Using an activation-tagging genetic screen, this work identified a basic helix-loop-helix–containing protein, named TCP1, as a positive regulator of the transcription of a key brassinosteroid biosynthesis enzyme DWARF4.
Abstract
Brassinosteroids (BRs) are essential phytohormones regulating normal plant growth and development. TCP1, a gene thought to be involved in floral organ symmetric control, was identified as a genetic suppressor of a weak BR receptor mutant, bri1-5, in an activation-tagging genetic screen. TCP1 encodes a putative transcription factor possessing a basic helix-loop-helix domain. The dominant allele of TCP1, tcp1-1D, suppresses the defective phenotypes of bri1-5. Overexpression of a dominant-negative form of TCP1, TCP1-SRDX, with a 12–amino acid repressor sequence fused to TCP1 at its C terminus, results in dwarfed plants resembling BR-deficient or insensitive mutants. The defective phenotypes can be rescued by exogenously applied brassinolide but cannot be recovered by auxins, gibberellins, or cytokinins. BR profile assay (quantitative analysis of BR biosynthetic intermediates) strongly suggests that TCP1 expression level positively coordinates with the function of DWARF4 (DWF4), a key enzyme in BR biosynthesis. Real-time RT-PCR analysis further demonstrated that TCP1 regulates the transcription levels of DWF4, and chromatin immunoprecipitation experiments showed that TCP1 indeed interacts with the DWF4 promoter. Confocal microscopy indicated that TCP1 is mainly confined to the nucleus. The expression of TCP1 appears to be regulated by BR levels. These studies demonstrate another level of regulation through which BRs mediate plant growth and development.
INTRODUCTION
Brassinosteroids (BRs) are a class of polyhydroxyl steroidal hormones naturally found in almost all plant species examined (Clouse and Sasse, 1998). BRs play critical roles in multiple physiological processes during normal plant growth and development, from seed germination to leaf senescence. Mutant plants unable to biosynthesize or perceive BRs exhibit typical phenotypes, including extremely dwarfed statures, shortened leaf petioles, rounded and curled leaves, prolonged life spans, reduced male fertility, and deetiolated hypocotyls when grown in darkness. Within the last two decades, detailed information regarding BR signal transduction and BR biosynthesis has been uncovered (Kim and Wang, 2010). A number of BR signaling regulators, such as the cell surface BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) (Li and Chory, 1997) and its coreceptor BRI1-ASSOCIATED PROTEIN KINASE1 (BAK1), were identified via genetic and biochemical methods (Li et al., 2002; Nam and Li, 2002). Other important regulatory proteins, such as a secreted Ser carboxypeptidase designated as bri1 SUPPRESSOR1 (Li et al., 2001a), a BRI1 inhibitory protein BKI1 (Wang and Chory, 2006), several putative BRI1 substrates, including an Arabidopsis thaliana paralog of TGF-BETA RECEPTOR-INTERACTING PROTEIN-1 (Jiang and Clouse, 2001; Ehsan et al., 2005), a TRANSTHYRETIN-LIKE protein (Nam and Li, 2004), and three homologous BR SIGNALING KINASES (Tang et al., 2008), a negative regulator called BRASSINOSTEROID INSENSITIVE2 (Li et al., 2001b; Li and Nam, 2002), a protein phosphatase (BSU1) (Mora-Garcia et al., 2004), 14-3-3 proteins (Bai et al., 2007; Gampala et al., 2007; Ryu et al., 2007), and two novel transcription factors, BZR1 (Wang et al., 2002) and BES1 (Yin et al., 2002), have also been identified using various approaches. Although a more detailed mechanistic understanding as to how the aforementioned proteins coordinate various steps in BR signaling is needed, evidence to date strongly indicates that they are key signaling components in BR signal transduction that relay information from the cell surface to nuclear transcription factors. A proposed pathway for BR signal transduction starts with the ligand (BR) binding to the extracellular domain of BRI1, which triggers a sequential phosphorylation between BRI1 and BAK1 (Wang et al., 2008). Activated receptor/coreceptor complex initiates a phosphorylation/dephosphorylation cascade that can transduce the BR signal from the cell surface to the cytoplasm and eventually to the nucleus where gene expression patterns are altered through the action of two transcription factors BZR1 and BES1 (Kim et al., 2009). As a consequence of these events, the plant is able to fine-tune its growth and development.
The entire BR biosynthetic pathway was initially elucidated using cultured Catharanthus roseus cells (Fujioka and Yokota, 2003). Several genes encoding key BR biosynthetic enzymes have also been cloned using BR-deficient mutants identified from a number of plant species, such as Arabidopsis, pea (Pisum sativum), tomato (Solanum lycopersicum), and rice (Orzya sativa; Fujioka and Yokota, 2003). For example, de-etiolated2 (det2) was identified as a deetiolated mutant from Arabidopsis (Chory et al., 1991). DET2 encodes a protein sharing sequence similarity with the mammalian steroid 5α-reductase (Li et al., 1997). Feeding experiments revealed that DET2 is involved in a 5α-reduction step of multiple related sterols during BR biosynthesis (Fujioka et al., 1997). An ortholog of Arabidopsis det2, named lk, was also identified from pea as an extremely dwarfed mutant (Nomura et al., 2004). dwarf4 (dwf4) is another BR-deficient mutant isolated in Arabidopsis (Choe et al., 1998). The dwarfed stature of dwf4 can be rescued by brassinolide (BL), the final product of the BR biosynthetic pathway and the most active form of BRs. DWF4 encodes a 22-hydroxylase and is responsible for multiple 22-hydroxylation steps during BR biosynthesis. It was proposed that DWF4 catalyzes a rate-limiting step during BR biosynthesis (Kim et al., 2006). constitutive photomorphogenesis and dwarfism (cpd) is another dwarf mutant isolated by T-DNA insertion analysis. It was shown that CPD encodes a 23α-hydroxylase and participates in a critical 23α-hydroxylation step in BR biosynthesis (Szekeres et al., 1996). However, recent feeding and biochemical analyses indicated that two P450 proteins, CYP90C1 and CYP90D1, act as true 23α-hydroxylases (Ohnishi et al., 2006). The severe phenotype of the cpd mutant indicated that it should be involved in a step earlier than the 23α-hydroxylation reaction. Another gene involved in BR biosynthesis is BR6ox, which was first identified in tomato by transposon tagging (Bishop et al., 1996). BR6ox catalyzed the C-6 oxidation of a number of different 6-deoxoBRs (Bishop et al., 1999). BR6ox orthologs from Arabidopsis and rice have conserved functions, which are responsible for linking the early and late C-6 oxidation pathways (Bishop et al., 2006).
Unlike other phytohormones, BRs are unable to be transported through a long distance mechanism in a plant (Symons and Reid, 2004). This suggests that homeostasis of bioactive BRs must be precisely controlled at tissue or even at cellular levels to ensure normal growth and development. There are a number of mechanisms for a plant to maintain adequate levels of bioactive BRs. For instance, excessive amounts of BRs can be inactivated by modifications of BRs (Fujioka and Yokota, 2003). Mechanisms for the inactivation of BRs include sulfonation at a 22-OH group by a steroid sulfotransferase named BNST3, identified in Brassica napus (Rouleau et al., 1999); 26-hydroxylation by BAS1, found in Arabidopsis (Neff et al., 1999); conjugation by a UDP-glycosyltransferase named UGT73C5 (Poppenberger et al., 2005); and a putative reduction step catalyzed by BEN1 (Yuan et al., 2007). Plants also use a feedback mechanism to monitor the BR biosynthetic rate, which is tightly linked with the BR signaling pathway (Mathur et al., 1998; He et al., 2005; Kim et al., 2006). If BRs are available, they can trigger a series of cellular processes, resulting in the accumulation of unphosphorylated BZR1 and BES1 in nuclei. Unphosphorylated BZR1 and BES1 have dual roles: repressing biosynthetic gene expression to slow down the biosynthetic rate and activating BR response genes to promote growth. How BR biosynthesis is positively regulated, however, is still poorly understood. In this article, we describe the identification of a transcription factor, which plays a positive role in regulating BR biosynthesis.
Using a gain-of-function genetic approach, we identified a number of suppressors for a weak BRI1 mutant allele, bri1-5 (Noguchi et al., 1999; Li et al., 2001a, 2002; Zhou et al., 2004; Yuan et al., 2007). One of these suppressors is tcp1-1D. TCP1 encodes a TCP transcription factor, which contains a basic helix-loop-helix (bHLH) domain. Previous studies suggested that TCP1 may play a role in regulating flower organ symmetry (Cubas et al., 2001; Busch and Zachgo, 2007). The activation-tagged locus, tcp1-1D, can suppress the defective phenotypes of bri1-5. Overexpression of a dominant-negative mutant TCP1-SRDX in wild-type plants, conversely, resulted in dwarfed transgenic plants similar to typical BR deficient mutants such as det2 (Li et al., 1996) or signaling defective mutants such as bri1-5 (Noguchi et al., 1999). Our detailed genetic, biochemical, and molecular analyses demonstrated that TCP1 positively regulates the expression of the key BR biosynthetic gene, DWF4, via a direct or indirect interaction with the promoter region of DWF4. Thus, our findings provide a molecular pathway in the regulation of BR biosynthesis under certain endogenous and external stimuli. The discoveries will significantly advance our knowledge about the functions of BR in regulating normal plant growth, development, and adaptation to various environments.
RESULTS
tcp1-1D Was Identified as a Gain-of-Function Genetic Suppressor of bri1-5
Activation tagging is a gain-of-function genetic approach that activates gene expression via inserting strong enhancers in the genome. The enhancers are commonly engineered in the T-DNA region of the transformation construct and inserted arbitrarily in the genome by Agrobacterium tumefaciens–mediated gene transformation (Clough and Bent, 1998). Usually only the genes in the vicinity of the enhancers can be transcriptionally activated (Weigel et al., 2000). This investigation takes advantage of bri1-5, a weak BR receptor mutant, in which a single Cys has been substituted by a Tyr at the N terminus (C69Y) (Noguchi et al., 1999). The bri1-5 protein is largely retained in endoplasmic reticulum and degraded through a proteosome-mediated degradation pathway (Hong et al., 2008). bri1-5 plants show a semidwarfed but fertile phenotype. The activation tagging–based bri1-5 genetic modifier screen has become an effective approach to identify components regulating BR signaling, catabolism, and biosynthetic pathways (Li et al., 2001a, 2002; Zhou et al., 2004; Yuan et al., 2007). One of the bri1-5 suppressors we identified by this method is called tcp1-1D.
The bri1-5 tcp1-1D double mutant shows partially suppressed phenotypes compared with the bri1-5 single mutant (Figure 1A). A bri1-5 mutant exhibits characteristic BR mutant phenotypes, such as rounded leaves, shortened petioles, and delayed flowering time. Although the leaf shapes of the double mutant plants remain unaltered, the petioles of bri1-5 tcp1-1D are significantly elongated. The inflorescences of the double mutant plants are twice as tall as those of bri1-5 plants at maturity. In addition, the delayed flowering time of bri1-5 was also significantly suppressed. Genetic analysis indicated that the double mutant phenotype was caused by a single dominant locus because the bri1-5 suppression phenotype is closely linked with the basta resistance gene from the T-DNA of the activation-tagging construct, pBASTA-AT2 (Yuan et al., 2007).
Figure 1.
tcp1-1D Was Identified as a Dominant Genetic Suppressor of bri1-5 by an Activation-Tagging Screen.
(A) Phenotypes of 16-d-old WS2, bri1-5, bri1-5 tcp1-1D, and bri1-5 35S-TCP1-GFP plants.
(B) In bri1-5 tcp1-1D, four copies of 35S enhancer were inserted at 2281 bp upstream of the start codon of At1G67260 (TCP1).
(C) The expression of TCP1 was elevated in bri1-5 tcp1-1D as well as in bri1-5 35S-TCP1-GFP plants as shown by real-time RT-PCR. The data were the average of three replicates. The error bars represent the sd. TCP1 expression level in the wild type was set to 1. TCP1 expression levels in other genotypes were determined relative to that of the wild type.
[See online article for color version of this figure.]
We cloned the flanking sequences of the T-DNA insertion by thermal asymmetric interlaced PCR (Liu and Whittier, 1995) and found that the T-DNA is inserted at 2281 bp upstream of the initiation codon of TCP1 (At1G67260) (Figure 1B). To determine whether TCP1 is the gene responsible for the suppression phenotype in the double mutant, we cloned the full-length cDNA of TCP1 and overexpressed it in bri1-5 driven by a constitutive cauliflower mosaic virus 35S promoter. Over 50% of the transgenic plants showed elongated petiole phenotypes similar to the originally identified activation tagging line (Figure 1A). Real-time RT-PCR analysis confirmed that the TCP1 expression levels in both bri1-5 tcp1-1D and bri1-5 35S-TCP1-GFP (for green fluorescent protein) were elevated by at least 10-fold compared with those in WS2 and bri1-5 (Figure 1C). These results demonstrated that increased expression of TCP1 is the cause of the suppression phenotype seen in the double mutant.
A Functional BRI1 Is Required for tcp1-1D to Regulate Plant Growth and Development
To determine whether tcp1-1D affects general growth or has a specific role in BR-related pathways, we conducted a series of genetic crosses. We first backcrossed the bri1-5 tcp1-1D with its background ecotype WS2 and segregated out bri1-5 after self-pollination. The tcp1-1D single mutant showed an elongated leaf phenotype reminiscent of BRI1-overexpressed or DWF4-overexpressed transgenic plants (Choe et al., 2001; Wang et al., 2001) (Figure 2A). A similar suppression outcome was observed when tcp1-1D was crossed with det2 (Figure 2B). det2 was first identified as a deetiolated mutant when grown in the dark (Chory et al., 1991). DET2 encodes a steroid reductase responsible for a reduction step from campesterol to campestanol during BR biosynthesis (Li et al., 1996, 1997; Fujioka et al., 1997). It was found that det2 mutants were still able to synthesize ∼5 to 10% wild-type levels of BRs. Therefore, the det2 null mutant was considered as an intermediate biosynthetic mutant (Fujioka et al., 1997; Fujioka and Yokota, 2003). To further test whether BR signaling is necessary for the function of tcp1-1D in regulating plant growth, we crossed tcp1-1D with bri1-4. bri1-4 has a 10–amino acid deletion at the N terminus of BRI1 resulting from an T-DNA insertion, which causes a premature stop codon after amino acid 153 (Noguchi et al., 1999). Therefore, this mutant is regarded as a null mutant of BRI1. The bri1-4 tcp1-1D double mutant did not show any leaf suppression phenotypes (Figure 2C). Thus, our genetic analyses clearly indicated that the role of tcp1-1D in regulating leaf growth is dependent on the presence of BRs and the BR signaling pathway.
Figure 2.
tcp1-1D Can Suppress Weak BR Signaling and Biosynthetic Mutants but Cannot Suppress a Null Allele of BRI1.
(A) WS2 tcp1-1D showed a phenotype reminiscent of the phenotypes of BRI1- or DWF4-overexpressed plants. The plants were photographed 4 weeks after germination.
(B) tcp1-1D can partially suppress the defective phenotypes of a BR biosynthetic mutant det2. The plants were photographed 5 weeks after germination.
(C) tcp1-1D cannot suppress a BRI1 null allele mutant bri1-4. The plants were photographed 3 weeks after germination.
[See online article for color version of this figure.]
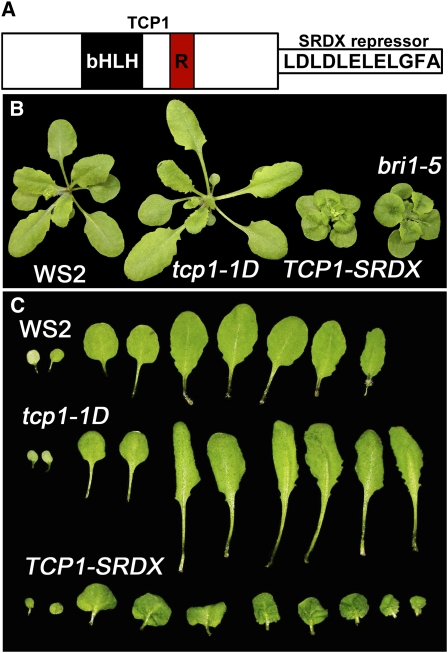
Expression of a TCP1-SRDX Chimeric Repressor Gene in Wild-Type Plants Results in a Typical BR Mutant Phenotype
Our genetic data suggested that TCP1 is involved in BR-related pathways. To further understand this role of TCP1, we searched for T-DNA null mutants of TCP1 from various resources. Unfortunately, no T-DNA alleles were identified from the available databases. We then tried to generate TCP1 knockdown mutants using an artificial microRNA strategy (Schwab et al., 2006). All the resulting plants did not show any obvious phenotypes, possibly due to gene redundancy. Therefore, we employed a gene silencing system, named chimeric repressor gene-silencing technology, in which TCP1 was fused with a 12–amino acid EAR motif repressor domain (SRDX) (Figure 3A) (Hiratsu et al., 2003). Previous experiments indicated that the chimeric version can effectively repress the expression of the target genes of a number of transcription factors, including TCP genes (Koyama et al., 2007). Expression of TCP1-SRDX driven by the constitutive 35S promoter resulted in dwarfed transgenic plants similar to BR-deficient or signaling mutants, such as det2, dwf4, and bri1-5 (Figure 3B). The dominant-negative plants showed phenotypes opposite to those of tcp1-1D plants. Whereas tcp1-1D plants showed elongated leaves and petioles, TCP1-SRDX plants displayed rounded and epinastic leaves, shortened petioles, and reduced statures (Figures 3B and 3C). When grown under darkness, TCP1-SRDX plants exhibited a typical deetiolated phenotype with opened cotyledons resembling that of det2 and bri1-4 mutants (see Supplemental Figure 1 online). The dominant-negative phenotypes were likely caused by the competitive binding of TCP1-SRDX with native TCP1 and its paralogs to the target gene(s) via the bHLH domain. Overexpression of TCP1ΔbHLH (in which bHLH domain sequence was deleted) or TCP1-SRDXΔbHLH failed to alter the growth of the transgenic plants (see Supplemental Figure 2 online), suggesting that the bHLH domain of TCP1 is prerequisite to the functions of TCP1 and TCP1-SRDX in transgenic plants. To ensure that the phenotypes of TCP1-SRDX transgenic plants were caused by the overexpression of TCP1-SRDX, we crossed the transgenic plants with tcp1-1D and isolated homozygous plants for both TCP1-SRDX and tcp1-1D loci. We found that tcp1-1D can partially complement the dominant-negative effect of TCP1-SRDX; this suggests that the phenotypes were indeed caused by TCP1-SRDX overexpression (see Supplemental Figure 3 online). These results suggested that the target gene(s) of the TCP1 transcription factor plays a significant role in regulating BR biosynthesis or signal transduction.
Figure 3.
Transgenic Plants Overexpressing a TCP1 Dominant-Negative Construct (TCP1-SRDX) Show a Typical BR Mutant Phenotype.
(A) Protein structures of the 359–amino acid TCP1. TCP1 contains a typical bHLH domain and an R domain. For the dominant-negative version, a 12–amino acid SRDX repressor sequence was fused at the C terminus of TCP1.
(B) Phenotypes of WS2, tcp1-1D, TCP1-SRDX, and bri1-5. The plants were photographed 3 weeks after germination.
(C) Leaf phenotypes of 3-week-old WS2, tcp1-1D, and TCP1-SRDX plants.
[See online article for color version of this figure.]
TCP1 Regulates BR Biosynthesis
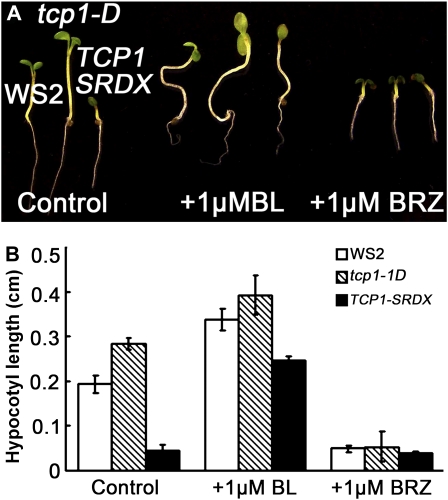
The phenotypes of BR-deficient and signaling mutants are morphologically similar. To determine whether the dwarfed phenotype of TCP1-SRDX plants was caused by the failure of the transgenic plants to respond to endogenous BRs or by disruptions in specific steps of the BR biosynthetic pathway, we thus tested their response to exogenous BRs in the form of BL. If BR signaling is altered, the mutant should show reduced response to exogenous BL. Conversely, if BR biosynthesis is impeded, the dwarf mutant should be rescued by exogenous applied BL. Our results show that the hypocotyl growth of the TCP1-SRDX plants can be greatly recovered by the addition of BL. In half-strength Murashige and Skoog (MS) medium supplemented with 1% sucrose and 1 μM BL, the hypocotyl length of the TCP1-SRDX seedlings was increased by fourfold, whereas those of wild-type WS2 and tcp1-1D seedlings were increased only by 20 to 40% (Figures 4A and 4B). Comparison analysis indicated that the growth of the TCP1-SRDX transgenic plants respond to exogenously applied BL in a similar manner as other BR-deficient mutants, such as det2 (see Supplemental Figure 4 online). Application of 1 μM BRZ, a specific BR biosynthetic inhibitor (Asami and Yoshida, 1999; Asami et al., 2000), did not further reduce the growth of TCP1-SRDX plants. Under the same treatment, the lengths of WS2 and tcp1-1D seedlings were reduced by four- to eightfold, respectively (Figures 4A and 4B). Furthermore, exogenous application of other known growth hormones, such as gibberellic acid 3, kinetin, and indole-3-acetic acid, showed no rescuing effects (see Supplemental Figure 5 online). These results suggest that expression of TCP1-SRDX specifically blocked the BR biosynthetic pathway.
Figure 4.
BL Treatment Rescues the Shortened Hypocotyl Phenotype of TCP1-SRDX.
(A) Phenotypes of representative WS2, tcp1-1D, and TCP1-SRDX seedlings grown on half-strength MS medium under light condition with no BL treatment, 1 μM BL treatment, or 1 μM BRZ treatment, respectively. The seedlings were photographed 5 d after germination.
(B) The measurements corresponding to the treatments shown in (A). Twenty seedlings per genotype were used for the measurements. The error bars represent the sd.
[See online article for color version of this figure.]
TCP1 Expression Levels Are Positively Correlated with the Catalytic Ability of DWF4, a Key Enzyme in the BR Biosynthesis Pathway
To investigate how TCP1 is involved in regulating BR biosynthesis, we performed BR profile analyses (quantitative analyses of BR biosynthetic intermediates) using 4-week-old WS2, tcp1-1D, and TCP1-SRDX plants. It was apparent that tcp1-1D enhanced the catalytic capability of DWF4, whereas TCP1-SRDX greatly reduced the catalytic ability of DWF4. For example, one of the substrates of DWF4, campestanol, is quantitatively alike in three different genotypes. Although the product of DWF4, 6-deoxocathasterone, is increased by approximately twofold in tcp1-1D seedlings, it decreased ∼10-fold in TCP1-SRDX seedlings in comparison with that of wild-type seedlings (Figure 5). Similar results were observed for the reaction from campesterol to (22S)-22-hydroxycampesterol, which is also catalyzed by DWF4 (Figure 5). None of the other reactions were affected in such a dramatic way. Because TCP proteins are transcription factors, TCP1 likely regulates DWF4-catalyzed reactions via the control of DWF4 expression.
Figure 5.
BR Profile Analyses of Four-Week-Old Soil-Grown WS2, tcp1-1D, and TCP1-SRDX Plants.
Levels (ng/g fresh weight) for each of the indicated molecules in the BR synthesis pathway were determined in WS, tcp1-1D, and TCP1-SRDX plants (top, middle, and bottom numbers, respectively). The numbers shown are the averages from three independent replicas. The ± represents sd. nd, not detected (below detection limit).
The Expression Level of DWF4 Is Positively Regulated by TCP1
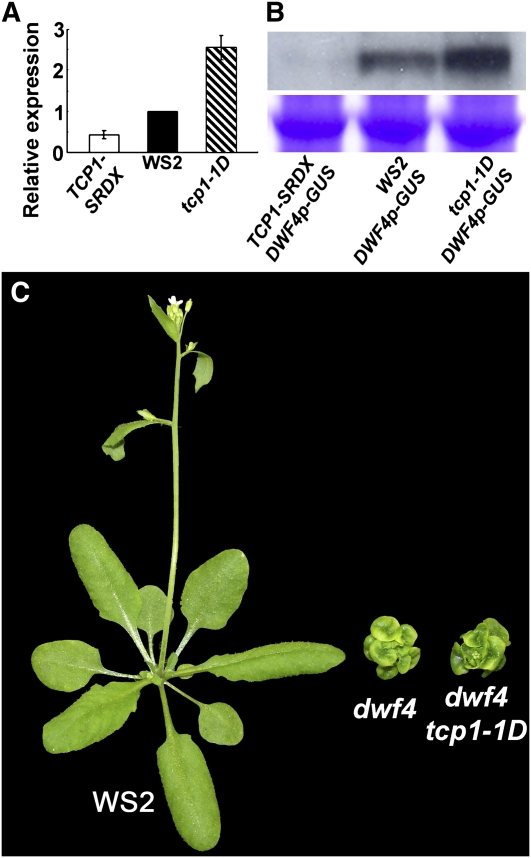
To test our hypothesis that TCP1 transcriptionally regulates DWF4 expression, we first generated TCP1p (TCP1 promoter)-β-glucuronidase (GUS) and DWF4p-GUS transgenic plants and examined the expression patterns of TCP1 and DWF4. If TCP1 directly regulates the expression of DWF4, we would expect to see overlapping expression patterns of the two genes. We analyzed six independent transgenic lines for each construct. Each transgenic event showed consistent GUS staining results, which clearly indicated that both genes have overlapped expression patterns (see Supplemental Figure 6 online). Both genes are expressed relatively strongly in the roots and at the junctions of roots and hypocotyls. In the leaves, both genes are mainly expressed in vascular tissues. The expression level of DWF4 in the leaves, however, is apparently much less than that of TCP1. The net DWF4 expression level is determined by both the positive effect of TCP1 and negative action of BZR1/BES1. To examine whether changing expression levels of TCP1 can affect the transcription of DWF4, we isolated total RNA from WS2, tcp1-1D, and TCP1-SRDX entire seedlings and performed real-time RT-PCR analyses. Our results clearly indicated that the expression level of DWF4 is elevated in the tcp1-1D gain-of-function mutant seedlings but reduced in the TCP1-SRDX dominant-negative mutant seedlings (Figure 6A). The expression levels of all other known Arabidopsis BR biosynthetic genes were not significantly altered in tcp1-1D and TCP1-SRDX seedlings (data not shown). We also crossed the homozygous lines of tcp1-1D and TCP1-SRDX with homozygous lines of DWF41p-GUS plants, respectively, and obtained F1 plants. These F1 plants, only containing one copy of each of the transgene, were used to test whether overexpression of TCP1 (tcp1-1D) or TCP1-SRDX can influence the expression levels of GUS by an immunoblotting analysis using anti-GUS antibody. If TCP1 controls DWF4 transcription via regulating the function of DWF4 promoter, we expect to see that overexpression of TCP1 upregulates the level of GUS and overexpression of TCP1-SRDX downregulates the level of GUS. Our immunoblotting results indeed confirmed these predictions (Figure 6B). These results confirmed that TCP1 functions as a positive regulator in controlling DWF4 transcription. In addition, when tcp1-1D was crossed into a null DWF4 mutant background, dwf4-1, the resulting dwf4-1 tcp1-1D double mutant plants showed the same phenotype as dwf4-1 single mutant plants. (Figure 6C). dwf4-1 is a T-DNA insertion mutant originally used to clone DWF4 (Choe et al., 1998). The null dwf4-1 mutant is more severe than a null det2 mutant but much milder than a cpd null mutant (Szekeres et al., 1996; Fujioka et al., 1997; Choe et al., 1998). BR profile analysis suggested that dwf4-1 still makes trace amount of BRs (Choe et al., 2001). The inability of tcp1-1D to suppress dwf4-1 is consistent with our notion about TCP1's role in regulating DWF4 expression. Without a functional target gene, overexpression of TCP1 becomes inconsequential as it would not be able to regulate the expression of the target gene and the subsequent downstream events leading to modified plant growth.
Figure 6.
TCP1 Positively Regulate the Expression of DWF4.
(A) Real-time RT-PCR results indicate that the expression of DWF4 is elevated in tcp1-1D but reduced in TCP1-SRDX plants. DWF4 expression level in WS2 was set to 1. DWF4 expression levels in other genotypes were relative to that in WS2. The data were the average of three replicates. The error bars represent the sd.
(B) The top panel is the immunoblotting data indicating that tcp1-1D positively, whereas TCP1-SRDX negatively regulates the expression of DWF4p-GUS. The bottom panel is a duplicate polyacrylamide gel stained with Coomassie blue to show equal loading.
(C) tcp1-1D does not rescue the phenotypes of a null dwf4 mutant. The plants were photographed 4 weeks after germination.
[See online article for color version of this figure.]
TCP1 Associates with the Promoter Region of DWF4
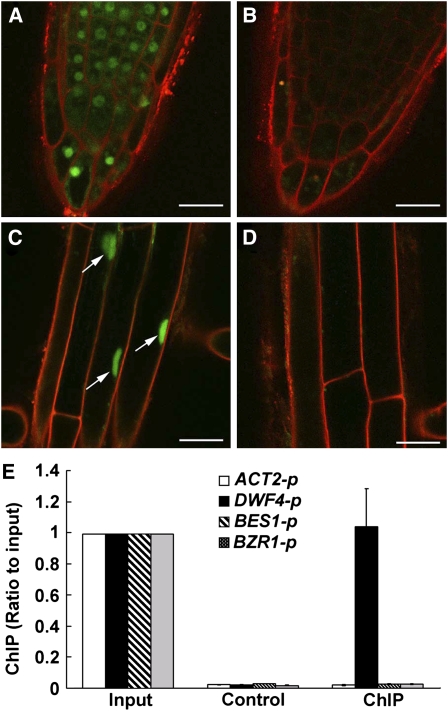
Our genetic and biochemical analysis data suggested that TCP1 is a transcription factor positively regulating DWF4 expression. To further examine TCP1 function, we conducted a TCP1 subcellular localization analysis to examine whether TCP1 is localized in the nucleus and a chromatin immunoprecipitation (ChIP) assay to determine whether TCP1 associates with the promoter of DWF4. We overexpressed a TCP1-GFP fusion gene in WS2 background, selected multiple transgenic lines, and identified homozygous plants. Confocal microscopy of primary roots from 4-d-old TCP1-GFP seedlings indicated that fluorescent signals originate exclusively from the nuclei. Representative results are shown in Figures 7A to 7D. These TCP1-GFP homozygous transgenic lines were then used to do ChIP experiments. According to predicted DNA binding sites of TCP family proteins from previous reports in rice, consensus TCP binding sites were found in the promoter regions of both BES1 and BRZ1 but not in the promoter region of DWF4. So we first checked whether the TCP1 protein binds to the promoter regions of BZR1 or BES1 through ChIP analysis. As shown in Figure 7E, TCP1 does not seem to associate with either the BES1 or BZR1 promoter. ChIP analysis, however, did demonstrate that the TCP1 protein is associated with the DWF4 promoter sequences, either via a novel binding sequence on the DWF4 promoter (Figure 7E) or via its interaction with another transcription factor that directly binds to the promoter sequence of DWF4. Our ChIP analysis results were consistent with our real-time RT-PCR results because we did not find any altered expression of BES1 and BZR1 in tcp1-1D or TCP1-SRDX plants compared with that in wild-type plants (data not shown). We did find, however, that DWF4 was upregulated in tcp1-1D and downregulated in TCP1-SRDX transgenic plants (Figures 6A and 6B).
Figure 7.
TCP1 Is Localized in the Nucleus, and ChIP Results Suggest That TCP1 Associates with DWF4 Promoter.
(A) and (C) Confocal microscopy shows that the TCP1-GFP fusion is localized in the nucleus. Homozygous transgenic plants harboring 35S-TCP1-GFP were used for the analysis.
(B) and (D) Wild-type plants were used as controls. No fluorescent signals were detected in the analysis. Roots were counterstained with propidium iodide to visualize the outline of the cells. Bars = 20 μm.
(E) CHIP results show that anti-GFP antibody can coimmunoprecipitate the DWF4 promoter region but not the promoter regions of ACT2, BES1, and BZR1. Input: using the samples collected before immunoprecipitation as real-time PCR templates. ChIP: using diluted immunoprecipitated (with anti-GFP antibody) samples as real-time PCR templates. Control: using diluted immunoprecipitated (without adding any antibody) samples as real-time PCR templates. Each input was set to 1. Error bars represent the sd of three replicates.
TCP1 Expression Can Be Regulated by BR
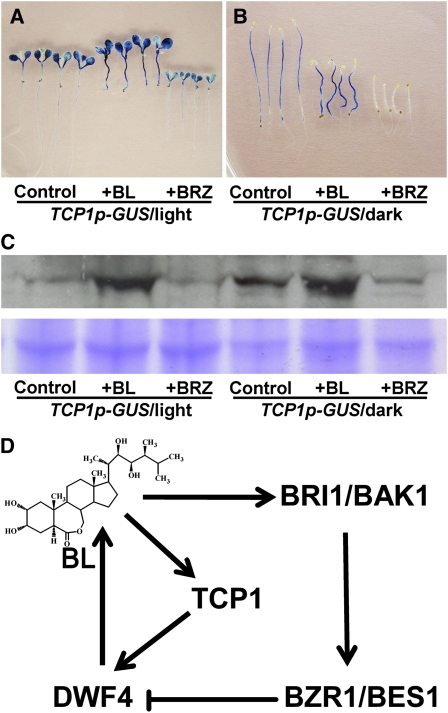
To examine whether BR levels affect the expression of TCP1, seeds from a representative homozygous TCP1p-GUS line were germinated in half-strength MS medium supplemented with 1% sucrose and 0 μM BL or BRZ, 1 μM BL, and 1 μM BRZ. The plants were grown under either complete darkness or continuous light conditions for 5 d. The seedlings were then stained for GUS activity and were also subjected to immunoblotting analyses using an anti-GUS antibody (Figures 8A to 8C). Our results indicated that application of 1 μM BL can significantly stimulate TCP1 expression as revealed by both GUS staining and immunoblotting results, whereas depletion of endogenous BRs by BRZ treatment can greatly reduce TCP1 expression (Figures 8A to 8C). We also grew WS2, det2-28 (in WS2 ecotype), and bri1-4 (in WS2 background) for 5 d in half-strength MS medium supplemented with 1% sucrose and 1 μM BL. Real-time RT-PCR analysis indicated that TCP1 expression levels were upregulated in WS2 and det2-28 but not in bri1-4 in the presence of 1 μM BL (see Supplemental Figure 7 online). The increasing level was quite dramatic in det2-28. These data suggest that the expression of TCP1 can be regulated by exogenous and endogenous levels of BL, and the regulation may rely on a functional BRI1 receptor (Figure 8D).
Figure 8.
The Expression of TCP1 Is Upregulated by BL.
(A) GUS-stained light-grown transgenic seedlings harboring TCP1 promoter-GUS shows that the expression levels of TCP1 can be positively regulated by the BL treatment and negatively regulated by the BRZ treatment.
(B) Dark-grown seedlings were used for the same experiments shown in (A).
(C) Immunoblotting analysis to confirm that GUS expression is positively regulated by BL. The top panel shows the immunoblotting results. The orders are the same as in (A) and (B). The bottom panel shows a duplicate polyacrylamide gel stained with Coomassie blue, indicating the equal amounts of total proteins were used in each lane.
(D) A model suggesting that the expression of DWF4 can be negatively regulated by BZR1/BES1 via a feedback loop and also can be positively regulated by TCP1.
DISCUSSION
BR homeostasis is critical for normal plant growth and development. BR-deficient mutants show a severely retarded growth phenotype mainly due to lack of cell elongation. Excessive amounts of BR, on the other hand, greatly inhibit root growth and trigger leaf senescence (Clouse, 1996; Clouse and Sasse, 1998). Bioactive levels of BR are mainly balanced by the rates of biosynthesis and inactivation. Within the last a few years, a feedback inhibitory regulation has been elucidated, which involves two novel transcription factors, BZR1 and BES1 (Mathur et al., 1998; He et al., 2005). Previous studies indicated that the expression levels of five BR-specific biosynthetic genes, including DET2, DWF4, CPD, BR6ox1, and ROT3, are upregulated in plants treated with the inhibitor BRZ (Bancos et al., 2002; Tanaka et al., 2005). The expression levels of four of the five genes (DWF4, CPD, BR6ox1, and ROT3) are significantly downregulated in response to exogenous BL treatment. Previous studies also revealed a number of biochemical reactions that plants use to inactivate BRs if an excessive amount of BR is present. How BR biosynthesis is positively regulated, however, is poorly understood. When additional BR is needed at certain developmental stages or under various biotic/abiotic stresses, how plants perceive internal or external signals to trigger the production of more BR is poorly understood. Generally, it is thought that the plants can accumulate additional BR via two different mechanisms, including acceleration of the biosynthetic rate and deceleration of inactivation speed or slowing down the feedback regulation. The most effective way is to speed up BR biosynthesis by elevating the expression of key BR biosynthetic genes.
Among the BR biosynthetic genes isolated, DWF4 was believed to be one of the key genes in the BR-specific biosynthetic pathway from campesterol to BL (Kim et al., 2006). DWF4 encodes a P450 protein that catalyzes multiple 22α-hydroxylation steps in BR biosynthesis (Choe et al., 1998). Relative to other BR biosynthetic genes, such as CPD and DET2, DWF4 is expressed at an extremely low level (Kim et al., 2006). Although BR profile analysis indicated that DWF4 substrates are always plentiful in plants, the products of the DWF4-catalyzed reactions are considerably low or even undetectable. Previous studies also indicated that DWF4 is mainly expressed in actively growing tissues, such as root and shoot apices, where BRs are significantly enriched (Kim et al., 2006). In addition, overexpression of DWF4 in wild-type Arabidopsis or tobacco (Nicotiana tabacum) plants can significantly increase biomass and seed production (Choe et al., 2001). These results suggest that DWF4 catalyzes a flux-determining step in the BR biosynthesis pathway (Kim et al., 2006). Elucidating how DWF4 expression is regulated will greatly advance our knowledge about the mechanisms controlling BR homeostasis in a plant.
Our detailed analyses clearly demonstrated that TCP1 is an important transcription factor positively regulating the BR biosynthesis by regulating the expression of DWF4. This notion is supported by several key observations. First, wild-type plants harboring tcp1-1D allele showed a phenotype similar to that of the DWF4-overexpressing plants (Choe et al., 2001). The transgenic plants overexpressing a dominant-negative chimeric gene, TCP1-SRDX, on the other hand, exhibited a typical BR deficiency or signaling mutant phenotype. The dwarfed phenotypes of the transgenic seedlings can be rescued by supplementing the culture medium with BL but not with any other growth-promoting hormones, such as auxins, gibberellins, or cytokinins, suggesting that TCP1-SRDX transgenic plants are BR-specific deficient mutants, rather than BR signaling mutants. Secondly, our BR profile analysis results clearly indicated that the expression levels of TCP1 affect the function of DWF4 but not other known BR biosynthetic enzymes. Our real-time RT-PCR results showed that overexpression of TCP1 upregulates DWF4 expression, whereas overexpression of TCP1-SRDX downregulates the expression of DWF4. Finally, our ChIP analysis using transgenic plants overexpressing TCP1-GFP demonstrated that TCP1 can interact with the promoter of DWF4 directly or indirectly, but not with the promoters of BZR1, BES1, and ACTIN2.
It is worth noting, however, that although tcp1-1D showed an organ elongation phenotype resembling plants overexpressing DWF4, BR profile analysis did not reveal any accumulation of castasterone (CS)/BL in tcp1-1D plants. As a matter of fact, several intermediates in the later BR biosynthetic pathway, such as CS, were reduced in both tcp1-1D and TCP1-SRDX plants. However, the phenotypes were opposite. BR profile analysis was also conducted in plants overexpressing DWF4 by Choe et al. (2001), and their results also showed there was no CS or BL accumulation in DWF4-overexpressing plants. Choe et al. (2001) proposed that BL, CS, or other bioactive BR in DWF4-overexpressing plants could be turned over rapidly after triggering the BR signal transduction chain. This idea is supported by the fact that in mutants impaired in BR signaling, such as in bri1, BL levels became elevated possibly because the signaling pathway has been disrupted in bri1 mutants (Noguchi et al., 1999). The similarity of the BR profiles in tcp1-1D and DWF4-overexpressing plants in comparison with wild-type plants is another piece of evidence linking TCP1 function to the regulation of DWF4 expression.
Although significant progress has been made in elucidating both the BR biosynthesis and signal transduction pathways, very little is known about how the expression of BR biosynthetic and signaling genes is positively regulated. Our identification of TCP1 as a positive regulator for DWF4 expression reveals a mechanism through which plants are able to modulate BR biosynthesis during normal plant growth and development. TCP proteins are plant-specific transcription factors regulating a number of processes during plant growth and development, such as floral symmetry (Luo et al., 1999; Broholm et al., 2008), embryonic growth (Tatematsu et al., 2008), morphology of shoot lateral organs (Koyama et al., 2007), and jasmonate biosynthesis and senescence (Schommer et al., 2008). The name TCP was used after TEOSINTE BRANCHED1 from maize (Zea mays), CYCLOIDEA from Anthirrinum majus, and PCF from rice (Cubas et al., 1999). These proteins contain a bHLH domain, which is thought to be involved in DNA binding. There is also an R-domain whose function is not yet known. In Arabidopsis, there are 24 TCP transcription factors. Based on similarity in the amino acid sequence of the TCP domain, 13 were grouped into class I, and 11 were group into class II. It was found that the two classes have distinct but overlapping binding sequences. For example, rice class I TCP proteins prefer to bind GGNCCCAC, whereas class II TCP proteins favor binding to GTGGNCCC (Kosugi and Ohashi, 2002). They share the core sequence GGNCCC. It is not known, however, whether Arabidopsis TCP transcription factors bind to the same DNA sequences. Since TCP1 belongs to class I of TCP transcription factors, we searched the promoter region sequence of DWF4 to see whether it contains a GGNCCCAC motif. We failed to identify the consensus sequence for either class I or class II TCPs in the DWF4 promoter region. This suggested that the possible binding sequence for TCP1 in DWF4 promoter may not be strictly identical to what was reported in rice. Alternatively, even though we have determined that TCP1 and DWF4 promoters are in the same complex as assayed by ChIP experiments, we cannot rule out the possibility that TCP1 is indirectly involved in activating DWF4 transcription by its association with another true DWF4 promoter binding transcriptional factor. Interestingly, in the promoters of both BZR1 and BES1, there are typical TCP binding sequences revealed from rice TCP orthologs. For instance, ∼400 bp upstream of the translation initiation codon of BES1, there are two putative TCP binding motifs, GGACCCAC and GGCCCCAC. Similar sequences have also been detected in the promoter region of BZR1. However, it appears that the expression of BES1 and BZR1 is not directly regulated by TCP1, as the expression levels of these two transcription factors are not altered in tcp1-1D or TCP1-SRDX plants (data not shown). It is possible that these transcription factors are regulated by other members of the TCP family. Previous studies suggested that TCP1 is involved in floral symmetric regulation (Cubas et al., 2001; Busch and Zachgo, 2007). The detailed molecular mechanism controlling organ mono- or polysymmetry is poorly understood. Our identification of TCP1 in regulating BR biosynthesis suggests that unequal expression of TCP1 may result in uneven distribution of BRs in floral meristems, which may contribute to the asymmetric regulation of floral organs. Future studies will focus on understanding how the expression of TCP1 is regulated by internal and external factors. The information about how TCP1 expression is regulated will facilitate our understanding of how BR biosynthesis is controlled.
METHODS
Plant Materials and Activation Tagging for bri1-5 Genetic Modifiers
All plants used in these studies, including bri1-5, dwf4-1, and bri1-4, were in Arabidopsis thaliana ecotype WS2 background. Unless specified, plants were grown under 16 h light (150 to 200 μmol m−2 s−1) and 8-h dark condition. The bri1-5 tcp1-1D double mutant was obtained by a large-scale activation-tagging screen for bri1-5 suppressors as described previously (Li et al., 2001a, 2002; Zhou et al., 2004; Yuan et al., 2007). Briefly, a homemade activation-tagging construct, pBASTA-AT2 (Yuan et al., 2007), was transformed into bri1-5 to generate transgenic plants with resistance to Basta herbicide. bri1-5 tcp1-1D was identified as one of the bri1-5 suppressors.
Identification of the tcp1-1D Locus
Thermal asymmetric interlaced PCR was used to amplify the flanking genomic sequence of the T-DNA of pBASTA-AT2 as described previously (Liu and Whittier, 1995; Terauchi and Kahl, 2000). The T-DNA insertion site was determined by sequencing the flanking genomic DNA. The T-DNA was inserted in the promoter of TCP1 (At1g67260), 2281 bp upstream from its start codon.
Construction of Expression Vectors
Full-length TCP1 cDNA was amplified with primers TCP1-attb1 and TCP1-attb2, and TCP1-SRDX was amplified with primers TCP1-attb1 and TCP1-SRDX-attb2 (see Supplemental Table 1 online). Both sequences were cloned into gateway vector pB35GWG as described previously (Gou et al., 2010) and overexpressed in bri1-5 and WS2 plants. Transgenic lines were analyzed for phenotypes.
Real-Time RT-PCR Analyses
Total RNA was extracted using RNeasy plant mini kits with on-column DNase-treatment (Qiagen). Total RNA (2 μg) was reversely transcribed to the first strand of the cDNA in a 20-μL volume using the SuperScript III first-strand synthesis system (Invitrogen). For non-real-time PCR, 1 to 2 μL RT product was used as PCR template in a 20-μL volume reaction. Different PCR cycles were conducted to get better results. PCR products were separated by 1% agarose gel electrophoresis and visualized under a UV scanner. For quantitative RT-PCR experiments, 10 μg total RNA was reverse transcribed into first-strand cDNAs using the SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instruction. The resulting cDNAs corresponding to 100 ng total RNAs (4 μL) were then used as templates for real-time PCR analyses of transcripts of interest using a MyIQ system (Bio-Rad) with gene-specific primers (see Supplemental Table 1 online). The relative expression level was calculated from three replicates using the comparative Ct method after normalized to an ACT2 control.
Hypocotyl Measurements
All of the seeds used for hypocotyl analysis were surface-sterilized as described previously (Yuan et al., 2007) and planted on half-strength MS medium supplemented with 1% sucrose, 0.8% agar, and 24-epiBL or BRZ. The plates were kept at 4°C for 2 d and then were grown vertically in white light at room temperature. Hypocotyl length of seedlings was measured after 5 d. All measurements were obtained from three independent experiments, and at least 20 seedlings were measured each time.
Immunoblotting Analysis
Immunoblotting analysis was performed as previously described (Li et al., 2002; Wang et al., 2005). Both anti-GFP (A11120) and anti-GUS (A-5790) antibodies were from Invitrogen.
BR Profile Analysis
Aerial parts of 4-week-old WS2, tcp1-1D, and TCP1-SRDX seedlings were harvested and lyophilized. The tissues (15 g of lyophilized tissues per assay) were extracted twice with 250 mL of methanol. Deuterium-labeled internal standards were added to the extracts. Purification and quantification of BRs and sterols were performed according to the method described previously (Fujioka et al., 2002).
Confocal Microscopy Analysis
Seeds of transgenic plants overexpressing TCP1-GFP were surface sterilized in 95% ethanol and 20% bleach and washed with sterile double deionized water prior to planting. Sterilized seeds were germinated on 62 × 48-mm glass cover slips coated with 0.5% agar supplemented with half-strength MS salts, 0.5 mg/mL pyridoxine-HCL, 0.5 mg/mL nicotinic acid, 1 mg/mL thiamine, 0.10 g/L myo-inositol, 0.5 g/L MES, and 1% sucrose. The pH of the agar nutrient medium was adjusted to 5.7 with 10 M KOH. After 3 to 4 d, the primary roots were imaged with a Leica TCS SP2 AOBS laser scanning confocal microscope equipped with a ×63 water immersion objective (Leica Microsystems). Seedlings were counterstained with 5 μM propidium iodide for simultaneous visualization of GFP and the cell wall. Propidium iodide and GFP were excited with the 488-nm line of the argon laser, and emission was detected at 510 and 620 nm, respectively.
CHIP Analysis
The EpiQuik Plant ChIP kit (Epigentek) was used to conduct ChIP analysis. The experiments were performed based on the specifications from the manufacturer. Briefly, seedlings (1 to 2 g) were used to get chromatin pellets. The chromatin pellet was resuspended in 500 μL CP3F containing protease inhibitor cocktail (Roche). DNA was sheared to ∼500-bp fragments by sonicating and then centrifuged to get supernatants. The supernatant was diluted, transferred to an antibody (anti-GFP)-bound strip well, and incubated at room temperature for 60 to 90 min with gentle shaking. TCP1-GFP specifically associated DNA fragments were purified and eluted from the column. The obtained DNA fragments were tested with real-time PCR. Primers for amplifying DWF4 were DWF4pF and DWF4pR; for amplifying BES1, they were BES1pF and BES1pR; for amplifying BZR1, they were BZR1pF and BZR1pR; and for amplifying ACT2, they were ACT2pF and ACT2pR (see Supplemental Table 1 online).
Generation of Promoter-GUS Transgenic Plants
A 1.5-kb promoter fragment of TCP1 was amplified from the Arabidopsis genomic DNA by PCR with primers TCP1p1 and TCP1p2 (see Supplemental Table 1 online). A 1.1-kb promoter fragment of DWF4 was amplified from genomic DNA by PCR with primers DWF4p1 and DWF4p2 (see Supplemental Table 1 online). Then, they were cloned into gateway vector pBIB-BASTA-GUS-GWR (Gou et al., 2010) and were transformed into Arabidopsis ecotype WS2. Homozygote transgenic plants were planted on soil or half-strength MS medium with 1% sucrose for GUS activity assay. TCP1p-GUS plants were planted on half-strength MS medium containing 1% sucrose with or without 1 μM epi-BL (Sigma-Aldrich) or 1 μM BRZ (Asami et al., 2000) for 5 d under light or darkness before determining for GUS activity. Seedlings or tissues were vacuum infiltrated in X-Gluc solution, followed by overnight incubation at room temperature, then destained with 70% ethanol and visualized for blue color.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TCP1 (At1G67260), ACT2 (AT3G18780), DWF4 (AT3G50660), BZR1 (AT1G75080), and BES1 (AT1G19350).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TCP1-SRDX Transgenic Plants Show a Typical Deetiolated Phenotype When Grown in Darkness.
Supplemental Figure 2. bHLH Domain in TCP1 Is Critical for the Functions of TCP1 and TCP1-SRDX in Regulating Plant Growth.
Supplemental Figure 3. The Dwarfed Phenotype of TCP1-SRDX Plants Can Be Partially Rescued by an Additional tcp1-1D Allele.
Supplemental Figure 4. Growth of TCP1-SRDX Plants in Response to BL Treatment Is Similar to a BR Deficiency Mutant det2.
Supplemental Figure 5. The Dwarfed Phenotype of TCP-SRDX Plants Can Be Rescued by the Treatment of BL but Not by Other Major Plant Hormones.
Supplemental Figure 6. Representative Expression Patterns of Transgenes, DWF4p-GUS and TCP1p-GUS, by GUS Staining.
Supplemental Figure 7. TCP1 Transcription Level Regulated by BL in Different Mutant Backgrounds.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Frans Tax for providing the dwf4-1 and det2 mutants. This work is partially supported by grants from the National Science Foundation (IBN-0317729 to J.L.) and a National Natural Science Foundation of China grant (90917019 to J.L.), as well as a National Science Foundation Arabidopsis 2010 grant (MCB-0419819 to S.D. Clouse, J.L., S. Huber, and M. Goshe). This work was also supported in part by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to S.F. (Grant 19380069). We also thank Suguru Takatsuto (Joetsu University of Education) for supplying deuterium-labeled internal standards.
References
- Asami T., Min Y.K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T., Yoshida S. (1999). Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 4: 348–353 [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos S., Nomura T., Sato T., Molnar G., Bishop G.J., Koncz C., Yokota T., Nagy F., Szekeres M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G., Nomura T., Yokota T., Montoya T., Castle J., Harrison K., Kushiro T., Kamiya Y., Yamaguchi S., Bancos S., Szatmari A.M., Szekeres M. (2006). Dwarfism and cytochrome P450-mediated C-6 oxidation of plant steroid hormones. Biochem. Soc. Trans. 34: 1199–1201 [DOI] [PubMed] [Google Scholar]
- Bishop G.J., Harrison K., Jones J.D. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8: 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G.J., Nomura T., Yokota T., Harrison K., Noguchi T., Fujioka S., Takatsuto S., Jones J.D., Kamiya Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm S.K., Tahtiharju S., Laitinen R.A., Albert V.A., Teeri T.H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Zachgo S. (2007). Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. USA 104: 16714–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., Feldmann K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (1996). Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M. (1998). BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Cubas P., Coen E., Zapater J.M.M. (2001). Ancient asymmetries in the evolution of flowers. Curr. Biol. 11: 1050–1052 [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Ehsan H., Ray W.K., Phinney B., Wang X., Huber S.C., Clouse S.D. (2005). Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J. 43: 251–261 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Li J., Choi Y.H., Seto H., Takatsuto S., Noguchi T., Watanabe T., Kuriyama H., Yokota T., Chory J., Sakurai A. (1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S., Takatsuto S., Yoshida S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., Clouse S.D., Li J. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Clouse S.D. (2001). Expression of a plant gene with sequence similarity to animal TGF-beta receptor interacting protein is regulated by brassinosteroids and required for normal plant development. Plant J. 26: 35–45 [DOI] [PubMed] [Google Scholar]
- Kim H.B., Kwon M., Ryu H., Fujioka S., Takatsuto S., Yoshida S., An C.S., Lee I., Hwang I., Choe S. (2006). The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis. Plant Physiol. 140: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol., in press. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Biswas M.G., Chao A., Russell D.W., Chory J. (1997). Conservation of function between mammalian and plant steroid 5alpha-reductases. Proc. Natl. Acad. Sci. USA 94: 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Lease K.A., Tax F.E., Walker J.C. (2001a). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001b). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Whittier R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Mathur J., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Mora-Garcia S., Vert G., Yin Y., Cano-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2004). The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE1. Plant Cell 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Nguyen S.M., Malancharuvil E.J., Fujioka S., Noguchi T., Seto H., Tsubuki M., Honda T., Takatsuto S., Yoshida S., Chory J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., et al. (2004). Brassinosteroid deficiency due to truncated steroid 5alpha-reductase causes dwarfism in the lk mutant of pea. Plant Physiol. 135: 2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Szatmari A.M., Watanabe B., Fujita S., Bancos S., Koncz C., Lafos M., Shibata K., Yokota T., Sakata K., Szekeres M., Mizutani M. (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G.L., Vaistij F.E., Hiranuma S., Seto H., Takatsuto S., Adam G., Yoshida S., Bowles D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M., Marsolais F., Richard M., Nicolle L., Voigt B., Adam G., Varin L. (1999). Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 274: 20925–20930 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chetelat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Reid J.B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 135: 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Nemeth K., Koncz-Kalman Z., Mathur J., Kauschmann A., Altmann T., Redei G.P., Nagy F., Schell J., Koncz C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Asami T., Yoshida S., Nakamura Y., Matsuo T., Okamoto S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Terauchi R., Kahl G. (2000). Rapid isolation of promoter sequences by TAIL-PCR: The 5′-flanking regions of Pal and Pgi genes from yams (Dioscorea). Mol. Gen. Genet. 263: 554–560 [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yuan T., Fujioka S., Takatsuto S., Matsumoto S., Gou X., He K., Russell S.D., Li J. (2007). BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 51: 220–233 [DOI] [PubMed] [Google Scholar]
- Zhou A., Wang H., Walker J.C., Li J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 40: 399–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.