996). Recent studies in budding yeast also demonstrated the existence of checkpoints monitoring morphogenesis events, like the assembly of the actin cytoskeleton (morphogenesis checkpoint) (Lew and Reed, 1995) and cell wall growth (cell wall integrity checkpoint) (Hoyt, 2004; Suzuki et al., 2004; Levin, 2005). Although genomic studies on yeasts have integrated cell wall synthesis into a global network of different cellular functions, including cell cycle control (Firon et al., 2004; Oliva et al., 2005), very little is known about the roles of degradation during cell cycle progression. By examining the effects of cellulase inhibitors on cell cycle progression in the dinoflagellate Crypthecodinium cohnii, our work demonstrates that cellulase activity plays a role during cell cycle progression.
RESULTS
Cellulase Inhibitors Delay Cell Cycle Progression at G2/M
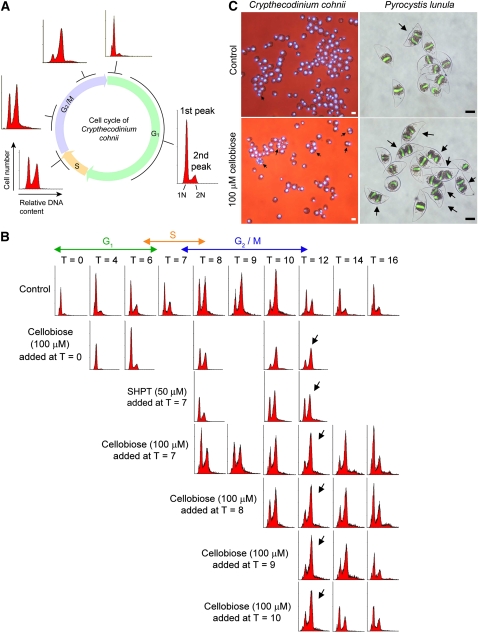
Flow cytometry provides a simultaneous, multiparametric analysis of every single cell or particle analyzed by the instrument. It is a common technique routinely used for the measurement of cellular DNA content to give a picture of the cell cycle status of a sample. By comparing the proportion of cells in the first and second peaks with the control, we can tell if there is a cell cycle delay/advancement in a population of synchronized cells (Figure 1A). For C. cohnii, we can obtain a highly synchronized population of cells at G1 (see Methods) and follow its progression in a cell cycle (10 to 12 h). To investigate if cellulase activities are required for cell cycle progression in C. cohnii, the cellulase inhibitors cellobiose (100 μM) and sodium hexachloropalladate (IV) tetrahydrate (SHPT) (50 μM) were added at T = 0 (early G1) or at T = 7 (early G2) to a synchronized population of cells. As shown in the flow cytograms (Figure 1B), it took 12 h for the control cells to complete the cell cycle. However, when cellobiose was added at T = 0 and T = 7, most of the cells were still in the G2/M phase (second peak) even at T = 12 (indicated by the arrows), indicating a cell cycle delay at the G2/M phase. Similar results were obtained when SHPT was added at T = 7. These suggest that cellulase activity is probably required for G2 progression in the C. cohnii cell cycle.
Figure 1.
Cellobiose Delayed the C. cohnii Cell Cycle at the G2/M Phase.
(A) A schematic diagram of DNA histograms showing the progression of the C. cohnii cell cycle from a population of synchronized G1 cells. The x axis of a DNA histogram represents the relative DNA content (propidium iodide [PI] fluorescence intensity), while the y axis represents the cell number (number of events). The first peak corresponds to the cells with the G1 DNA content (1N) and the second corresponds to the cells with the G2/M content (2N). In the G1 phase, the first peak (1N DNA content) appears as the major peak in a histogram. As the cells progress to the S phase, the first (G1, 1N) peak drops, and the second (G2/M, 2N) increases. During the G2/M phase, the DNA content doubles. Eventually, the cells complete mitosis and reenter the next G1 phase (the second peak drops and the first peak increases again).
(B) Flow cytograms of PI-stained synchronous C. cohnii. T refers to the time (hours) at which the samples were harvested after cell cycle synchronization at early G1. Cellulase inhibitors, cellobiose and SHPT, were added to synchronized cells at different time points to investigate the possible involvement of cellulase in cell cycle progression. Cellobiose (100 μM) was added at T = 0 (early G1), T = 7 (early G2), T = 8, T = 9, or T = 10 (late G2/M). SHPT (50 μM) was added at T = 7 (early G2). For control cells, the cells finished a cell cycle within 12 h (the first peak becomes the major peak again at T = 12). A G2/M delay, indicated by the arrows, was observed in cells treated with both cellobiose (added at T =0, 7, 8, 9, and 10) and SHPT (added at T = 7). It took longer for the cells to leave the G2/M phase and return to the G1 phase. This was revealed by the histogram with a higher second peak at T = 12 (indicated by the arrows) when compared with the control.
(C) Merged images of bright-field and fluorescence photomicrographs showing the effects of cellobiose (100 μM) on asynchronous C. cohnii and P. lunula. Cells were incubated with cellobiose for 24 h. DNA was stained blue with 4',6-diamidino-2-phenylindole for C. cohnii and green with SYTOX Green for P. lunula. Arrows indicate the multinucleated cells. For C. cohnii, bars = 10 μm; for P. lunula, bars = 50 μm.
[See online article for color version of this figure.]
To further examine if cellulase activity is required for the progression of early G2/M, middle G2/M or late G2/M, cellobiose was added at different time points within the G2/M phase (T = 7, 8, 9, and 10) (Figure 1B). The cell cycle was delayed (cells were not able to finish the cell cycle within 12 h) when cellobiose was added at any time point within the G2/M phase. When cellobiose was added at T = 10 (very late stage of the G2/M phase), a shorter delay was observed than when cellobiose was added earlier in the G2/M phase. These observations suggest that cellulase activity is required for the progression of the whole G2 phase, leading to mitosis.
Effects of cellulase inhibition on dinoflagellates were also studied with asynchronous cells of C. cohnii and Pyrocystis lunula with the assistance of fluorescent DNA stains (Kwok and Wong, 2003). Fluorescent photomicrographs suggest that the addition of cellobiose (100 μM) resulted in the appearance of more multinucleated cells in both species (Figure 1C).
Control experiments using other small oligosaccharides that do not inhibit cellulase activity (e.g., cellotriose and maltose) were performed to verify that the observed cell cycle effects were due to the specific inhibition of cellulase activity. Cellotriose and maltose were individually added to the synchronized cells (at T = 7) at concentrations essentially the same as for the cellobiose. Unlike cellobiose, neither cellotriose nor maltose delayed the cell cycle when compared with the control (see Supplemental Figure 1 online). This confirms that the observable cell cycle delay following the addition of cellobiose was a consequence of cellulase activity inhibition.
Cellulase Activity Peaks at G2/M
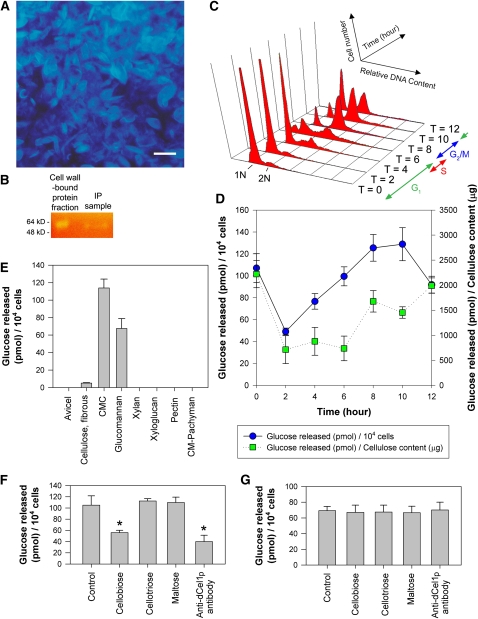
To determine if there is any cell wall–associated cellulase activity in the C. cohnii cells, cell walls were isolated and purified, and cell wall–bound proteins were extracted. Cellulosic cell walls in the cell wall fraction were stained blue with Calcofluor White (Figure 2A). To detect the presence of cellulase activity in the cell wall–bound protein sample, a Congo red-carboxymethylcellulose (CMC) staining method was employed that locates bands of cellulase activity in a polyacrylamide gel following electrophoresis (Schwarz et al., 1987). By staining the CMC-containing gel replica with Congo red, light-yellow bands (of ∼60 kD, corresponding to the molecular mass of dCel1p) against a red background demonstrated the breakdown of CMC substrate (Figure 2B) and suggested the presence of cellulase activity in the samples.
Figure 2.
Cellulase Activity in the C. cohnii Cell Cycle.
(A) Fluorescence photomicrograph of cellulosic cell walls prepared for the isolation of the cellulase-containing cell wall–bound protein fraction. Cellulose in the cell wall was stained blue with Calcofluor White. Bar = 10 μm.
(B) Cellulase activity in CMC-containing agarose replica was stained with Congo red following gel electrophoresis. Yellow bands (∼60 kD) against an orange background were observed in both the cell wall–bound protein fraction and immunoprecipitation (IP) sample.
(C) Flow cytograms of PI-stained synchronous C. cohnii cells were plotted, with the x axis representing the relative DNA amount, the y axis representing the cell number, and the z axis representing the time (hours) after synchronization.
(D) Cellulase activity of the C. cohnii cell wall protein fraction on the CMC substrate was determined by measuring the increase in reducing power of the reaction solution. Cell wall–bound protein fractions were harvested at different time points after cell cycle synchronization. Activity was expressed as pmole glucose released per 104 cells or per cellulose content (μg).
(E) Substrate specificities of C. cohnii cell wall–bound protein sample (harvested at the G2 phase, T = 7) toward different polysaccharides. Data represent means ± se of three replicate experiments.
(F) Effects of different small oligosaccharides and anti-dCel1p antibody on cell wall–bound CMCase activity. Cellobiose (100 μM), cellotriose (100 μM), maltose (100 μM), and anti-dCel1p antibody (3 μg mL−1) were tested for their ability to affect cell wall–bound CMCase activity. Cellobiose and anti-dCel1p antibody significantly (P < 0.01) reduced the CMCase activity, indicated by the asterisks, when compared with the control. Data represent means ± se of three replicate experiments.
(G) Effects of different small oligosaccharides and anti-dCel1p antibody on cell wall–bound glucomannanase activity. Cellobiose (100 μM), cellotriose (100 μM), maltose (100 μM), and anti-dCel1p antibody (3 μg mL−1) were tested for their ability to affect cell wall–bound glucomannanase activity. No significant (P < 0.05) difference was found in any of the treatments compared with the control. Data represent means ± se of three replicate experiments.
[See online article for color version of this figure.]
During the C. cohnii cell cycle (Figure 2C), cellulase activity per cell (glucose released per 104 cells) increased in the G1 phase (T = 2 to T = 6) (Figure 2D). Cellulase activity per cell peaked at G2/M phase (T = 10) and dropped when the cells entered the G1 phase of the next cell cycle (T = 12). At the very beginning of the G1 phase (T = 0), when daughter cells had just shed their mother cell walls (Bhaud et al., 1991, 1994), relatively high cellulase activity was detected when compared with that at T = 2. This suggests that cellulase activity might also be involved during the daughter cell break-off process.
Because the cellulase activity assayed above was derived from the cell wall, we also normalized the cellulase activity data to the cellulose content in the cell wall samples (Figure 2D). By accounting for the changes in cellulosic content, cellulase activity (glucose released per cellulose content) appeared to be maintained at different levels within the G1 and G2/M phase. Cellulase activity per cellulose content doubled when the cells proceeded to the early G2 phase (T = 6 to T = 8), which is in agreement with the significant drop in cellulosic content reported previously (Kwok and Wong, 2003). A doubling in cellulase activity per unit cellulose content was observed in G2/M (T = 8 to T = 12) when compared with that in G1 (T = 2 to T = 6), implicating cellulase activity in the G2/M phase.
The activity of cell wall–bound endo-1,4-β-glucanase was demonstrated in suspension-cultured poplar cells by detecting the release of cello-oligosaccharides in culture medium (Ohmiya et al., 2000). If the observable increase in cellulase activity at the G2 phase is responsible for causing cellulose degradation in the C. cohnii cell wall, one would expect cello-oligosaccharide degradation products to be released into the culture medium. To test this hypothesis, cells were collected at T = 8 (G2) and the cell-free medium was tested for the presence of short cello-oligosaccharides. Degradation of potential cello-oligosaccharides was detected upon addition of commercial cellulase. An increase in the release of reducing sugars (resulting from the breakdown of cello-oligosaccharides) was detected in the cell-free medium harvested at T = 8 at a rate of 234.6 ± 6.5 pmol glucose per 104 cells but not in the sample not treated with commercial cellulase. Similarly, there was no detectable increase in the release of reducing sugars in the cellulase only control.
Substrate Specificity of the Cell Wall–Bound CMCase
The substrate specificity of the cell wall–bound CMCase (T = 7, early G2) was determined by performing the enzyme assay with different substrates. Besides cellulose substrates (Avicel, fibrous cotton cellulose, and CMC), other polysaccharides, such as xyloglucan, xylan, glucomannan, pectin, and pachyman (β-1,3-glucans), were also tested. The cell wall–bound CMCase mainly degraded CMC and glucomannan and showed a limited ability to degrade fibrous cotton cellulose (Figure 2E). There was no detectable activity against Avicel (microcrystalline cellulose) and all other forms of polysaccharides tested.
To verify that cellobiose and anti-dCel1p antibody can be used as cellulase inhibitors in cell cycle studies, in vitro CMCase and glucomannanase assays using the cell wall–bound protein sample (harvested at G2 phase, T = 7) were performed in the presence of different small oligosaccharides (cellobiose, cellotriose, and maltose) or the antibody. For the CMCase assay, a significant (P < 0.01) reduction in activity, by ∼40 and 50%, was observed with cellobiose and anti-dCel1p antibody, respectively, but not in the small oligosaccharide controls (cellotriose and maltose) (Figure 2F). For the glucomannanase assay, no significant (P < 0.05) difference in activity was observed between the control and all the treatments (Figure 2G).
Bioinformatic Analysis of dCel1
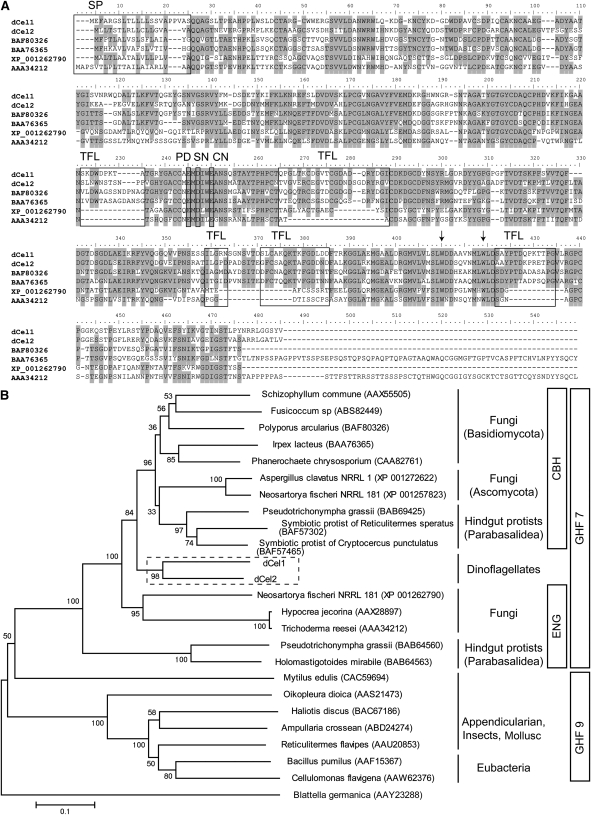
We identified a dinoflagellate cellulase that was reported to be highly expressed in the dark phase of P. lunula (Okamoto and Hastings, 2003). By performing inverse PCR with self-ligated single-stranded cDNA, full-length cDNA of the P. lunula cellulase was cloned (see Supplemental Figure 2A online). The cDNA encoding the P. lunula cellulase, dCel1, contained an open reading frame (ORF) 1392 bp (464 amino acids), starting with an ATG codon at position 67 and ending with a TGA codon at position 1458. BLAST searches using the deduced amino acid sequence of dCel1 showed high sequence homology to a glycosyl hydrolase family 7 (GHF7) cellulase (∼60% similarity). GHF7 includes endoglucanase (EC 3.2.1.4) and reducing end-acting cellobiohydrolase (EC 3.2.1.-), and members are mainly found in fungi and some symbiotic protists living in the hindgut of termites (http://www.cazy.org/Glycoside-Hydrolases.html). The predicted molecular mass of dCel1p was calculated to be 50 kD, and the first 22 amino acids were predicted to be a signal peptide sequence (with a predicted cleavage site between amino acids 22 and 23; AS-QQ). The dCel1p protein contains a GHF7 domain (24 to 454 amino acids), but no known cellulose or carbohydrate binding module could be identified (Figure 3A). A potential N-glycosylation site was found at the C terminus (449 amino acids; NSTL).
Figure 3.
Structural and Evolutionary Relationships between Cellulases from Dinoflagellates and Other Organisms.
(A) Comparison of the deduced amino acid sequences of P. lunula dCel1 and L. polyedrum dCel2 with other cellulase protein sequences from GHF7. Multiple sequence alignment was performed with ClustalX 2.0 (Larkin et al., 2007). Identical amino acids are shaded. As reported by Zhou et al. (2007), conserved amino acids, putative domains and structures, including signal peptide (SP), tunnel-forming loops (TFLs), and catalytic domain (PD = proton donor, SN = probable secondary nucleophile, and CN = catalytic nucleophile), are identified and framed. Arrows indicate the probable substrate binding Trp (W) residues.
(B) Possible evolutionary relationships between cellulases from dinoflagellates (boxed area), fungi, metazoans and bacteria. Phylogenetic analyses were conducted in MEGA4 (Saitou and Nei, 1987). The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). Genbank accession numbers for all sequences are shown in parentheses. CBH, cellobiohydrolases; ENG, endoglucanases. Bar represents the phylogenetic distance of 0.1 amino acid substitution per site.
BLAST searches using the deduced amino acid sequence of dCel1 returned several dinoflagellate EST sequences from the GenBank EST database. These allowed us to clone another cellulase, dCel2, from the dinoflagellate Lingulodinium polyedrum (see Supplemental Figure 2B online). The ORF of dCel2 encodes polypeptides consisting of 475 amino acids. dCel2 exhibits 59% identity (71% similarity) at the whole deduced amino acid level to dCel1. By performing a multiple sequence alignment of P. lunula dCel1 and L. polyedrum dCel2 with other cellulase protein sequences from GHF7 (BAF80326: cellobiohydrolase I from Polyporus arcularius, BAA76365: cellulase from Irpex lacteus, XP_001262790: endo-1,4-β-glucanase from Neosartorya fischeri, AAA34212: endoglucanase I from Trichoderma reesei), some conserved amino acids, putative domains, and structures of GHF7 cellulases (Zhou et al., 2007), including signal peptide, tunnel-forming loops, and catalytic domain, were identified in both dCel1 and dCel2 (Figure 3A). It is noteworthy that the probable substrate binding Trp residues of GHF7 cellulases (indicated by the arrows) were also conserved in both dCel1 and dCel2.
A phylogenetic tree was constructed with dinoflagellate cellulases and other GHF7 and GHF9 members from fungi, protozoa, eubacteria, and metazoa (Figure 3B; see Supplemental Data Set 1 online). The GHF7 clade was divided into two sister groups, cellobiohydrolase and endoglucanase. The phylogenetic tree aligned the dinoflagellate cellulases (dCel1 and dCel2) more closely with cellobiohydrolases than endoglucanases within GHF7. Within the group Alveolata, it should be noted that the apicomplexans, some members of which also carry relic plastids (Leander and Keeling, 2004), do not possess any cellulase gene. This suggests that dinoflagellates might have obtained the cellulase gene (probably together with cellulose synthase gene) after acquiring relic plastids in the evolutionary history. Although both the BLAST searches and the phylogenetic analyses using deduced amino acid sequence of dCel1 returned significant similarity to GHF7 cellobiohydrolases, the residues involved in the putative tunnel-forming loops were very different from those of other GHF7 members (Figure 3A).
Abundance of Immunodetectable dCel1p per Cell Curface Area Peaks at Both the G1 and G2/M Phase
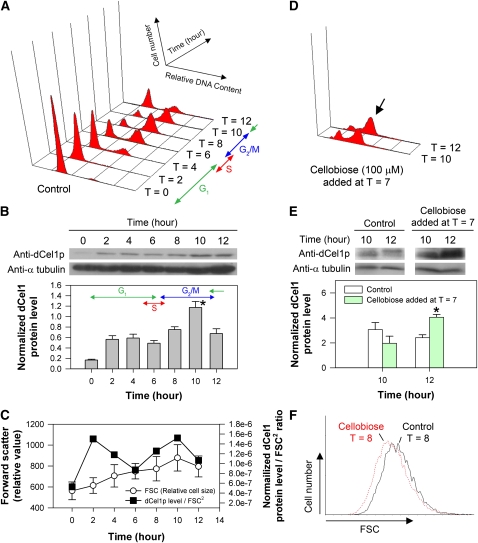
Immunoblotting was performed for studying the expression level of dCel1p during the C. cohnii cell cycle (Figure 4A). The affinity-purified anti-dCel1p antibody recognized a 60-kD protein band in the C. cohnii total protein sample, which is consistent with the CMCase activity staining results. At T = 10 (G2/M), the abundance of immunodetectable dCel1p reached a maximum and was significantly higher when compared with G1 cells at T = 4 (P < 0.05) (Figure 4B). This is in agreement with the observable increase in cellulase (CMCase) activity in the G2/M phase of the C. cohnii cell cycle.
Figure 4.
Immunodetection of dCel1p in the Cell Cycle and Effects of Cellobiose on dCel1p Expression and G2/M Cell Size.
(A) Flow cytograms of PI-stained synchronous C. cohnii cells harvested at different time points after cell cycle synchronization.
(B) Immunodetection of dCel1p in the normal cell cycle as shown in (A). Protein blots of C. cohnii protein samples harvested at different time points after cell cycle synchronization. SDS-PAGE–separated C. cohnii protein was probed with polyclonal antibodies raised against dCel1p. Anti-α-tubulin was also probed as a loading control. Immunodetectable signals for dCel1p were determined using ImageJ and normalized to the corresponding α-tubulin signals. Blots are representative of three independent experiments. During the cell cycle, the abundance of immunodetectable dCel1p peaked at the G2/M phase (T = 10) and was significantly higher (P < 0.05) when compared with that in the G1 phase (T = 4). Data represent means ± se of three replicate experiments. Asterisk indicates significant (P < 0.05) difference as compared with T = 4.
(C) FSC, which is a flow cytometric measurement related to the relative cell size, of the samples harvested at different time points during a cell cycle was plotted. Note that the cell size varied throughout the cell cycle. Data represent means ± sd (n = 10,000). By normalizing the abundance of immunodetectable dCel1p to FSC2 (proportional to the cell surface area), the data revealed that dCel1p level peaked at both the G1 (T = 2) and G2/M (T = 10) phase.
(D) When cellobiose was added at T = 7, a G2/M delay, as indicated by the arrow, was observed.
(E) Immunodetection of dCel1p in cells treated with cellobiose (100 μM). Protein blots of protein samples extracted from control and cellobiose-treated cells. Blots are representative of three independent experiments. Upon cellobiose treatment, the abundance of immunodetectable dCel1p increased significantly at T = 12 (P < 0.05) when compared with that of the control. Data represent means ± se of three replicate experiments. Asterisk indicates significant (P < 0.05) difference compared with the control.
(F) Cell size of the cellobiose-treated cells (T = 8) were generally smaller than the control at T = 8. The FSC data set (10,000 events) of control cells and cellobiose-treated cells were overlaid on a histogram using the software WinMDI. The x axis represents the FSC, while the y axis represents the cell number (number of events).
[See online article for color version of this figure.]
Forward scatter (FSC) is a flow cytometric measurement related to relative cell size. A greater FSC corresponds to a cell with a larger cross-sectional area, which can refract a larger amount of light onto the photosensor positioned parallel to the laser beam. Based on the relationship between FSC and cell size, the mean cell surface area could be estimated roughly from FSC2 (Kwok and Wong, 2003; Macey, 2007). For walled organisms, the cell wall poses constraints on cell size increase. In a normal cell cycle, C. cohnii cell size increased continuously from the G1 to G2/M phase and reached a maximum at T = 10 (Figure 4C). To investigate if there is any relationship between dCel1p level and cell size, we also plotted the abundance of imunodetectable dCel1p per cell surface area (FSC2) against time (Figure 4C). The results revealed that the dCel1p level per cell surface area indeed peaked in both the G1 and G2/M phases (lowest at S phase), possibly implicating a role for dCel1p in cell growth.
To investigate if dCel1p expression is regulated by a feedback mechanism during G2/M, cellobiose was added to synchronized cells at T = 7 (early G2). Cellobiose treatment resulted in a cell cycle delay at G2/M (Figure 4D), and the abundance of immunodetectable dCel1p at T = 12 increased significantly (P < 0.05, increased by ∼60%) when compared with the control (Figure 4E). During the G2/M delay, cellobiose-treated cells (at T = 8) were smaller in size than the control (Figure 4F).
To further investigate if cellulase activity was required for cell cycle progression at G1, cellobiose (100 and 500 μM) was added to synchronous C. cohnii cells at T = 0. For cellobiose-treated cells, a G1 cell cycle delay (as reflected by the percentage of G1 cells in the DNA histogram) (see Supplemental Figure 3A online) and a decrease in cell size were observed when compared with the control (see Supplemental Figure 3B online). However, there was no significant difference (P > 0.1) in immunodetectable dCel1p abundance at T = 4 between the control and cells treated with cellobiose (100 and 500 μM) (see Supplemental Figure 3C online).
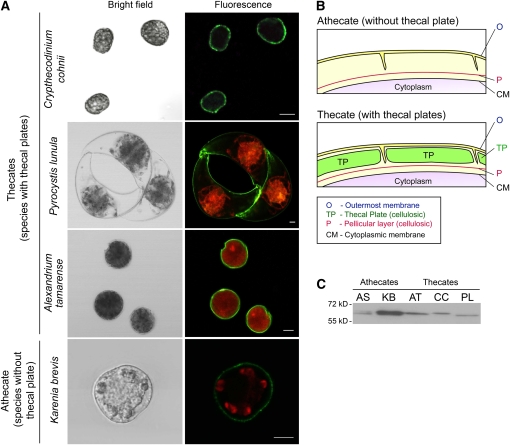
dCel1p Is Present in Both Thecate and Athecate Dinoflagellates and Is Mainly Localized to the Cell Wall of Dinoflagellates
Immunolocalization experiments on different dinofagellate cells were performed to validate the target of dCel1p in dinoflagellates. As shown in the confocal micrographs (Figure 5A), immunofluorescent signals (green) of dCel1p were observed on all of the dinoflagellate cells tested, including C. cohnii, P. lunula, Alexandrium tamarense, and the athecate Karenia brevis. The green immunofluorescence signals of dCel1p were essentially found only in the cell wall of dinoflagellate cells. No immunofluorescence and autofluorescence signals were found on the cell wall in the secondary antibody control (see Supplemental Figure 4 online).
Figure 5.
Subcellular Localization of dCel1p.
(A) Confocal images showing the immunofluorescence pattern in thecate (C. cohnii, P. lunula, and A. tamarense) and athecate (K. brevis) dinoflagellate species labeled with anti-dCel1p antibody. Green immunofluorescence signals (Alexa Fluor 488 goat anti-rabbit IgG), corresponding to dCel1p, were mainly localized in the cell wall. Red fluorescence signals correspond to the autofluorescence of chloroplast pigments. Bars = 10 μm.
(B) Schematic diagram showing the difference in cell wall between a typical thecate and athecate dinoflagellate. Cellulose can be found in two layers of the cell wall of dinoflagellates: (1) in the continuous pellicle layer in both the thecate and athecate species; and (2) in the thecal plates among the thecate species (Morrill and Loeblich, 1983).
(C) Immunoblot analysis detecting putative cellulase in different dinoflagellates. The anti-dCel1p antibody recognized bands of around 60 kD in all dinoflagellate species tested. AS, A. sanguine; KB, K. brevis; AT, A. tamarense; CC, C. cohnii; PL, P. lunula.
The dinoflagellate cellulose can be deposited on both cellulosic thecal plates as well as the pellicular layer (pellicle) in the dinoflagellate cell wall (Loeblich, 1970; Morrill and Loeblich, 1983). Based on the presence or absence of cellulosic thecal plates, dinoflagellates are classified as thecate (with cellulosic thecal plates and pellicle) or athecate (cellulosic pellicle only) species (Loeblich, 1970) (Figure 5B). Therefore, we further verified the subcellular target of dCel1p using thecate and athecate dinoflagellate species. Protein gel blot analysis using the anti-dCel1p antibody recognized protein bands of ∼60 kD in all the thecate (C. cohnii, P. lunula, and A. tamarense) and athecate (K. brevis and Akashiwo sanguine) dinoflagellate species tested (Figure 5C). This suggests that dCel1p is likely present in all dinoflagellate species and also implies that the common target for dCel1p is the cellulosic pellicle.
Cellulase Is Required for and Is Coupled to Cell Cycle Progression
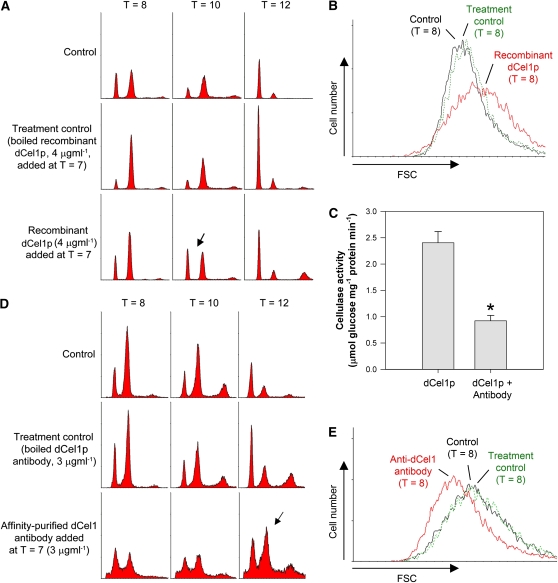
Since immunolocalization results suggest that dCel1p is mainly localized in the cell wall (Figure 5A), it is possible that the addition of exogenous dCel1p might affect the cell cycle. Recombinant dCel1p was purified under native conditions (see Supplemental Figure 5 online), and its CMCase activity was confirmed by Congo red CMC staining (see Supplemental Figure 5C online). The addition of recombinant dCel1p at the early G2 phase (T = 7) advanced the cell cycle (by ∼2 h) (Figure 6A) and increased cell size (Figure 6B) when compared with the control. Cell cycle progression for G1 cells was not affected when exogenous dCel1p was added in the G1 phase (see Supplemental Figure 6 online). This result was not surprising, as surface coverings are intact at this stage.
Figure 6.
Effects of Exogenous dCel1p and Purified Anti-dCel1p Antibody on Cell Cycle Progression.
(A) The addition of exogenous dCel1p to the cells at T = 7 (early G2) advanced the cell cycle. In the presence of exogenous dCel1p, the cells completed the cell cycle and returned to the G1 phase earlier (as shown by the emergence of first peak at T = 10 in the flow cytogram; indicated by the arrow) than in the controls. An equal amount of boiled recombinant dCel1p was added to the culture as control.
(B) Cells treated with exogenously added dCel1p were generally larger in size (at T = 8, G2/M phase) than the control cells.
(C) Purified anti-dCel1p antibody suppressed the activity of recombinant dCel1p. Recombinant dCel1p was incubated with purified anti-dCel1p antibody for 1 h at 4°C prior to cellulase activity assay. Data represent means ± se of four replicate experiments. Asterisk indicates significant (P < 0.01) difference compared with the control.
(D) The addition of affinity-purified anti-dCel1p antibody to the cells at T = 7 (early G2) resulted in a G2/M delay. Most of the cells treated with the affinity-purified antibody were still in the G2/M phase (the second peak at T = 12 was higher than the first peak, indicated by the arrow) at T = 12. An equal amount of boiled antibody was added to the culture as a control.
(E) Cells treated with the anti-dCel1p antibody were generally smaller (at T = 8, G2/M phase) than the control cells.
[See online article for color version of this figure.]
Because the affinity-purified anti-dCel1p antibody was able to reduce cellulase activity of recombinant dCel1p by ∼60% (Figure 6C), we also tested the effects of the purified antibody on cell cycle progression. The addition of antibody, conceptually similar to a knockdown experiment, inhibits cellulase activity and simulates the situation where cell wall expansion is slower than cell growth. By adding the antibody to the culture at T = 7 (early G2), a delay at the G2/M phase of ∼2 h was observed (Figure 6D). The cells treated with the anti-dCel1p antibody were smaller than control cells (Figure 6E). Similar to the results of adding exogenous dCel1p to cells at the G1 phase, the addition of anti-dCel1p antibody at G1 did not delay the cell cycle (see Supplemental Figure 6 online).
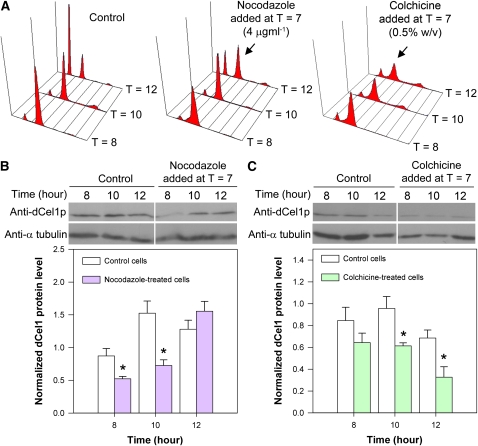
To further investigate the relationship between cell cycle progression at G2/M and dCel1p expression level, a reversible microtubule inhibitor, nocodazole, was used to independently prolong the G2/M phase as described previously (Yeung et al., 2000). When compared with the control, the addition of nocodazole (4 μg mL−1) to synchronous cells at T = 7 resulted in a G2/M delay (Figure 7A) and caused a temporary drop in immunodetectable dCel1p abundance at T = 8 and T = 10 (Figure 7B). It took a longer time, ∼2 more hours, for the dCel1p to reach its maximum level when compared with control cells. We further confirmed the effects on dCel1p level by delaying G2/M phase with an irreversible microtubule inhibitor, colchicine. When compared with nocodazole-treated cells, a longer G2/M delay was observed in colchicine-treated cells (Figure 7A). This resulted in a decrease in immunodetectable dCel1p abundance in the colchicine-treated cells (Figure 7C). These results suggest that the dCel1p expression level is tightly regulated by cell cycle progression at least in G2/M.
Figure 7.
Effects of G2/M Delay on dCel1p Level.
(A) Flow cytograms of PI-stained synchronous C. cohnii cells harvested at different time points after cell cycle synchronization. Microtubule inhibitors, nocodazole and colchicine, delayed the cell cycle at the G2/M phase. The arrows indicate the G2/M delay (the second peak was higher than the first peak at T = 12 when compared with the control) upon nocodazole and colchicine treatments.
(B) and (C) Immunodetection of dCel1p in cells treated with nocodazole and colchicine, respectively. Anti-α-tubulin was used as a loading control. Immunodetectable signals for dCel1p were determined using ImageJ and normalized to the corresponding α-tubulin signals. Blots are representative of three independent experiments. Both nocodazole and colchicines led to a significant decrease (P < 0.05) in the abundance of immunodetectable dCel1p at T = 10 when compared with the control. Data represent means ± se of three replicate experiments. Asterisk indicates significant (P < 0.05) difference compared with the corresponding control.
[See online article for color version of this figure.]
DISCUSSION
Is cell wall degradation required for and coordinated with cell cycle progression? Although there is some documentation about the regulation of cellulase expression levels in plants and fungi (Beguin and Aubert, 1994; Ilmen et al., 1997; Nicol et al., 1998; Park et al., 2003; Ohnishi et al., 2007), no data suggests a cell cycle–regulated cellulase level. Interestingly, the dCel1p level was upregulated upon addition of cellobiose (Figure 4E) and dropped with the independent arrest of the cell cycle at G2/M (Figures 7B and 7C). This apparent feedback and coupling mechanism reinforces the view that the balance between synthesis and degradation is tightly regulated in the cell cycle.
Cellobiohydrolases form a tunnel-like catalytic site (containing many substrate binding subsites) and are able to hydrolyze crystalline cellulose to release cellobiose (Teeri, 1997). Because of having a more open substrate binding groove than cellobiohydrolases, endoglucanases exhibit comparatively poor activity toward crystalline cellulose and presumably act on amorphous cellulose (Teeri, 1997). Among the different types of cellulases (e.g., endoglucanase, cellobiohydrolase, and cellobiase), only endoglucanases actively degrade CMC (Bisaria and Mishra, 1989). Our substrate specificity assays revealed that the preferred substrate for C. cohnii cell wall–bound cellulase was CMC (Figure 2E), implying that the C. cohnii cell wall–bound cellulase (recognized by the anti-dCel1p antibody) is probably an endoglucanase. Similar to C. cohnii cell wall–bound cellulase, some of the T. reesei endoglucanases (which also belong to GHF7) are able to hydrolyze glucomannan other than cellulose substrates (Biely and Tenkanen, 1998). However, further studies will be required to determine whether glucomannan is present in dinoflagellate cell walls and if the glucomannanase activity, together with cellulase activity, is involved in cell cycle progression.
Cellulose can be found in two layers of the cell wall of dinoflagellates: (1) in the continuous pellicle layer in both the thecate and athecate species; and (2) in the thecal plates among the thecate species (Morrill and Loeblich, 1983) (Figure 5B). The cellulosic thecal plates of many dinoflagellates, including C. cohnii, will be shed after the S phase (early G2) (Morrill, 1984). During the G2/M phase, the daughter cells are enclosed by the continuous parental pellicle until the division process is complete, although the broken thecal plate layer can still be partially attached in some cells. The addition of cellobiose to synchronized populations of C. cohnii cells demonstrated that dCel1p activities were required throughout the cell cycle, consistent with its expression pattern and the requirement of its activities for the pellicle layer. If dCel1p were required only to break the discontinuous thecal plate, a burst of activity at the beginning of G2 would be sufficient. However, the dCel1p expression level is lowest at early G2 (T = 6; Figure 4B). This is not consistent with the requirement of dCel1p for the detachment of the thecal plate layer. The detached nature of the thecal plate layer at the beginning of G2 would account for the ability of exogenously added recombinant dCel1p and anti-dCel1p antibodies to advance and delay cell cycle progression, respectively, only at the G2/M phase. Since the pellicle layer is continuous, any increase in cell size will require its ordered loosening throughout the cell cycle. The expression pattern of dCel1p throughout the cell cycle is consistent with dCel1p being a requirement of cell growth. Cellobiose-delayed cells are generally smaller than controls cells (Figure 4F; see Supplemental Figure 3B online), consistent with a role of dCel1p in cell growth during the G1 and G2 phases.
Cellulose is found in a wide range of different phylogenetic groups, including bacteria, slime mold, algae, plants, and animals (Delmer and Amor, 1995). It is believed that cellulose-synthesizing eukaryotes, including plants and algae, acquired the ability to synthesize cellulose (cellulose synthase) from cynaobacteria during the primary or secondary endosymbiosis (Nobles et al., 2001; Niklas, 2004). Dinoflagellates have acquired plastids from red algae through secondary or tertiary endosymbiosis (Yoon et al., 2002; Falkowski et al., 2004; Keeling et al., 2004; Saldarriaga et al., 2004). It is possible that the cellulose-synthesizing activities, and the regulation of these activities, were acquired from the ancestral plastid host.
As for cell walls in other species, plant cell walls require integration with other cellular processes (e.g., cell size control) during development. In this work, we demonstrated that cellulase activity acts to link cell wall homeostasis to cell cycle progression at the G2/M phase. Cumulatively, we propose that cellulose synthesis and cellulase activity (required for cell wall degradation), collectively defining cell wall homeostasis, are coupled to cell growth and cell cycle progression Recently, a cell size–mediated negative feedback mechanism to the cell cycle regulation engine has been reported for fission yeast (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). The cell size signal, in terms of Pom1 gradients localized at cell ends, provides a feedback mechanism for a cell to continuously monitor its size by responding to the concentration changes in the center of the cell. Cell wall proteins, which are on the cell boundary, are likely to play a role upstream of the cell size–sensing mechanism. Feedback mechanisms of cell size on cellulase expression are possibly present in the cells, all plants and other organisms containing a cell wall to perceive changes in the cell wall during cell growth and to coordinate cell wall degradation with cell cycle progression.
METHODS
Cell Culture and Cell Cycle Synchronization
Crypthecodinium cohnii Biecheler strain 1649 was obtained from the Culture Collection of Algae at the University of Texas at Austin, maintained in MLH liquid medium (Tuttle and Loeblich, 1975) and cultured at 28°C in the dark. Cell growth occurred in both G1 and G2, but mainly in G1. The duration of different cell cycle phases were as follows: G1 phase, 6 h; S phase, 1 to 1.5 h; G2/M phase, 1.5 to 2 h (Bhaud et al., 1991). Highly synchronized C. cohnii cells were obtained through colony release and filtration (Wong and Whiteley, 1996). Pyrocystis lunula, Lingulodinium polyedrum, Alexandrium tamarense, Karenia brevis, and Akashiwo sanguine (Gymnodinium sanguineum) were obtained from The Provasoli-Guillard National Center for Culture of Marine Phytoplankton and grown in f/2 medium on 12-h-light/12-h-dark cycles at 18°C.
Chemical Inhibitors and Flow Cytometric Analysis
Cellobiose (C12H22O11) is the structural unit of cellulose molecules and also the main final product of cellulose hydrolysis by both cellobiohydrolases (Beguin and Aubert, 1994) and endoglucanases (Van Tilbeurgh et al., 1989; Beguin and Aubert, 1994). For both cellobiohydrolase and endoglucanse, strong product inhibition (competitive inhibition) in cellulose hydrolysis by cellobiose has been reported based on in vitro cellulase activity assays (Hsu et al., 1980; Holtzapple et al., 1990; Gusakov and Sinitsyn, 1992; Kruus et al., 1995; Christakopoulos et al., 1999). Experimental data of FT-IR spectra, fluorescence spectrum, and circular dichroism suggested that cellobiose binds to the Trp residue of the substrate binding site and causes steric hindrance that prevents cellulose molecule chains from diffusing into the active site of cellulase (Zhao et al., 2004). Similarly, sodium hexachloropalladate (IV) tetrahydrate (Na2PdCl6.4H2O) (SHPT) has been shown to strongly inhibit cellulase activity, including endoglucanase and cellobiohydrolase, in the fungus Trichoderma reesei (Lassig et al., 1995; Shultz et al., 1995). However, the mechanism of cellulase inhibition by SHPT is still unknown. The inhibition is irreversible, which means that the cellulase activity could not be restored by the removal of the inhibitor. A 0.1 M stock was prepared by dissolving SHPT in distilled water.
Nocodazole and colchicine are microtubule-depolymerizing agents (Margolis and Wilson, 1977; Saoudi et al., 1998) and are known to cause a cell cycle delay at G2/M in dinoflagellates (Yeung et al., 2000; Kwok and Wong, 2003). Nocodazole acts by inactivating free tubulin dimers and can induce microtubule depolymerization from plus ends (Saoudi et al., 1998). Unlike nocodazole, colchicine forms stable complexes with free tubulin dimers, stabilizes the plus ends, and blocks further polymerization of the microtubule (Margolis and Wilson, 1977). Nocodazole and colchicine were first dissolved in DMSO and added to synchronized cells at T = 7 with a final concentration of 4 μg mL−1 and 0.5% (w/v), respectively.
At defined time intervals, cells were harvested by low-speed centrifugation (1000g, 10 min), fixed in 70% (v/v) ethanol (for DNA and cellulosic contents analyses), and kept at 4°C until analysis. The ethanol was replaced with PBS containing 5 μg mL−1 RNase and incubated at 37°C for 60 min. The cells were stained with 25 μg mL−1 PI and 0.1% (w/v) Calcofluor White M2R (CFW) for DNA and cellulosic content analyses, respectively (Kwok and Wong, 2003). All flow cytometric analyses were performed in a Becton-Dickinson Vantage fluorescence-activated cell sorter. All flow cytometric data, including DNA content histograms and FSC, were analyzed using the software WinMDI (version 2.8; The Scripps Research Institute) running “total” events (10,000). To estimate the percentage of G1 and G2/M cells in a sample, the specific regions corresponding to the G1 and G2/M peaks were gated on a flow cytogram using the “Marker” function available in the WinMDI software.
Preparation of the Cell Wall–Bound Protein Fraction and Determination of Total Cellulose Content in the Cell Wall Fraction
C. cohnii cells were first collected by low-speed centrifugation (1000g, 10 min) and resuspended in 25 mM MOPS buffer, pH 7.0, supplemented with 20 μg mL−1 aproptinin, 20 μg mL−1 leupetin, 20 μg mL−1 pepstain A, and 1 mM phenylmethylsulfonyl fluoride. The cells were disrupted by sonication on ice. Cell lysate was centrifuged briefly at 400g for 1 min to remove cell debris (other than the cellulosic cell wall). Various speeds (0g to 1000g) of centrifugation have been tested and the percentage recovery of cellulosic content from the lysate was estimated by Calcofluor white staining. Most of the cell debris was removed by centrifugation at 400g, with a percentage recovery of 75% cellulose content. The supernatant was recovered and constituted the soluble fraction. Cell wall proteins were then extracted with 25 mM MOPS buffer, pH 7.0, and 200 mM calcium chloride (CaCl2) under vigorous shaking at 4°C for 1 h (Goujon et al., 2003). The sample was then centrifuged at 1000g for 10 min at 4°C, and the supernatant (cell wall–bound protein fraction) was maintained. The supernatant was further salt purified and concentrated using Centricon YM-30 (Centricon Centrifugal Filter Unit with Ultracel YM-30 membrane; Mr cutoff, 30 kD) (Millipore), according to the supplier's instructions. The insoluble cell wall fractions were used to determine the total cellulose content by the Updegraff cellulose assay (Updegraff, 1969).
Cellulase Activity Staining
The Congo red CMC staining method, which provides a reliable and sensitive way for measuring endoglucanase activity (Wood, 1981; Gilkes et al., 1984), was used for locating bands of cellulase activity in polyacrylamide or agarose gels following electrophoresis (Schwarz et al., 1987). Detection of CMCase using Congo red–stained agar replicas was a sensitive assay and 6 × 10−4 units of Clostridium thermocellum cellulase could be detected in the CMC-agar (Beguin, 1983). Protein samples (100 μg per lane) were first separated on 12% polyacrylamide gel. After completion of electrophoresis, the gel was washed at room temperature in two changes of a solution containing 0.5 M sodium acetate buffer, pH 5.0, and 25% isopropanol to remove the SDS. The proteins were renatured in a 50 mM sodium acetate buffer, pH 5.0, containing 5 mM β-mercaptoethanol overnight at 4°C (Coral et al., 2002; Qin et al., 2008). Then, the separation gel was overlaid with a cellulase detection gel (1% agarose and 1% carboxymethylcellulose, ∼1 mm in thickness) (Lamed et al., 1983; Murao et al., 1988), wrapped with cling wrap, and incubated at 28°C for 5 h. For activity staining of cellulase, the gel was briefly rinsed with distilled water, stained with 1% (w/v) Congo red in water for 30 min, and exposed to gentle shaking in 1 M NaCl solution to remove excess dye (Yuan et al., 2001; Sugimura et al., 2003). Light-yellow bands or orange halos on a red background indicate breakdown of CMC and, thereby, cellulase activity (Wood and Weisz, 1987; Saul et al., 1990).
Cellulase Activity and Substrate Specificity Assays
Measurement of reducing sugars released form soluble CMC and insoluble Avicel (crystalline cellulose) is widely used for assessing endo-β-(1,4)-glucanase (or carboxymethyl cellulase) and cellobiohydrolase activity, respectively (Murao et al., 1988). CMCase activity was determined using a 1-mL assay mixture that contained 1% (w/v) CMC (Fluka) dissolved in 50 mM phosphate buffer, pH 6.9. To this mixture, an equivalent amount of protein sample was added prior to incubation at 28°C (temperature for growing C. cohnii) for 24 h. Samples were examined for the presence of reducing sugar by the dinitrosalicylic acid (DNSA) method (Miller, 1959; Coughlan, 1988). Controls were performed in an identical manner, except that the protein samples were boiled for 10 min prior to incubation. The amount of reducing sugar released was determined spectrophotometrically by measuring the absorbance of the solution at 530 nm. Absorbance readings were compared with the glucose standard curve. Each sample was assayed three times and averaged. Using 1% (w/v) Avicel 101 (crystalline cellulose) (Fluka) as substrate during incubation, Avicelase activity (Murao et al., 1988; Han et al., 1995) was detected using the DNSA method after removal of remaining Avicel by brief centrifugation.
Substrate specificity assays for cell wall–bound protein samples were performed essentially the same as for the CMCase activity and Avicelase activity assay mentioned above. Besides CMC and Avicel, the following substrates (1%, w/v) were also tested: fibrous cotton cellulose (Sigma-aldrich), Konjac glucomannan (Megazyme), xylan from oat spelled (Sigma-Aldrich), xyloglucan (Megazyme), pectin from apple (Sigma-Aldrich), and carboxymethyl-pachyman (Megazyme).
To confirm the specificity of affinity-purified anti-dCel1p antibody and cellobiose on inhibiting cellulase activity of cell wall–bound protein sample, CMCase and glucomannanase activity assays were performed in reactions containing the following: cellobiose (100 μM), cellotriose (100 μM; Seikagaku), maltose (100 μM; Sigma-Aldrich), boiled anti-dCel1p antibody (3 μg mL−1), and anti-dCel1p antibody (3 μg mL−1). Instead of using CMC as substrate, 1% (w/v) glucomanan was used for the detection of glucomannanase activity in the samples. The effects of anti-dCel1p antibody (3 μg mL−1) on recombinant dCel1p (0.1 μg mL−1) were also tested by incubating the antibody with the recombinant dCel1p for 1 h at 4°C prior to performing the cellulase activity assay.
Determination of Cello-Oligosaccharides
To test for the presence of short cello-oligosaccharides during the C. cohnii cell cycle, cell-free medium was harvested at the G2 phase (T = 8) for the cellulase activity assay. First, synchronous culture was concentrated by low-speed centrifugation (1000g, 10 min) at T = 6.5 and then resuspended in a small volume (2 mL) of low glucose-MLH medium (0.22 mM). Because MLH medium is a glucose-rich medium (22 mM glucose), low glucose medium was used to minimize the sugar background for the cellulase activity assay. At T = 8, cells were pelleted (1000g, 10 min) and discarded. The culturing medium, potentially containing the short cello-oligosaccharides, was harvested and boiled for 5 min to equilibrate the anomer configuration of the reducing sugars (Ohmiya et al., 2000). The amount of cello-oligosaccharides was determined using commercial cellulase from Humicola insolens (Sigma-aldrich). The reaction mixture contained 1 unit of cellulase and sample solution in 330 μL of 50 mM sodium phosphate buffer, pH 6.9. After incubation for 24 h at 28°C, the release of reducing sugars was detected by the DNSA method (Miller, 1959). Controls were performed in the identical manner, except that the cellulase was excluded in the reaction mixture. The amount of reducing sugar released was determined spectrophotometrically by measuring the absorbance of the solution at 530 nm. Absorbance readings were compared with the glucose standard curve. Each sample was assayed three times and averaged.
Cloning of the Dinoflagellate Cellulase and Plasmid Construction for Protein Expression
The partial nucleotide sequence of a putative dinoflagellate (P. lunula) cellulase was obtained from GenBank. Total RNA was isolated by grinding frozen dinoflagellate cells into fine powder in liquid nitrogen with a mortar and pestle. The frozen powder was resuspended in 20 volumes of Trizol reagent (Invitrogen), and total RNA was isolated according to the manufacturer's instructions. The poly(A)-containing RNA was isolated using the GenElute mRNA Miniprep kit (Sigma-Aldrich). First-strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen), according to the manufacturer's instructions. A DNA fragment encoding the N terminus of the dinoflagellate (P. lunula) cellulase was amplified by PCR using P. lunula cDNA as a template. The PCR was run as follows: 94°C for 2 min, 30 cycles of 94°C for 1 min, 67°C for 1.5 min, 72°C for 2 min, and 72°C for 10 min. The sense primer (Cello_pQE1-F3) consisted of the sequence 5′-ATAGCATGCGAGACCCCATCCAGTGCGCCA-3′. The antisense primer (Cello_pQE1-R3) consisted of the sequence 5′-TAAAGCTTGCCCGTGCCGTACGTGGCACC-3′. The PCR product was digested with restriction enzymes HindIII and SphI and cloned into a HindIII/SphI-predigested protein expression vector, pQE-1 (Qiagen). Using CircLigase single-stranded DNA ligase (Epicentre Biotechnologies), self-ligated single-stranded-cDNA was obtained (see Supplemental Figure 2A online) (Polidoros et al., 2006). By performing inverse PCR with primers Cello-Inv A (5′-TTGGATCCATTCCCGTCCAGATTCAC-3′) and Cello-Inv B (5′-GGTGTCTTCGTCGAGATGG-3′), we cloned the full-length cDNA of P. lunula cellulase. The inverse PCR was run as follows: 94°C for 2 min, 30 cycles of 94°C for 1 min, 52°C for 1.5 min, 72°C for 2 min, and 72°C for 10 min. DNA sequences and orientation of inserts were confirmed using the ABI Prism BigDye Terminator v3.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems). The full-length cDNA sequence of the P. lunula cellulase, dCel1, was deposited in GenBank (accession number GQ258704).
The nucleotide sequences of another putative dinoflagellate (L. polyedrum) cellulase were obtained from GenBank. By performing PCR with primers (5′-ATGACGATGCTGCTTACCAGC-3′) and (5′-AGTTGACCGCTACAACGAACG-3′), full-length cDNA of L. polyedrum cellulase, dCel2, was cloned (see Supplemental Figure 2B online).
For investigating the roles of dCel1p in the cell cycle, full-length dCel1 was cloned into the pQE-1 vector (excluding the putative signal peptide, the first 24 amino acids, because signal peptide might not be recognized by Escherichia coli). The plasmid was introduced into E. coli SG13009, and overexpression of His-tagged recombinant protein was induced by 1 mM isopropyl-β-d-thiogalactopyranoside. Recombinant dCel1p was purified from bacteria using a Ni-NTA column under native conditions as described in the manufacturer's manual (QIAexpressionist; Qiagen).
Bioinformatic Analyses
Searches for sequence homology were conducted using BLAST (Altschul et al., 1990) at NCBI. Possible ORFs were identified with the ORF finder at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/projects/gorf/). SignalP 3.0 Server (http://www.cbs.dtu.dk /services/SignalP/) was used to predict the presence of a signal peptide. Conserved domains in the deduced amino acid sequence were analyzed using Conserved Domain Server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Conserved Domain Architecture Retrieval Tool (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi) at NCBI. Determination of the predicted isoelectric point (pI) was performed using the Expasy website (http://www.expasy.ch/tools/pi_tool.html). Potential glycosylation sites were predicted using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk-/services/NetNGlyc/).
Amino acid sequences of dinoflagellate cellulases (dCel1 and dCel12) and some GHF7 and GHF9 members were used for phylogenetic analyses. An α-amylase from the cockroach (Blattella germanica) was used as the outgroup. Multiple alignments were performed using ClustalX 2.0 (NCBI) (Thompson et al., 1997; Larkin et al., 2007) under default settings (gaps and matches were equally weighted). Alignments were further adjusted by eye to minimize the effects of insertion/deletion events on the analysis. A neighbor-joining tree was constructed using MEGA4 (Tamura et al., 2007) with a 1000-replicate bootstrap analysis. A maximum parsimony consensus tree was also constructed using the same procedure to confirm the topology of the inferred tree.
Dinoflagellate Protein Extraction for SDS-PAGE
Cells were harvested by centrifugation at 1000g for 10 min. The cell pellet was resuspended in lysis buffer (50 mM Tris, pH 7.4, 1% [w/v] SDS, 5 mM EDTA, 10 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 20 μg mL−1 aproptinin, 20 μg mL−1 leupetin, and 20 μg mL−1 pepstain A). Cells were disrupted using a high-pressure cell disruption machine (Constant Systems) at 30 k.p.s.i. Insoluble substances were removed by centrifuging at 10,000g for 10 min. The supernatant fraction was used for SDS-PAGE analysis and immunoblotting. Gel electrophoresis was performed according to Laemmi (1970). Protein concentration was determined using the DC protein assay kit (Bio-Rad) with BSA as standard. Protein bands were visualized by Coomassie Brilliant Blue and silver staining (Berkelman and Stenstedt, 1998).
Preparation of Anti-dCel1p Antibody and Immunoblot Analysis
Production and purification of recombinant dCel1p from E. coli was performed under denaturing conditions according to the manufacturer's manual (QIAexpressionist; Qiagen). The purified protein was dialyzed against PBS and used to immunize rabbit (Harlow and Lane, 1988). The antibody was affinity purified using antigen on a blot as described by Rybicki et al. (1990). Gel electrophoresis and immunoblot analysis were performed according to Laemmi (1970) and the manufacturer's manual (ECL protein gel blotting protocols; Amersham). Whole-cell lysates were prepared from cells harvested at specific time points. Eighty micrograms of protein from the lysates was resolved by SDS-PAGE and probed with affinity-purified rabbit polyclonal anti-dCel1p antibody (1:1000 dilution). Horseradish peroxidase–conjugated anti-rabbit IgG (Zymax) was used as secondary antibody (1:1000 dilution). Protein loading was verified by stripping the blot and reprobing with mouse monoclonal anti-α-tubulin (1:3000 dilution) (Sigma-Aldrich) and horseradish peroxidase–conjugated anti-mouse IgG (1:5000 dilution). Stripping of the membrane was performed as described by Gallagher et al. (2008). Labeled bands were detected with the ECL chemiluminescence system (Amersham) according to the manufacturer's manual. Band intensities were determined using ImageJ software (NIH) (Sheffield, 2008).
Immunostaining
Cells were fixed with 2% (w/v) paraformaldehyde overnight at 4°C. The control was cells incubated with Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen; A11008) only. After fixation, cells were washed three times with PBS and incubated with anti-dCel1p antibody (1:100 dilution) overnight at 4°C. Cells were washed three times with PBS before incubating with Alexa Fluor 488 goat anti-rabbit IgG (1:1000 dilution) at room temperature for 1 h. For confocal microscopy, all images were obtained with a Nikon C1 confocal microscope system and EZ-C1 software (Nikon).
Immunoprecipitation
Proteins were extracted from C. cohnii cells as mentioned above. Immunoprecipitation was performed as described by Bonifacino and Dell'Angelica (1998). For the preparation of antibody-conjugated beads, ∼1 μg of affinity-purified antibody was added to 100 μL of 50% (v/v) protein A-sepharose bead slurry in ice-cold PBS. The mixture was then incubated at 4°C in a tube rotator for 1 h (Bonifacino and Dell'Angelica, 1998).
Statistical Analysis
The Student's t test was used to analyze data, and the level of significance was set at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: partial nucleotide sequence of a putative cellulase from dinoflagellate L. polyedrum (BU582415), dCel1 from dinoflagellate P. lunula (GQ258704), dCel2 from dinoflagellate L. polyedrum (GQ258705), cellobiohydrolase I from Polyporus arcularius (BAF80326), cellulase from Irpex lacteus (BAA76365), endo-1,4-β-glucanase from Neosartorya fischeri (XP_001262790), endoglucanase I from Trichoderma reesei (AAA34212), α-amylase from Blattella germanica (AY945930), and EST sequences used for assembling the dCel2 contig (BP743767, CD810733, CD810109, BP743542, CD810238, BP742910, CD809782, CD810792, CD809750, BP744088, CD809923, CD809734, BP743695, CD809732, BP743622, BP742342, and BP743519).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effects of Small Oligosaccharides on Cell Cycle Progression.
Supplemental Figure 2. Cloning of Dinoflagellate Cellulases.
Supplemental Figure 3. Effects of Cellobiose on G1 Cell Size and dCel1p Expression.
Supplemental Figure 4. Control Experiment for Immunolocalization of dCel1p.
Supplemental Figure 5. Expression of Recombinant dCel1p.
Supplemental Figure 6. Effects of Exogenous dCel1p and Purified anti-dCel1p Antibody on G1 Progression.
Supplemental Data Set 1. Amino Acid Sequences Used to Generate the Phylogeny Presented in Figure 3B.
Acknowledgments
This work was partly supported by a The Competitive Earmarked Research Grants (HKUST662707) from the Research Grant Council of Hong Kong to J.T.Y.W. A.C.M.K. was partly supported by a Hong Kong University of Science and Technology Postdoctoral Fellowship Matching Fund during the course of the study. We thank Michael Bennett for his help with grammar and revision of the manuscript.
References
- Adams D.J. (2004). Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Baladron V., Ufano S., Duenas E., Martin-Cuadrado A.B., del Rey F., Vazquez de Aldana C.R. (2002). Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1: 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras D.R., Stone B.A. (1969). Beta-1,3-glucan hydrolases from Euglena gracilis. II. Purification and properties of the beta-1,3-glucan exo-hydrolase. Biochim. Biophys. Acta 191: 342–353 [DOI] [PubMed] [Google Scholar]
- Beguin P. (1983). Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal. Biochem. 131: 333–336 [DOI] [PubMed] [Google Scholar]
- Beguin P., Aubert J.P. (1994). The biological degradation of cellulose. FEMS Microbiol. Rev. 13: 25–58 [DOI] [PubMed] [Google Scholar]
- Berkelman T., Stenstedt T. (1998). 2-D Electrophoresis Using Immoblized pH Gradients: Principles and Methods. (Sweden: Amersham Biosciences; ). [Google Scholar]
- Bhaud Y., Barbier M., Soyer-Gobillard M.O. (1994). A detailed study of the complex cell cycle of the dinoflagellate Crypthecodinium cohnii Biecheler and evidence for variation in histone H1 kinase activity. J. Eukaryot. Microbiol. 41: 519–526 [Google Scholar]
- Bhaud Y., Salmon J.M., Soyer-Gobillard M.O. (1991). The complex cell cycle of the dinoflagellate protoctist Crypthecodinium cohnii as studied in vivo and by cytofluorimetry. J. Cell Sci. 100: 675–682 [Google Scholar]
- Biely P., Tenkanen M. (1998). Enzymology of hemicellulose degradation. Trichoderma and Gliocladium, Harman G.E., Kubicek C., (London: Taylor and Francis; ), pp. 25–47 [Google Scholar]
- Bisaria V.S., Mishra S.J. (1989). Regulatory aspects of cellulase biosynthesis and secretion. Crit. Rev. Biotechnol. 9: 61–103 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Dell'Angelica E.C. (1998). Immunoprecipitation. In Current Protocols in Cell Biology, Bonifacino J.S., Dasso M., Harford J.B., Lippincott-Schwartz J., Yamada K.M., (New York: John Wiley & Sons; ), pp. 7.2–7.2.21 [Google Scholar]
- Cabib E., Roh D.H., Schmidt M., Crotti L.B., Varma A. (2001). The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276: 19679–19682 [DOI] [PubMed] [Google Scholar]
- Carpita N., McCann M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, Buchanan B., Gruissem W., Jones R., (Rockville, MD: American Society of Plants Physiologists; ), pp. 52–108 [Google Scholar]
- Carpita N.C., Gibeaut D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Christakopoulos P., Hatzinikolaou D.G., Fountoukidis G., Kekos D., Claeyssens M., Macris B.J. (1999). Purification and mode of action of an alkali-resistant endo-1, 4-beta-glucanase from Bacillus pumilus. Arch. Biochem. Biophys. 364: 61–66 [DOI] [PubMed] [Google Scholar]
- Cid V.J., Duran A., del Rey F., Snyder M.P., Nombela C., Sanchez M. (1995). Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59: 345–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coral G., Arikan B., Unaldi M.N., Guvenmez H. (2002). Some properties of crude carboxymethyl cellulase of Aspergillus niger Z10 wild-type strain. Turk. J. Biol. 26: 209–213 [Google Scholar]
- Cosgrove D.J. (1998). Cell wall loosening by expansins. Plant Physiol. 118: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 391–417 [DOI] [PubMed] [Google Scholar]
- Coughlan M.P. (1988). Staining techniques for the detection of the individual components of cellulolytic enzyme systems. Methods in Enzymology, Wood W.A., Kellogg S.T., (New York: Academic Press; ), pp. 135–144 [Google Scholar]
- del Campillo E. (1999). Multiple endo-1,4-beta-D-glucanase (cellulase) genes in Arabidopsis. Curr. Top. Dev. Biol. 46: 39–61 [DOI] [PubMed] [Google Scholar]
- del Campillo E., Bennett A.B. (1996). Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 111: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer D.P., Amor Y. (1995). Cellulose biosynthesis. Plant Cell 7: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J. (1996). Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Falkowski P.G., Katz M.E., Knoll A.H., Quigg A., Raven J.A., Schofield O., Taylor F.J. (2004). The evolution of modern eukaryotic phytoplankton. Science 305: 354–360 [DOI] [PubMed] [Google Scholar]
- Firon A., Lesage G., Bussey H. (2004). Integrative studies put cell wall synthesis on the yeast functional map. Curr. Opin. Microbiol. 7: 617–623 [DOI] [PubMed] [Google Scholar]
- Fischer R.L., Bennett A.B. (1991). Role of cell wall hydrolased in fruit ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 675–703 [Google Scholar]
- Gallagher S., Winston S.E., Fuller S.A., Hurrell J.G. (2008). Immunoblotting and immunodetection. Curr. Protoc. Immunol. 8: 10. [DOI] [PubMed] [Google Scholar]
- Gilkes N.R., Langsford M.L., Kilburn D.G., Miller R.C., Jr., Warren R.A. (1984). Mode of action and substrate specificities of cellulases from cloned bacterial genes. J. Biol. Chem. 259: 10455–10459 [PubMed] [Google Scholar]
- Goujon T., Minic Z., El Amrani A., Lerouxel O., Aletti E., Lapierre C., Joseleau J.P., Jouanin L. (2003). AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J. 33: 677–690 [DOI] [PubMed] [Google Scholar]
- Gusakov A.V., Sinitsyn A.P. (1992). A theoretical analysis of cellulase product inhibition: Effect of cellulase binding constant, enzyme/substrate ratio, and beta-glucosidase activity on the inhibition pattern. Biotechnol. Bioeng. 40: 663–671 [DOI] [PubMed] [Google Scholar]
- Han S.J., Yoo Y.J., Kang H.S. (1995). Characterization of a bifunctional cellulase and its structural gene. The cell gene of Bacillus sp. D04 has exo- and endoglucanase activity. J. Biol. Chem. 270: 26012–26019 [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. (New York: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Hartwell L.H., Weinert T.A. (1989). Checkpoints: Controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Holtzapple M., Cognata M., Shu Y., Hendrickson C. (1990). Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol. Bioeng. 36: 275–287 [DOI] [PubMed] [Google Scholar]
- Hoson T. (1993). Regulation of polysaccharide breakdown during auxin-induced cell wall loosening. J. Plant Res. 103: 369–381 [Google Scholar]
- Hoyt M.A. (2004). A new checkpoint takes shape. Nat. Cell Biol. 6: 801–803 [DOI] [PubMed] [Google Scholar]
- Hsu T.-A., Gong C.-S., Tsao G.T. (1980). Kinetic studies of cellodextrins hydrolysis by exocellulase from Trichoderma reesei. Biotechnol. Bioeng. 22: 2305–2320 [Google Scholar]
- Ilmen M., Saloheimo A., Onnela M.L., Penttila M.E. (1997). Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J., Archibald J.M., Fast N.M., Palmer J.D. (2004). Comment on “The evolution of modern eukaryotic phytoplankton.” Science 306: 2191. [DOI] [PubMed] [Google Scholar]
- Kemmerer E.C., Tucker M.L. (1994). Comparative study of cellulases associated with adventitious root initiation, apical buds, and leaf, flower, and pod abscission zones in soybean. Plant Physiol. 104: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruus K., Andreacchi A., Wang W.K., Wu J.H. (1995). Product inhibition of the recombinant CelS, an exoglucanase component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 44: 399–404 [DOI] [PubMed] [Google Scholar]
- Kuranda M.J., Robbins P.W. (1991). Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266: 19758–19767 [PubMed] [Google Scholar]
- Kwok A.C.M., Wong J.T.Y. (2003). Cellulose synthesis is coupled to cell cycle progression at G1 in the dinoflagellate Crypthecodinium cohnii. Plant Physiol. 131: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmi U.K. (1970). Clevage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lamed R., Setter E., Bayer E.A. (1983). Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156: 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.R., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lassig J.P., Shultz M.D., Gooch M.G., Evans B.R., Woodward J. (1995). Inhibition of cellobiohydrolase I from Trichoderma reesei by palladium. Arch. Biochem. Biophys. 322: 119–126 [DOI] [PubMed] [Google Scholar]
- Leander B.S., Keeling P.J. (2004). Early evolutionary history of dinoflagellates and apicomplexans (Alveolata) as inferred from hsp90 abd actin phylogenies. J. Phycol. 40: 341–350 [Google Scholar]
- Levin D.E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Reed S.I. (1995). A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129: 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeblich A.R., III (1970). The amphiesma or dinoflagellate cell covering. In Proceedings of the North American Paleontology Convention, Chicago 1969, Yochelson E.L., ed (Lawrence, KS: Allen Press; ), pp. 867–929 [Google Scholar]
- Macey M.G. (2007). Principles of flow cytometry. Flow Cytometry: Principles and Applications, Macey M.G., (Totowa, NJ: Humana Press; ), pp. 1–15 [Google Scholar]
- Margolis R.L., Wilson L. (1977). Addition of colchicine–tubulin complex to microtubule ends: The mechanism of substoichiometric colchicine poisoning. Proc. Natl. Acad. Sci. USA 74: 3466–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Berthelot-Grosjean M. (2009). Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 459: 852–856 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S.J., Cosgrove D.J. (1995). Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G.L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31: 426–428 [Google Scholar]
- Molhoj M., Pagant S., Hofte H. (2002). Towards understanding the role of membrane-bound endo-beta-1,4-glucanases in cellulose biosynthesis. Plant Cell Physiol. 43: 1399–1406 [DOI] [PubMed] [Google Scholar]
- Morrill L.C. (1984). Ecdysis and the location of the plasma membrane in the dinoflagellate Heterocapsa niei. Protoplasma 119: 8–20 [Google Scholar]
- Morrill L.C., Loeblich A.R., III (1983). Ultrastructure of the dinoflagellate amphiesma. Int. Rev. Cytol. 82: 151–180 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Mayeux A., Paoletti A., Nurse P. (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459: 857–860 [DOI] [PubMed] [Google Scholar]
- Murao S., Sakamoto R., Arai M. (1988). Cellulases of Aspergillus aculeatus. Methods in Enzymology, Wood W.A., Kellogg S.T., (New York: Academic Press; ), pp. 274–299 [Google Scholar]
- Nicol F., His I., Jauneau A., Vernhettes S., Canut H., Hofte H. (1998). A plasma membrane-bound putative endo-1,4-b-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas K.J. (2004). The cell walls that bind the tree of life. Bioscience 54: 831–841 [Google Scholar]
- Nobles D.R., Romanovicz D.K., Brown R.M., Jr (2001). Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 127: 529–542 [PMC free article] [PubMed] [Google Scholar]
- Ohmiya Y., Nakai T., Park Y.W., Aoyama T., Oka A., Sakai F., Hayashi T. (2003). The role of PopCel1 and PopCel2 in poplar leaf growth and cellulose biosynthesis. Plant J. 33: 1087–1097 [DOI] [PubMed] [Google Scholar]
- Ohmiya Y., Samejima M., Shiroishi M., Amano Y., Kanda T., Sakai F., Hayashi T. (2000). Evidence that endo-1,4-beta-glucanases act on cellulose in suspension-cultured poplar cells. Plant J. 24: 147–158 [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Nagase M., Ichiyanagi T., Kitamoto Y., Aimi T. (2007). Transcriptional regulation of two cellobiohydrolase encoding genes (cel1 and cel2) from the wood-degrading basidiomycete Polyporus arcularius. Appl. Microbiol. Biotechnol. 76: 1069–1078 [DOI] [PubMed] [Google Scholar]
- Okamoto O.K., Hastings J.W. (2003). Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 321: 73–81 [DOI] [PubMed] [Google Scholar]
- Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J. (2005). The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 3: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.W., Tominaga R., Sugiyama J., Furuta Y., Tanimoto E., Samejima M., Sakai F., Hayashi T. (2003). Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J. 33: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Peng L., Kawagoe Y., Hogan P., Delmer D. (2002). Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150 [DOI] [PubMed] [Google Scholar]
- Polidoros A.N., Pasentsis K., Tsaftaris A.S. (2006). Rolling circle amplification-RACE: a method for simultaneous isolation of 5′ and 3′ cDNA ends from amplified cDNA templates. Biotechniques 41: 35–36, 38, 40 passim [DOI] [PubMed] [Google Scholar]
- Qin Y., Wei X., Liu X., Wang T., Qu Y. (2008). Purification and characterization of recombinant endoglucanase of Trichoderma reesei expressed in Saccharomyces cerevisiae with higher glycosylation and stability. Protein Expr. Purif. 58: 162–167 [DOI] [PubMed] [Google Scholar]
- Robert S., Bichet A., Grandjean O., Kierzkowski D., Satiat-Jeunemaitre B., Pelletier S., Hauser M.T., Hofte H., Vernhettes S. (2005). An Arabidopsis endo-1,4-beta-D-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17: 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki E.P., von Wechmar M.B., Burger J.T. (1990). Monospecific antibody preparation for use in the detection of viruses. World Perspectives on Barley Yellow Dwarf, Burnett P.A., (Mexico: CIMMYT; ), pp. 149–153 [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Saldarriaga J.F., Taylor F.J.R., Cavalier-Smith T., Menden-Deuer S., Keeling P.J. (2004). Molecular data and the evolutionary history of dinoflagellates. Eur. J. Protistol. 40: 85–111 [Google Scholar]
- Saoudi Y., Fotedar R., Abrieu A., Doree M., Wehland J., Margolis R.L., Job D. (1998). Stepwise reconstitution of interphase microtubule dynamics in permeabilized cells and comparison to dynamic mechanisms in intact cells. J. Cell Biol. 142: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul D.J., Williams L.C., Grayling R.A., Chamley L.W., Love D.R., Bergquist P.L. (1990). celB, a gene coding for a bifunctional cellulase from the extreme thermophile “Caldocellum saccharolyticum”. Appl. Environ. Microbiol. 56: 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz W.H., Bronnenmeier K., Grabnitz F., Staudenbauer W.L. (1987). Activity staining of cellulases in polyacrylamide gels containing mixed linkage beta-glucans. Anal. Biochem. 164: 72–77 [DOI] [PubMed] [Google Scholar]
- Sheffield J. (2008). An introduction to ImageJ: A useful tool for biological image processing and analysis. Microsc. Microanal. 14 (suppl. 2): 898–899 [Google Scholar]
- Shultz M.D., Lassig J.P., Gooch M.G., Evans B.R., Woodward J. (1995). Palladium–A new inhibitor of cellulase activities. Biochem. Biophys. Res. Commun. 209: 1046–1052 [DOI] [PubMed] [Google Scholar]
- Smits G.J., van den Ende H., Klis F.M. (2001). Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147: 781–794 [DOI] [PubMed] [Google Scholar]
- Sugimura M., Watanabe H., Lo N., Saito H. (2003). Purification, characterization, cDNA cloning and nucleotide sequencing of a cellulase from the yellow-spotted longicorn beetle, Psacothea hilaris. Eur. J. Biochem. 270: 3455–3460 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Igarashi R., Sekiya M., Utsugi T., Morishita S., Yukawa M., Ohya Y. (2004). Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6: 861–871 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Teeri T. (1997). Crystalline cellulose degradation: New insight into the function of cellobiohydrolases. Trends Biotechnol. 15: 160–167 [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.L., Sexton R., del Campliio E., Lewis L.N. (1988). Bean abscission cellulase. Plant Physiol. 88: 1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle R.C., Loeblich A.R., III (1975). An optimal growth medium for the dinoflagellate Crypthecodinium cohnii. Phycologia 14: 1–8 [Google Scholar]
- Updegraff D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Van Tilbeurgh H., Loontiens F.G., Engelborgs Y., Claeyssens M. (1989). Studies of the cellulolytic system of Trichoderma reesei QM 9414. Binding of small ligands to the 1,4-beta-glucan cellobiohydrolase II and influence of glucose on their affinity. Eur. J. Biochem. 184: 553–559 [DOI] [PubMed] [Google Scholar]
- Wong J.T.Y., Whiteley A. (1996). An improved method for the cell cycle synchronization of the heterotrophic dinoflagellate Crypthecodinium cohnii. J. Exp. Mar. Biol. Ecol. 197: 91–99 [Google Scholar]
- Wood P.J. (1981). Specificity in the interaction of direct dyes with polysaccharides. Carbohydr. Res. 94: c19–c23 [Google Scholar]
- Wood P.J., Weisz J. (1987). Detection and assay of (1-4)-beta-D-glucanase, (1-3)-beta-D-glucanase, (1-3)(1-4)-beta-D-glucanase, and xylanase based on complex formation of substrate with Congo Red. Cereal Chem. 64: 8–15 [Google Scholar]
- Yeung P.K.K., New D.C., Leveson A., Yam C.H., Poon R.Y.C., Wong J.T.Y. (2000). The spindle checkpoint in the dinoflagellate Crypthecodinium cohnii. Exp. Cell Res. 254: 120–129 [DOI] [PubMed] [Google Scholar]
- Yoon H.S., Hackett J.D., Bhattacharya D. (2002). A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. USA 99: 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Wu Y., Cosgrove D.J. (2001). A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 127: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Smith J.A., Oi F.M., Koehler P.G., Bennett G.W., Scharf M.E. (2007). Correlation of cellulase gene expression and cellulolytic activity throughout the gut of the termite Reticulitermes flavipes. Gene 395: 29–39 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wu B., Yan B.X., Gao P.J. (2004). Mechanism of cellobiose inhibition in cellulose hydrolysis by cellobiohydrolase. Sci China C Life Sci 47: 18–24 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Nishizawa N., Wu Y., Kost B., Chua N.H. (2000). KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]