Abstract
The Arabidopsis (Arabidopsis thaliana) trichome birefringence (tbr) mutant has severely reduced crystalline cellulose in trichomes, but the molecular nature of TBR was unknown. We determined TBR to belong to the plant-specific DUF231 domain gene family comprising 46 members of unknown function in Arabidopsis. The genes harbor another plant-specific domain, called the TBL domain, which contains a conserved GDSL motif known from some esterases/lipases. TBR and TBR-like3 (TBL3) are transcriptionally coordinated with primary and secondary CELLULOSE SYNTHASE (CESA) genes, respectively. The tbr and tbl3 mutants hold lower levels of crystalline cellulose and have altered pectin composition in trichomes and stems, respectively, tissues generally thought to contain mainly secondary wall crystalline cellulose. In contrast, primary wall cellulose levels remain unchanged in both mutants as measured in etiolated tbr and tbl3 hypocotyls, while the amount of esterified pectins is reduced and pectin methylesterase activity is increased in this tissue. Furthermore, etiolated tbr hypocotyls have reduced length with swollen epidermal cells, a phenotype characteristic for primary cesa mutants or the wild type treated with cellulose synthesis inhibitors. Taken together, we show that two TBL genes contribute to the synthesis and deposition of secondary wall cellulose, presumably by influencing the esterification state of pectic polymers.
As the major component of the plant cell wall, cellulose has diverse functions. In the primary wall, cellulose is important for the production of the cell plate during cell division, for anisotropic cell expansion, and for turgor pressure distribution (Shedletzky et al., 1992). Thus, the cellulose microfibrils largely determine cell shape and patterns of development (Carpita and McCann, 2000). Once plant cells have stopped expanding, some cell types deposit a secondary cell wall (Taylor et al., 2000) that is mainly composed of highly aligned, crystalline cellulose microfibrils, noncellulosic polysaccharides, such as xylans and mannans (Carpita and McCann, 2000; Ebringerova and Heinze, 2000; Brown et al., 2005), and lignin. These polymers provide a framework that gives further strength to cells that have to sustain enhanced mechanical stress. Secondary cell walls are the major components in wood and plant fibers, underlining their economical importance (Brown et al., 2005). Cell walls are also important for protecting cells against pathogens, dehydration, or other environmental factors (Braam, 1999; Jones and Takemoto, 2004; Vorwerk et al., 2004).
More than 1,000 genes in the Arabidopsis (Arabidopsis thaliana) genome are estimated to encode cell wall-related proteins, but the specific biological contexts and the biochemical functions of most of these proteins are largely unknown (Carpita et al., 2001; Somerville et al., 2004). The first plant CELLULOSE SYNTHASE (CESA) genes were identified in cotton (Gossypium hirsutum) through sequence homology to conserved regions of bacterial cellulose synthases and high expression levels coinciding with high rates of cellulose synthesis (Pear et al., 1996). A family of CESA-related genes was rapidly identified in Arabidopsis, and their function as cellulose synthases was subsequently corroborated by classical genetic approaches (Arioli et al., 1998; Taylor et al., 1999, 2000; Fagard et al., 2000). Mutants in primary CESA genes generally contain strongly reduced levels of cellulose and exhibit dwarfed growth phenotypes (Arioli et al., 1998; Fagard et al., 2000; Burn et al., 2002). Mutants in secondary CESA genes, on the other hand, show characteristic irregular xylem vessels (Taylor et al., 1999, 2000), a phenotype that subsequently also has been reported for other secondary cell wall mutants (Brown et al., 2005). Besides the CESAs, several other components are believed to participate in cellulose deposition, such as the endo-1,4-β-d-glucanase KORRIGAN (Nicol et al., 1998), the glycophosphatidyl inositol-anchored plant-specific COBRA protein (Schindelman, et al., 2001), and the plasma membrane-localized KOBITO1 protein (Pagant et al., 2002; Lertpiriyapong and Sung, 2003). Analysis of Arabidopsis fragile fiber (fra) mutants, in which interfascicular fibers exhibit reduced mechanical strength, resulted in the identification of mutant alleles for the secondary CESA genes (fra5 and fra6; Zhong et al., 2003) but also yielded novel components, some of which presumably are indirectly required for secondary wall synthesis. These include the kinesin-like protein FRA1, which is essential for oriented deposition of cellulose microfibrils and cell wall strength (Zhong et al., 2002), the katanin-like protein FRA2 involved in regulating microtubule disassembly by severing microtubules (Burk et al., 2001), a type II inositol polyphosphate 5-phosphatase (Zhong and Ye, 2004), and the GTP-binding protein RHD3 (Wang et al., 1997; Hu et al., 2003), both required for actin organization in fiber cells. Identification of FRA8 as a putative xylan glucuronyltransferase (Zhong et al., 2005) from the glycosyl transferase (GT) 47 family of carbohydrate-active enzymes (www.cazy.org) points to the importance of acidic xylan (i.e. glucuronoxylan) modifications for normal cellulose deposition during secondary wall formation in Arabidopsis (Reis and Vian, 2004; Peña et al., 2007).
Coexpression analyses of microarray data with CESA genes as bait have been used to identify genes associated with cellulose synthesis (Brown et al., 2005; Persson et al., 2005). Mutations in several such genes display cell wall phenotypes characteristic of cellulose deficiency, for example the COBRA-like gene IRREGULAR XYLEM6 (IRX6) that contains reduced levels of secondary wall cellulose (Brown et al., 2005). Likewise, the irx8 and irx9 mutants display slightly reduced levels in stem secondary wall cellulose (Brown et al., 2005) but appear to be associated with the synthesis of xylans. IRX8 (AT5G54690) and IRX9 (AT2G37090) encode putative glycosyl transferase genes from the GT8 and GT43 families, respectively (www.cazy.org), and were found to be involved in glucuroxylan (GX) synthesis (Bauer et al., 2006; Peña et al., 2007; Persson et al., 2007), suggesting a requirement for normal hemicellulosic polysaccharide synthesis in order for normal secondary wall synthesis and cellulose deposition to occur.
Another important component thought to be required for secondary wall cellulose synthesis is the gene that controls a trait referred to as TRICHOME BIREFRINGENCE (TBR; Potikha and Delmer, 1995). The highly ordered cellulose found in the cell walls of Arabidopsis trichomes displays strong birefringence under polarized light, whereas the Arabidopsis tbr mutant displays no such birefringence (Fig. 1), and the cellulose content in tbr mutant trichomes is strongly reduced (Potikha and Delmer, 1995). In this work, we report the identification of the gene responsible for the TBR trait and a further characterization of the Arabidopsis tbr mutant. We show that TBR belongs to a plant-specific, poorly described gene family (TBR-like [TBL]) with 46 members in Arabidopsis. TBR and other gene family members are strongly coexpressed with primary and secondary CESA genes, respectively. We further provide evidence that TBR and an additional member of the family influence secondary wall cellulose deposition. Our results also suggest the involvement of the latter genes in pectin modification, more specifically methylesterification, which may affect the deposition of secondary walls.
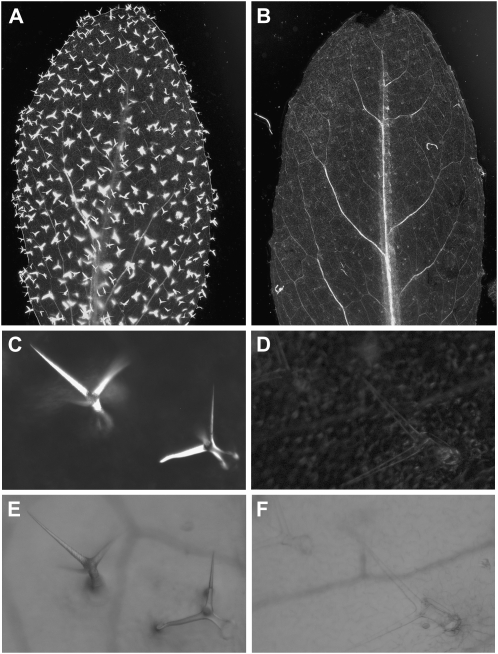
Figure 1.
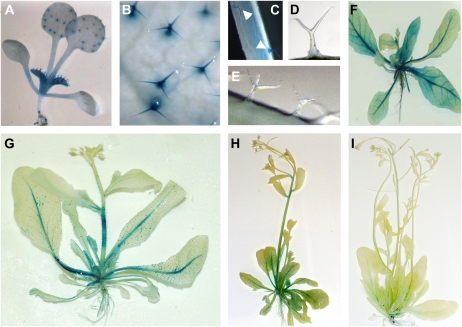
Lack of trichome birefringence in tbr mutants. A and B, Birefringence phenotypes of a wild-type Col-0 leaf (A) and a tbr mutant leaf (B). C to F, Appearance of individual wild-type and tbr mutant trichomes under polarized (C and D) and nonpolarized (E and F) white light.
RESULTS
Novel Phenotypes of the tbr Mutant
The tbr mutant of Arabidopsis was previously characterized as lacking leaf and stem trichome birefringence (Potikha and Delmer, 1995; Fig. 1) characteristic of plant cells that contain highly ordered cellulose in their secondary walls. Consistently, cellulose levels were decreased in tbr trichomes by greater than 80%. In addition, the cellulose content in the leaf vasculature was decreased approximately 30%, whereas cellulose contents in stems, roots, and callus tissue were unaffected (Potikha and Delmer, 1995). Other tbr phenotypes reported by Potikha and Delmer (1995) included educed trichome density, altered trichome shape and surface appearance, lack of trichome papillae and basal cells, altered stomata shape, and altered patterns of callose deposition.
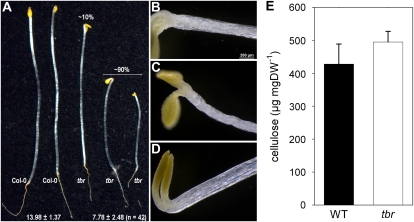
An additional cell wall-related phenotype was detected when we grew tbr mutant seedlings in the dark. Mutant seedlings frequently displayed a marked reduction in hypocotyl length (Fig. 2A), reminiscent of etiolated primary cesa mutant seedlings like prc1-8, ixr1-2, and rsw1-10 (Fagard et al., 2000; Mouille et al., 2003) and dark-grown wild-type seedlings treated with cellulose synthesis inhibitors like thaxtomin A (Scheible et al., 2003; Bischoff et al., 2009) or dichlobenil (Robert et al., 2004). In addition, young etiolated tbr mutant seedlings occasionally showed slight isotropic cell expansion symptoms in the upper part of the hypocotyl (Fig. 2, B and C), a phenotype that was reported previously for thaxtomin A-treated seedlings (Scheible et al., 2003). Such symptoms were never observed in wild-type seedlings (Fig. 2D). These results point to a function of TBR in primary cell wall synthesis in etiolated seedlings. To assess whether this is due to reduced levels of primary wall cellulose, we measured the amount of crystalline cellulose in tbr and wild-type hypocotyls (Fig. 2E). We did not detect any differences in cellulose levels between tbr and the wild-type control, indicating that alterations in other cell wall polymers may be the cause of the reduced hypocotyl elongation.
Figure 2.
Hypocotyl phenotypes of etiolated tbr mutants. A, Aspects of 4-d-old etiolated wild-type and tbr mutant seedlings. The length distribution among tbr seedlings is indicated by percentage numbers. Numbers at the bottom [13.98 ± 1.37 and 7.78 ± 2.48 (n = 42)] indicate mean values (in millimeters) of hypocotyl length in wild-type and tbr populations. B to D, Cell-swelling phenotype occasionally observed at the top of tbr mutant hypocotyls (B and C) and the invariant, regular wild-type phenotype (D). E, Crystalline cellulose determination as measured by the Updegraff (1969) protocol. Mean values ± se (n = 3) are given. DW, Dry weight; WT, wild type.
Greenhouse-grown tbr mutants (i.e. progeny with five backcrosses to ecotype Columbia [Col-0] wild type) also repeatedly displayed a marked growth variation that is unrelated to variation in germination time. The size of tbr mutants ranged between wild type like and considerably dwarfed, with the frequency of the latter being less than 10% (Supplemental Fig. S1). Additional visible phenotypes of tbr mutants included an approximately 40% reduced stem thickness at the base (0.82 ± 0.12 mm in tbr versus 1.47 ± 0.21 mm in the wild type) and reduced leaf size (Supplemental Fig. S1).
Cell Wall Composition of tbr Trichomes
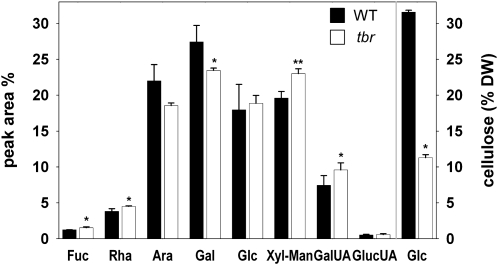
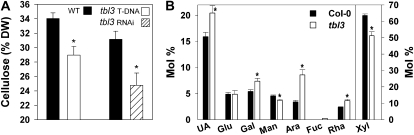
To investigate the effects on cell wall composition of the TBR gene product, we isolated mature trichomes from tbr and wild-type rosette leaves. Cellulose measurements in the trichome preparations revealed an approximately 70% decrease in tbr relative to the wild type (Fig. 3), confirming previous results (Potikha and Delmer, 1995). We also analyzed the noncellulosic sugar composition and uronic acid content in trichomes by high-performance anion-exchange chromatography (Fig. 3). Compared with the wild type, tbr trichomes displayed a 25% increase of the pectic component GalUA. Similarly, another prominent pectic component, Rha, was increased by approximately 15%. In contrast, the relative Gal and Ara contents were reduced by approximately 15%, while GlcUA, a minor but, for biomineralization processes, important compound found in plant cell walls, was unaltered. Neutral sugars (i.e. Xyl and Fuc), apparent in the side chains of hemicelluloses such as xyloglucans, were increased by around 20% (Fig. 3). A clear separation of Xyl and Man could not be achieved by the high-performance anion-exchange chromatography method used, but additional data obtained from analysis of alditol acetates by gas chromatography-mass spectrometry (data not shown) suggested that the Xyl content was increased in tbr mutant trichomes, whereas the Man content was unchanged.
Figure 3.
Altered cellulose and noncellulosic sugar composition in tbr mutant trichomes. Individual sugars and uronic acids (UA) are expressed as a percentage of the total noncellulosic cell wall sugar. Cellulose-derived Glc (right side) is expressed as a percentage of dry weight (DW). Typical results (means ± se; n = 4) from one of several experiments are shown. Significant changes as deduced from Student's t test (P ≤ 0.05) are marked with asterisks. WT, Wild type.
Identification of TBR
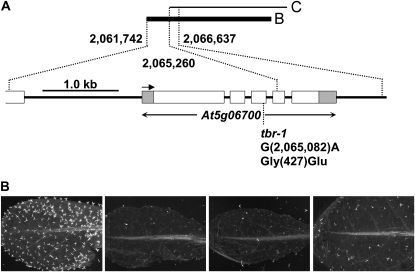
The morphological changes and the reduction in cellulose in tbr mutant trichomes are robust and transmitted to the progeny in a pattern typical for recessive mutants (Potikha and Delmer, 1995). Since the genetic lesion in the ethyl methanesulfonate-induced tbr mutant was unknown, we identified the TBR gene by map-based cloning (Lukowitz et al., 2000) and cosmid complementation (Fig. 4; Supplemental Fig. S2; Supplemental Table S1). This led to the identification of a complementing clone B and an overlapping noncomplementing clone C (Fig. 4; Supplemental Fig. S2). End sequencing of the Arabidopsis genomic sequence integrated in clone B and C revealed that the only TBR candidate gene was AT5G06700. This gene is completely contained in B, whereas the first two-thirds of the gene is absent in C. Sequencing of AT5G06700 from tbr mutant DNA subsequently revealed a G→A transition in the third exon of the annotated coding region, resulting in replacement of Gly-427 by Glu in the predicted protein (Fig. 4A). A cleaved-amplified polymorphic sequence (CAPS) marker was developed based on the single base change found in AT5G06700. Cosegregation of strong trichome birefringence and the heterozygous CAPS genotype was found in the progeny of tbr mutants transformed with clone B, as expected (Supplemental Fig. S2D).
Figure 4.
Identification of TBR. A, View of the approximately 5-kb region responsible for complementation of the tbr phenotype. The structure of the one annotated gene (AT5G06700) in that region is given. Exons are depicted as white boxes, and untranslated regions of AT5G06700 (i.e. TBR) are shown as gray boxes. The arrow indicates the direction of transcription. The point mutation in the tbr-1 allele (G→A) leads to a predicted Gly-to-Glu exchange at position 427 of the encoded protein. B, Knockdown of TBR by RNAi (the three photographs to the right show leaves of different RNAi lines) leads to strongly reduced trichome birefringence as compared with the wild type (the photograph to the left).
To provide further and independent confirmation of the identity of TBR, we investigated additional reduction/loss-of-function alleles for AT5G06700. To this end, we first tested several T-DNA insertion lines (SALK_134006, SALK_134014, SALK_058509, and SAIL_707_D07) but were unable to (1) identify homozygous mutants by PCR or (2) detect plants with reduced or lacking trichome birefringence. Therefore, we next produced an RNA interference (RNAi) construct to AT5G06700 and introduced it into the wild type. Many of the resulting RNAi lines displayed considerable or complete loss of trichome birefringence (Fig. 4B). In addition, complementation of the tbr mutant was achieved by expression of the annotated AT5G06700 coding sequence under control of the cauliflower mosaic virus 35S promoter (Supplemental Fig. S3).
TBR and Its Homologs Are Plant-Specific DUF231 Proteins
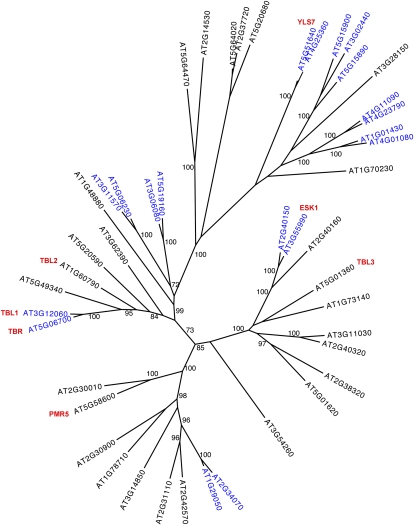
According to the annotation provided by The Arabidopsis Information Resource (TAIR), TBR spans approximately 2.173 kb (from start to stop codon; Fig. 4A), contains five exons, and encodes a 2.207-kb transcript that yields a predicted 608-amino acid 67.9-kD protein with an pI of 9.12. BLASTP and TBLASTN searches with the TBR protein sequence revealed that TBR is plant specific and has 45 homologs (e < 10−29) in the Arabidopsis genome. We aligned the protein sequences (Supplemental Table S2) and created an unrooted phylogenetic tree (Fig. 5). The proteins cluster into three major branches consistent with previous analyses (Xin et al., 2007), with TBR (AT5G06700) being part of the smallest clade (Fig. 5). Within the family, The Institute for Genomic Research database lists eight pairs of segmental duplicated genes and one triplet that arose through segmental and tandem duplication (highlighted in blue in Fig. 5).
Figure 5.
Unrooted, bootstrapped tree of the TBL gene family in Arabidopsis. MAFFT (version 5) was used to create an alignment of the protein sequences available at TAIR. Highly variable, gapped N-terminal regions were removed, and a maximum likelihood phylogeny was generated with RAxML model PROTCATWAG (Stamatakis, 2006) and bootstrapped (n = 1,000 trials) to generate the final tree. Bootstrap values greater than 70 are shown. Segmental/tandem gene pairs are highlighted in blue, and genes mentioned in the text are shown in red.
TBR, its duplicated twin AT3G12060 (TBL1), and the closely related AT1G60790 (TBL2) encode the longest annotated proteins in the family (608, 556, and 541 amino acids, respectively), while AT2G31110 and AT3G14850 appear to encode the shortest TBL proteins, with 364 and 356 amino acids, respectively (Supplemental Fig. S4A). All of the TBR and TBL homologs harbor a putative transmembrane domain at the N terminus (TMH) and contain a domain of unknown function (DUF231; PFAM 3005; IPR004253) near their C terminus (Supplemental Fig. S4A). The predicted amino acid exchange (Gly-427 → Glu) in the tbr mutant introduces a negative charge just in front of the DUF231 domain (asterisk in Supplemental Fig. S4B). This Gly residue is conserved in TBR homologs from other plant species (Supplemental Fig. S4B) as well as 26 of the Arabidopsis TBLs (Supplemental Table S2). Three additional Arabidopsis TBLs have Ala and the residual 16 have an aromatic amino acid (Tyr, Trp, or Phe) in this position.
Furthermore, the 46 proteins contain another plant-specific region, typically 87 to 89 residues long, that has not been recognized previously and that we termed the TBL domain (Supplemental Fig. S4). Thirteen amino acids in this domain, including five unequally spaced Cys residues, two Trp residues, and a Gly-Asp-Ser (GDS) signature, are fully conserved among the 46 family members. In addition, similar amino acids are found in another 25 positions of the TBL domains (Supplemental Fig. S4B; Supplemental Table S2). The GDS signature is followed by a Leu in over half of the proteins (24 proteins) or by a similar nonaromatic, aliphatic amino acid like Ile, Val, or Met, which in turn is preceded by several hydrophobic amino acids. GDS in the same amino acid context (i.e. GDSL) has previously been found to be a conserved motif in some esterases/lipases (Upton and Buckley, 1995; Akoh et al., 2004).
Within the 150- to 180-residue-long DUF231 domain, we detected another 13 residues with absolute identity (including another three Cys, three His, and two Trp residues) in all 46 proteins and 27 residues with conserved similarity (Supplemental Fig. S4B; Supplemental Table S2). There is a remarkable stretch of highly conserved amino acids toward the end of the DUF231 domain, including a DxxH motif (Supplemental Fig. S4B; Supplemental Table S2), which is also found in the same class of GDSL motif-containing esterases/lipases. Interestingly, the Asp and His residues in the DxxH motif together with the Ser residue in the distant GDS motif were determined to form the catalytic triad in the 2.5-Å crystal structure of a fungal rhamnogalacturonan acetylesterase (Mølgaard and Larsen, 2004).
TBR/TBLs have highly similar DUF231 protein homologs in other plant species. Within the C-terminal part starting with the TBL domain, TBR, for example, shares 77% identity (90% similarity) with grapevine (Vitis vinifera) protein CAO23412, 69% identity (82% similarity) with rice (Oryza sativa) lustrin A-like protein BAD35858 (Os06g0207500), and 61% identity (80% similarity) with Medicago truncatula protein ABE91344 (Supplemental Fig. S4B).
Expression Pattern of TBR
To investigate where the TBR gene is active, we created promoter:GUS gene reporter lines. When expressed under the control of a 1.6-kb sequence upstream of the annotated start codon (which includes the 140-bp annotated 5′ untranslated region), GUS activity in young plantlets was prominent in leaf and stem trichomes (Fig. 6, A–C). However, similar GUS constructs for the close TBR homologs AT3G12060 (TBL1) and AT1G60790 (TBL2) showed that these genes were not active in trichomes (Fig. 6, D and E). Interestingly, the TBL1 coding sequence expressed under control of the TBR promoter complemented the trichome birefringence phenotype of tbr mutants (data not shown). These results suggest that TBL1 is functionally equivalent to TBR but may work in different tissues or cell types.
Figure 6.
GUS expression of TBR. A, Staining of a 9-d-old seedling transformed with GUS driven by the TBR promoter. B, Magnified view of stained leaf trichomes of a 9-d-old seedling. C to E, Staining of stem trichomes of seedlings transformed with TBR:GUS (C), TBL1:GUS (D), or TBL2:GUS (E). F to I, Typical staining of 3-week-old (F), 4-week-old (G), 5-week-old (H), and 6-week-old (I) TBR:GUS transgenic plants.
In 3-week-old TBR:GUS gene reporter plants, GUS activity was mainly associated with the leaf vasculature and was also present in younger expanding/maturating rosette leaves (Fig. 6F) as well as rapidly growing parts of the root (e.g. lateral root tips; data not shown). In 4-week-old plants, the signal persisted in the vasculature and trichomes and was also strong in rapidly expanding, fortifying inflorescence stems (Fig. 6G), where primary and secondary wall cellulose is deposited. With increasing age and organ maturation, GUS activity continuously decreased (Fig. 6H) until it was hardly detectable in 6-week-old plants (Fig. 6I). This expression pattern is in agreement with the developmental AtGenExpress ATH1 gene chip data (Schmid et al., 2005; for visualization, see http://www.bar.utoronto.ca/efp) and is also consistent with the one expected for a component required during cellulose deposition.
Coexpression of TBR and TBLs with CESA and Other Cell Wall Genes
The strong reduction in crystalline cellulose in tbr trichomes (Potikha and Delmer, 1995) led us to investigate whether the gene is coexpressed with CESA genes. Analyses using the GeneCAT coexpression tool (http://genecat.mpg.de; Mutwil et al., 2008) revealed that CESA5, CESA6, and CESA3 rank at positions 2, 6, and 12 on the list of genes coexpressed with TBR (Supplemental Table S3). We also found the endochitinase ELP/POM/CTL1 (AT1G05850) and the glycophosphatidyl inositol-anchored protein-encoding COBRA gene (AT5G60920) on this list. These genes are well-known components for primary wall cellulose production (Hauser et al., 1995; Schindelman et al., 2001; Zhong et al., 2002). Similar results were also obtained using other coexpression tools, including ATTED-II (Obayashi et al., 2009) and AraGenNet (Mutwil et al., 2010). Vice versa, TBR also appears in the list of coexpressed genes when primary CESA genes are used as bait (data not shown; Persson et al., 2005).
To see if other members of the family also may be transcriptionally coordinated with cellulose-related processes, we extended the analysis to include the additional 35 DUF231 family members represented on ATH1 gene chips. Interestingly, the TBL gene AT5G01360 was tightly coexpressed with the secondary CESA genes; conversely, the analysis of genes coexpressed with AT5G01360 in GeneCAT revealed many of the known or suspected genes important for secondary wall cellulose synthesis (Brown et al., 2005), including all three secondary CESA genes, IRX8, IRX9, IRX12, and COBL4 (Supplemental Table S3).
Finally, several other DUF231 family members, including AT1G78710, AT2G37720, AT2G40160, AT3G28150, AT3G62390, AT5G01620, AT5G15890, and AT5G58600, displayed remarkable coexpression with other cell wall-related genes, like cellulose synthase-like, endo-1,4-β-glucanase, xyloglucan:xyloglucosyl transferase, laccase, polygalacturonase, and pectinesterase genes (http://genecat.mpg.de). Taken together, the coexpression analyses suggest that, besides TBR, at least AT5G01360 and possibly several other members of the plant-specific TBL/DUF231 domain gene family might have a function in cell wall biology.
Isolation and Phenotypic Characterization of tbl3 Mutants
To investigate the potential involvement of AT5G01360/TBL3 in cellulose/cell wall synthesis, we obtained seeds from a corresponding T-DNA insertion line (SALK_065959) available through the European Arabidopsis Stock Centre. Homozygous mutant plants were identified by PCR screening of genomic DNA samples and were further characterized for expression of AT5G01360 (subsequently named TBL3) at the cDNA level. No PCR amplification was possible from mutant cDNA with primers spanning the T-DNA insertion site (Supplemental Fig. S5, A and B), and quantitative real-time PCR revealed that the cDNA template corresponding to the last exon of AT5G01360 was more than 1,000-fold less abundant in the mutant compared with the wild-type cDNA pool (Supplemental Fig. S5, A and C). These results suggest that the isolated homozygous mutant is a null.
Growth analysis revealed that the homozygous mutant progeny displayed a variety of growth phenotypes, ranging from mild to severe reductions in inflorescence stem elongation (Supplemental Fig. S6), whereas rosette leaf or root growth was only mildly affected (Supplemental Fig. S6; data not shown). The variation in growth and stem size is similar to what was observed for the tbr mutants (Supplemental Fig. S1). However, TBR gene expression was not changed in the tbl3 mutants (data not shown). The tbl3 mutants also frequently displayed significantly (10%–20%) reduced stem diameter (data not shown), as found for tbr mutants and for other secondary wall mutants (Turner and Somerville, 1997). To corroborate the mutant phenotype, we also produced gene-specific RNAi lines for AT5G01360 and found the tbl3 growth phenotypes recapitulated in these (data not shown). These results suggest that TBL3 is required for normal stem development in Arabidopsis.
Biochemical analysis of tbl3 T-DNA mutant stems revealed a 15% to 20% reduction in cellulose content independent of stem age (Fig. 7A). These data were reproduced using the tbl3 RNAi lines (Fig. 7A). We also detected changes in the noncellulosic carbohydrate composition in stem material sampled 20 d after bolting (Fig. 7B). In agreement with previous studies, wild-type stems showed a high Xyl content characteristic of secondary cell walls (Turner and Somerville, 1997; Brown et al., 2005). Cell wall material from tbl3 mutant stems displayed a 20% relative reduction in Xyl content and strong relative increases in Ara, Gal, Rha, and Fuc as well as uronic acids (Fig. 7B). These changes resemble the sugar compositions measured in stems of irx7 and irx8 mutants and of mutants in other genes coexpressed with secondary CESAs (Brown et al., 2005; Persson et al., 2007). Finally, stem sections were cut from the base of the mature inflorescence stem to investigate xylem morphology of tbl3 mutant and the wild type. The xylem vessels of tbl3 mutants were indistinguishable from wild-type vessels displaying open xylem elements with relatively round shape (data not shown). These data suggest that while the constituents for several important secondary wall polymers were decreased in tbl3, the integrity of the wall still is sufficient for normal xylem morphology.
Figure 7.
Biochemical cell wall phenotypes of tbl3 T-DNA mutants. A, Cellulose-derived Glc is expressed as percentage of dry weight (DW) and shows the typical result from one of several experiments (means ± se; n = 5). B, Individual sugars and uronic acids (UA) of wild-type and tbl3 stems are expressed as percentage of the total noncellulosic cell wall sugar (means ± se; n = 4). Significant changes as deduced from Student's t test (P ≤ 0.05) are marked with asterisks.
Additional Hypocotyl Phenotypes of tbr and tbl3
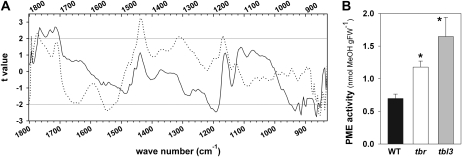
To determine possible structural changes in the cell walls of tbr hypocotyls, we performed Fourier transform infrared (FTIR) microspectroscopy (Fig. 8A). This analysis revealed, according to Student's t test, significant changes at wave number 1,168 cm−1 and also at wave number 1,774 cm−1, which most likely correspond to decreases of ester linkages, presumably of methyl-esterified pectins (Sene et al., 1994; Mouille et al., 2003). The peak at 1,546 cm−1 may represent changes in not further specified cell wall proteins. These results indicate that the tbr hypocotyls are not holding lower levels of cellulose (Fig. 2E) but that there may be structural changes in the cell wall framework involving esterified pectins. In agreement with this observation, a significantly elevated pectin methylesterase (PME) activity was detected in tbr protein extracts derived from etiolated seedlings (Fig. 8B).
Figure 8.
FTIR spectra and PME activity in tbr and tbl3 mutants. A, FTIR spectrum from 4-d-old, dark-grown tbr (dotted line) and tbl3 (solid line) mutant hypocotyls. Student's t test values for the comparison between the wild type and tbr or tbl3 (x axis) are plotted against the wave numbers (y axis). Horizontal gray lines refer to the significance threshold (P = 0.95). Several highly significant maxima can be assigned to pectin ester linkages and alterations in cell wall crystallinity. B, Measurement of PME activity isolated from etiolated seedlings. The mean values of PME activity are expressed as release of methanol in nmol g−1 fresh weight (FW) derived from wild-type (WT), tbr, and tbl3 seedlings. Mean values ± se are shown (n = 5). Significant changes (P ≤ 0.01) as deduced from Student's t test are marked with asterisks.
In contrast to tbr (Fig. 2), etiolated tbl3 seedlings did not show a significant reduction in hypocotyl length (data not shown). The level of crystalline cellulose in these hypocotyls tended to be slightly increased on a dry weight basis, and FTIR analysis revealed similar changes in cell wall structures of etiolated tbl3 hypocotyls as seen for tbr (Fig. 8A). We found significant changes at wave numbers 852, 917, 933, and 1,191 cm−1 and at wave numbers 1,762, 1,731, and 1,712 cm−1 (Fig. 8A), indicating structural changes of cellulose and esterified pectins in tbl3 compared with wild-type seedlings. In addition and similar to tbr, PME activity was significantly increased in tbl3 (Fig. 8B). These data suggest that the tbr and tbl3 seedling hypocotyls hold lower levels of methylesterified pectins due to enhanced PME activities.
DISCUSSION
In this study, we applied forward and reverse genetic approaches to identify two members of the undescribed DUF231 gene family, TBR (AT5G06700) and TBL3 (AT5G01360), as novel components contributing to secondary wall cellulose synthesis in higher plants. The tbr mutant was previously reported as a mutant that lacks crystalline cellulose in trichomes and to lesser extent also in the vasculature (Potikha and Delmer, 1995). The involvement of TBR in cellulose synthesis is underpinned by the expression pattern of the gene. TBR displays extraordinary coexpression with primary CESA genes, such as CESA3, CESA5, or CESA6, while coexpression with secondary CESA genes is inconspicuous. Several genes coexpressed with the primary and secondary CESA genes were previously shown to be associated with primary and secondary wall cellulose production, respectively (see introduction and “Results”). The expression patterns obtained from the microarray data sets were mirrored in the TBR promoter:GUS staining patterns, corroborating that TBR is similarly expressed as the primary wall CESA genes. Considering the substantial reduction in trichome-associated crystalline cellulose levels in tbr, it is perhaps surprising that no decrease in primary wall cellulose levels were observed. However, it is possible that other TBR-related gene products may functionally compensate for the loss of TBR in these tissues. In support of such scenario, TBL1 could rescue the trichome birefringence phenotype when expressed under the control of the TBR promoter.
We showed that the reduction in cellulose content in tbr trichomes was accompanied by an increased amount of pectic compounds (i.e. GalUAs), whereas the relative Gal and Ara contents were reduced by approximately 15%. These results imply increased amounts of HG and decreased abundance of the Gal- and Ara-containing side chains of the pectin rhamnogalacturonan I in the mutant. The amount of GlcUA remained stable in trichomes, in turn. This result confirms that the changes in uronic acids are associated with pectic structures and rules out the idea that loss of trichome birefringence is a result of reduced uronic acid-dependent biomineralization/crystallization. The amount of Xyl-Man was increased, indicating elevated amounts of xyloglucans. Since the effects on pectin/xyloglucan content, and/or modification, were significant, we propose that tbr directly interferes with cellulose biosynthesis in trichomes and directly or indirectly influences pectin composition in this organ. In support of this, several mutants deficient in cellulose production display increased levels of pectins, most likely homogalacturonans (His et al., 2001).
tbr hypocotyls contained wild-type levels of primary wall cellulose, less methylesterified pectins, and hold modified cell wall structures, as assessed by FTIR analysis. In addition, results of the PME assay confirm that the lower degree of pectin methylesterification comes along with a higher methylesterase activity in tbr compared with wild-type seedlings. Many primary cell wall mutants (e.g. prc1-1, rsw1-10, kor1-1, and kob1-1) have less crystalline cellulose and short hypocotyls (Desnos et al., 1996; Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000; Pagant et al., 2002). However, dwarfed hypocotyls are not necessarily linked to the level of crystalline cellulose but can also result from altered levels of methylesterified pectins (Derbyshire et al., 2007). In addition, it is interesting that the tbr hypocotyl phenotypes described partially resemble the mutant quasimodo2 (qua2), affecting a putative pectin methyltransferase. qua2 has higher levels of crystalline cellulose, a shorter hypocotyl, as well as altered pectin composition (Mouille et al., 2003, 2007). Our results suggest that TBR, besides its role in secondary wall cellulose deposition in trichomes, is involved in pectin modifications in etiolated hypocotyls, a tissue mainly containing primary wall cellulose. Alternatively, the changes described could reflect a feedback loop from secondary to primary cell wall biosynthesis, as has been described for mur10 (Bosca et al., 2006). However, TBR has a largely nonredundant/critical biological function in trichomes, as suggested by the missing trichome expression of its close homologs (Fig. 6, D and E). In this context, it is interesting (1) that Arabidopsis trichomes do not appear to express secondary CESA genes (CESA4, CESA7, and CESA8) while primary CESA genes (CESA1, CESA3, CESA5, and CESA6) and KORRIGAN are expressed (Jakoby et al., 2008; Marks et al., 2008), and (2) that the Arabidopsis trichome cell wall contains polysaccharides that are more like those of typical primary walls (Marks et al., 2008). This may suggest that the “secondary wall” crystalline cellulose deposited in trichomes is a product of the primary CESA complex. Alternatively, the secondary CESA complex may be recruited from other cell types, such as neighboring trichome companion or basal cells. However, both interpretations seem to be rejected by the observation that irx3 (cesa7) mutants as well as primary cesa mutants (cesa3, cesa2, cesa5, cesa6, cesa2/cesa5, and cesa2/cesa6) display wild-type-like trichome birefringence (V. Bischoff, S. Nita, and W. Scheible, unpublished data). Another possibility would be that cellulose synthase-like proteins synthesize cellulose in trichomes, similar to the situation described for pollen tubes of Nicotiana alata (Doblin et al., 2001).
The second TBL/DUF231 domain gene family member that we functionally implicated in secondary cell wall and cellulose synthesis is TBL3 (AT5G01360). This gene was strongly coexpressed with CESA4, CESA7, and CESA8, and analysis of a T-DNA insertion null allele and RNAi lines confirmed its function in secondary cell wall cellulose deposition. The T-DNA mutants have shorter and weaker stems at high frequency, contained significantly less cellulose in the inflorescence stems, and displayed a cell wall monosaccharide pattern that highly resembles those of several irx mutants (Brown et al., 2005). tbl3 stems appear to be enriched in pectin, as judged from increased levels of uronic acids, Ara, Gal, and Rha. While Xyl levels decreased by approximately 20%, this neutral sugar pattern is reminiscent of those found in irx7, irx8, and irx9 mutants (Brown et al., 2005). The irregular xylem structure is a known criterion for mutants involved in secondary wall cellulose biosynthesis (Turner and Somerville, 1997). However, the reduction of cellulose and/or the structural changes in the secondary cell wall of tbl3 were not sufficient to yield an irregular xylem phenotype with collapsed xylem vessels (data not shown). This is in agreement with several other mutants notably involved in secondary wall cellulose biosynthesis. Some of the latter showed normal or modestly disturbed xylem vessels (Brown et al., 2005, 2009; Persson et al., 2005), while others developed the irregular xylem phenotype only in a double irx background (Brown et al., 2009). Additional analyses showed that etiolated tbl3 hypocotyls displayed wild-type-like phenotypes but, as seen for tbr hypocotyls, had slightly elevated crystalline cellulose levels and significantly reduced amounts of methylesterified pectins. Along with the PME assay showing higher PME activity in tbl3, our results suggest that TBL3, similar to TBR, is involved in secondary wall cellulose biosynthesis in specific tissues and also has an influence on the pectic structures in tissue containing mainly primary cell wall. A role of AT5G01360 in secondary cell wall biosynthesis was previously suggested, as it turned up as a member of a core xylem-specific gene set resulting from global comparative transcriptome analysis (Ko et al., 2006).
What might be the biochemical functions of the plant-specific DUF231 domain genes? One report (Yoshida et al., 2001) describes YLS7 (AT5G51640) as highly expressed in senescent leaves, but beyond this the gene remained anonymous. Another member of the gene family (ESK1; AT3G55990) was found to act as a negative regulator of cold acclimation, as mutations in the ESK1 gene provide strong freezing tolerance (Xin et al., 2007). In a third report, it was noted that a null mutation in the PMR5 gene (i.e. AT5G58600) rendered Arabidopsis resistant to powdery mildew species and that, based on cell wall and FTIR analyses, pmr5 cell walls were enriched in pectin and displayed a reduced degree of pectin methylesterifiation relative to wild-type cell walls, suggesting that the gene affects pectin composition (Vogel et al., 2004). This observation is in line with our results, as we also noted changes in pectin composition in various tissues of tbr and tbl3. Although the alterations in pectin composition in tbr trichomes and tbl3 stems seem to be due to compensatory effects following the reduction in cellulose levels, our results from etiolated tbr and tbl3 hypocotyls suggest a more direct impact of these gene products on pectin modification. FTIR analysis revealed less esterified pectins, and this could be confirmed by an elevated PME activity. These results indicate that at least some DUF231 gene family members, including PMR5, TBR, and TBL3, are required to maintain higher pectin methylesterification states, for example by inhibiting PME activity, rather than being PMEs by themselves, as might be deduced from the conserved GDSL and DxxH motifs known from some esterases (see “Results”).
Whatever the biochemical function of the proteins, loss of TBR or TBL3 appears to increase PME activity, reduce pectin esterification, and decrease cellulose deposition in trichomes and stems, respectively. There is existing evidence indicating that correct modifications of noncellulosic cell wall polysaccharides are important for growth of cells undergoing secondary wall thickening, the synthesis of secondary wall cellulose, and secondary wall integrity. For example, the dwarfed Arabidopsis irx8/gaut12 mutant is deficient in GX and HG (Persson et al., 2007), has alterations of the glycosyl sequence at the GX reducing end (Peña et al., 2007), and shows a significant reduction in secondary cell wall thickness and cellulose content (Persson et al., 2007). The degree of pectin methylesterification can limit cell growth and hypocotyl elongation in Arabidopsis, as shown by ectopic expression of a fungal PME (Derbyshire et al., 2007). Similarly, PME prevents correct growth of developing wood cells/fibers in poplar (Populus species; Siedlecka et al., 2008), suggesting that proper pectin esterification is likely to be essential for xylem development and lignification (Pelloux et al., 2007). Furthermore, there is evidence for potent binding of pectins, which are enriched in neutral side chains, to cellulose (Zykwinska et al., 2005), thus leading to the assumption that pectin-cellulose interactions are significant for cell wall assembly and normal cellulose deposition during primary and secondary cell wall formation. In tobacco leaf explants, oligogalacturonides derived from esterified HG were shown to stimulate cellulose deposition and cell wall thickening (Altamura et al., 1998), and such oligogalacturonides were also shown to elicit the production of hydrogen peroxide (H2O2; Legendre et al., 1993; Svalheim and Robertsen, 1993), probably via induction of small Rac-type GTPases, which activate H2O2-producing plasma membrane NADPH oxidases (Keller et al., 1998; Potikha et al., 1999; Wong et al., 2007). In this respect, it should be noted that the plasma membrane-localized small GTPase AtRAC2/ROP7 (AT5G45970) is specifically induced during later stages of xylem differentiation in Arabidopsis (Brembu et al., 2005), and strong coexpression with TBL3 was detected in our study (Supplemental Table S3). Constant production of low levels of H2O2 stimulates the onset of secondary wall cellulose synthesis and secondary wall differentiation, as shown in cotton fiber cells (Potikha et al., 1999; Karlsson et al., 2005; Hovav et al., 2008), which from the botanical point of view represent seed trichomes. It is tempting, therefore, to speculate that (1) TBR and/or TBL3 function in maintaining HG esterification, as suggested by our results, and that (2) this is required for triggering secondary wall cellulose synthesis in a manner similar to the suite of events outlined above. Therefore, the esterification state of pectic polymers might influence the synthesis and deposition of cellulose in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in environmental chambers (120 μE, 16 h of light/8 h of dark, 60% relative humidity, 20°C) or in the greenhouses of the Max Planck Institute of Molecular Plant Physiology (150 μE, 12–14 h of light, approximately 70% relative humidity, 20°C–22°C) on a commercial Arabidopsis substrate (Stender). Etiolated seedlings were grown on vertical agar plates (0.7% [w/v] agar, half-strength Murashige and Skoog medium supplemented with 0.5% [w/v] Suc) wrapped in aluminum foil. Arabidopsis wild-type (Col-0) and tbr mutant seeds were obtained from an in-house collection. The tbl3 T-DNA insertion line SALK_065959 was obtained from the European Arabidopsis Stock Centre.
Positional Cloning of the TBR Locus
See Supplemental Protocol S1.
Cell Wall Analyses and PME Activity Assay
See Supplemental Protocols S2 and S3.
Analysis of Trichome Birefringence
Trichome birefringence of old rosette leaves was analyzed by incubation in methanol for 20 min followed by discoloration for 1 h in boiling 85% lactic acid and rinsing with water (three times). The leaves were then observed with a stereomicroscope (Leica MZ 12.5) equipped with a polarizer.
Stem Cross-Sections
The 50-μm cross-sections were cut with a vibratome (Leica). Sections were stained with 0.1% toluidine blue (analysis of irregular xylem structures) and observed with a stereomicroscope (Olympus BX41 and Leica MZ 125).
FTIR Microspectrometry
Four-day-old seedlings were squashed between two barium fluoride windows and rinsed abundantly with distilled water for 2 min before drying at 37°C for 20 min. For each mutant, 20 spectra were collected from individual hypocotyls of seedlings from four independent cultures (five seedlings from each culture), as described by Mouille et al. (2003). Normalization of the data and statistical analyses were performed as described by Mouille et al. (2003). Normalization of the data set and statistical analyses were performed using the statistical language R version 2.6. (R Development Core Team, 2006). To normalize the spectra, the baseline, estimated using a linear regression involving 10 points at each end of the spectrum, was subtracted from each absorbance value, and the area was set to 1 by dividing each absorbance value by the sum of all absorbance values. To determine the difference of the composition and the structure between mutants and the wild type, Student's t test was performed.
RNAi Constructs for TBR and TBL3
A 381-bp TBR-specific PCR fragment was amplified using primers 5′-CACCAACCACTCGTCACGCGCC-3′ and 5′-GGTTTGGTTGAAGTGACATTGGG-3′, subcloned into pENTR/SD/D-TOPO (Invitrogen), and recombined into pK7GWIWG2(II) (Karimi et al., 2002). A 177-bp TBL3-specific PCR fragment was amplified with primers 5′-CACCAATTACCAGATTTGCCACTATGAGC-3′ and 5′-TTGAAGAAAGATGAAGACGAAGAGG-3′ and shuttled into pK7GWIWG2(II) with the same strategy. The constructs were transferred into Col-0 plants by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998), and T1 transformants were selected on kanamycin and scored for trichome birefringence as described above.
Promoter:GUS Constructs and GUS Staining
To obtain a TBR promoter:GUS gene fusion construct, a PCR product was amplified from genomic DNA of Arabidopsis wild-type Col-0 using PfuTurbo polymerase (Stratagene) and primers 5′-CAATGTCGACGGAAGATGAGTTGGACAATGC-3′ and 5′-CAATGGATCCGCATATACTTAACGGCGTCTG-3′. The 1.64-kb BamHI/SalI promoter fragment was subsequently fused to the GUS gene in vector pBI101.1 (Clontech); after sequence verification, the recombinant vector was introduced into Arabidopsis wild-type Col-0 via A. tumefaciens (GV3101) according to Clough and Bent (1998). Detection of GUS activity was performed according to Jefferson et al. (1987) by incubation in staining buffer at 37°C overnight. Promoter:GUS lines for the close TBR homologs AT3G12060 (TBL1) and AT1G60790 (TBL2) were produced likewise using primers 5′-CAATGTCGACTTTCGGAGTTTCTAGTCTGGA-3′ and 5′-CAATGGATCCTTCAGGCAATGCGTAATCTAT-3′ and primers 5′-CAATGTCGACCCATTTGTATCAATCTCCCCC-3′ and 5′-CAATCCCGGGAATGAAGCACACGGAAAGTGA-3′, respectively.
Molecular Analysis of tbl3 Mutants
A T-DNA insertion line (SALK_065959) for TBL3 (AT5G01360) was obtained from the European Arabidopsis Stock Centre. Plant DNA was extracted as described previously (Lukowitz et al., 2000). Mutant lines were confirmed for T-DNA insertion using spanning primers LP (5′-GTATGAAAAACGGACAGCCAGAAACT-3′) and RP (5′-TCCGAAAGAACCCCACAGAGC-3′; designed on http://signal.salk.edu/tdnaprimers.html) and a primer from the left T-DNA border, LBc1 (5′-CCGCAATGTGTTATTAAGTTG-3′). Primer combinations LBC1/LP and LP/RP were used. PCR conditions were 96°C for 5 min; 96°C for 30 s, 58°C for 40 s, and 72°C for 1 min; 4°C hold. RNA was isolated from tbl3 mutant stems using the Plant RNeasy Mini Kit (Qiagen). RNA quality checks, DNase I treatment, and reverse transcription were performed as described previously (Czechowski et al., 2005). PCR with the primers 5′-GTATGAAAAACGGACAGCCAGAAACT-3′ and 5′-TCCGAAAGAACCCCACAGAGC-3′ spanning the insertion site (Supplemental Fig. S5A) was performed on wild-type and tbl3 cDNA to confirm the absence of a PCR product when using the mutant template (Supplemental Fig. S5B). Quantitative real-time PCR was used to analyze the expression level of AT5G01360 using an Applied Biosystems HT7900 sequence detection system, SYBR Green reagent (Applied Biosystems), and primers 5′-ATCGCATCGACGCTCACAC-3′ and 5′-TCAGCGGTTAGGATCTTGCC-3′. For further details and subsequent data normalization procedures, see Czechowski et al. (2005).
Sequence Alignment and Phylogenetic Analysis
MAFFT version 5 (Katoh and Toh, 2005) was used to create an alignment of the Arabidopsis DUF231 protein sequences available at TAIR and that of other plant TBR homologs available at the National Center for Biotechnology Information. The alignment was processed with Boxshade 3.21 (http://www.ch.embnet.org) and arranged manually into the final layout (Supplemental Fig. S4B; Supplemental Table S2). Prior to computing a phylogeny, the highly variable and gapped N-terminal region was removed manually, and all columns with 50% or more gaps were removed with the software REAP (Hartmann and Vision, 2008). RAxML version 7 was then used to compute an unrooted maximum likelihood phylogeny from the masked alignment using the PROTCATWAG model (Stamatakis, 2006). The phylogenetic tree was subsequently bootstrapped (n = 1,000 trials) to create the final tree in FigTree version 1.1.2 (http://tree.bio.ed.ac.uk/software/figtree/) as shown in Figure 5.
Coexpression Analysis
Coexpression analyses were performed using the Web-based tool GeneCAT (Mutwil et al., 2008) and ATTED-II (Obayashi et al., 2009).
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following locus identifiers: AT5G06700 (TBR), AT3G12060 (TBL1), AT1G60790 (TBL2), and AT5G01360 (TBL3). The mutant tbr sequence is available from GenBank (HM120873).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Growth phenotype of tbr mutants.
Supplemental Figure S2. Identification of TBR by recombinational mapping and cosmid complementation.
Supplemental Figure S3. Complementation of the tbr mutant by 35S::TBR.
Supplemental Figure S4. Structure and sequence alignment of DUF231 domain proteins.
Supplemental Figure S5. Molecular characterization of tbl3 T-DNA insertion mutants.
Supplemental Figure S6. Growth phenotype of tbl3 mutants.
Supplemental Table S1. Specifications and primer sequences of mapping markers and the tbr CAPS marker.
Supplemental Table S2. Partial alignment of the 46 Arabidopsis DUF231 proteins.
Supplemental Table S3. Genes coexpressed with TBR or TBL3.
Supplemental Protocol S1. Positional cloning of the tbr locus.
Supplemental Protocol S2. Biochemical analyses of tbr and tbl3 cell walls.
Supplemental Protocol S3. PME extraction and activity assay.
Supplementary Material
Acknowledgments
We are grateful to Kian Hematy and Gregory Mouille (both INRA), and Eugenia Maximova (Max Planck Institute of Molecular Plant Physiology) for excellent advice and help with respect to microscopy and to Joachim Selbig and Stefanie Hartmann (both University of Potsdam) for help with phylogenetic analysis. We also acknowledge Erwin Grill (Technical University Munich) for providing the Col-0 cosmid library, CEREON Genomics for supplying information on Col-0/Landsberg erecta polymorphisms, and the European Arabidopsis Stock Centre for seeds of the T-DNA insertion line SALK_065959.
References
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534–552 [DOI] [PubMed] [Google Scholar]
- Altamura MM, Zaghi D, Salvi G, De Lorenzo G, Bellincampi D. (1998) Oligogalacturonides stimulate pericycle cell wall thickening and cell divisions leading to stoma formation in tobacco leaf explants. Planta 204: 429–436 [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. (2006) Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA 103: 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Cookson SJ, Wu S, Scheible WR. (2009) Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J. (1999) If walls could talk. Curr Opin Plant Biol 2: 521–524 [DOI] [PubMed] [Google Scholar]
- Brembu T, Winge P, Bones AM. (2005) The small GTPase AtRAC2/ROP7 is specifically expressed during late stages of xylem differentiation in Arabidopsis. J Exp Bot 56: 2465–2476 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N, McCann M. (2000) The cell wall. Buchanan B, Gruissem W, Jones R, , Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 52–108 [Google Scholar]
- Carpita NC, Tierney M, Campbell M. (2001) Molecular biology of the plant cell wall: searching for the genes that define structure, architecture, and dynamics. Plant Mol Biol 47: 1–5 [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, McCann MC, Roberts K. (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Orbović V, Bellini C, Kronenberger J, Caboche M, Traas J, Höfte H. (1996) Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsis seedlings. Development 122: 683–693 [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125: 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebringerova A, Heinze T. (2000) Naturally occurring xylans: structures, isolation procedures and properties. Macromol Rapid Commun 21: 542–556 [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann S, Vision TJ. (2008) Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol Biol 8: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. (1995) Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Höfte H. (2001) Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-β-glucanase. Planta 212: 348–358 [DOI] [PubMed] [Google Scholar]
- Hovav R, Udall JA, Chaudhary B, Hovav E, Flagel L, Hu G, Wendel JF. (2008) The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet 4: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong R, Morrison WH, Ye ZH. (2003) The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217: 912–921 [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148: 1583–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Takemoto D. (2004) Plant innate immunity: direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol 16: 48–62 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Melzer M, Prokhorenko I, Johansson T, Wingsle G. (2005) Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J Exp Bot 56: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh M. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. (1998) A plant homolog of the neutrophil NADPH oxidase gp91-phox subunit gene encodes a plasma membrane protein with Ca21-binding motifs. Plant Cell 10: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Beers EP, Han KH. (2006) Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol Genet Genomics 276: 517–531 [DOI] [PubMed] [Google Scholar]
- Legendre L, Rueter S, Heinstein PF, Low PS. (1993) Characterization of the oligogalacturonide-induced oxidative burst in cultured soybean (Glycine max) cells. Plant Physiol 102: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertpiriyapong K, Sung Z. (2003) The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Mol Biol 53: 581–595 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH. (2008) A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J 56: 483–492 [DOI] [PubMed] [Google Scholar]
- Mølgaard A, Larsen S. (2004) Crystal packing in two pH-dependent crystal forms of rhamnogalacturonan acetylesterase. Acta Crystallogr Sect D 60: 472–478 [DOI] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al. (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Mutwil M, Obro J, Willats WG, Persson S. (2008) GeneCAT: novel Webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Usadel B, Schütte M, Loraine A, Ebenhöh O, Persson S. (2010) Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. (1998) A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Höfte H. (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha T, Delmer DP. (1995) A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J 7: 453–460 [Google Scholar]
- Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A. (1999) The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 119: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2006) R: a language and environment for statistical computing. http://www.R-project.org
- Reis D, Vian B. (2004) Helicoidal pattern in secondary cell walls and possible role of xylans in their construction. C R Biol 327: 785–790 [DOI] [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H. (2004) The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11: 351–364 [Google Scholar]
- Scheible WR, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR. (2003) An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell 15: 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sene C, McCann MC, Wilson RH, Grinter R. (1994) Fourier-transform raman and Fourier-transform infrared spectroscopy (an investigation of five higher plant cell walls and their components). Plant Physiol 106: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Trainin T, Kalman S, Delmer D. (1992) Cell wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2,6-dichlorobenzonitrile: a comparison between two dicotyledonous plants and a graminaceous monocot. Plant Physiol 100: 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlecka A, Wiklund S, Péronne MA, Micheli F, Lesniewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ. (2008) Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol 146: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Svalheim O, Robertsen B. (1993) Elicitation of hydrogen peroxide production in cucumber hypocotyl segments by oligo-1,4-alpha d-galacturonides and an oligo-beta-glucan preparation from cell walls of Phytophthora megasperma f. sp. glycinea. Physiol Plant 88: 675–681 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Upton C, Buckley JT. (1995) A new family of lipolytic enzymes? Trends Biochem Sci 20: 178–179 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C. (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, et al. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Mandaokar A, Chen J, Last RL, Browse J. (2007) Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J 49: 786–799 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. (2001) Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol 42: 170–178 [DOI] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH. (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2004) Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol 45: 1720–1728 [DOI] [PubMed] [Google Scholar]
- Zhong RQ, Morrison WH, Freshour GD, Hahn MG, Ye ZH. (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong RQ, Pena MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH, Darvill AG, York WS, Ye ZH. (2005) Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykwinska AW, Ralet MCJ, Garnier CD, Thibault JFJ. (2005) Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol 139: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.