Abstract
The hemicellulose glucuronoxylan (GX) is a major component of plant secondary cell walls. However, our understanding of GX synthesis remains limited. Here, we identify and analyze two new genes from Arabidopsis (Arabidopsis thaliana), IRREGULAR XYLEM9-LIKE (IRX9-L) and IRX14-LIKE (IRX14-L) that encode glycosyltransferase family 43 members proposed to function during xylan backbone elongation. We place IRX9-L and IRX14-L in a genetic framework with six previously described glycosyltransferase genes (IRX9, IRX10, IRX10-L, IRX14, FRAGILE FIBER8 [FRA8], and FRA8 HOMOLOG [F8H]) and investigate their function in GX synthesis. Double-mutant analysis identifies IRX9-L and IRX14-L as functional homologs of IRX9 and IRX14, respectively. Characterization of irx9 irx10 irx14 fra8 and irx9-L irx10-L irx14-L f8h quadruple mutants allows definition of a set of genes comprising IRX9, IRX10, IRX14, and FRA8 that perform the main role in GX synthesis during vegetative development. The IRX9-L, IRX10-L, IRX14-L, and F8H genes are able to partially substitute for their respective homologs and normally perform a minor function. The irx14 irx14-L double mutant virtually lacks xylan, whereas irx9 irx9-L and fra8 f8h double mutants form lowered amounts of GX displaying a greatly reduced degree of backbone polymerization. Our findings reveal two distinct sets of four genes each differentially contributing to GX biosynthesis.
Secondary cell walls constitute the major form of biomass produced by plants and represent a material that provides an important source of renewable and sustainable energy. The major components of the secondary cell wall are the polysaccharides cellulose and hemicellulose together with the more complex lignin polymer that is made up of phenylpropanoid subunits. Although considerable understanding has been gained about the enzymes required for cellulose and lignin synthesis (Boerjan et al., 2003; Scheible and Pauly, 2004; Lerouxel et al., 2006; Somerville, 2006), little is currently understood about hemicellulose. Although the plant cell wall represents a rich source of stored energy in the form of polysaccharides, the presence of lignin and hemicellulose in the secondary cell wall reduces the accessibility of cellulases to cellulose and hence negatively affects the net energy yield from plant biomass. One potential route to improve the energy yield is to alter the cell wall structure or composition via modulation of hemicellulose biosynthesis. It has been found that down-regulation of the poplar (Populus spp.) PoGT47C gene that is a homolog of the Arabidopsis (Arabidopsis thaliana) FRAGILE FIBER8 (FRA8) can lead to an increase in the saccharification yield from woody material (Lee et al., 2009b).
Glucuronoxylan (GX) represents the predominant form of xylan in plant secondary cell walls and is comprised of a linear backbone of β(1-4)-linked Xyl subunits with α-linked side branches of GlcUA and 4-O-methyl GlcUA (Me-GlcUA). The Xyl residues can also be substituted with arabinosyl or acetyl groups (Ebringerova and Heinze, 2000). Analysis of xylan isolated from Arabidopsis indicates that only GlcUA and Me-GlcUA side branches are present (Peña et al., 2007), and that they occur on average once every eight Xyl residues (Brown et al., 2007). In addition, a tetrasaccharide sequence, β-d-Xyl-(1,4)-β-d-Xyl-(1,3)-α-l-Rha-(1,2)-α-d-GalUA-(1,4)-d-Xyl is found at the reducing end of the xylan chain. This tetrasaccharide is conserved in a range of plant species, suggesting that it plays an important role either in the synthesis or function of the xylan (Johansson and Samuelson, 1977; Andersson et al., 1983; Peña et al., 2007). Several genes encoding enzymes that appear to be required for xylan synthesis have been identified either via analysis of Arabidopsis mutants that display collapsed xylem vessels in stem tissues (Turner and Somerville, 1997) or by gene identification using transcriptomic-based approaches (Brown et al., 2005; Persson et al., 2005). The genes identified include members of the GT47 (FRA8/IRX7 and IRX10), glycosyltransferase family 43 (GT43; IRX9 and IRX14), and GT8 families (IRX8 and PARVUS) of glycosyltransferases (Zhong et al., 2005; Bauer et al., 2006; Brown et al., 2007, 2009; Lee et al., 2007b; Peña et al., 2007; Persson et al., 2007; Wu et al., 2009). Analysis of mutants in each of these genes indicates that IRX9, IRX10, and IRX14 encode enzymes that function as xylosyltransferases in the synthesis of the β-1-4-xylan backbone while IRX8, FRA8, and PARVUS appear to be involved in synthesis of the reducing end tetrasaccharide structure (Bauer et al., 2006; Brown et al., 2007; Lee et al., 2007a; Peña et al., 2007). Additionally, it has been shown that the poplar homolog of IRX9, PoGT43B, can complement the Arabidopsis irx9 mutant and that it is specifically expressed in the secondary xylem in wood (Zhou et al., 2007), supporting a role for PoGT43B during xylan biosynthesis in poplar wood. Similar results have also been found for two poplar homologs of the PARVUS gene that are able to complement the Arabidopsis parvus mutant (Kong et al., 2009). However, to date there have been no direct demonstrations of enzyme activity for any of the enzymes, leaving their specific functions open to question.
Although genes that putatively function in different phases of GX synthesis have now been identified, there is little understood about the biosynthesis mechanism. One possibility is that the β-1-4-xylan backbone is synthesized first and the reducing end tetrasaccharide added subsequently to terminate chain elongation. Alternatively the reducing end tetrasaccharide could act as a primer for elongation of the xylan backbone via addition of Xyl residues at the nonreducing end (York and O'Neill, 2008). Currently available data supports the first model as it has been found from NMR analysis of xylan isolated from the irx8 and fra8 mutants that β-1-4-xylan chains can be made that apparently lack the reducing end tetrasaccharide (Peña et al., 2007). Furthermore, the irx9, irx10, and irx14 mutants have a reduced backbone length but retain the reducing-end tetrasaccharide. Attempts to demonstrate xylosyltransferase activity for IRX9 by heterologous expression in yeast (Saccharomyces cerevisiae) were not successful, leading to the suggestion that multiple-subunit enzyme complexes may function in GX synthesis (Brown et al., 2007; Peña et al., 2007; York and O'Neill, 2008). These models cannot be fully substantiated until the full complement of necessary synthesis enzymes is identified and a functional assay is established.
Recently, two independent studies have identified IRX10-L as a homolog of the Arabidopsis IRX10 gene (Brown et al., 2009; Wu et al., 2009). Double mutants between irx10 and irx10-L displayed a strongly enhanced phenotype, suggesting functional redundancy between these genes. A similar finding was also made for FRA8 and its paralog FRA8 HOMOLOG (F8H; Lee et al., 2009a). Analysis of the double-mutant combinations has greatly improved our understanding of the importance of xylan synthesis for the secondary cell wall development.
This study aimed to gain a better understanding about the respective functions and interactions of the complete set of genes that function in GX biosynthesis by initially characterizing the IRREGULAR XYLEM9-LIKE (IRX9-L) and IRX14-LIKE (IRX14-L) homologs of the IRX9 and IRX14 Arabidopsis genes. The severe phenotypes of the irx9 irx9-L and irx14 irx14-L double mutants demonstrate previously unrecognized important roles for IRX9-L and IRX14-L. The double mutants further revealed effects on the synthesis of GX with consequences for the structure and function of the secondary cell wall. Additional analysis of the fra8 and f8h mutants questions whether their sole function is synthesis of the GX reducing-end tetrasaccharide, raising new questions that need to be addressed if hemicellulose synthesis is to be understood fully.
RESULTS
Duplication of the GX Biosynthesis Machinery in Arabidopsis
The partial functional redundancy exhibited by the IRX10/IRX10-L and FRA8/F8H gene pairs (Brown et al., 2009; Lee et al., 2009a; Wu et al., 2009) from the GT47 subfamily I (Zhong and Ye, 2003; Supplemental Fig. S1) has highlighted the importance of studying both single- and double-mutant combinations to gain a complete understanding of the enzyme function. To establish whether homologs also exist for other putative GX synthesis enzymes, full-length IRX9 and IRX14 sequences were used to BlastP search against the Arabidopsis protein database. A single closely related sequence exists for IRX14 encoded by At5g67230 (IRX14-L) that displays 72% amino acid sequence identity and 83% similarity. The closest IRX9-related sequence is encoded by At1g27600 (IRX9-L) and exhibits 44% sequence identity and 64% similarity with IRX9. The IRX9, IRX9-L, IRX14, and IRX14-L genes belong to the glycosyltransferase 43 family (GT43; http://www.cazy.org/fam/GT43.html), which has members from vertebrates, invertebrates, and plants and is defined by a conserved GlcAT domain (Fondeur-Gelinotte et al., 2006). A phylogenetic tree including sequences from rice (Oryza sativa), Medicago truncatulata, poplar, and Arabidopsis places IRX14 and IRX14-L together in a subbranch separate from IRX9 and IRX9-L (Supplemental Fig. S2).
Homologous Pairs of Genes Encoding Putative GX Synthesis Enzymes Show Partial Functional Redundancy and Dosage Dependency
Two T-DNA insertion alleles were isolated for irx9 within exon 1 (Salk_058238; irx9-1) and exon 2 (Salk_057033; irx9-2) and two for irx9-L located in close proximity within intron 1 (Salk_037323, irx9-L1 and Salk_037330, irx9-L2; Fig. 1A). Reverse transcription-PCR showed that there was no detectable message for either the irx9-L1 or irx9 alleles (Supplemental Fig. S3). Single T-DNA insertion alleles were isolated for irx14 in exon 1 (Salk_038212) and for irx14-L in exon 2 (Salk_066961; Fig. 1C). Both irx9 single-mutant alleles displayed reduced stature in 4-week-old plants grown on soil, although under our conditions the phenotype was somewhat weaker than previously reported (Brown et al., 2005). The irx9-L1 and irx9-L2 alleles in contrast appeared similar to wild type (Fig. 1B; data not shown for irx9-L2). Neither the irx14 nor the irx14-L alleles displayed a visible phenotype (Fig. 1D; Supplemental Table S1) as found previously for irx14 (Brown et al., 2007). To test for genetic redundancy, crosses were made to create the irx9-1 irx9-L2, irx9-2 irx9-L1, and irx14 irx14-L double-mutant combinations. All double mutants exhibited a stunted growth habit, forming a small rosette with reduced leaf size that did not form an inflorescence stem after 6 weeks (Fig. 1, B and D; Supplemental Table S1), reminiscent of the phenotypes shown by irx10 irx10-L (Brown et al., 2009; Wu et al., 2009) and fra8 f8h (Lee et al., 2009a). Since both irx9 irx9-L combinations showed similar phenotypes, subsequent work used the irx9-2 and irx9-L1 alleles.
Figure 1.
The irx9 irx9-L, irx14 irx14-L, and fra8 f8h double mutants show similar, severe phenotypes. A, Diagram of the Arabidopsis IRX9 and IRX9-L genes showing positions of exons (black boxes), introns (lines), 5′ and 3′ untranslated regions (white boxes), and the T-DNA insertion sites indicated by black triangles. Primer annealing sites used for genotyping insertion lines are indicated by arrows. B, 30-d-old soil-grown wild-type (WT), irx9-L2, irx9-1, irx9-1/+ irx9-L2, irx9-1 irx9-L2/+, and irx9-1 irx9-L2 double-mutant plants (from left to right). The inset section shows a 10× magnification of the irx9-1 irx9-L2 double mutant. C, Diagram of the Arabidopsis IRX14 and IRX14-L genes showing positions of gene features as annotated in A. D, 30-d-old soil-grown wild-type, irx14-L, irx14, irx14-L irx14/+, irx14-L/+ irx14, and irx14 irx14-L double-mutant plants. The inset section shows a 10× magnification of the irx14 irx14-L double mutant. E, Diagram of the Arabidopsis FRA8 and F8H genes showing positions of positions of gene features as annotated in A. F, 30-d-old soil-grown wild-type, f8h, fra8, fra8/+ f8h, fra8 f8h/+, and fra8 f8h double-mutant plants. The inset section shows a 10× magnification of the fra8 f8h double mutant.
Both the irx9 irx9-L/+ and irx14 irx14-L/+ combinations exhibited an intermediate phenotype similar to previously described irx10 irx10-L/+ plants (Brown et al., 2009; Wu et al., 2009). In contrast irx9/+ irx9-L and irx14/+ irx14-L combinations appeared similar to the wild type (Fig. 1, B and D; Supplemental Table S1). To establish whether fra8 f8h/+ or fra8/+ f8h plants also exhibited an intermediate phenotype, T-DNA insertion alleles were identified for FRA8 (Salk120296) and F8H (f8h1, GK-052G08 and f8h2, GK-298C10; Fig. 1E). The single mutants were crossed and homozygotes selected in the F2 generation. In agreement with previous findings, both of the fra8 f8h double-mutant combinations showed a reduced growth habit (Lee et al., 2009a; Fig. 1F). The fra8 f8h/+ plants displayed an intermediate phenotype with reduced stem height and leaf size while fra8/+ f8h plants were indistinguishable from the wild type (Fig. 1F; Supplemental Table S1).
Double Mutants between Homologous Pairs of Putative GX Synthesis Genes Show Defects in Secondary Cell Wall Formation
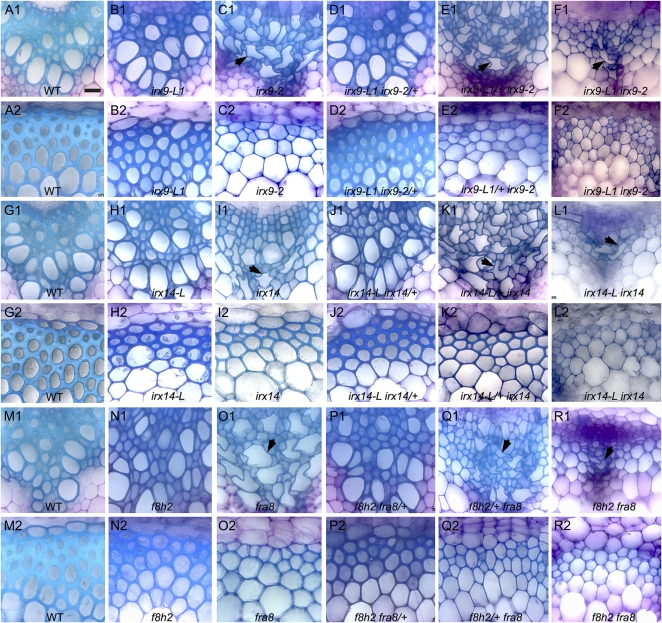
The effect of the irx9 irx9-L and irx14 irx14-L mutant combinations on secondary cell wall formation and xylem vessel morphology was examined in transverse sections taken from the basal region of the stem. The fra8 f8h double mutant was also examined as previous work has focused on root and petiole tissues (Lee et al., 2009a). Each of the double mutants exhibited little if any evidence of secondary cell wall formation in either the xylem or interfascicular regions and xylem vessels were collapsed and misshapen (Fig. 2, F1, F2, L1, L2, R1, and R2). The cell wall diameter of interfascicular fibers and xylem vessels was significantly reduced compared to the wild type for all of the double mutants (Supplemental Table S2). The irx9 irx9-L/+, irx14 irx14-L/+, and fra8 f8h/+ combinations also had reduced secondary cell wall diameters compared to the wild type and had collapsed xylem vessels (Fig. 2, E1, E2, K1, K2, Q1, and Q2; Supplemental Table S2). In contrast, the irx9-L, irx14-L, f8h, irx9/+ irx9-L, irx14/+ irx14-L, and fra8/+ f8h stem sections all appeared similar to the wild type with similar cell wall diameters (Fig. 2, B1, B2, D1, D2, H1, H2, J1, J2, N1, N2, P1, and P2; Supplemental Table S2). The irx9, irx14, and fra8 single mutants all displayed collapsed xylem vessels as previously reported (Fig. 2, C1, I1, and O1; Zhong et al., 2005; Brown et al., 2007; Peña et al., 2007).
Figure 2.
Transverse sections of stem tissue from the irx9 irx9L, irx14 irx14L, and fra8 f8h mutant combinations reveal collapsed xylem vessels and reduced secondary cell wall formation. Transverse sections of stem tissues from wild-type (A, G, M), irx9-L (B), irx9 (C), irx9/+ irx9-L (D), irx9 irx9-L/+ (E), irx9 irx9-L (F), irx14-L (H), irx14 (I), irx14/+ irx14-L (J), irx14 irx14-L/+ (K), irx14 irx14-L (L), f8h (N), fra8 (O), fra8/+ f8h (P), fra8 f8h/+ (Q), and fra8 f8h (R) plants. Xylem (A1–R1), interfascicular region (A2–R2). Arrows indicate collapsed xylem elements. Scale bar, 20 μm.
The IRX9/IRX9-L, IRX14/IRX14-L, and FRA8/F8H Pairs of Genes Show Partially Overlapping Expression Patterns
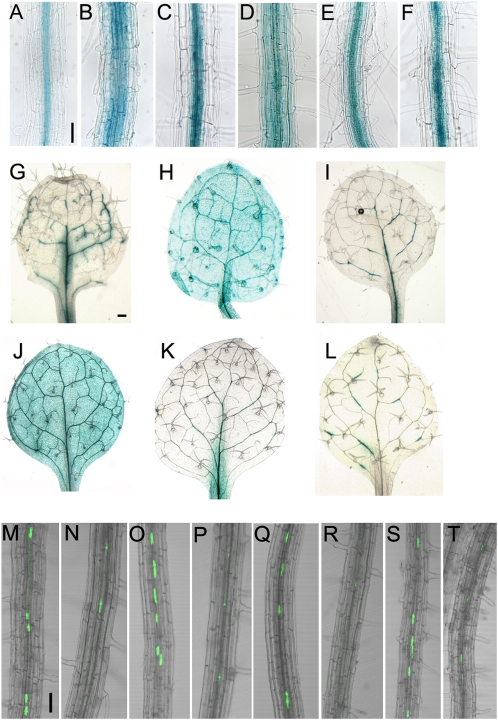
The partial functional redundancy exhibited between the IRX9/IRX9-L, IRX14/IRX14-L, and FRA8/F8H gene pairs indicates that their expression patterns are likely to overlap. Promoter:GUS fusions were made for each of the six genes and introduced into wild-type Arabidopsis. Within root tissues IRX9, IRX14, IRX14-L, and FRA8 expression is predominantly limited to the central stele tissues whereas IRX9-L and F8H expression is found in the peripheral cell layers in addition to the stele (Fig. 3, A–F). Within leaves IRX9, IRX14, IRX14-L, and FRA8 are expressed discontinuously within the vasculature, while IRX9-L and F8H are found more generally expressed throughout leaf tissues (Fig. 3, G–L). To further compare the expression of the pairs of related genes including IRX10 and IRX10-L, the respective promoters were used to drive expression of yellow fluorescent protein (YFP) fused to a nuclear localization signal (YFP-NLS; Kubo et al., 2005). The IRX9, IRX10, IRX14, and FRA8 promoters led to a strong YFP signal within the root xylem tissues whereas the signal from the IRX9-L, IRX10-L, IRX14-L, and F8H promoters was much weaker or absent (Fig. 3, M–T). The Genevestigator database supports the predominantly vascular associated expression patterns for IRX9, IRX10, IRX14, IRX14-L, and FRA8 with the highest levels being found in stem, node, and hypocotyl tissues whereas IRX9-L, IRX10-L, and F8H exhibit a more widespread expression profile (Supplemental Fig. S4; https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004; Hruz et al., 2008).
Figure 3.
Overlapping expression patterns of the FRA8, F8H, IRX9, IRX9-L, IRX14, and IRX14-L genes. GUS expression in root tissues of plants transformed with proFRA8:GUS (A), proF8H:GUS (B), proIRX9:GUS (C), proIRX9-L:GUS (D), proIRX14:GUS (E), proIRX14-L:GUS (F). GUS expression in leaves of plants transformed with proFRA8:GUS (G), proF8H:GUS (H), proIRX9:GUS (I), proIRX9-L:GUS (J), proIRX14:GUS (K), proIRX14-L:GUS (L). Expression of YFP-NLS driven by the IRX10 (M), IRX10-L (N), FRA8 (O), F8H (P), IRX9 (Q), IRX9-L (R), IRX14 (S), and IRX14-L (T) promoters, respectively. Scale bar, 50 μm.
Complementation of the irx9 irx9-L, irx14 irx14-L, and fra8 f8h Mutants
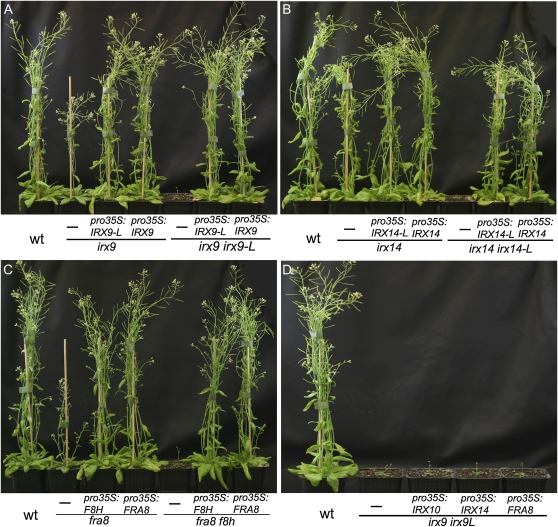
Complementation experiments were carried out to confirm that the irx9 irx9-L, irx14 irx14-L, and fra8 f8h double-mutant phenotypes resulted from T-DNA insertions in the respective genes. The IRX9 and IRX9-L genes were placed under the control of the 35S cauliflower mosaic virus (CaMV) promoter and transformed into the irx9 or irx9-L irx9/+ backgrounds. Plants with a wild-type appearance carrying the 35S:IRX9 transgene either in an irx9 single-mutant or irx9 irx9L double-mutant background were selected in the T1 generation by PCR genotyping, thereby confirming that the double-mutant phenotype is due to T-DNA insertions in the IRX9 and IRX9-L genes (Fig. 4A). A single copy of the 35S:IRX9-L transgene fully rescued both the irx9 single-mutant, and irx9 irx9-L double-mutant phenotype in T1 plants (Fig. 4A; Supplemental Fig. S5, B and C), demonstrating that IRX9-L can fulfil the same enzymatic function as IRX9. Similar results were found for 35S:IRX14 and 35S:IRX14-L that fully complemented both the irx14 irx14-L double mutant and irx14 single mutant (Fig. 4B; Supplemental Fig. S5, D and E) and for 35S:FRA8 and 35S:F8H that complemented the fra8 f8h and fra8 mutants (Fig. 4C; Supplemental Fig. S5, F and G). Similarly, expression of IRX10-L under control of the 35S CaMV promoter rescued the irx10 single-mutant phenotype (Supplemental Fig. S6).
Figure 4.
Complementation of the irx9 irx9L, irx14 irx14-L, and fra8 f8h double-mutant combinations demonstrates conservation of function between homologs. A, Complementation of irx9 mutant and irx9 irx9-L double mutant with 35S:IRX9 and 35S:IRX9-L. B, Complementation of irx14 mutant and irx14 irx14-L double mutant with 35S:IRX14 and 35S:IRX14-L. C, Complementation of fra8 mutant and fra8 f8h double mutant with 35S:FRA8 and 35S:F8H. D, Overexpression of IRX10, IRX14, or FRA8 does not rescue the irx9 irx9-L double mutant. For each plant the transgene is written above the horizontal line and the background genotype below the line. wt, Wild type. [See online article for color version of this figure.]
Attempts to rescue the irx9 irx9-L double mutant with 35S:IRX10, 35S:IRX14, or 35S:FRA8 did not result in complementation (Fig. 4D). Similarly, expressing IRX9, IRX10, or FRA8 in the irx14 irx14-L double mutant or IRX9, IRX14, or FRA8 in the irx10 irx10-L double-mutant background did not rescue the mutant phenotypes (data not shown). Thus despite IRX9, IRX10, and IRX14 genes all encoding enzymes proposed to function in xylan backbone elongation, they are not functionally interchangeable.
To establish whether the intermediate phenotype of the fra8 f8h/+ plants is due to a difference in the activity of the two proteins or differences in expression patterns, promoter swap constructs were made and introduced into the fra8 f8h double mutant. Expression of pFRA8:FRA8 rescued the fra8 f8h double mutant but both proF8H:F8H and proFRA8:F8H showed only a weak partial rescue (Fig. 5). These results indicate that FRA8 is functionally more important than F8H.
Figure 5.
Promoter swaps between FRA8 and F8H show that the function of FRA8 rather than its specific expression pattern is critical for normal development. The proF8H:F8H, proFRA8:FRA8, proFRA8:F8H, proIRX10-L:FRA8, and proIRX10:FRA8 transgenes were introduced into the fra8 f8h double mutant and the plants grown for 5 weeks on soil together with wild type (wt). For each plant the transgene is written above the horizontal line and the background genotype below the line. [See online article for color version of this figure.]
The GX Synthesis Machinery Can Be Divided into Sets of Major Function and Minor Function Genes
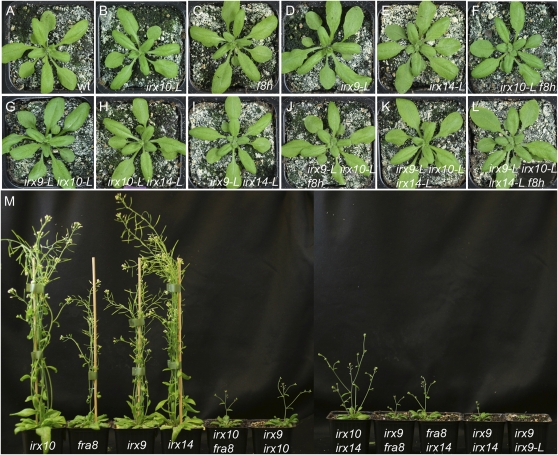
The dosage dependency exhibited by the homozygous/heterozygous combinations of the different gene pairs, together with the rescues of the double mutants by overexpression of either gene and analysis of promoter swaps has allowed a distinction to be made between a set of major function genes comprising IRX9, IRX10, IRX14, and FRA8 and a set of minor function genes that includes IRX9-L, IRX10-L, IRX14-L, and F8H. To further test the validity of this distinction, all possible double-, triple-, and quadruple-mutant combinations were made for the four minor function genes. All combinations were visually indistinguishable from the wild type, indicating that IRX9-L, IRX10-L, IRX14-L, and F8H do not perform an essential redundant function (Fig. 6, A–L). The six double-mutant combinations between genes belonging to the major function set show a more severe phenotype compared with either single mutant (Fig. 6M) though all can form stem material and show more growth in comparison to the double mutants between homologous pairs of genes (irx10 irx10-L, irx9 irx9-L, irx14 irx14-L, and fra8 f8h). The irx10 irx14-L, irx14 irx9-L, irx14 irx10-L, irx9 irx10-L, and irx9 irx14-L double-mutant combinations between one major gene and one minor gene, showed the same phenotype as the corresponding single mutant in the major gene (Supplemental Fig. S7, A–G). An additive effect on silique development was found for the irx10 f8h, fra8 irx10-L, fra8 irx9-L, and irx9 f8h double-mutant combinations in comparison to the fra8 or irx9 single mutants (Supplemental Fig. S7I; data not shown).
Figure 6.
Genetic analysis allows distinction of the GX synthesis genes into a major and minor function sets. A to L, The double, triple, and quadruple mutant combinations between irx9-L, irx10-L, irx14-L, and f8h do not exhibit a visible phenotype. A, Wild type (wt). B, irx10-L. C, f8h. D, irx9-L. E, irx14-L. F, irx10-L f8h. G, irx9-L irx10-L. H, irx10-L irx14-L. I, irx9-L irx14-L. J, irx9-L irx10-L f8h. K, irx9-L irx10-L irx14-L. L, irx9-L irx10-L irx14-L f8h. M, Double-mutant combinations between irx9, irx10, irx14, and fra8 show an enhanced phenotype and are infertile. From left to right are: irx10, fra8, irx9, irx14, irx10 fra8, irx9 irx10, irx10 irx14, irx9 fra8, fra8 irx14, irx9 irx14, and irx9 irx9-L. [See online article for color version of this figure.]
Reduced Cell Wall Xylose Content Correlates with the Severity of the Mutant Phenotypes
The Xyl compositions of the noncellulosic carbohydrate fraction isolated from stem material of the irx9 irx9-L, irx14 irx14-L, and fra8 f8h double mutants and each of the single mutants were determined using gas chromatography of alditol acetates. In agreement with previously published results, the irx9, irx14, and fra8 single mutants had a 50% or greater reduction in Xyl content compared to the wild type (Brown et al., 2007; Supplemental Fig. S8) whereas irx9-L, irx14-L, and f8h mutants showed a reduction of about 20%. The Xyl content of irx9 irx9-L, irx14 irx14-L, and fra8 f8h double mutants was reduced by around 85% compared to wild type (Supplemental Fig. S8), a value that is similar to that found for irx10 irx10-L (Wu et al., 2009). Thus there is a high degree of correlation between the severity of the mutant phenotype and the stem Xyl content.
The Abundance and Structure of Xylan Isolated from Stems Is Affected in the irx9 irx9-L, irx14 irx14-L, and fra8 f8h Mutants
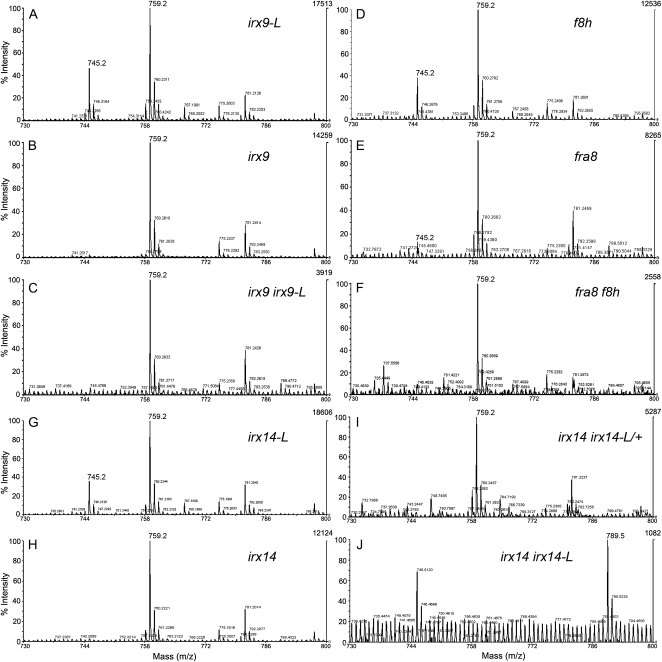
Xylan was isolated from stem material of all single- and double-mutant combinations of the irx9 irx9-L, irx14 irx14-L, and fra8 f8h pairs to establish whether a change in the abundance or structure can explain the reduction in Xyl content of the cell walls. Mass spectrometric analysis of xylanase-treated GX samples showed that irx9, irx14, and fra8 retain the 4-O-Me-GlcUA side chain (mass-to-charge ratio [m/z] 759) but lack the nonmethylated form (m/z 745) consistent with previously published results (Fig. 7, B, H, and E; Zhong et al., 2005; Brown et al., 2007; Peña et al., 2007). The irx9-L, irx14-L, and f8h samples showed a similar profile of methylated and nonmethylated GlcUA to the wild type (Wu et al., 2009), consistent with the lack of a visible phenotype exhibited by these mutants (Figs. 1, B and D and 7, A, G, and D). The amount of xylan in irx9 irx9-L and fra8 f8h double mutants was significantly reduced compared to the respective single mutants though it was still possible to detect the 4-O-Me-GlcUA signal (Fig. 7, C and F). However, there was no detectable xylan in the irx14 irx14-L double-mutant sample as previously found for irx10 irx10-L stems (Wu et al., 2009; Fig. 7J).
Figure 7.
Gas chromatography-mass spectrometry analysis of GX from single- and double-mutant combinations of irx9, irx9-L, irx14, irx14-L, fra8, and f8h. Mass spectrometry of acidic endoxylanase-generated GX fragments isolated from stems of the irx9-L (A), irx9 (B), irx9 irx9-L (C), f8h (D), fra8 (E), fra8 f8h (F), irx14-L (G), irx14 (H), irx14 irx14-L/+ (I), and irx14 irx14-L (J). The signal at m/z 745 refers to Xyl4GlcUA and at m/z 759 to Xyl4 4-O-Me-GlcUA fragments.
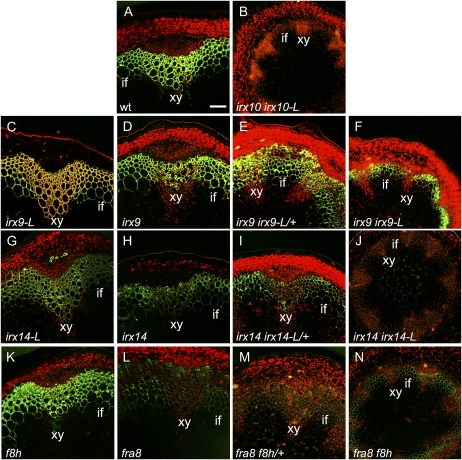
The anti-xylan LM10 monoclonal antibody (McCartney et al., 2005) was used to determine whether the different single- and double-mutant combinations affected the xylan distribution or abundance. The biggest changes were seen in the irx14 irx14-L double mutant, the fra8, fra8 f8h/+, and fra8 f8h combinations with either a strong reduction or absence of a signal (Fig. 8, J and L–N). In contrast the single- and double-mutant combinations of irx9 and irx9-L all retained a strong LM10 signal though the irx9 irx9-L double mutant had a reduced intensity and lacked a signal in the xylem cells (Fig. 8, C–F). The possibility that reduced signals are due to access of the antibody being affected in the mutants cannot be discounted.
Figure 8.
Immunolocalization of xylan in transverse stem sections using the LM10 antibody reveals changes in abundance. Transverse stem sections of basal stem material from wild type (wt; A), irx10 irx10-L (B), irx9-L (C), irx9 (D), irx9 irx9-L/+ (E), irx9 irx9-L (F), irx14-L (G), irx14 (H), irx14 irx14-L/+ (I), irx14 irx14-L (J), f8h (K), fra8 (L), fra8 f8h/+ (M), and fra8 f8h (N). Xylem (xy) and the interfascicular region (if) are indicated. Scale bar, 50 μm.
irx9 irx9-L, irx14 irx14-L, and fra8 f8h Are All Affected in Their Xylan Backbone Chain Length
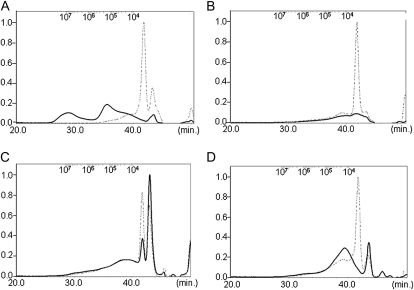
Size-exclusion chromatography analysis was performed on xylan isolated from stem material of the irx9 irx9-L, irx14 irx14-L/+, and fra8 f8h double-mutant combinations to test whether the degree of backbone polymerization was altered. Following digestion with xylanase only short oligosaccharides with a similar size distribution of less than 104 g−1 mol−1 were present in all samples, confirming that the preparations were comprised predominantly of xylan. Prior to digestion, the wild-type xylan size was 105 g−1 mol−1 or larger (Fig. 9A). In contrast, the major xylan peaks isolated from the irx9 irx9-L, irx14 irx14-L/+, and fra8 f8h double mutants showed a greater than 10-fold reduction in size compared to the wild type (Fig. 9, B–D), demonstrating that polymerization of the β-1-4-xylan backbone was affected.
Figure 9.
The degree of polymerization of the GX backbone is reduced in irx9 irx9-L, irx14 irx14-L, fra8 f8h, and irx10 irx10-L double mutants. high-pressure size-exclusion chromatography analysis of xylan isolated from stem material of wild type (A), fra8 f8h (B), irx9 irx9-L (C), and irx14 irx14-L/+ (D). The dashed lines indicate the size distribution following digestion of the xylan with xylanase, while the solid lines show the size distribution of the undigested xylan.
DISCUSSION
IRX9 and IRX14 and Their Homologs Play Critical Roles during GX Synthesis in Arabidopsis
Despite the fact that xylan constitutes up to 30% of the secondary cell wall biomass, relatively little is understood about how this polymer is synthesized. Although a number of genes have been identified that are proposed to function in synthesizing either the xylan backbone or the reducing-end tetrasaccharide, the lack of a working assay to demonstrate the function of the enzymes has hindered progress.
Our study demonstrates that both IRX9 and IRX14 have functional homologs that result in partial redundancy in irx9 and irx14 single mutants. This places new emphasis on the roles of the pairs of enzymes during GX synthesis. The phenotypes of the irx9 irx9-L and irx14 irx14-L double mutants are similar to those previously described for irx10 irx10-L and fra8 f8h, supporting the conclusion that they function in a common pathway or in synthesis of the same polymer. It is apparent that xylan synthesis is not abolished in the irx9 irx9-L double mutant (Figs. 7, A–C and 8, C–F) although the degree of backbone polymerization is greatly reduced (Fig. 9C). This indicates that IRX9/IRX9-L activity may not be absolutely required for GX synthesis. In contrast, it was not possible to isolate xylan from either irx14 irx14-L (Fig. 7J) or irx10 irx10-L stem tissues (Wu et al., 2009) or to detect it using the LM10 antibody (Fig. 8J), demonstrating that these pairs of genes play a critical and indispensable role. Furthermore, the low-Mr xylan formed in irx14 irx14-L/+ indicates that the activity of IRX14/IRX14-L represents an important rate-limiting component of the GX backbone synthesis machinery.
What Role Do FRA8 and F8H Play in Xylan Backbone Elongation?
Although proposed mechanisms for GX synthesis remain hypothetical at present, it has been suggested that formation of the reducing-end tetrasaccharide plays a role in regulating the degree of polymerization of the xylan backbone. This conclusion was based on the observation that fra8 and irx8 single mutants had increased xylan backbone lengths coupled with an apparent loss of the reducing-end tetrasaccharide. However, it is now apparent that the fra8 mutant retains F8H activity, making interpretation of the single-mutant data more difficult (Lee et al., 2009a). Work describing identification of F8H did not analyze xylan from the fra8 f8h double mutant but by growing fra8 f8h plants under a clear plastic cover it has been possible to obtain sufficient stem material to perform the analysis. Surprisingly, although fra8 single mutants displayed an increase in xylan backbone polymerization (Peña et al., 2007), the fra8 f8h double mutant only formed very short xylan chains (Fig. 9B). This implies that, contrary to conclusions based on the fra8 single-mutant analysis, FRA8/F8H function is critical in some aspect of xylan chain elongation other than, or in addition to, the proposed function in regulating the degree of backbone polymerization indirectly via synthesis of the reducing-end tetrasaccharide (Peña et al., 2007). It will be informative to establish whether FRA8 and/or F8H form a functional interaction with other GX synthesis enzymes and the precise activity of the individual enzymes or complex.
Xylan Biosynthesis Genes Can Be Divided into Major and Minor Function Sets
It is thought that Arabidopsis has undergone two rounds of genome duplication (Blanc et al., 2000; Simillion et al., 2002) so it is likely that the IRX10/IRX10-L, FRA8/F8H, IRX9/IRX9-L, and IRX14/IRX14-L sets of genes arose from duplication and diversification from common GT47 and GT43 ancestor genes. Detailed genetic analysis has revealed that all four gene pairs (IRX9/IRX9-L, IRX10/IRX10-L, IRX14/IRX14-L, and FRA8/F8H) exhibit a gene-dosage influence on plant development with one gene from each pair being functionally more important. This has led to the definition of a set of major function genes comprising IRX9, IRX10, IRX14, and FRA8 and a second set of minor function genes consisting of the four homologs IRX9-L, IRX10-L, IRX14-L, and F8H. The set of four major function genes as well as IRX14-L have very similar expression patterns, supporting the idea that they function in a common process and that some or all of them could interact to form a functional complex. Currently there is no identified functional homolog of IRX8 and double mutants between irx8 and gaut13 (At3g01040), gaut14 (At5g15470), or gaut15 (At3g58790) that belong to a branch of the GT8 family (Persson et al., 2007; Caffall et al., 2009) do not result in additive phenotypes (A.-M. Wu and A. Marchant, unpublished data). Thus, existence of major and minor function pairs of genes involved in GX synthesis does not extend to IRX8.
The reason for persistence in the genome of closely related pairs of genes encoding proteins with partially redundant function in GX synthesis is currently unclear. Although the catalytic functions of the related genes are likely to be similar or the same, their respective roles during development are distinct. This is evidenced by the intermediate phenotypes shown by the homozygous/heterozygous combinations of major and minor gene pairs (Fig. 1; Brown et al., 2009; Wu et al., 2009) and the ability of the minor function genes to complement the double mutant when overexpressed. This has allowed a simple mechanism to be proposed whereby the set of major function genes provide the predominant enzymatic activity required for GX synthesis. However, mutation of any one of the genes from the major function set can be compensated for by the homologous gene from the minor function set. It is possible that the rate of GX synthesis or degree of backbone polymerization could be modulated by substitution of major function genes with the minor function homologs. This mechanism is speculative at present in the absence of a functional assay and demonstration of the formation of a complex of GX synthesis enzymes but does provide the basis for future work leading to strategies to modify GX synthesis in planta.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) seed sterilization, growth condition, medium, and resistance screening were carried out as previously described (Wu et al., 2009). In brief, seed was surface sterilized (Forsthoefel et al., 1992) and sown in vitro on 1× Murashige and Skoog medium, 1% Suc, and 0.8% plant agar (Duchefa), buffered to pH 5.8 with 0.5 m KOH. The seeds on plates were incubated at 4°C for 48 h prior to germination under a 16-h light/8-h dark cycle. Soil-grown plants were maintained in controlled growth rooms with a light intensity of 120 μmol m−2 s−1 at 23°C under a 16-h light period.
Isolation of Mutants
All T-DNA insertion lines were obtained from the Nottingham Arabidopsis Stock Centre and were as follows: SALK_120296 (fra8, first exon), GABI 052G08 (f8h-1, first exon), GABI 298C10 (f8h-2, third exon), SALK_058238 (irx9-1, first exon), SALK_057033 (irx9-2, second exon), SALK_037323 (irx9-L-1, first intron), SALK_037330 (irx9-L-2, first intron), SALK_038212 (irx14, first exon), and SALK_066961 (irx14-L, second exon). PCR was performed using gene-specific primers designed to anneal 5′ and 3′ to the T-DNA insertion site together with insert-specific primers (primers listed in Supplemental Table S3). All mutant T-DNA insertion lines and the wild type were in the Columbia ecotype.
GUS Staining and YFP-NLS Fluorescence
Promoter fragments of 2.5, 2.5, 2.0, 1.4, 2.8, and 1.5 kb upstream of the ATG start codons of FRA8, F8H, IRX9, IRX9-L, IRX14, and IRX14-L, respectively, were amplified by PCR using GATEWAY-compatible primers (Supplemental Table S3). The promoter fragments were cloned into the pDONR207 vector, sequenced, and then recombined into either pGWB3 (Nakagawa et al., 2007) to create promoter:GUS constructs or pBGYN YFP-NLS (Kubo et al., 2005) to create promoter:YFP-NLS binary vectors according to the manufacturer's instructions (Invitrogen). Finally, constructs were transformed into wild-type Arabidopsis using the floral-dip procedure (Clough and Bent, 1998) and transgenic plants selected on Murashige and Skoog agar plates containing 50 μg/mL kanamycin. Tissues were stained for GUS activity by incubating in staining solution (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.5 mm ferricyanide, 0.5 mm ferrocyanide, and 1 mm 5-bromo-4-chloro-3-indolyl β-d-GlcUA) at 37°C for up to 12 h. Tissues were cleared in 70% ethanol, and were examined using a light microscope. YFP-NLS-expressing seedlings (1-week-old) were mounted in microtubule-stabilizing buffer (50 mm PIPES, 5 mm EGTA, 5 mm MgSO4; pH 7.0; Lauber et al., 1997) and the YFP fluorescence was observed using a Zeiss LSM510 confocal microscope.
Sectioning of Stems
Segments were cut from the basal third of stems of 6-week-old soil-grown plants and immediately fixed in buffer (1.6% [v/v] paraformaldehyde and 0.2% [w/v] glutaraldehyde in 25 mm sodium phosphate, pH 7.2). Sections (50 μm) were cut using a Leica VT1000S vibratome using a 3% agarose as support. The sections were stained for 1 to 2 min in 0.02% toluidine blue O (Sigma-Aldrich), rinsed in water, and then mounted in 50% glycerol before observing using a light microscope (Axioplan 2 microscope equipped with Axiovision software; Zeiss).
Complementation
The gene sequence incorporating the region from the ATG start codon to the stop codon were amplified by PCR using gene-specific gateway-compatible primers (Supplemental Table S1). The products were cloned into the pDONR207 vector and sequenced. The gene sequences were transferred into the pEarleyGate 100 destination vector (Earley et al., 2006) to produce the 35S CaMV promoter fusion constructs that were transformed into one of the f8h fra8/+, irx9-L2 irx9-1/+, or irx14-L irx14/+ mutant combinations using the floral-dip procedure (Clough and Bent, 1998). Transgenic plants were selected by spraying 120 mg/L BASTA solution onto 1-week-old seedlings in soil. Transformed seedlings were genotyped using gene-specific primers 5′ and 3′ to the T-DNA insertion site to identify those carrying the transgene in an otherwise double-mutant background.
Analysis of the Sugar Composition of the Cell Wall
Stem samples were collected from 6-week-old plants except in the case of double mutants where plants were grown for 9 weeks, and put into 80% ethanol before freeze drying. The material was treated and fractionated and the sugar composition of alcohol-insoluble residues were analyzed using alditol acetate derivatives as described previously (Englyst and Cummings, 1984) with modifications (Wu et al., 2009).
GX Extraction and Analysis
Stems were heated at 70°C for 30 min in 70% (v/v) ethanol. The samples were ground into fine powder in a Potter homogenizer. Then the samples were extracted and analysis by mass spectrometry performed as described previously (Wu et al., 2009).
High-Pressure Size-Exclusion Chromatography
The determination of the average Mr and Mr distribution were performed by coupling a high-pressure size-exclusion chromatography column to a multiangle laser light scattering and a Shimadzu RID-10A (Shimadzu) differential refractive index (DRI) detector. The light-scattering signal is proportional to the product concentration and Mr and the DRI signal is only proportional to the concentration. The size-exclusion chromatography line consisted of an OHPAK SB-G guard column and two Shodex OHPAK SB 804 and 806 HQ columns (Showa Denko K.K.) with a polyhydroxymethylmethacrylate gel used to pack the column. The flow carrier (0.1 m LiNO3) was degassed and eluted at a flow rate of 0.5 mL min−1 using a Shimadzu HPLC pump LC10 AI. Multiangle laser light scattering was carried out as previously reported (Picton et al., 2000) using a DAWN enhanced optical system multiangle laser light scattering photometer (Wyatt Technology Inc.) fitted to a K5 cell with 18 photodiodes and a InGasAs 30 mW laser (λ = 690 nm). The collected data were analyzed using the Astra V-5.3.2 software package. The concentrations of each eluted fraction have been determined with the DRI. Before injection, the samples were dissolved in 0.1 m LiNO3 containing 0.02% NaN3 at a 2 g L−1 concentration and filtered on a 0.45 μm membrane (Millipore).
Xylan Immunolocalization
The basal regions of inflorescence stems were collected from 6-week-old plants, except in the case of double mutants where 8-week-old plants were used. All samples were fixed overnight at 4°C using 2% glutaraldehyde in phosphate-buffered saline (0.5 m Na2HPO4/NaH2PO4, pH 7.2). Tissues were embedded in 3% agarose, and 40-μm-thick sections cut using a vibratome (Leica Microsystems) and used for immunolocalizations (Freshour et al., 1996, 2003; Wu et al., 2009).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At2g37090 (IRX9), At1g27600 (IRX9-L), At4g36890 (IRX14), At5g67230 (IRX14-L), At2g28110 (FRA8), and At5g22940 (F8H).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of subgroup I the GT47 family.
Supplemental Figure S2. Phylogenetic tree of the GT43 family members from Arabidopsis and other species.
Supplemental Figure S3. Reverse transcription-PCR analysis of the irx9 and irx9-L1 alleles.
Supplemental Figure S4. Gene expression profiles obtained from the Genevestigator resource.
Supplemental Figure S5. Overexpression of minor function GX synthesis genes is able to restore the phenotype of the mutant in the homologous major function gene.
Supplemental Figure S6. Complementation of the irx10 mutant by overexpression of IRX10-L.
Supplemental Figure S7. Phenotypes of double-mutant combinations between major and minor function genes.
Supplemental Figure S8. Reduction in cell wall Xyl content is correlated with the severity of the mutant phenotype.
Supplemental Table S1. Measurements of growth parameters for wild type and different mutant combinations.
Supplemental Table S2. Cell wall thickness of fibers and vessels in the stems of wild type and different mutant combinations.
Supplemental Table S3. Sequences of primers used for analyses.
Supplementary Material
Acknowledgments
We thank Taku Demura (RIKEN Plant Science Center, Yokohama, Japan) for the pBGYN YFP-NLS vector. The pGWB vectors were supplied by Tsuyoshi Nakagawa (Shimane University, Matsue, Japan).
References
- Andersson SI, Samuelson O, Ishihara M, Shimizu K. (1983) Structure of the reducing end-groups in spruce xylan. Carbohydr Res 111: 283–288 [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. (2006) Development and application of a suite of polysaccharide-degrading enzymes for analysing plant cell walls. Proc Natl Acad Sci USA 103: 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Vicky WWA, Goodacre R, Stephens E, Dupree P, Turner SR. (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D. (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ebringerova A, Heinze T. (2000) Naturally occurring xylans: structures, isolation procedures and properties. Macromol Rapid Commun 21: 542–556 [Google Scholar]
- Englyst HN, Cummings JH. (1984) Simplified method for the measurement of total non-starch polysaccharides by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst (Lond) 109: 937–942 [DOI] [PubMed] [Google Scholar]
- Fondeur-Gelinotte M, Lattard V, Oriol R, Mollicone R, Jacquinet JC, Mulliert G, Gulberti S, Netter P, Magdalou J, Ouzzine M, et al. (2006) Phylogenetic and mutational analyses reveal key residues for UDP-glucuronic acid binding and activity of b1,3-glucuronosyltransferase I (GlcAT-I). Protein Sci 15: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel NR, Wu Y, Schultz B, Bennett MJ, Feldmann KA. (1992) T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust J Plant Physiol 19: 353–366 [Google Scholar]
- Freshour G, Bonin CP, Reiter WD, Albersheim P, Darvill AG, Hahn MG. (2003) Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol 131: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MH, Samuelson O. (1977) Reducing end groups in birch xylan and their alkaline degradation. Wood Sci Technol 11: 251–263 [Google Scholar]
- Kong Y, Zhou G, Avic U, Gu X, Jones C, Yin Y, Xu Y, Hahn MG. (2009) Two poplar glycosyltransferase genes, PdGATL1.1 and PdGATL1.2 are functional orthologs to PARVUS/AtGATL1 in Arabidopsis. Mol Plant 2: 1040–1050 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, O'Neill MA, Tsumuraya Y, Darvill AG, Ye ZH. (2007a) The irregular xylem 9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 48: 1624–1634 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. (2009a) The F8H glycosyltransferase is a functional paralog of FRA8 involved in glucuronoxylan biosynthesis in Arabidopsis. Plant Cell Physiol 50: 812–827 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. (2009b) Downregulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol 50: 1075–1089 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH. (2007b) The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48: 1659–1672 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. (2006) Biosynthesis of plant cell wall polysaccharides—a complex process. Curr Opin Plant Biol 9: 621–630 [DOI] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP. (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall HK, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton L, Bataille I, Muller G. (2000) Analysis of a complex polysaccharide (gum arabic) by multi-angle laser light scattering coupled on-line to size exclusion chromatography and flow field flow fractionation. Carbohydr Polym 42: 23–31 [Google Scholar]
- Scheible WR, Pauly M. (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7: 285–295 [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- York WS, O'Neill MA. (2008) Biochemical control of xylan biosynthesis—which end is up? Curr Opin Plant Biol 11: 258–265 [DOI] [PubMed] [Google Scholar]
- Zhong R, Peña MJ, Zhou GK, Naim CJ, Wood-Jones A, Richardson EA, Morrison WH, Darvill AG, York WS, Ye ZH. (2005) The FRA8 gene which encodes a putative glucuronyltransferase is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2003) Unraveling the functions of glycosyltransferase family 47 in plants. Trends Plant Sci 8: 565–568 [DOI] [PubMed] [Google Scholar]
- Zhou GK, Zhong R, Himmelsbach DS, McPhail BT, Ye ZH. (2007) Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiol 48: 689–699 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.