Abstract
Arabidopsis (Arabidopsis thaliana) RAP2.2 (At3g14230) is an APETALA2/ethylene response factor-type transcription factor that belongs to the same subfamily as the rice (Oryza sativa) submergence tolerance gene SUB1A. RAP2.2 is expressed at constitutively high levels in the roots and at lower levels in the shoots, where it is induced by darkness. Effector studies and analysis of ethylene signal transduction mutants indicate that RAP2.2 is induced in shoots by ethylene and functions in an ethylene-controlled signal transduction pathway. Overexpression of RAP2.2 resulted in improved plant survival under hypoxia (low-oxygen) stress, whereas lines containing T-DNA knockouts of the gene had poorer survival rates than the wild type. This indicates that RAP2.2 is important in a plant's ability to resist hypoxia stress. Observation of the expression pattern of 32 low-oxygen and ethylene-associated genes showed that RAP2.2 affects only part of the low-oxygen response, particularly the induction of genes encoding sugar metabolism and fermentation pathway enzymes, as well as ethylene biosynthesis genes. Our results provide a new insight on the regulation of gene expression under low-oxygen conditions. Lighting plays an important regulatory role and is intertwined with hypoxia conditions; both stimuli may act collaboratively to regulate the hypoxic response.
Flooding is one of the most common stresses affecting plant growth and development. Many important crop plants are sensitive to flooding or waterlogging conditions caused by heavy rain or irrigation. Flooding and submergence conditions impose a variety of challenges on the plant. Most critical is maintaining an energy supply for continued metabolism and growth. To cope with the reduction in oxygen supply, plants have developed a number of metabolic and morphological adaptations that enable them to survive transient periods of complete or partial submergence (Kende et al., 1998; Drew et al., 2000; Bailey-Serres and Voesenek, 2008). Escape from hypoxia involves shoot elongation, development of aerenchyma, and adventitious root formation (Drew et al., 2000; Sauter, 2000; Voesenek et al., 2006; Perata and Voesenek, 2007). Nonetheless, oxygen may ultimately become limiting, necessitating a switch from aerobic respiration to anaerobic fermentation, with a variety of electron acceptors such as pyruvate and acetaldehyde replacing oxygen. This metabolic adaptation helps maintain ATP production and NAD+ regeneration (Dennis et al., 2000).
Anaerobic fermentation is far less efficient in producing energy than aerobic respiration. Carbohydrate availability, either from photosynthesis or from stored reserves, becomes an important issue the longer the plant remains submerged. As a reaction to rising water levels, some plants extend their shoots and leaf blades to reestablish gas exchange (Kende et al., 1998; Voesenek et al., 2004; Pierik et al., 2009). However, this extension growth is at the expense of increased carbohydrate consumption and is detrimental when submergence is too deep or lasts for too long and carbohydrate reserves become depleted. Some plants, therefore, have adapted an alternative strategy that aims to conserve energy and carbohydrate consumption by limiting growth under low-oxygen conditions. Plant survival under flooding conditions is controlled by an intricate balance between escape and endurance (Fukao and Bailey-Serres, 2004).
The molecular basis of the morphological, anatomical, and metabolic adaptations to flooding and the signaling events leading to these responses remain to be elucidated. Recent research has focused on ethylene as a signal for the regulation of the early response to flooding. Expression levels of ethylene biosynthetic genes are up-regulated under hypoxic conditions (Van der Straeten et al., 1997; Peng et al., 2005). In submerged plants, ethylene levels increase rapidly due to physical entrapment of this volatile hormone (Métraux and Kende, 1983; Peng et al., 2001; Voesenek et al., 2004, 2006). The programmed cell death response that results in lysigenous aerenchyma formation is also controlled by ethylene (Drew et al., 2000), as is epidermal cell death above adventitious root primordia in rice (Oryza sativa; Mergemann and Sauter, 2000; Steffens and Sauter, 2009) and adventitious root growth in Rumex palustris and rice (Visser et al., 1996; Steffens and Sauter, 2005; Steffens et al., 2006). The internode elongation response, which serves as an important adaptation response in deepwater rice, is also controlled by ethylene (Hattori et al., 2009). In Arabidopsis (Arabidopsis thaliana), ethylene induces alcohol dehydrogenase gene expression (ADH1; Peng et al., 2001, 2005). Ethanolic fermentation through ADH1 activity contributes substantially to metabolic adaptation to low-oxygen stress; an adh1 null mutant that showed lower survival rates when exposed to low oxygen (Ellis et al., 1999) and overexpression of pyruvate decarboxylase (PDC1 and PDC2) in Arabidopsis results in improved survival under low-oxygen conditions (Ismond et al., 2003).
A major quantitative trait locus responsible for submergence tolerance, Submergence1 (SUB1), was identified in lowland indica rice (Fukao et al., 2006; Xu et al., 2006). The locus consists of a cluster of three ethylene response factor (ERF) genes, SUB1A, SUB1B, and SUB1C. The SUB1A gene is present only in indica and not in japonica cultivars. Although SUB1A is present in several, but not all, indica cultivars, only the SUB1A-1 allele from the submergence-tolerant indica cv FR13A was able to confer the flooding tolerance phenotype. Overexpression of SUB1A-1 in a flooding-sensitive japonica cultivar resulted in increased ADH1 expression and flooding tolerance (Fukao et al., 2006; Xu et al., 2006). The SUB1A-1 allele appears to confer submergence tolerance via a complex regulatory pathway that reduces elongation growth and carbohydrate consumption (Fukao et al., 2006; Xu et al., 2006; Perata and Voesenek, 2007, Jung et al., 2010). A role of the SUB1A-2 allele in submergence tolerance remains to be analyzed. However, more recently, it was shown that the opposite response in deepwater rice is also controlled by ethylene and ERF factors. The capacity of deepwater rice to elongate stem internodes and extend the hollow stem to the water surface requires the SNORKEL1 (SK1) and SK2 ERF transcription factors (Hattori et al., 2009). A detailed analysis of cell type-specific changes in translated mRNA in response to short-term hypoxic treatment of Arabidopsis seedlings revealed a ubiquitous increase in translated ERF71 and ERF73 mRNA (Mustroph et al., 2009). Increased expression of these two ERFs by low-oxygen conditions was recently confirmed, but induction of these factors by ethylene was not analyzed (Licausi et al., 2010).
ERF-type transcription factors belong to the plant-specific multigene family of APETALA2 (AP2)/ERF transcription factors. The AP2/ERF superfamily of proteins is characterized by having either one or two AP2 domains: the ERF or EREBP (for ethylene-responsive element-binding proteins) family has one AP2 domain, the RAV family has one AP2 domain and a B3 domain, and the AP2 family has two AP2 domains (Okamuro et al., 1997; Riechmann and Meyerowitz, 1998; Nakano et al., 2006). Some ERF factors can bind as activators or repressors to the GCC box (AGCCGCC) elements in the promoters of ethylene-responsive genes (Ohme-Takagi and Shinshi, 1995), while others mediate the response to cytokinin (Rashotte et al., 2006). ERF factors also play a role in a variety of developmental processes such as flower and seed development (Riechmann and Meyerowitz, 1998) and abiotic and biotic stress responses (Büttner and Singh, 1997; Stockinger et al., 1997; Fujimoto et al., 2000; Thomashow, 2001; Sakuma et al., 2002).
We studied the role of the ERF transcription factor, RAP2.2 (At3g14230), in the Arabidopsis response to hypoxia. RAP2.2 shows structural and phylogenetic relationships to the rice SUB1A gene (Nakano et al., 2006). Our results show that ethylene and RAP2.2, together with oxygen-dependent signal transduction, play an important role in the response to hypoxia.
RESULTS
Relationship between Arabidopsis RAP2.2 and the Rice SUB1 Proteins
BLASTP searches revealed that Arabidopsis RAP2.2 is related to the rice SUB1A protein. RAP2.2, however, is longer than SUB1 and shares only 37% amino acid identity over the aligned regions relative to SUB1A-1. The protein alignment of RAP2.2 with the rice SUB1 proteins (Fig. 1A) shows only short stretches of high homology, particularly the AP2 domain and two other domains, one with unknown function located between amino acids 49 and 70 (rich in Asp) and the other at the very N-terminal end (MCGG motif; Fig. 1A). The rice SK1 and SK2 proteins that are responsible for elongation growth have a different AP2 domain and therefore were not included in the alignment (Hattori et al., 2009). An extensive classification of the Arabidopsis and rice AP2/ERF transcription factors based on the AP2 domain structure grouped RAP2.2 and the rice SUB1B and SUB1C proteins together in subfamily VII (Nakano et al., 2006); rice SK1 and SK2 have a different AP2 domain structure and belong to subfamily XI. The group VII subfamily contains 15 members in japonica rice but only five in Arabidopsis. A phylogenetic analysis with the entire protein sequences of the Arabidopsis and rice group VII members and closely related AP2/ERF proteins from other plants was carried out (Fig. 1B; for the sequence alignment, see Supplemental Fig. S1). RAP2.2 was found to be most closely related to Arabidopsis RAP2.12 (At1g53910) and two other dicot ERF factors, tomato (Solanum lycopersicum) JERF3 and pepper (Capsicum annuum) CaPF1, which play roles in salt and freezing tolerance, respectively (Wang et al., 2004; Yi et al., 2004; Fig. 1B). No rice group VII proteins clustered with the clade containing RAP2.2. The only rice proteins that cluster with the dicot proteins are Os02g54160, Os06g09390, and Os09g26420 (Fig. 1B). Os02g54160 (OSEREBP1) plays a role in pathogenesis-related gene expression (Cheong et al., 2003). This alignment of the amino acid sequences shows that the rice SUB1A, -B, and -C proteins cluster separately from the other rice group VII ERF factors and that their closest Arabidopsis relatives are At1g72360 (ERF73) and At2g47520 (ERF71; Fig. 1B) but not RAP2.2. But, as is the case for RAP2.2, ERF73, and ERF71, homology with SUB1A is mainly confined to the AP2 domain (overall amino acid identity of 26% and 33% for ERF73 and ERF71, respectively). These results indicate that Arabidopsis does not have a close relative of the rice SUB1 proteins and that the rice SUB1 genes may have evolved independently after the separation of monocots and dicots (Fukao et al., 2009).
Figure 1.
Phylogenetic tree of group VII AP2/ERF transcription factors from Arabidopsis and rice. A, Protein sequence alignment of RAP2-2 with the rice Sub1A, Sub1B, and Sub1C proteins. Alignment shows homology especially in the AP2 domain (amino acids 130–186). Another rather highly conserved region of unknown function is present between amino acids 49 and 70 (2). The N-terminal MCGG motif (1) is strictly conserved between RAP2.2, Sub1A, Sub1B, and Sub1C and is also found in barley (Hordeum vulgare) HvRAF (Jung et al., 2007). A potential nuclear localization signal (NLS) is present between positions 113 and 128. B, A phylogenetic tree of group VII AP2/ERF transcription factors from Arabidopsis and rice was constructed using the entire protein sequence of only the group VII (Nakano et al., 2006) ERF transcription factors of japonica rice (prefix Os) and Arabidopsis (prefix At). Bootstrap figures are given at the branch points of the tree. For the SUB1 proteins from indica rice, the equivalent japonica locus is given for SUB1B and -C; the GenBank entry is given for the SUB1A locus. Subgroup VII corresponds to the B-2 subgroup according to the analysis of Sakuma et al. (2002). Known gene names are mentioned in parentheses. The phylogenetic tree also includes group VII AP2/ERF transcription factors from bell pepper (CaPF1) and tomato (JERF3; Nakano et al., 2006). Barley HvRAF is also included as a close relative of the group VII ERF subfamily. The bar indicates the number of amino acid substitutions per site.
RAP2.2 Is Induced in Dark-Grown Shoots
RAP2.2 was originally identified as a gene that was induced 2- to 3-fold on microarrays after 1 h of low-oxygen treatment (0.5% oxygen) of Arabidopsis hairy root cultures (Klok et al., 2002). Induction of RAP2.2 by hypoxia was not observed on RNA blots of Arabidopsis root mRNA in our experimental system; expression of RAP2.2 in roots was high in the dark under both aerobic and hypoxic conditions (Fig. 2A). In aerobic shoots, RAP2.2 expression was much lower than in aerobic roots; expression was strongly induced when plants were transferred to hypoxic conditions in the dark, up to levels similar to those in roots (Fig. 2, A and B). ADH1 expression was strongly induced by low-oxygen conditions in the roots and shoots, but overall expression levels were much lower in the shoots compared with the roots (Fig. 2, A and B). RAP2.2 induction by low-oxygen stress was not observed in roots from dark-grown hairy root cultures, but overall expression levels were already very high in untreated root cultures (Fig. 2B). When low-oxygen stress was carried out in the light, RAP2.2 induction by 5% oxygen was not observed, but expression levels of RAP2.2 increased in shoots in the dark, even under aerobic conditions; the induction is transient and peaks after 2 h of darkness (Fig. 2C). In contrast, expression of ADH1, a well-characterized low-oxygen-responsive gene, was strongly induced by hypoxia in roots, hairy root cultures, and shoots (Fig. 2, A and B) and responded specifically to low oxygen but not to darkness (Fig. 2C). These results indicate that RAP2.2 responds only to darkness and not to low-oxygen conditions in shoots.
Figure 2.
Expression analysis of RAP2.2 and ADH1 in roots and shoots during low-oxygen and dark treatments of Arabidopsis plants. A, RNA blots showing expression of RAP2.2 and ADH1 in roots and shoots of 17-d-old plants treated with 5% oxygen (hypoxia treatment) or 21% oxygen (aerobic control). B, RNA-blot analysis of RAP2.2 and ADH1 expression in hairy root cultures and shoots of plants treated with hypoxia or kept in aerobic conditions. C, RNA-blot analysis of RAP2.2 and ADH1 expression in the shoots treated with hypoxia (for 2 or 8 h) or kept in aerobic conditions (0 h) under normal lighting conditions and in the dark. Twenty micrograms of total RNA was used per lane, and ethidium bromide staining of rRNA was used as a loading control.
RAP2.2 Expression Is Regulated by Ethylene
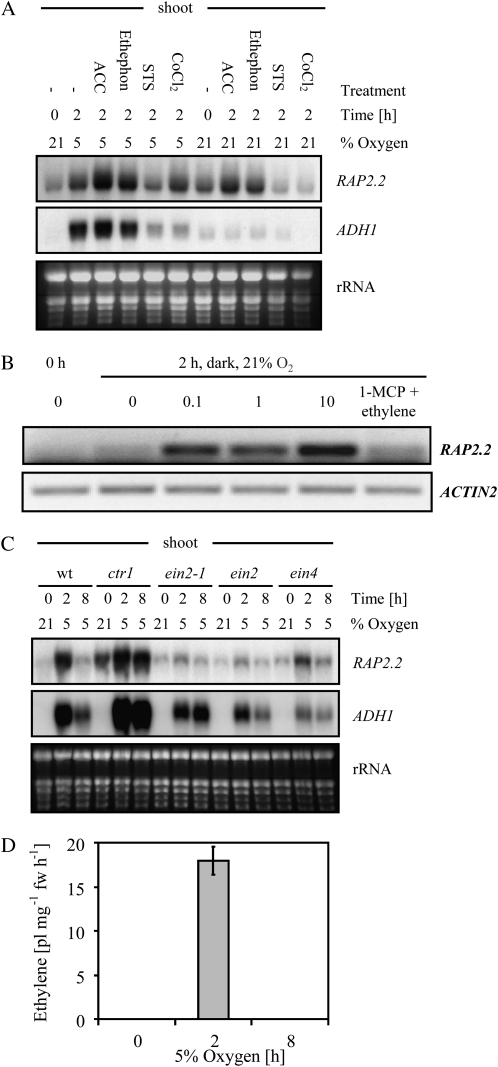
Treatments were carried out in the dark on shoot tissues under normoxic (21% oxygen) or hypoxic (5% oxygen) conditions to determine whether RAP2.2 is regulated by ethylene. The ethylene-releasing chemical ethephon, the natural ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), ethylene itself, and the inhibitor of ethylene biosynthesis CoCl2 were used to manipulate ethylene levels in the plant. In the presence of 50 μm ACC or 50 μm ethephon, RAP2.2 expression was enhanced under both normoxic and hypoxic conditions in the dark. The 50 μm CoCl2 treatment appeared to reduce expression only under normoxic conditions (Fig. 3A). We also applied the inhibitors of ethylene signaling silver thiosulfate (STS) and 1-methylcyclopropene (1-MCP). In the presence of 50 μm STS, RAP2.2 transcript levels were reduced under both normoxic and hypoxic conditions (Fig. 3A). Treatment of Arabidopsis plantlets with ethylene (0.1, 1, and 10 μL L−1) induced RAP2.2 expression; this induction was blocked by the simultaneous treatment with 1-MCP (ethylene at 1 μL L−1 and 1-MCP at 10 μL L−1; Fig. 3B). Treatments with the ethylene precursors ethephon and ACC increased ADH1 transcript abundance in shoots, while the inhibitors of ethylene synthesis or signaling reduced ADH1 expression (Fig. 3A).
Figure 3.
Effects of ethylene, ethylene-producing agents, inhibitors of ethylene synthesis, and ethylene signaling mutants on the expression of RAP2.2 and ADH1. A, RNA-blot analysis of 17-d-old plants that were preincubated for 2 h with 50 μm ethephon, 50 μm ACC, 50 μm STS (an ethylene perception inhibitor), or 50 μm CoCl2 (an ethylene synthesis inhibitor) and then subjected to hypoxic conditions in the dark. As a control, nontreated plants were left in the light. B, RT-PCR data showing the effects of ethylene treatment (0.1, 1, and 10 μL L−1) and the combined treatment of ethylene (1 μL L−1) and 1-MCP (10 μL L−1) on RAP2.2 gene expression. Amplification of Actin1 cDNA was used as a control for RNA input. C, RNA-blot analysis of RAP2.2 and ADH1 in shoots of the wild type (wt) and the ethylene signaling mutants ctr1, ein2-1, ein2, and ein4 under normoxic or hypoxic conditions in the dark. D, Ethylene production of 17-d-old plants kept at 21% oxygen (0 h) or at 5% oxygen for 2 or 8 h in the dark. Ethylene accumulation was determined by gas chromatography during the last hour of treatment. Average values ± sd were obtained from three independent biological experiments with three independent setups including 30 plants each. fw, Fresh weight.
To further study the role of ethylene signaling in RAP2.2 regulation, the constitutive triple response ctr1 mutant and three ethylene-insensitive mutants, ein2, ein3, and ein4, were employed (Olmedo et al., 2006). Expression of RAP2.2 in shoots was increased in ctr1 under both normoxic-light and hypoxic-dark conditions (Fig. 3C). In contrast, in ein2, ein2-1, ein3-1, and ein4, induction of RAP2.2 expression under hypoxic-dark conditions in shoots was reduced (Fig. 3C). RAP2.2 is induced weakly in ein4 after 2 h of hypoxic treatment; this could be due to the fact that ein4 is semidominant, as other ethylene receptors can partially complement ein4 (Hua and Meyerowitz, 1998). Similar to RAP2.2, ADH1 mRNA levels in shoots were enhanced in ctr1 but only under hypoxic-dark conditions (Fig. 3C), suggesting that ADH1 is under dual control by low-oxygen signaling and ethylene signaling.
Direct measurement of ethylene production by Arabidopsis plantlets showed that ethylene levels increased strongly after 2 h of oxygen deprivation in the dark and returned to nondetectable levels after 8 h (Fig. 3D). The time of maximal ethylene synthesis coincided with the time of maximal expression of RAP2.2 (Fig. 3B).
In conclusion, both ethylene and ethylene signaling affect RAP2.2 expression under normoxic-dark and hypoxic-dark conditions, but ADH1 expression is affected by ethylene signaling only when hypoxic conditions are present at the same time.
Spatial Expression Pattern of the RAP2.2 Promoter
Seventeen-day-old Arabidopsis plants transformed with a RAP2.2 promoter-driven GUS reporter gene showed GUS activity in both shoots and roots. Expression was weak in shoots but high in the roots (Fig. 4, A and B). When plants were transferred to the dark for 2 h, GUS staining increased in the shoot (Fig. 4C). In 4-d-old light-grown seedlings, GUS activity was high in the stele of the root and in the hypocotyl but weaker in the vasculature of the cotyledon (Fig. 4D). Expression was highest at the junction between the root and hypocotyl (Fig. 4, E and F) and the hypocotyl and cotyledons (Fig. 4, E and G). In light-grown seedling roots, GUS staining was present in the stele but not in the root tip (Fig. 4H). In 4-d-old dark-grown seedlings, GUS activity was high in the roots and the root-hypocotyl junction (Fig. 4, I–K), but in contrast to light-grown seedlings, expression in the root was also observed in the root tip (Fig. 4L). In the hypocotyl, overall expression appeared to be lower compared with light-grown seedlings of the same age and was mostly vascular (Fig. 4I). However, high levels of expression were seen in the subapical hook region (Fig. 4, I and M). The spatial distribution of RAP2.2 promoter-driven GUS expression in whole plantlets showed quantitative differences in expression but no qualitative differences, nor did we observe any qualitative differences in the expression pattern following darkness or low-oxygen treatment (data not shown).
Figure 4.
Localization of RAP2.2 expression using promoter:GUS transgenic plants. A to C, Seventeen-day-old plants subjected to either normal lighting conditions (A and B) or dark conditions for 2 h (C). GUS staining for both light-grown and dark-treated plantlets was 30 min. D to H, Light-grown 4-d-old Arabidopsis seedlings. D, Complete seedling. E, Enlargement of the root-hypocotyl junction. F, Further enlargement of the root-hypocotyl junction. G, The vascular tissue leading from the hypocotyl to the cotyledons. H, Root tip. I to M, Four-day-old etiolated Arabidopsis seedlings. I, Intact seedling. J, Root-hypocotyl junction. K, Staining in the vascular tissue at the root-hypocotyl junction. L, Root tip. M, Staining in the apical hook region.
RAP2.2 Overexpression Increases ADH1 and PDC1 Expression under Hypoxic Conditions
RAP2.2 under the control of the 35S promoter was transformed into ecotype Columbia (Col-0), and the ox1, ox2, and ox3 lines were recovered. Strong RAP2.2 overexpression in shoots of light-grown aerobic plants was evident for ox1, ox2, and ox3, and mRNA levels were further increased in all lines following hypoxic treatment in the dark (Fig. 5).
Figure 5.
RAP2.2, ADH1, and PDC1 expression in the wild type (wt) and RAP2.2-overexpressing lines. Seventeen-day-old wild-type plants and the 35S::RAP2.2 ox transgenic lines ox1, ox2, and ox3 were treated with 21% or 5% oxygen in the dark, and relative transcript levels in shoots of RAP2.2, ADH1, and PDC1 were determined by RNA-blot analysis.
Overexpression of RAP2.2 resulted in overexpression of ADH1 and, to a lesser degree, of PDC1 in shoots following hypoxic-dark treatment but not under normoxic-dark conditions (Fig. 5). This increase was reflected in increased ADH and PDC enzyme activities in shoots (Fig. 6). In shoots of Col-0 wild type, ADH activity increased after 8 and 24 h of hypoxic-dark treatment. The same time course experiment for the RAP2.2-overexpressing lines ox1 and ox2 showed significantly higher ADH and PDC activities than the wild type after 24 h of hypoxia treatment (Fig. 6; P < 0.01, n = 6). In the ox2 line but not the ox1 line, ADH and PDC activities were already increased significantly compared with the wild type after 8 h of hypoxia treatment (Fig. 6). Similar results were obtained for a third RAP2.2-overexpressing line, ox3 (Supplemental Fig. S2). The increased ADH activity of the RAP2.2-overexpressing lines was significant under hypoxic-dark conditions and mirrors the observations from the RNA blots (Fig. 5). These results clearly indicate that overexpression of RAP2.2 leads to increased levels of gene expression and enzyme activity levels of the two ethanol fermentation pathway enzymes, ADH and PDC. This overexpression is mainly evident under a hypoxic environment in the dark. Similarly, expression of ERF71 (HRE2) and ERF73 (HRE1) in roots of Arabidopsis seedlings is induced by treatment with 1% oxygen in the dark. Overexpression of ERF73 improved survival of Arabidopsis seedlings exposed to anaerobic treatment in the dark and resulted in up-regulation of ADH1 expression (Licausi et al., 2010). Since anaerobic treatments were carried out in the dark, it is possible that darkness or hypoxia or both treatments is required as signals.
Figure 6.
ADH and PDC enzyme activity measurements in shoots of wild-type Col-0 and the RAP2.2-overexpressing lines ox1 and ox2. A, ADH enzyme activity measurements following 0 (t0), 8, and 24 h of hypoxia (5% oxygen) treatment, showing significantly increased levels of ADH activity in the overexpressing lines. B, PDC enzyme activity measurements following 0 (t0), 8, and 24 h of hypoxia (5% oxygen) treatment, showing overexpression of PDC activity in the two overexpressing lines. The data represent averages ± se of three biological repeats with three measurements per sample.
T-DNA Insertions in RAP2.2 Reduce ADH1 and PDC1 Hypoxic Induction in Shoots
Two independent T-DNA insertion lines of RAP2.2, rap2.2-1 (CS871911) and rap2.2-2 (CS876942), segregated as single locus insertions. The rap2.2-1 and rap2.2-2 T-DNA insertions are located in the first and second exons of RAP2.2, respectively, and are expected to eliminate protein function. Expression levels were reduced more strongly in the rap2.2-2 insertion line compared with rap2.2-1, but both lines still showed induction of mRNA levels following hypoxia in the shoots (Fig. 7). T-DNA insertions in RAP2.2 resulted in slightly lower shoot ADH1 and PDC1 mRNA induction levels under hypoxia in the dark. The fact that RAP2.2 knockdown lines only partially reduced ADH1 and PDC1 expression may be an indication that other redundant ERF factors can compensate for lower RAP2.2 expression levels.
Figure 7.
RNA-blot analysis of RAP2.2, ADH1, and PDC1 expression in the wild type (wt) and the RAP2.2 T-DNA insertion lines rap2.2-1 and rap2.2-2. Seventeen-day-old wild-type plants and plants of the T-DNA insertion lines rap2.2-1 and rap2.2-2 were treated with 21% oxygen or 5% oxygen in the dark for 2 or 8 h before isolation of RNA from the shoot.
RAP2.2 Is Important for Survival of Hypoxia
To determine whether RAP2.2 is important for plant survival under low oxygen, the two highest overexpressing lines (ox2 and ox3) and the two insertion knockout lines (rap2.2-1 and rap2.2-2) were subjected to low-oxygen survival assays as described by Christianson et al. (2009). Different survival assay conditions had to be established for overexpressing and knockout lines of RAP2.2 because of their different behavior in terms of low-oxygen survival relative to the wild type. The insertion line treatment consisted of 24 h of 5% oxygen pretreatment followed by 40 h of 0.1% oxygen treatment, both in the dark. The overexpressing lines were subjected to 24 h of 5% oxygen pretreatment followed by 48 h of 0.1% oxygen treatment in the dark. Control aerobic plants were also kept in the dark. Each assay consisted of three replicates of 12 plants, using Col-0 as the wild-type control, and results were averaged from at least three independent experiments. Plant survival in the light was scored 1 week after the stress treatment. The results show that rap2.2-1 and rap2.2-2 had significantly lower survival than Col-0 (P < 0.01; Figs. 8A and 9A), whereas the ox2 and ox3 overexpressing lines possessed significantly higher survival levels than the wild type (P < 0.01; Figs. 8B and 9B). These results indicate that there is a correlation between RAP2.2 expression and survival under the low-oxygen conditions imposed in this assay. Root and shoot fresh weights were determined from rap2.2-2 and ox3 plants recovered from the survival assays shown in Figure 9, A and B, and expressed relative to the fresh weights of their respective untreated controls (Fig. 9C). Results indicate that knockout plants have in general a lower fresh weight than the wild type, while plants of the ox3 line tend to have higher shoot and root fresh weights.
Figure 8.
Survival of plants with elevated and reduced expression of RAP2.2 as compared with the wild type (wt). A, Images of wild-type seedlings and the T-DNA insertion lines rap2.2-1 and rap2.2-2 at 1 week after recovery from 24 h of 5% oxygen and 40 h of 0.1% oxygen in the dark. B, Images of wild-type seedlings and the overexpression lines ox2 and ox3 at 1 week after recovery from 24 h of 5% oxygen and 48 h of 0.1% oxygen in the dark. The control plants in this experiment were also kept in the dark.
Figure 9.
Effects of low-oxygen treatment on the percentage of plant survival. A, Survival percentage of the wild type (wt) and the T-DNA insertion lines rap2.2-1 and rap2.2-2 measured 1 week after recovery from 24 h of 5% oxygen and 40 h of 0.1% oxygen (±se; n = 5). Asterisks denote line survival percentages significantly different from each other as determined by t test (P < 0.01). B, Survival percentage of the wild type and the overexpression lines ox2 and ox3 measured 1 week after recovery from 24 h of 5% oxygen and 48 h of 0.1% oxygen (±se; n = 3). Asterisks denote line survival percentages significantly different from each other as determined by t test (P < 0.01). C, Fresh weight (FW) of roots and shoots of 4-week-old light-grown plants of the wild type, the T-DNA knockout line rap2.2-2, and the RAP2.2-overexpressing line ox3 measured after the recovery period of the survival assays. rap2.2-2 has consistently lower fresh weight. The opposite was observed for the ox3 overexpression line. Data points with asterisks denote fresh weights significantly different from the wild type as determined by t test (P < 0.01).
RAP2.2 Expression Affects Only a Subset of Low-Oxygen-Responsive Pathways
To understand the role of RAP2.2 under hypoxia conditions, we investigated what genes (apart from ADH1 and PDC1) are affected by RAP2.2 in darkness, hypoxia (5% oxygen), and the combination of darkness and hypoxia. We studied the effect of these three treatments in wild-type plants as well as in RAP2.2-overexpressing and knockout lines (ox3 and rap2.2-2) and investigated the effect of altering RAP2.2 expression levels on the expression of 32 genes that have previously been shown to be affected by hypoxia treatment in plants (Supplemental Table S1). The genes were chosen from two low-oxygen stress microarray experiments that were carried out in the dark (Klok et al., 2002; Loreti et al., 2005) and one carried out under light conditions (Liu et al., 2005; Supplemental Table S1). The choice was influenced by function under low-oxygen concentrations (e.g. sugar metabolism, fermentation pathway genes) or a potential role in senescence or ethylene-mediated responses. Also included were ethylene biosynthesis genes and other ERF genes, particularly members of the group VII ERF factors (Fig. 1B) and those ERF factors that were differentially expressed in the microarray experiments. Since RAP2.2 expression levels were shown to reach a maximum around 2 h after induction in the shoots, a treatment length of 5 h was chosen for the darkness and hypoxia treatments to analyze the effect of changing RAP2.2 expression on downstream genes. Quantitative reverse transcription (QRT)-PCR expression profiling was carried out for three individual repeat samples; all expression data are provided in Supplemental Figure S3, and a heat map summarizing the expression data is provided in Supplemental Figure S4.
Hierarchical clustering of the expression profiles of the 32 genes and treatments/genotypes revealed that the combined hypoxia-darkness treatment responses were closer to the darkness-only treatment under normoxic conditions than they were to hypoxia treatment under light conditions (Fig. 10). Of the 21 genes that were induced by at least 2-fold under hypoxia in the dark in wild-type plants, only eight were induced by hypoxia in the light, whereas 11 were induced by darkness alone (Fig. 10; Supplemental Figs. S3 and S4). Those genes that were induced by hypoxia in the light had a much stronger induction by hypoxia in darkness (Supplemental Figs. S3 and S4). Only three genes appeared to have a weak induction by hypoxia in the light and did not further increase their expression under hypoxia in darkness (RopGAP4, RAP2.12, and ACO; Supplemental Fig. S3). Many genes thought to be “low-oxygen-induced” genes are in fact induced by darkness during the assay. Changing RAP2.2 expression caused changes to specific genes, but overall expression profiles were similar to wild-type responses and clustered with them. Overexpression of RAP2.2 resulted in up-regulation of ADH1 and down-regulation of ACS9 under normoxia in the light. When hypoxia-darkness was imposed on ox3, increases in expression of AlaAT1 and RAP2.3 and decreases in AtMYB2 and ACS2 were observed (Fig. 10). Loss of a functional RAP2.2 resulted in up-regulation of a number of genes under normoxia-light, including the ethylene biosynthesis genes ASC2, ACS7, ASC9, and AOX1 as well as NIP2;1 and AtMYB2. Loss of hypoxia-darkness induction in AtMYB2, ACS2, and ETR2 was also observed in rap2.2-2 (Fig. 10).
Figure 10.
Hierarchical clustering of expression data for 32 candidate genes. Expression analysis was carried out on RNA extracted from whole seedlings of the wild type, RAP2.2-overexpressing line ox3 (ox), and RAP2.2 knockout line rap2.2-2 (ko). Plant treatments consisted of 5 h in the dark under normoxia (dark), 5 h with 5% oxygen in the light (o2), or a combination of 5 h in the dark with 5% oxygen (o2 + dark). Expression ratios for wild-type plants are treatment divided by the value obtained under normoxia-light conditions. ox and ko values are ox3 and rap2.2-2 expression under normoxia-light compared with the wild type under normoxia-light. Treatment values for ox and ko are treatments divided by ox and ko values obtained under normoxia-light. The full description of the genes used (locus, gene name, and function) is listed in Supplemental Table S1. Hierarchical clustering was used to divide the gene set in three clusters: group I consists of genes induced by darkness and hypoxia-darkness but repressed under hypoxia-light; group II consists of predominantly genes induced by darkness, hypoxia-darkness, and hypoxia-light treatments and contains the best known low-oxygen-induced genes that are involved in fermentation and sugar metabolism as well as ethylene biosynthesis genes and the ethylene receptor ETR2; group III consists of genes that were normally not induced by any of the three treatments in wild-type plants but whose expression was altered by changing RAP2.2 expression levels.
Overall, there were three major clusters in expression profiles (Fig. 10). Group I consisted of eight genes that were mainly darkness and hypoxia-darkness induced but repressed under hypoxia-light conditions. This group contained the senescence-associated genes SEN1 and SEN5 as well as a number of other ethylene-associated genes, such as ACS2 and AOX1. Group II is the largest cluster (16 members) and contains predominantly genes induced by darkness, hypoxia-darkness, and hypoxia-light treatments. All of the known low-oxygen-induced genes involved in fermentation and sugar metabolism (ADH1, PDC1, LDH1, AtSUS1, and AtSUS4) as well as four transcription factors (ANR1, AtERF4, ERF71, and ERF73), the ethylene biosynthesis genes ASC7 and ACO1, and the ethylene receptor ETR2 were present in this group. Group III was made up of eight genes that were normally not induced by any of the three treatments in wild-type plants but whose expression was altered by changing RAP2.2 expression levels. This group contains RAP2.12, a close relative of RAP2.2.
The expression data demonstrate that RAP2.2 affects only part of the response to low-oxygen stress. Specific genes important for metabolic adaptations such as fermentation and sugar metabolism, as well as some ethylene biosynthetic genes, are affected by RAP2.2.
DISCUSSION
Ethylene plays a major role in hypoxia-induced gene expression in plants: ethylene biosynthesis genes are up-regulated under low-oxygen conditions, ethylene induces ADH1 in Arabidopsis, and, more recently, quantitative trait locus mapping revealed that both submergence tolerance and elongation growth in deepwater rice are controlled by transcription factors of the ERF family (Xu et al., 2006; Hattori et al., 2009). Ethylene regulates several morphological adaptation responses to flooding and submergence, such as shoot elongation, aerenchyma formation, and adventitious root growth (for review, see Bailey-Serres and Voesenek, 2008). Both tolerance to submergence (SUB1A-1) and the escape reaction of elongation growth in deepwater rice (SK1 and SK2) are controlled by particular alleles of ERF genes.
ERFs that play a role in low-oxygen stress conditions have recently also been identified in dicots. The Arabidopsis ERF71 and ERF73 genes (HRE2 and HRE1, respectively) are induced by low-oxygen stress in the dark, and overexpression results in improved hypoxia survival (Licausi et al., 2010). We studied Arabidopsis RAP2.2 (At3g14230), originally thought to be responsive to hypoxia in roots (Klok et al., 2002) but shown here to be constitutively expressed under hypoxia in that tissue. RAP2.2 belongs to the same group VII ERF family as SUB1A (Fig. 1; Nakano et al., 2006) and was recently shown to activate two Arabidopsis genes encoding the carotenoid biosynthesis pathway enzymes phytoene synthase and phytoene desaturase (Welsch et al., 2007). These genes are also differentially expressed under hypoxic conditions (Loreti et al., 2005). Our phylogenetic analysis of the group VII ERF factors from rice and Arabidopsis shows that the dicot ERF transcription factors have evolved independently. This is reflected by the relatively high degree of sequence divergence outside the highly conserved AP2 domain. This complicates finding a true SUB1 homolog in Arabidopsis based on sequence similarity only. But RAP2.2, according to AP2 domain structure, is certainly more related to SUB1 from rice than to the rice SK1 and SK2 genes, which belong to a different ERF subfamily (Hattori et al., 2009). However, two other Arabidopsis group VII proteins, ERF73 and ERF71, are even more closely related to SUB1A than RAP2.2 (Fig. 1B). These transcription factors also play a role in low-oxygen stress in the dark (Licausi et al., 2010).
We show that RAP2.2 does play a significant role in metabolic adaptation to flooding stress in Arabidopsis, suggesting that the group VII ERF factors are specialized in abiotic stress responses. RAP2.12, the closest relative of RAP2.2 (Fig. 1), was recently shown to regulate ADH1 expression (Papdi et al., 2008). Other related dicot ERFs, tomato JERF3 and pepper CaPF1, play a role in salt and freezing tolerance, respectively (Wang et al., 2004; Yi et al., 2004). RAP2.2 was expressed at high levels in roots and at low levels in the shoots, a distribution similar to fermentation pathway genes like ADH1 (Dolferus et al., 1994), PDC1 (R. Dolferus, unpublished data), LDH1 (Dolferus et al., 2008), and AlaAT1 (Miyashita et al., 2007). While RAP2.2 expression in roots is constitutively high and not affected by hypoxia, in shoots its expression appeared to be induced by darkness but not by hypoxia. The gene is also regulated by ethylene and ethylene-generating compounds, and the fact that mutations in ethylene signal transduction components affect its expression clearly indicates that RAP2.2 functions in an ethylene-responsive signaling pathway. Low-light conditions trigger leaf senescence, which involves ethylene accumulation and the ethylene signaling pathway (for review, see Gan and Amasino, 1997; Quirino et al., 2000; Wingler and Roitsch, 2008; Lin et al., 2009). Darkness may trigger ethylene biosynthesis, as Arabidopsis ethylene biosynthesis genes ACS4 and ACS7 are induced by darkness (Wang et al., 2005). It is not clear why RAP2.2 expression in roots is so high and no superinduction can be seen by hypoxia or darkness. It is possible that roots grown in soil (darkness) or in liquid medium (entrapment) naturally accumulate higher ethylene levels.
A variety of experimental setups have been used to study low-oxygen responses in plants. Some of the more recent studies have used anaerobiosis (0% oxygen; Mustroph et al., 2006, 2009), while others used hypoxic conditions (0.1%–5% oxygen; Van Dongen et al., 2009; Licausi et al., 2010). Most of these experiments use darkness or low-light conditions, but the effect of light has so far not been investigated. We have routinely carried out our low-oxygen treatments on Arabidopsis plantlets in sealed tanks with a defined gas composition and in the dark to stop complications arising from altered oxygen concentrations due to photosynthesis or the release of trapped oxygen from plant tissues. These treatments resemble complete submergence, which in nature also leads to reduced photosynthetic activity due to a combination of restricted light and CO2 availability. It is conceivable that low light and CO2 availability, induction of ethylene synthesis, and induction and promotion of senescence are naturally intertwined and therefore inherent to natural submergence conditions. ADH1 expression in roots is low and is strongly induced by hypoxic conditions (Dolferus et al., 1994). Previous work on Arabidopsis seedlings indicated that ADH1 is induced within 12 h by ethylene, but this induction is only strong under hypoxic conditions (Peng et al., 2001). These studies were carried out on intact seedlings, likely in the light, but nonetheless support the conclusion that ethylene is not the only trigger required for inducing ADH1 expression. Hypoxic conditions are essential to fully induce ADH1 expression. The rice SUB1A gene is induced by submergence conditions, but it remains to be shown whether SUB1A also responds to darkness.
Overexpressing RAP2.2 using a strong constitutive promoter did not cause any noticeable phenotypic changes under normal growth conditions, but both ADH1 and PDC1 expression in shoots was increased following hypoxic-dark treatment. The overexpressing lines also had higher ADH and PDC enzyme activity in shoots, suggesting that RAP2.2 is a regulator of the alcohol fermentation pathway genes. The overexpression of ADH1 and PDC1 in the RAP2.2-overexpressing lines was only evident under hypoxic conditions in the dark but not under normoxic-dark conditions. This suggests that ethylene accumulation and RAP2.2 induction are not sufficient for ADH1 and PDC1 activation and that hypoxic conditions are also required.
The RAP2.2-overexpressing lines showed significantly higher hypoxia plant survival rates in the dark, while the knockout lines showed the opposite phenotype. This demonstrates that RAP2.2 can act as a regulator of the plant's response to hypoxia in the dark and that either expression of RAP2.2 is rate limiting for regulating genes that lead to hypoxia tolerance or that higher preexisting levels of RAP2.2 enable a quicker response to hypoxia, resulting in enhanced hypoxia tolerance. Submergence-tolerant rice plantlets produced highly elevated levels of ethanol during anaerobiosis in the dark with sufficient ATP synthesis to ensure plantlet survival, whereas flooding-sensitive wheat (Triticum aestivum) seedlings were much less efficient in fermentative ATP production. Fermentation occurred mainly in the shoot, indicating a vital role of anaerobic shoot metabolism for plantlet survival in the dark (Mustroph et al., 2006).
We investigated the overlap between hypoxic treatment under light and darkness as well as the effect of RAP2.2 overexpression and knockout lines on these responses using 32 candidate genes that play a role in hypoxia and ethylene responses. The clustering of the treatments indicates that hypoxia-darkness treatment responses were closer to the darkness-only treatment under normoxic conditions than they were to hypoxia treatment under light conditions. Darkness alone generally causes a lower induction than that observed when both darkness and hypoxia are combined. This suggests that there is a clear link between hypoxia and darkness responses in shoots. Observing the effect of altering levels of RAP2.2 showed that it affects only a subset of these pathways. The major effect observed was on fermentation genes such as ADH1 and PDC1 and ethylene biosynthesis and receptor genes such as ACS2, ASC7, ASC9, and ETR2. It is possible that RAP2.2 may affect other genes; however, they would have to be involved in pathways not yet associated with hypoxia tolerance.
The importance of fermentation and glycolysis for surviving low oxygen has been well established. ADH1, PDC1, and the Suc synthase genes AtSUS1 and AtSUS4 are vital in Arabidopsis for tolerance to low oxygen (Ellis et al., 1999; Rahman et al., 2001; Ismond et al., 2003; Kürsteiner et al., 2003; Bieniawska et al., 2007). Increased root survival was also observed in LDH1-overexpressing Arabidopsis lines (Dolferus et al., 2008). Expression of PDC1 is considered to be the rate-limiting step in ethanol fermentation, and overexpression has been shown to increase Arabidopsis survival (Ellis et al., 1999; Ismond et al., 2003). The observation that in the shoots, overexpression of RAP2.2 resulted in higher ADH and PDC activity indicates that the improved hypoxia survival of plants overexpressing RAP2.2 is in part via enhancement of fermentation. The poor hypoxia tolerance of the RAP2.2 knockouts may only be partly related or even unrelated to deficiencies in fermentation, as ADH1 and PDC1 expression is only partly reduced. Knocking out RAP2.2 does result in changes to ACS2, ACS7, and ACS9 that indicate negative feedback regulation of ethylene biosynthesis by RAP2.2. It may be hypothesized that altered ethylene signaling in rap2-2 plants contributes to poor hypoxia tolerance.
Although ERF factors were shown to interact with the GCC box or AP2/ERF-binding motif (Ohme-Takagi and Shinshi, 1995; Lin et al., 2008), RAP2.2 was shown to bind to an ATCTA promoter element (Welsch et al., 2007). The ADH1 promoter contains an ATCTA element proximal to the TATA box (positions −57 to −53; data not shown). A GCC box-like motif, the GC motif, is together with the GT motif part of the anaerobic response element (Olive et al., 1991; Dolferus et al., 1994). Transactivation assays with wild-type and deleted ADH1 promoter constructs are required to investigate whether RAP2.2 interacts directly with the GC motif or the ATCTA element. RAP2.2 may also act indirectly on ADH1 expression via other transcription factors. ERF transcription factors are known to interact with each other in complicated networks (Oñate-Sánchez et al., 2007). ERF71, ERF73, and ERF4 are all expressed in the same group of genes as ADH1; it is possible that these ERF factors function downstream of RAP2.2 to activate group II genes. A hypoxic signal is required to activate ADH1 expression, suggesting that there are two regulatory pathways that act on the ADH1 promoter. The ethylene-dependent pathway that involves RAP2.2 alone is not able to induce ADH1, but hypoxic conditions are also essential. The fact that hypoxia is able to induce ethylene biosynthesis suggests that the ethylene pathway is not completely independent of hypoxic conditions. On the other hand, AtMYB2, which binds to the GT motif (Hoeren et al., 1998), appears to be affected by darkness and ethylene (Fig. 10); this suggests that there is cross talk between the two regulatory pathways. Oxygen-dependent regulatory mechanisms that trigger anaerobic gene expression have been identified in bacteria, yeast, and mammals but remain so far elusive in plants (Poyton, 1999; Gilles-Gonsalez, 2001; Schofield and Ratcliffe, 2004; Bailey-Serres and Chang, 2005). To further unravel how gene regulation works under hypoxic conditions and to identify the components of the ethylene pathway and especially the hypoxic pathway will require more carefully designed experiments that take into account lighting conditions during hypoxic treatment. It will also be important to further establish the importance of lighting conditions and the difference between real submergence conditions and artificial assays that use gas mixtures in combination with light or dark conditions.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Low-Oxygen Treatments
Experiments were carried out on Arabidopsis (Arabidopsis thaliana ecotype Col-0). Col-0 wild-type seeds were originally obtained through GABI-Kat (Max-Planck-Institut), the T-DNA insertion lines SAIL_184_G12 (CS871911) and SAIL_799_D10 (CS876942) were from the SALK Institute, and the ethylene signaling mutants ctr1, ein2, and ein3 were from the Arabidopsis Biological Resource Center. Seeds were surface sterilized for 20 min in 1 mL of 0.5% (w/v) sodium hypochlorite and washed five times with autoclaved water. Five or 10 seeds were sown per plate (60 mm × 15 mm or 120 mm × 120 mm; height, 17 mm) on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), sealed with 3M micropore tape (Eydam), and kept at 4°C in the dark for 2 d. Plates were then transferred to a growth chamber, and plants were grown at 22°C in a photoperiod of 16 h of light/8 h of dark for the times indicated. Arabidopsis root cultures were generated by incubating leaf discs on callus induction medium (Valvekens et al., 1988) for 3 to 4 d in the dark. Leaf discs were then infected with Agrobacterium rhizogenes strain A4RS (Vilaine et al., 1987). When hairy roots appeared, the leaf discs were transferred to liquid MS medium and incubated at 50 rpm on a rotary shaker platform in the dark.
For RNA isolation for northern-blot analysis, promoter:GUS, and enzyme activity assays, plants were grown on half-strength MS medium containing 1.5% (w/v) Suc and 0.8% (w/v) agar at pH 5.7 (70 μE m−2 s−1). Hypoxic treatment of 17-d-old plants was carried out in an atmosphere containing 5% (v/v) oxygen, 0.04% (v/v) CO2, and 94.96% (v/v) N2 in the dark for the duration indicated. For ethylene measurements, plantlets were transferred after 1 week to MS medium containing 2% (w/v) agarose to prevent root growth into the agarose and subsequent wounding during the transfer of plants to the vials that were used for ethylene measurement, as described previously (Peng et al., 2001). For treatment with the effectors ACC, ethephon, STS (freshly prepared from 100 mm sodium thiosulfate and 100 mm silver nitrate at a ratio of 1:1), and CoCl2, plants were first grown on solid medium and transferred 2 d before treatment to liquid MS medium to improve uptake of the effectors. After preincubation for 2 h with ethephon, ACC, STS, or CoCl2 at the concentrations indicated, the plants were exposed to hypoxic treatment as described above.
For RNA isolation for QRT-PCR, plants were grown on half-strength MS medium containing 1.5% (w/v) Suc and 0.8% (w/v) agar at pH 5.7 (70 μE m−2 s−1). Plants were divided into one of four treatments (five plants per treatment): normoxia in the light (70 μE m−2 s−1) for 5 h, normoxia in the dark for 5 h, hypoxia (5% oxygen) in the light for 5 h, or hypoxia (5% oxygen) in the dark for 5 h.
Hypoxic Assays
Low-oxygen survival assays were performed as outlined by Christianson et al. (2009). Briefly, 2.5-week-old plants were grown under a 16-h light cycle (100 μE m−2 s−1) on MS medium containing 3% (w/v) Suc and 0.8% (w/v) agar at pH 5.7 and then transferred to MS liquid medium containing 3% (w/v) Suc for 1 d prior to treatment. Immediately prior to treatment, plants were transferred to liquid medium that had been sparged with 5% (v/v) oxygen and then placed in 3.5-L anaerobic chambers (Oxoid) and purged with 5% (v/v) oxygen at a flow rate of approximately 10 L min−1 for 20 min. The plants were left in 5% (v/v) oxygen in the dark with gentle shaking for 24 h, then purged with 0.1% (v/v) oxygen and left for 40 to 48 h. Following treatment, plants were given fresh liquid medium and allowed to recover on an orbital shaker in normal growth cabinet conditions for 1 week before survival scoring. Survival scores were averaged from at least three independent experiments. Each experiment contained three technical replicates, with each replicate consisting of 12 plants.
Generation and Characterization of Transgenic Plants
To constitutively overexpress RAP2.2, a 1.9-kb genomic fragment including the open reading frame, 5′ and 3′ untranslated regions, and two introns was amplified from Col-0 using the gene specific primers 5′-GAATTCGAGTAGAGCTTTCGTGAAGCCACCAT-3′ (forward) and 5′-GGATCCAGAAGATTCATTGAACAGATA-3′ (reverse). The fragment was restricted with EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pART7 (Gleave, 1992). The resulting 35S-RAP2.2-OCS fragment was then subcloned as a NotI fragment into the binary vector pART27 and transformed into the Arabidopsis ecotype Col-0 using the floral dip method (Clough and Bent, 1998). The presence and integrity of the RAP2.2 genomic DNA sequence was verified by sequencing the insertion in pART27.
To generate a RAP2.2 promoter construct, a 1,112-bp RAP2.2 promoter region was amplified from genomic DNA of ecotype Col-0 using the gene-specific primers 5′-GAATTCCAATATTGCCACTCCATGATAA-3′ (forward) and 5′-ATAGCTCCTCCACCCATGGTGGCTTCACGA-3′ (reverse). The resultant PCR fragment, which contained an EcoRI site and an NcoI site, was ligated in front of the GUS-NOS cassette of pHW9 (Dolferus et al., 1994) and subsequently cloned into the binary vector pBIN19 using the restriction sites EcoRI and XbaI (Bevan, 1984). Arabidopsis ecotype Col-0 plants were transformed, and GUS-expressing lines were analyzed.
Phylogenetic Analysis of ERF Sequences
Full-length ERF protein sequences were retrieved from the GenBank database and analyzed using the MEGA 4.0 software (Tamura et al., 2007). Sequences were aligned using the ClustalW progressive alignment algorithm (Higgins et al., 1994) and the neighbor-joining method (Saitou and Nei, 1987). Bootstrap figures are indicated in the phylogenetic tree.
RNA Extraction, Northern Blotting, and QRT-PCR
Total RNA was extracted by grinding the frozen tissue in liquid nitrogen in a mortar with a pestle. The root and shoot sections of the rosette-stage plants were separated by butting the base of the rosette with a scalpel. Tri-Reagent (Sigma-Aldrich) was added to the ground tissue, and RNA was isolated according to the manufacturer's instructions. For all gels, 20 μg of RNA each per sample was separated on a denaturing agarose gel (1% [w/v] agarose, 40 mm MOPS buffer, and 6% [v/v] formaldehyde). The ribosomal RNA staining pattern (ethidium bromide) was used as a loading control. RNA was transferred to a nylon membrane (Hybond N+; GE Healthcare) and hybridized to 32P-labeled probes obtained using a Ready-To-Go dCTP labeling kit (GE Healthcare). Hybridization was performed overnight at 68°C in hybridization solution (5% [w/v] dextran sulfate, 1 m NaCl, 1% [w/v] SDS, and 0.1 mg mL−1 heat-denatured salmon sperm DNA). Blots were washed two times in 2× SSC/1% (w/v) SDS for 1 and 10 min, once in 1× SSC/1% (w/v) SDS for 10 min, and once in 0.5× SSC/1% (w/v) SDS for 10 min. The gene-specific primer pairs used to generate the PCR probes from genomic DNA were 5′-CCTAGCGTCGTATCCCAGAA-3′ and 5′-CAAGGCGTTGTCAAGGTATGC-3′ for RAP2.2, 5′-CGAGACACCTTACAACAACAC-3′ and 5′-CAGATGAGAGGTTCAAACACAT-3′ for RAP2.12, 5′-GATTGTTGAGAGTGTTGGAG-3′ and 5′-CTTGGTCGAATCTTTTAGAGT-3′ for ADH1, and 5′-TCTCTCACACACATACACAAAC-3′ and 5′-CAAGCAAAGTGAGGTTGAAATC-3′ for PDC1. For RAP2.2 and ADH1, probes were generated from the coding region. For RAP2.12, a probe including the coding region and part of the 3′ untranslated region was generated. For PDC1, a probe including part of the 5′ untranslated region and the coding region was used. The probes for RAP2.2 and RAP2.12 shared only 23% sequence identity.
QRT-PCR was performed as described previously (Wilson et al., 2005). The RT product was amplified using gene-specific primers (Supplemental Table S2). Reactions were performed in triplicate on a Rotor-Gene 6000 (Qiagen). Data were normalized to At5g08290 (Czechowski et al., 2005) and analyzed using a comparative quantification procedure (Wilson et al., 2005). Hierarchical clustering of log-transformed expression data was carried out using the Cluster and Treeview (Eisen et al., 1998) programs. Heat maps were constructed using the University of Toronto BAR Heatmapper tool (http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi).
Expression Analysis of RAP2.2:GUS Plants
For RAP2.2:GUS studies on etiolated seedlings, seeds were exposed to light for 4 h to synchronize germination and then incubated in the dark for 4 d. In addition, RAP2.2:GUS seedlings were grown for 4 d and plants were grown for 17 d on half-strength MS medium containing 1.5% (w/v) Suc and 0.8% (w/v) agar at pH 5.7 and 70 μE m−2 s−1 in a light/dark cycle as described above. To analyze RAP.2.2 promoter activity under 5% (v/v) oxygen and in the dark, 17-d-old plants were grown under a 16-h light cycle at 100 μE m−2 s−1 on MS medium containing 3% (w/v) Suc and 0.8% (w/v) agar at pH 5.7 and then transferred to MS liquid medium containing 3% (w/v) Suc for 1 d prior to treatment. Immediately prior to treatment, plants were transferred to liquid medium. For the low-oxygen-treated plants, the medium was sparged with 5% (v/v) oxygen whereas the control was not sparged. Plants were then placed in 3.5-L anaerobic chambers. The low-oxygen-treated plants were purged with 5% (v/v) oxygen at a flow rate of approximately 10 L min−1 for 20 min. The plants were left in the dark with gentle shaking before harvest. Two independent transgenic RAP2.2:GUS transgenic lines were analyzed for GUS activity at the seedling and vegetative stages (Blázquez et al., 1997; Bomblies, 2000).
ADH and PDC Activity Assays
For ADH enzyme activity measurements, soluble protein was extracted in cold extraction buffer (100 mm Tris-HCl, pH 8.0, 25% [v/v] glycerol, 0.8% [v/v] β-mercaptoethanol, 2% [w/v] polyvinylpyrrolidone, and 5 mm dithiothreitol) and centrifuged at 15,000g for 15 min at 4°C. PDC enzyme activity was measured using a modified protocol by Rivoal et al. (1997). Soluble protein was extracted in cold extraction buffer (50 mm MES-NaOH, pH 6.2, 5 mm dithiothreitol, 1 mm MgCl2, and 1 mm thiamine pyrophosphate) and centrifuged at 15,000g for 15 min at 4°C. Protein concentration was determined with the Bradford assay (Roti-Quant; Roth). The enzymatic reaction was started by the addition of 800 μL of activation buffer (0.15 m Tris-HCl, pH 8.0, 0.3 mm NAD+, and 0.175% [v/v] ethanol for ADH activity measurements or 50 mm MES-NaOH, pH 6.2, 0.1 mm thiamine pyrophosphate, 0.5 mm MgCl2, 0.15 mm NADH, 3.3 mm sodium pyruvate, and 40 units mL−1 yeast ADH for PDC enzyme activity measurements). Activities were calculated either from the increase or decrease in A340 over time.
Ethylene Measurement
Thirty plants each were treated with 21% (v/v) or 5% (v/v) oxygen as described above, transferred to a 22-mL vial, and capped gas tight. Ethylene was measured after 1 h in the head space using a gas chromatograph (GC-14B; Shimadzu). Plant fresh weights were determined, and ethylene production per mg fresh weight was calculated.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_180251.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of RAP2.2-related ERF transcription factors.
Supplemental Figure S2. ADH and PDC activity measurements in shoots of wild-type Col-0 and the RAP2.2-overexpressing line ox3.
Supplemental Figure S3. QRT-PCR expression data for 32 candidate genes.
Supplemental Figure S4. Detailed heat map of QRT-PCR gene expression data for 32 candidate genes.
Supplemental Table S1. List of 32 candidate genes used for gene expression profiling studies by QRT-PCR.
Supplemental Table S2. List of PCR primers used in QRT-PCR experiments.
Supplementary Material
References
- Bailey-Serres J, Chang R. (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bevan M. (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2000) Whole mount GUS staining. Weigel D, Glazebrook J, , Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 243–245 [Google Scholar]
- Büttner M, Singh KB. (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al. (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis thaliana seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, Grover A, Ismond KP, Good AG, Peacock WJ. (2000) Molecular strategies for improving waterlogging tolerance in plants. J Exp Bot 51: 89–97 [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES. (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Wolansky M, Carroll R, Miyashita Y, Ismond K, Good AG. (2008) Functional analysis of lactate dehydrogenase during hypoxic stress in Arabidopsis. Funct Plant Biol 35: 131–140 [DOI] [PubMed] [Google Scholar]
- Drew MC, He CJ, Morgan PW. (2000) Programmed cell death and aerenchyma formation in roots. Trends Plant Sci 5: 123–127 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Paul T, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2004) Plant responses to hypoxia: Is survival a balancing act? Trends Plant Sci 9: 449–456 [DOI] [PubMed] [Google Scholar]
- Fukao T, Harris T, Bailey-Serres J. (2009) Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann Bot (Lond) 103: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1997) Making sense of senescence. Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA. (2001) Oxygen signal transduction. IUBMB Life 51: 165–173 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, Matsuoka M, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. (1998) Evidence for a role of AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, De Pauw M, Dennis ES, Good AG. (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M. (2007) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225: 575–588 [DOI] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho HT. (1998) Deepwater rice: a model plant to study stem elongation. Plant Physiol 118: 1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kürsteiner O, Dupuis I, Kuhlemeier C. (2003) The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol 132: 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Lin RC, Park HJ, Wang HY. (2008) Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant 1: 42–57 [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu F, VanToai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxia response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergemann H, Sauter M. (2000) Programmed death of epidermal cells facilitates emergence of adventitious roots in deepwater rice. Plant Physiol 124: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux JP, Kende H. (1983) The role of ethylene in the growth response of submerged deep water rice. Plant Physiol 72: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG. (2007) Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J 49: 1108–1121 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 18: 100–127 [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJJ, Harren FJM, Albrecht G, Grimm B. (2006) Organ-specific analysis of the anaerobic primary metabolism in rice and wheat seedlings: dark ethanol production is dominated by the shoots. Planta 225: 103–114 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive MR, Walker JC, Singh K, Dennis ES, Peacock WJ. (1991) Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol Biol 15: 593–604 [DOI] [PubMed] [Google Scholar]
- Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. (2006) ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exo-ribonuclease required for the regulation of the EIN3 targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA 103: 13286–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB. (2007) AtERF14, a member of the ERF family of transcription factors, plays a non-redundant role in plant defense. Plant Physiol 143: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L. (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HP, Chan CS, Shih MC, Yang SF. (2001) Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 126: 742–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HP, Lin TY, Wang NN, Shih MC. (2005) Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol Biol 58: 15–25 [DOI] [PubMed] [Google Scholar]
- Perata P, Voesenek LA. (2007) Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci 12: 43–46 [DOI] [PubMed] [Google Scholar]
- Pierik R, Van Aken JM, Voesenek LACJ. (2009) Is elongation-induced leaf emergence beneficial for submerged Rumex species? Ann Bot (Lond) 103: 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyton RO. (1999) Models for oxygen sensing in yeast: implications for oxygen-regulated gene expression in higher eucaryotes. Respir Physiol 115: 119–133 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Rahman M, Grover A, Peacock WJ, Dennis ES, Ellis MH. (2001) Effects of manipulation of pyruvate decarboxylase and alcohol dehydrogenase levels on the submergence tolerance of rice. Aust J Plant Physiol 28: 1231–1241 [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Rivoal J, Thind S, Pradet A, Ricard B. (1997) Differential induction of pyruvate decarboxylase subunits and transcripts in anoxic rice seedlings. Plant Physiol 114: 1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguch-Shinozaki K. (2002) DNA binding specificity of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sauter M. (2000) Rice in deep water: how to take heed against a sea of troubles. Naturwissenschaften 87: 289–303 [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ. (2004) Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol 5: 343–354 [DOI] [PubMed] [Google Scholar]
- Steffens B, Sauter M. (2005) Epidermal cell death in rice is regulated by ethylene, gibberellin, and abscisic acid. Plant Physiol 139: 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M. (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an auto-amplified signal pathway. Plant Cell 21: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Wang J, Sauter M. (2006) Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223: 604–612 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (2001) So what's new in the field of plant cold acclimation? Lots! Plant Physiol 125: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Straeten D, Anuntalabhochai S, Van Caeneghem W, Zhou Z, Gielen J, Van Montagu M. (1997) Expression of three members of the ACC synthase gene family in deepwater rice by submergence, wounding, and hormonal treatments. Plant Sci 124: 79–87 [Google Scholar]
- Van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaine F, Charbonnier C, Casse-Delbart F. (1987) Further insight concerning the TL region of the Ri plasmid of Agrobacterium rhizogenes strain A4: transfer of a 1.9 kb fragment is sufficient to induce transformed roots on tobacco leaf fragments. Mol Gen Genet 210: 111–115 [Google Scholar]
- Visser E, Cohen JD, Barendse G, Blom C, Voesenek L. (1996) An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiol 112: 1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LA, Colmer TD, Pierik R, Millenaar FF, Peeters AJ. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Rijnders JH, Peeters AJM, Van de Steeg HM, De Kroon H. (2004) Plant hormones regulate fast shoot elongation under water: from genes to communities. Ecology 85: 16–27 [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R. (2004) Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol 55: 183–192 [DOI] [PubMed] [Google Scholar]
- Wang NN, Shih MC, Li N. (2005) The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J Exp Bot 56: 909–920 [DOI] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. (2007) Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IW, Kennedy GC, Peacock JW, Dennis ES. (2005) Microarray analysis reveals vegetative molecular phenotypes of Arabidopsis flowering-time mutants. Plant Cell Physiol 46: 1190–1201 [DOI] [PubMed] [Google Scholar]
- Wingler A, Roitsch T. (2008) Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biol (Suppl) 1: 50–62 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D. (2004) The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol 136: 2862–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.