Abstract
Most cellular processes are enabled by cohorts of interacting proteins that form dynamic networks within the plant proteome. The study of these networks can provide insight into protein function and provide new avenues for research. This article informs the plant science community of the currently available sources of protein interaction data and discusses how they can be useful to researchers. Using our recently curated IntAct Arabidopsis thaliana protein–protein interaction data set as an example, we discuss potentials and limitations of the plant interactomes generated to date. In addition, we present our efforts to add value to the interaction data by using them to seed a proteome-wide map of predicted protein subcellular locations.
For well over two decades, plant scientists have studied protein interactions within plants using many different and evolving approaches. Their findings are represented by a large and growing corpus of peer-reviewed literature reflecting the increasing activity in this area of plant proteomic research. More recently, a number of predicted interactomes have been reported in plants and, while these predictions remain largely untested, they could act as a useful guide for future research. These studies have allowed researchers to better understand the function of protein complexes and to refine our understanding of protein function within the cell (Uhrig, 2006; Morsy et al., 2008). The extraction of protein interaction data from the literature and its standardized deposition and representation within publicly available databases remains a challenging task. Aggregating the data in databases allows researchers to leverage visualization, data mining, and integrative approaches to produce new insights that would be unachievable when the data are dispersed within largely inaccessible formats (Rodriguez et al., 2009).
Currently, there are three databases that act as repositories of plant protein interaction data. These are IntAct (http://www.ebi.ac.uk/intact/; Aranda et al., 2010), The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/; Poole, 2007), and BioGRID (http://www.thebiogrid.org/; Breitkreutz et al., 2008). These databases curate experimentally established interactions available from the peer-reviewed literature (as opposed to predicted interactions, which will be discussed below). Each repository takes its own approach to the capture, storage, and representation of protein interaction data. TAIR focuses on Arabidopsis thaliana protein–protein interaction data exclusively; BioGRID currently focuses on the plant species Arabidopsis and rice (Oryza sativa), while IntAct attempts to capture protein interaction data from any plant species. Unlike the other repositories, IntAct follows a deep curation strategy that captures detailed experimental and biophysical details, such as binding regions and subcellular locations of interactions using controlled vocabularies (Aranda et al., 2010). While the majority of plant interaction data held by IntAct concern protein–protein interaction data in Arabidopsis, there is a small but growing content of interaction data relating to protein–DNA, protein–RNA, and protein–small molecule interactions, as well as interaction data from other plant species.
Using the IntAct Arabidopsis data set as an example, we outline how the accumulating knowledge captured in these repositories can be used to further our understanding of the plant proteome. We compare the characteristics of predicted interactomes with the IntAct protein–protein interaction data set, which consists entirely of experimentally measured protein interactions, to gauge the predictive accuracy of these studies. Finally, we show how the IntAct data set can be used together with a recently developed Divide and Conquer k-Nearest Neighbors Method (DC-kNN; K. Lee et al., 2008) to predict the subcellular locations for most Arabidopsis proteins. This data set predicts high confidence subcellular locations for many unannotated Arabidopsis proteins and should act as a useful resource for future studies of protein function. Although this article focuses on the IntAct Arabidopsis protein–protein interaction data set, readers are also encouraged to explore the resources offered by our colleagues at TAIR and BioGRID.
Each database employs its own system to report molecular interactions, as represented in the referenced source publications, and each avoids making judgments on interaction reliability or whether two participants in a complex have a direct interaction. Thus, the user should carefully filter these data sets for their specific purpose based on the full annotation of the data sets. In particular, the user should consider the experimental methods and independent observation of the same interaction in different publications when assessing the reliability and type of interaction of the proteins (e.g., direct or indirect). Confidence scoring schemes for interaction data are discussed widely in the literature (Yu and Finley, 2009).
COMPOSITION OF THE INTACT AND OTHER ARABIDOPSIS INTERACTION RESOURCES
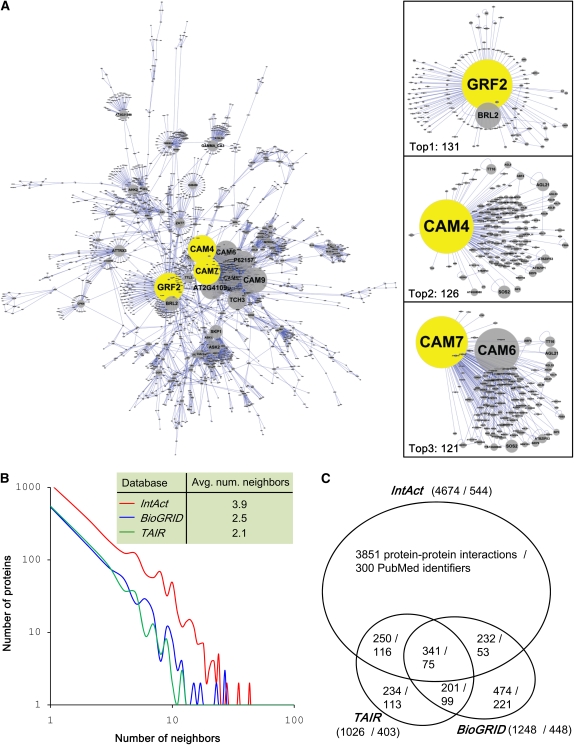
At the time of this writing, the IntAct team has curated 544 publications resulting in a data set of 4674 binary interactions among 2334 Arabidopsis proteins. The vast majority of these proteins have less than or equal to 15 interacting partners (2263 proteins; 97%). A few proteins, however, have a very large number of interactions, including GRF2 (14-3-3-like protein GF14 omega, TAIR locus identifier AT1g78300), CAM4 (Calmodulin-1/4 protein, TAIR locus identifier AT1g66410), and CAM7 (Calmodulin-7 protein, TAIR locus identifier AT3g43810) with 131, 126, and 121 partners, respectively (Figure 1A). The Arabidopsis interactome within IntAct shows a power law distribution, which implies a scale-free network (Figure 1B). We compared the Arabidopsis protein–protein interactions in IntAct to those in the TAIR and BioGRID databases (Figure 1C). We observed that there is a significant overlap of curated publications and protein interactions between the three data sets. To reduce this redundant effort, the International Molecular Exchange Consortium (IMEx) was established to encourage data deposition and sharing by all of the participating databases, including IntAct and BioGRID, to ensure maximal data availability to the scientific community (http://imex.sf.net; Orchard et al., 2007). TAIR also makes BioGRID and IntAct data available via its website.
Figure 1.
Characteristics of Curated Arabidopsis Interactions within IntAct and Comparison with Those of TAIR and BioGRID.
(A) The Arabidopsis interactions within IntAct. We have drawn the largest connected component (3949 interactions among 1848 Arabidopsis proteins) among Arabidopsis interactions within the IntAct database (a total of 4674 unique interactions among 2334 Arabidopsis proteins) using the Cytoscape tool (http://www.cytoscape.org/). Right three panels are the top three hub proteins, including GRF2 (AT1g78300; 131 interacting partners), CAM4 (AT5g37780; 126 partners), and CAM7 (AT3g43810; 121 partners).
(B) The distributions of numbers of interacting neighbors in the currently curated Arabidopsis interactomes within IntAct, TAIR, and BioGRID.
(C) Comparison of IntAct interactome to other well-known Arabidopsis interaction databases, including TAIR and BioGRID.
Central to the usefulness of these data sets is the accuracy of the database record for each experiment. This is dependent on curation accuracy, which has been a topic of debate recently. Two recent studies found very different levels of curation accuracy within the data sets sampled using different approaches (Cusick et al., 2009a; Salwinski et al., 2009). Salwinski et al. calculated an error rate of 2% for the IntAct Arabidopsis protein–protein interaction data set, while Cusick et al. calculated an error rate of 10.7% for the same data set (Cusick et al., 2009b; Salwinski et al., 2009). The conclusions that can be drawn from this debate are that expert appraisal of repositories’ veracity is central to quality assurance and that all participants in the production, capture, storage, and dissemination of the data can take steps to lower error rates. Some specific steps that researchers can take to assist curators to improve accuracy are discussed at the end of this article.
All three databases (IntAct, TAIR, and BioGRID) have developed versatile user interfaces and search engines, and each welcomes user feedback to improve its service. BioGRID and IntAct provide data and search results in a tabular or XML PSI-MI (Protein Standards Initiative Molecular Interaction) compliant format. TAIR makes tab-delimited file formats available, which also allows the user to programatically interrogate large data sets. All data sets mentioned in this article, including subcellular location prediction data files and Arabidopsis-specific data sets, are available at the following URL: ftp://ftp.ebi.ac.uk/pub/databases/IntAct/various/2010-LeeEtAl/.
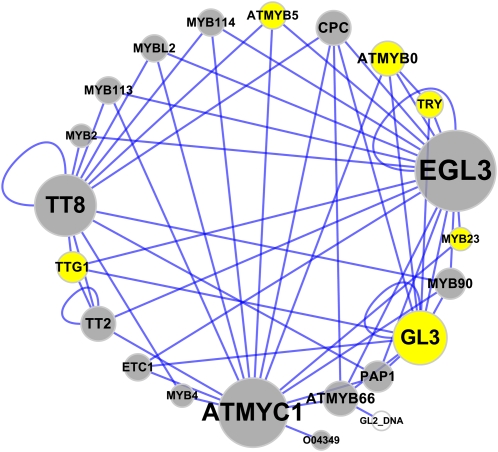
Once interaction data are downloaded, researchers typically will wish to explore the data visually. IntAct has now incorporated Cytoscape (http://www.cytoscape.org/; Cline et al., 2007), a popular open source network visualization tool that has a large number of plug-in packages that add extra functionality beyond that of the core software (Shannon et al., 2003; Cline et al., 2007). BioGRID has provided similar functionality using the Osprey visualization package (http://biodata.mshri.on.ca/osprey/servlet/Index). Figure 2 displays an aggregated data set derived from several publications curated within IntAct involving proteins that function in trichome differentiation (Gene Ontology term GO:0010026). The network is visualized using Cytoscape and the BINGO plug-in, which annotates GO process terms to each protein (Maere et al., 2005). It is apparent that GO-annotated proteins interact with many other proteins not annotated as playing a role in trichome differentiation. A recent report indicates that TT8 (transcription factor TT8, TAIR locus identifier AT4g09820), annotated by GO as involved in anthocyanin production, may also play a role in trichome formation (Maes et al., 2008). This finding provides an example of how protein interaction data can help refine our understanding of protein function. The integration of network data with existing biological information, such as GO terms or expression data, elevates the network from a static representation to a condition-dependent dynamic structure with added biological context and utility. The plug-in architecture of Cytoscape is one solution to the pressing challenge of studying the dynamic nature of protein networks, but other tools are widely available. Suderman and Hallett (2007) provide a recent and highly informative review of this topic.
Figure 2.
Protein Network of Proteins Involved in Trichome Differentiation from the IntAct Protein–Protein Interaction Data Set.
Interactions of proteins (in gray) annotated as being involved in trichome development (GO term GO:0010026) in Arabidopsis. Proteins currently not annotated as involved in trichome development are in yellow. Node and node font sizes are proportional to number of interactions. The GL2_DNA (GLABRA2 gene promoter MYB binding site) is not circled to indicate that it is a nucleic acid not a protein molecule.
COMPARISON OF LITERATURE-CURATED AND PREDICTED PROTEIN INTERACTION STUDIES
A recent study predicted 17,624 protein interactions in Arabidopsis, but the authors reported that of these, only 75 predicted interactions occurred in publicly available data sets of experimentally detected interactions (De Bodt et al., 2009). These authors also reported that of the 5840 predicted interactions from a study by Geisler-Lee et al. (2007), 37 appeared in the same publicly available data sets (De Bodt et al., 2009). Such low overlap likely occurs for two reasons: First, the interactions that have been curated from the literature to date represent only a fraction of the complete Arabidopsis interactome. Second, the predictive approaches developed so far have failed to detect many experimentally proven and highly probable interactions.
Both of the studies above (Geisler-Lee et al., 2007; De Bodt et al., 2009) were based on an interolog approach, which infers an interaction between two proteins if orthologs to both proteins have been shown to interact in a reference species. By contrast, in the IntAct data set, many Arabidopsis protein interactions involve a single orthologous protein (from at least one reference species) and a protein that does not have an ortholog in that reference species (Table 1). This phenomenon probably arises from a widespread evolutionary expansion of conserved protein roles within the plant lineage. An equally large number of protein interactions occur between proteins that both lack orthologs in the reference species (Table 1). If these trends hold for complete plant interactomes, predictive tools based solely on an interolog approach will have limited coverage of the plant interactome but should provide information on the most highly conserved protein networks.
Table 1.
Analysis of Orthologous Protein Interactions in the IntAct Arabidopsis Data Set
| Interaction Type | Number in Data Set |
| Ortholog interacting with ortholog | 424 |
| Ortholog interacting with nonortholog | 2016 |
| Nonortholog interacting with nonortholog | 1735 |
Reference species were Homo sapiens, Caenorhabditis elegans, Saccharomyces cerevisiae, and Drosophila melanogaster as used in the studies of Geisler-Lee et al. (2007) and De Bodt et al. (2009). The source ortholog database was InParanoid, as used by the two studies.
Two recent studies employed supervised learning approaches to predict protein interactions in Arabidopsis (Cui et al., 2008; Lin et al., 2009). These approaches used features such as shared GO annotation, orthology, and gene coexpression to predict protein interactions and predicted 28,062 (Cui et al., 2008) and 224,206 (Lin et al., 2009) protein interaction pairs, respectively. To estimate the false-negative rate of these predicted interactions, we downloaded a data set of 1418 experimentally measured protein interactions from IntAct that were not used by either of the predictive studies above. The study by Cui et al. (2008) correctly predicted 9% of the measured protein interactions, suggesting a false-negative rate of 91%. The study by Lin et al. (2009) correctly predicted 19% of the measured protein interactions, suggesting a false-negative rate of 81%. By way of comparison, random pairs of Arabidopsis proteins consistently returned false-negative rates of >99.8%. Although these false-negative rates indicate that many highly probable interactions were not predicted in either study (Cui et al., 2008; Lin et al., 2009), further development of supervised learning approaches may have potential as predictive tools. In contrast with false-negative rates, estimation of false positive rates is much more difficult given that the current literature-curated databases do not record negative protein interaction results. We therefore did not try to estimate false-positive rate here, although it is clear that IntAct and other literature-curated interaction databases would do well to record negative results when available.
ADDING VALUE TO PROTEIN INTERACTION DATA SETS
The determination of subcellular locations of proteins is not a trivial task, and, frequently, different experimental studies report contradictory locations for the same protein. While many proteins may occupy multiple cellular locations during their lifetime, it is becoming apparent that many protein locations reported to date should be viewed with caution unless supported by both targeting and accumulation studies (Millar et al., 2009). Millar et al. (2009) highlighted the importance of understanding the relationship between subcellular location, protein interaction, and protein function. As these authors commented, protein interaction data can provide useful information for predicting protein subcellular location, since interacting proteins typically represent colocalized functional units of a biological process. To illustrate how curated protein interaction data sets can be applied to address current problems in plant biology, we used a recently developed DC-kNN method (K. Lee et al., 2008) that uses protein interaction data to predict protein locations with high accuracy in multiple species.
We assembled a set of known locations based on the following sources: IntAct subcellular location data; green fluorescent protein data from the Arabidopsis Subcellular Database (Heazlewood et al., 2007); data from both ARAPerox (Reumann et al., 2004) and AtNoPDB (Brown et al., 2005); and localization data annotated by the GO project having evidence codes EXP (inferred from experiment), IDA (inferred from direct assay), or IPI (inferred from interaction) (Rhee et al., 2008). From these sources, we were able to assign 2032 proteins with 3144 annotated subcellular locations covering 15 specific cellular compartments (Table 2).
Table 2.
Subcellular Locations Considered in the Prediction of Protein Cellular Location Using Protein Interaction Data
| Location | Number of Known Proteins | Number of Predicted Locations among Unknown Test Proteins |
| Apoplast | 68 | 750 |
| Cell plate | 14 | 1,094 |
| Chloroplast | 399 | 506 |
| Cytoskeleton | 43 | 851 |
| Cytosol | 397 | 937 |
| Endoplasmic reticulum | 118 | 670 |
| Extracellular | 31 | 1,523 |
| Golgi apparatus | 79 | 822 |
| Mitochondrion | 332 | 1,523 |
| Nucleolus | 132 | 188 |
| Nucleus | 830 | 4,149 |
| Peroxisome | 138 | 2,164 |
| Plastid | 136 | 1,894 |
| Plasma membrane | 181 | 1,110 |
| Vacuole | 246 | 607 |
| All | 3,144 | 18,788 |
Next, we assembled a set of protein features for predicting a protein's subcellular location, including its sequence, chemical properties, motifs, and functions (so-called single protein features) along with characteristics of its network neighbors (capturing neighbors’ single protein features and their subcellular locations if known) (see Supplemental Methods online for detailed information on location prediction). Using the DC-kNN method (K. Lee et al., 2008) together with the known locations, we selected a subset of the protein features that were informative for predicting each subcellular location of Arabidopsis (see Supplemental Figures 1 to 3 online). Finally, based on the selected feature set for each location (see Supplemental Figure 2 online), we predicted the locations of all 25,497 Arabidopsis proteins for which subcellular locations had not yet been reported. It was possible to predict 18,788 locations for 13,749 proteins of these with high confidence (P value < 0.05 and corresponding to a false discovery rate of 0.35; see Supplemental Data Set 1 online for all predicted results).
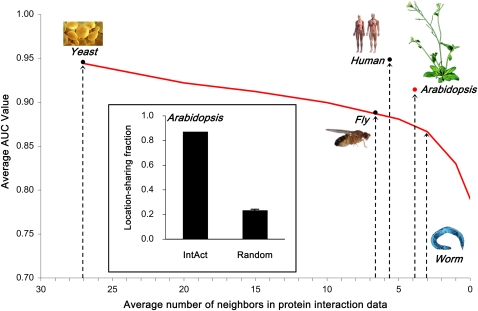
Using cross-validation on the known locations, we observed a very high accuracy of 0.914 average AUC (area under receiver operator characteristic curve) (Molodianovitch et al., 2006; Streiner and Cairney, 2007) for prediction of subcellular location (the red “x” marker in Figure 3; see Supplemental Figure 3 online for the receiver operator characteristic curves of individual locations). Given that the average number of interacting neighbors (referred to as average degree) of Arabidopsis interactome is 3.9, the performance is a little higher than the previous simulation result using yeast interactions (Figure 3). This might result from the higher fraction (87%) of location-sharing interactions of Arabidopsis proteins than that (59%) of yeast interactions that are used in the simulation (see the inset of Figure 3). As expected, the fraction was much higher than for random networks (z-score = 40 and P value ≈ 0 based on 100 location-permutated random networks). The high performance of location prediction mainly depends on the use of group features of interacting neighbors as well as characteristics of self proteins (K. Lee et al., 2008). For example, proteins in mitochondrion have a weak degree of self purity of location, which means that many of the proteins can reside at other locations, including chloroplast (see Supplemental Figure 4A online). However, many interacting partners of mitochondrial protein reside also in the mitochondrion, which means a high degree of neighbor purity (see Supplemental Figure 4A online). Location prediction using a DCkNN method thus showed relatively high performance on the mitochondrion (0.95 AUC; see Supplemental Figure 4C online). For the case of cytosol, however, both degrees of self and neighbor purities are low, which resulted in relatively low performance even using the DCkNN method with both kinds of features.
Figure 3.
Performance of Predicted Locations on Location-Known Arabidopsis Proteins.
The red “x” denotes the performance of predicted locations on 2032 location-known Arabidopsis proteins using IntAct interactions based on leave-one-out cross-validation. The x axis and y axis are the average degree (or number) of interacting partners in protein-protein interaction data and the average AUC value, respectively. Performance on yeast, fly, worm, and human were redrawn from our previous work (courtesy of Nucleic Acid Research). The inset denotes the fraction of location-sharing interactions among the Arabidopsis interactome within IntAct. In the calculation of location-sharing fraction, we used the 2032 known locations only. For a random case, we randomly selected 100 location-permutated random networks and averaged them.
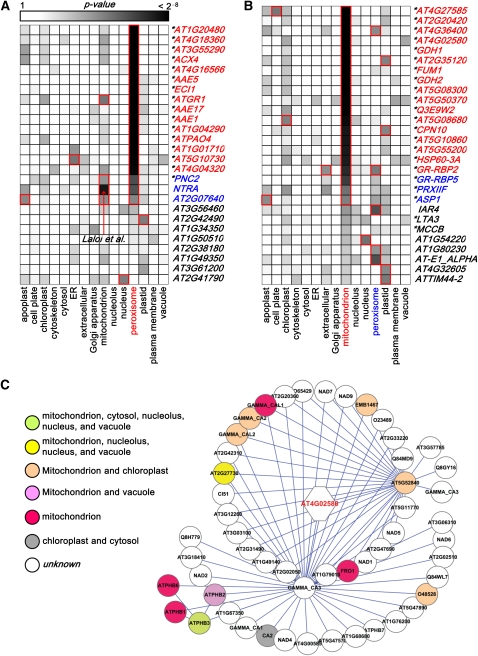
Next, we assessed the accuracy of the newly predicted locations using two recent studies of peroxisomal and mitochondrial proteomes that employed mass spectrometry (Eubel et al., 2008; C.P. Lee et al., 2008). The selected studies employed stringent purification protocols for peroxisomal and mitochondrial proteins, which would seem to offer the highest quality data of this type. These two data sets were reserved for testing prediction accuracy and thus were not included in the DC-kNN training set.
When the proteins detected in these studies were compared with the predicted locations, it was shown that the predictions based on the IntAct interactions performed quite well, with many agreements between the in vivo study and in silico predictions (Figure 4). For example, C.P. Lee et al. (2008) found peroxisomal locations for 26 proteins by mass spectrometry that were not included in the DC-kNN training set; of these, 18 proteins were predicted as peroxisomal with high confidence (Figure 4A; sensitivity = 0.69). Strikingly, our predictions also support the previously reported dual targeting of NTRA (AT2G17420), NADPH-dependent thioredoxin reductase A, to both mitochondria (highest score) and peroxisomes (second highest score) (Laloi et al., 2001). We suppose that the high accuracy of prediction results from the fact that the DC-kNN method uses known subcellular locations of the extended network neighborhood, in addition to single protein features of each neighbor (see Figure 4C for an example of AT4G02580, NADH-ubiquinone oxidoreductase 24-kD subunit, which is part of a protein network containing many characterized components of the mitochondrial electron transport chain). Thus, the method will be most accurate in large networks seeded with a sizable fraction of proteins whose locations are already known. A number of subcellular locations have very limited numbers of experimentally proven protein residents; therefore, our predictions in those locations will tend to be less robust than those subcellular locations with large cohorts of experimentally proven protein residents. By referring to the results section, the reader can make a judgment about the level of caution with which they should view the predictions for each location.
Figure 4.
Validation of Predicted Locations on Independent Test Data Sets.
(A) and (B) Heat maps of validation results on two independent test data sets for peroxisome (A) and mitochondrion (B), respectively. Each column corresponds to each test protein, and each row corresponds to one of 15 locations tested. Each cell shows the significance of prediction assigned to each protein and location (with grayscale representing the P value); darker gray means a higher significance. Proteins in red have <0.01 P value, and proteins in blue have <0.05. Predicted locations (P value <0.05) are marked in red rectangles, and asterisks indicate the proteins with the highest predicted scores in the peroxisome (A) or mitochondrion (B), respectively. Especially in the case of NTRA (AT2G17420) in (A), our prediction showed a high score on mitochondrion in addition to peroxisome. The mitochondrion prediction of NTRA is also confirmed by other literature (Laloi et al., 2001).
(C) Up to second network neighbors of AT4G02580 (NADH-ubiquinone oxidoreductase 24-kD subunit) in the IntAct interactome of Arabidopsis proteins. Different color on each protein means different locations of the protein. AT4G02580 has many network neighbors known to locate in mitochondrion (12 neighbors among 13 location-known proteins). Thus, our prediction of AT4G02580 is mitochondrion.
Subcellular localization is a prerequisite to interaction and function, but in order to discover biologically meaningful protein networks, these data need to be integrated with other data sets to build a robust and verifiable model to guide future research (Fukushima et al., 2009). Currently, gene coexpression data sets are widely available (Brady and Provart, 2009), and a commonly held assumption in system biology is that coexpression of genes is a strong indicator of possible interaction of their translated products. A recent study, however, demonstrated that in Arabidopsis, only a low proportion of experimentally determined interacting protein pairs were coexpressed (Lysenko et al., 2009). While the Arabidopsis interaction data set is limited and may be unrepresentative of the full interactome, this finding might indicate that other factors, such as posttranslational protein modification and protein disorder, may be of much more relevant to protein interaction than coexpression of the encoding genes (Stein et al., 2009). Although many studies have been performed in these areas, the integration and analysis of these data with protein interaction data sets to predict and verify protein interaction networks is challenging and in its early stages.
FUTURE CONSIDERATIONS
Unfortunately, there are few literature curators, while there are a growing army of researchers producing and publishing their results. This inevitably results in a backlog of data buried within the primary literature, resistant to modern analytical approaches. But it is widely agreed that the capture, storage, and dissemination of this information is vital to modern research (Jorrin et al., 2007). How can the plant science community resolve this dilemma and ensure that the efforts of hundreds of dedicated scientists are maximized to gain new insights into the complexity of plant biology?
The Human Proteome Organization–Protein Standards Initiative has published guidelines to outline the minimum information required for reporting a molecular interaction experiment (MIMIx) (Orchard et al., 2007). MIMIx outlines the minimum information required to describe all relevant aspects of the interaction experiment while minimizing the burden placed on the researchers generating the data. This standard does not dictate how to conduct research but, if followed, allows researchers to set out their findings in a manner that can be understood by the widest audience, including curators (Orchard and Taylor, 2009). Following standards like MIMIx is the most effective action a researcher can take to assist database curators and ensure the efficient and accurate deposition of their data into a relevant database, the content of which is often the raw materials for bioinformatic analysis. Currently, up to half of curators’ working hours are taken up with identifying missing information required to correctly represent the data in a curated paper, and this bottleneck would be removed if MIMIx guidelines were followed by the reporting authors. The IMEx website (http://imex.sourceforge.net/MIMIx/index.html) and a recent article by Orchard et al. (2007) document concrete entry points for efficient deposition and dissemination of molecular interaction data (Orchard et al., 2007). We contend that direct data deposition as part of the manuscript publication process will ensure a higher visibility of both the data set and the publication and will result in a higher data quality through direct author validation of the data representation in the database.
As protein interaction studies expand and evolve, the relationship between researcher, publisher, and repository will also have to evolve to allow researchers access to the bulk and breadth of data. It would appear that, although text mining can assist manual curation, it has yet to fulfill its obvious potential for reliable high-throughput literature curation (Winnenburg et al., 2008; Kabiljo et al., 2009). On the other hand, large-scale funding to create a cadre of dedicated database curators has not been forthcoming either and would seem unlikely in the near future if science budgets contract due to the recent economic downturn. It would appear that the increased use of standardized reporting and researcher-initiated deposition of data are our best hope of maintaining and improving critical resources for the future of plant research. By minimizing the burden of adopting standards and by working with the research community to streamline data deposition, we hope the full potential of the efforts of hundreds of scientists can be harnessed for the benefit of agriculture and plant biology.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Feature Set Selection for the Subcellular Locations of Arabidopsis Proteins Based on AUC Measure.
Supplemental Figure 2. Selected Feature Sets and Model for Each Subcellular Location of Arabidopsis Proteins.
Supplemental Figure 3. ROC Curves of Generated Models for Individual Locations of Arabidopsis.
Supplemental Figure 4. Location Purity of Self and Neighbor Proteins and Performance for Individual Locations of Arabidopsis.
Supplemental Methods. The Detailed Information on Location Prediction of Arabidopsis Proteins.
Supplemental Data Set 1. The Predicted Locations of Location-Untagged Arabidopsis Proteins.
Supplementary Material
Acknowledgments
K.L. was supported by the Brain Korea 21 Project for Medical Science, Ajou University and the Korea Food & Drug Administration in 2010 (Grant 10182KFDA992). T.I. was supported by Grant GM070743 from the National Institute of General Medical Sciences. D.T., P.A., and the IntAct database team are funded by EU Grant QLRI-CT-2001-00015 under the Research and Technological Development program “Quality of Life and Management of Living Resources” and EU Contract 21902 “Felics–Free European Life-Science Information and Computational Services.”
References
- Aranda B., et al. (2010). The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38: D525–D531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Provart N.J. (2009). Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz B.J., Stark C., Reguly T., Boucher L., Breitkreutz A., Livstone M., Oughtred R., Lackner D.H., Bahler J., Wood V., Dolinski K., Tyers M. (2008). The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 36: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W., Shaw P.J., Shaw P., Marshall D.F. (2005). Arabidopsis nucleolar protein database (AtNoPDB). Nucleic Acids Res. 33: D633–D636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M.S., et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li P., Li G., Xu F., Zhao C., Li Y., Yang Z., Wang G., Yu Q., Li Y., Shi T. (2008). AtPID: Arabidopsis thaliana protein interactome database–an integrative platform for plant systems biology. Nucleic Acids Res. 36: D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick M.E., et al. (2009a). Literature-curated protein interaction datasets. Nat. Methods 6: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick M.E., et al. (2009b). Addendum: Literature-curated protein interaction datasets. Nat. Methods 6: 934–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Proost S., Vandepoele K., Rouze P., Van de Peer Y. (2009). Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics 10: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Meyer E.H., Taylor N.L., Bussell J.D., O'Toole N., Heazlewood J.L., Castleden I., Small I.D., Smith S.M., Millar A.H. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Redestig H., Arita M., Saito K. (2009). Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13: 532–538 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J., O'Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrin J.V., Maldonado A.M., Castillejo M.A. (2007). Plant proteome analysis: A 2006 update. Proteomics 7: 2947–2962 [DOI] [PubMed] [Google Scholar]
- Kabiljo R., Clegg A.B., Shepherd A.J. (2009). A realistic assessment of methods for extracting gene/protein interactions from free text. BMC Bioinformatics 10: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Rayapuram N., Chartier Y., Grienenberger J.M., Bonnard G., Meyer Y. (2001). Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., O'Toole N., Millar A.H. (2008). Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell. Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Lee K., Chuang H.Y., Beyer A., Sung M.K., Huh W.K., Lee B., Ideker T. (2008). Protein networks markedly improve prediction of subcellular localization in multiple eukaryotic species. Nucleic Acids Res. 36: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Hu B., Chen L., Sun P., Fan Y., Wu P., Chen X. (2009). Computational identification of potential molecular interactions in Arabidopsis. Plant Physiol. 151: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko A., Hindle M.M., Taubert J., Saqi M., Rawlings C.J. (2009). Data integration for plant genomics–exemplars from the integration of Arabidopsis thaliana databases. Brief. Bioinform. 10: 676–693 [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Maes L., Inze D., Goossens A. (2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148: 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Carrie C., Pogson B., Whelan J. (2009). Exploring the function-location nexus: Using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodianovitch K., Faraggi D., Reiser B. (2006). Comparing the areas under two correlated ROC curves: Parametric and non-parametric approaches. Biom. J. 48: 745–757 [DOI] [PubMed] [Google Scholar]
- Morsy M., Gouthu S., Orchard S., Thorneycroft D., Harper J.F., Mittler R., Cushman J.C. (2008). Charting plant interactomes: Possibilities and challenges. Trends Plant Sci. 13: 183–191 [DOI] [PubMed] [Google Scholar]
- Orchard S., Kerrien S., Jones P., Ceol A., Chatr-Aryamontri A., Salwinski L., Nerothin J., Hermjakob H. (2007). Submit your interaction data the IMEx way: A step by step guide to trouble-free deposition. Proteomics 7(Suppl 1): 28–34 [DOI] [PubMed] [Google Scholar]
- Orchard S., Taylor C.F. (2009). Debunking minimum information myths: One hat need not fit all. N. Biotechnol 25: 171–172 [DOI] [PubMed] [Google Scholar]
- Poole R.L. (2007). The TAIR database. Methods Mol. Biol. 406: 179–212 [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.Y., Wood V., Dolinski K., Draghici S. (2008). Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 9: 509–515 [DOI] [PubMed] [Google Scholar]
- Rodriguez H., et al. (2009). Recommendations from the 2008 International Summit on Proteomics Data Release and Sharing Policy: The Amsterdam principles. J Proteome Res. 8: 3689–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L., et al. (2009). Recurated protein interaction datasets. Nat. Methods 6: 860–861 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Pache R.A., Bernado P., Pons M., Aloy P. (2009). Dynamic interactions of proteins in complex networks: A more structured view. FEBS J. 276: 5390–5405 [DOI] [PubMed] [Google Scholar]
- Streiner D.L., Cairney J. (2007). What's under the ROC? An introduction to receiver operating characteristics curves. Can. J. Psychiatry 52: 121–128 [DOI] [PubMed] [Google Scholar]
- Suderman M., Hallett M. (2007). Tools for visually exploring biological networks. Bioinformatics 23: 2651–2659 [DOI] [PubMed] [Google Scholar]
- Uhrig J.F. (2006). Protein interaction networks in plants. Planta 224: 771–781 [DOI] [PubMed] [Google Scholar]
- Winnenburg R., Wachter T., Plake C., Doms A., Schroeder M. (2008). Facts from text: Can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief. Bioinform. 9: 466–478 [DOI] [PubMed] [Google Scholar]
- Yu J., Finley R.L., Jr. (2009). Combining multiple positive training sets to generate confidence scores for protein-protein interactions. Bioinformatics 25: 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.