Abstract
Rapid Alkalinization Factors (RALFs) are plant peptides that rapidly increase the pH of plant suspension cell culture medium and inhibit root growth. A pollen-specific tomato (Solanum lycopersicum) RALF (SlPRALF) has been identified. The SlPRALF gene encodes a preproprotein that appears to be processed and released from the pollen tube as an active peptide. A synthetic SlPRALF peptide based on the putative active peptide did not affect pollen hydration or viability but inhibited the elongation of normal pollen tubes in an in vitro growth system. Inhibitory effects of SlPRALF were detectable at concentrations as low as 10 nm, and complete inhibition was observed at 1 μm peptide. At least 10-fold higher levels of alkSlPRALF, which lacks disulfide bonds, were required to see similar effects. A greater effect of peptide was observed in low-pH-buffered medium. Inhibition of pollen tube elongation was reversible if peptide was removed within 15 min of exposure. Addition of 100 nm SlPRALF to actively growing pollen tubes inhibited further elongation until tubes were 40 to 60 μm in length, after which pollen tubes became resistant to the peptide. The onset of resistance correlated with the timing of the exit of the male germ unit from the pollen grain into the tube. Thus, exogenous SlPRALF acts as a negative regulator of pollen tube elongation within a specific developmental window.
Peptide signaling regulates a variety of developmental processes and environmental responses in plants (Olsen et al., 2002; Ryan et al., 2002; Boller, 2005; Germain et al., 2006; Matsubayashi and Sakagami, 2006; Farrokhi et al., 2008). For example, the CLAVATA3 peptide regulates meristem size (Fletcher et al., 1999), and the SCR peptide is the pollen self-incompatibility recognition factor in the Brassicaceae (Schopfer et al., 1999; Takayama et al., 2000). The peptide systemin induces the systemic defense response (Ryan and Pearce, 2003), and defensins are small Cys-rich proteins that are involved in the innate immune system of plants (Lay and Anderson, 2005; Okuda et al., 2009).
The Rapid Alkalinization Factor (RALF) family of peptides was discovered as a factor that could rapidly cause alkalinization of suspension culture medium in a search for tobacco (Nicotiana tabacum) systemins (Pearce et al., 2001). Tomato (Solanum lycopersicum) RALF (now called SlRALF, previously LeRALF [Scheer et al., 2005]) is a 49-amino acid active peptide produced from a 115-amino acid propeptide. When Arabidopsis (Arabidopsis thaliana) or tomato seeds were germinated in the presence of 10 μm synthetic RALF, root growth and root hair initiation were inhibited, suggesting a negative growth regulatory function for this peptide. Subsequently, RALF-encoding genes have been identified in a wide variety of plant species, including dicots, monocots, and gymnosperms (Pearce et al., 2001). The 40 RALF or RALF-like (RALFL) genes encoded in Arabidopsis have variable expression patterns, and ovule-specific expression of a subset of RALF genes has been observed in Solanum chacoense (Olsen et al., 2002; Germain et al., 2005; Punwani et al., 2007). A study of NaRALF down-regulation in Nicotiana attenuata has demonstrated that this root-expressed RALF is required for normal root and root hair growth (Wu et al., 2007). Overexpression of either of two Arabidopsis RALF genes, AtRALF1 or AtRALF23, results in a dwarf phenotype (Matos et al., 2008; Srivastava et al., 2009). AtRALF1 was identified in a search for bioactive peptides using Arabidopsis seedlings expressing aequorin-a bioluminescent Ca2+ reporter as a factor that can rapidly (within 40 s) induce a transient internal Ca2+ spike (Haruta et al., 2008), suggesting that RALF peptides may act through a calcium signaling pathway. Recent experiments using exogenous RALF peptide purified from sugarcane (Saccharum officinarum) leaves (SacRALF) demonstrated that the peptide inhibits sugarcane microcalli development and that SacRALF transcripts were found in actively expanding zones of both roots and leaves. These studies also showed that exogenous recombinant AtRALF1 reduced Arabidopsis hypocotyl growth (Mingossi et al., 2010). RALF gene expression in turn appears to be regulated by other plant hormones: PtdRALF2 transcripts are down-regulated by methyl jasmonate in poplar (Populus spp.) cell suspension cultures (Haruta and Constabel, 2003), and AtRALF23 is significantly down-regulated by brassinolide (Srivastava et al., 2009).
Prohormone proteins in animals and yeast are typically processed at dibasic sites by Golgi-localized subtilisin-related proteinases called proprotein convertases, and the processed, active peptides are released into the extracellular matrix (Nakayama, 1997; Seidah and Chrétien, 1999). RALF precursors also possess a conserved dibasic site upstream to the active peptide, suggesting that the processing mechanism may be similar. Recent studies show that both AtRALF23 and AtRALF1 are processed at the dibasic site by Golgi-located plant subtilisin-like Ser proteases and that this processing step is required for the activation of the peptide (Matos et al., 2008; Srivastava et al., 2009).
Pollen tube germination and growth are autoregulated by several pollen-produced peptides. For example, phytosulfokines are responsible for the stimulatory “pollen population effect” for in vitro pollen tube germination (Chen et al., 2000). The small Cys-rich LAT52 protein is required for normal pollen hydration and germination (Muschietti et al., 1994). In both of these cases, the peptides can act in an “autocrine”-like manner to regulate pollen tube germination (Tang et al., 2002; Johnson and Preuss, 2003).
In this article, we describe the discovery of SlPRALF, a pollen-specific RALF peptide from tomato that does not affect pollen viability, hydration, or early germination events but does inhibit the elongation of pollen tubes within a specific developmental window.
RESULTS
Discovery of a Pollen-Expressed RALF Gene
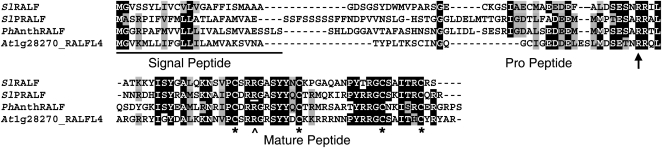
A yeast two-hybrid screen was conducted to identify potential proteins that interact with the recognition domain of the tomato pollen-specific cell wall-localized Leu-rich repeat extensin chimera (LRX) protein (Rubinstein et al., 1995a, 1995b; Stratford et al., 2001; Baumberger et al., 2003). One pollen cDNA identified by this screen (Pol2) encodes a peptide similar to SlRALF, a factor shown to rapidly increase the pH of plant tissue culture medium and to inhibit root growth (Pearce et al., 2001). While additional experiments to confirm and characterize pollen LRX-RALF interactions are in progress, the effects of the pollen-expressed RALF on pollen tube germination and growth became the focus of this study. The Pol2 cDNA sequence was used to query the Sol Genomics Network (SGN) unigene collection (http://www.sgn.cornell.edu/tools/blast/), and a single tomato unigene constructed from S. lycopersicum and Solanum pennellii ESTs was identified (SGN-U324197; SlPRALF in Fig. 1). Like the previously identified vegetative tissue-expressed SlRALF (Pearce et al., 2001), the Pol2 gene encodes a prepropeptide predicted to be targeted to the endomembrane system and then proteolytically processed near a conserved dibasic site that, in Arabidopsis, is required for propeptide processing (and activity) of both AtRALF1 and AtRALF23 (Matos et al., 2008; Srivastava et al., 2009). The predicted active Pol2 peptide includes four conserved Cys residues likely to be involved in disulfide bridges that have been shown to be required for both alkalinization of somatic suspension cell culture medium and root growth inhibition (Pearce et al., 2001). The predicted mature Pol2 peptide also possesses additional well-conserved sequences found in the original SlRALF and RALFL peptides, including an ISY motif near the mature N terminus, a GASYY motif between the first and second conserved Cys residues, and an YXRGCS motif that contains the third conserved Cys residue.
Figure 1.
Amino acid alignment of SlRALF with the pollen-expressed SlPRALF, PhanthRALF, and AtRALFL4. The underlined section of the alignment corresponds to the predicted signal peptide sequences, the dibasic site within the proregion is identified with an arrow, and the conserved Cys residues are indicated with asterisks. Identical residues are boxed in black, and similar residues are shaded gray. The predicted amino acid sequence encoded by the Pol2 cDNA differs from SlPRALF at a single Arg residue in the mature peptide (underlined and indicated by the caret), which in Pol2 is a His. Vegetatively expressed SlRALF corresponds to the translated tomato unigene SGN-U316452, SlPRALF corresponds to the translated pollen-expressed tomato unigene SGN-U324197, PhanthRALF corresponds to the translated P. hybrida anther-expressed unigene SGN-U207478, and AtRALFL4 corresponds to the translated Arabidopsis pollen-expressed At1g28270 gene.
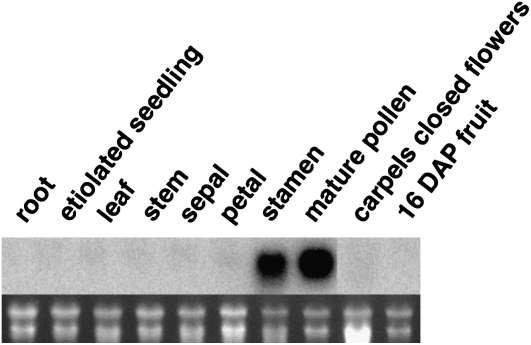
Pol2 Expression Is Pollen Specific
Pollen expression of the Pol2 gene could be inferred, since the Pol2 cDNA was identified using a pollen cDNA library and the corresponding SGN unigene was compiled mainly from S. pennellii pollen cDNA sequences. In order to assess expression in other tissues, an RNA blot with RNA from tomato root, etiolated seedlings, leaf, stem, sepal, petal, stamen, pollen, unpollinated carpels from closed flowers, and 16 d-after-pollination fruit was probed with Pol2 sequences. As seen in Figure 2, a strong signal was only detected in pollen and stamens, which contain pollen. Thus, the Pol2 sequence is most highly expressed in pollen. Therefore, we have named this tomato gene SlPRALF for S. lycopersicum pollen RALF. The Arabidopsis genome contains 40 genes encoding RALFL proteins (Olsen et al., 2002; www.arabidopsis.org), and at least six Arabidopsis RALFL genes are expressed in pollen (Honys and Twell, 2004; Pina et al., 2005; Schmid et al., 2005); At1g28270_RALFL4 is the Arabidopsis pollen-expressed gene most closely related to SlPRALF. The encoded amino acid sequence of this gene as well as a pollen/anther-expressed RALF from Petunia hybrida (PhanthRALF) are shown in the amino acid alignment in Figure 1. No clear pollen-specific motif is evident when pollen-expressed RALFs are compared with vegetative SlRALF, other than an enrichment of positively charged amino acids between the third and fourth Cys residues in the active peptide region.
Figure 2.
RNA gel-blot analysis of SlPRALF expression. The top panel shows the hybridization pattern of the Pol2 probe to RNA from designated tissues, and the bottom panel shows ethidium bromide-stained ribosomal RNA bands. DAP, Days after pollination.
Detection of SlPRALF in Growing Pollen Tubes and Germination Medium
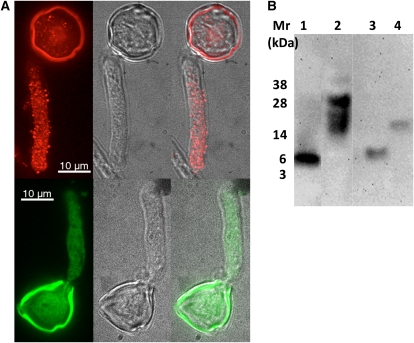
The SlPRALF gene encodes a predicted preproprotein of 129 amino acids with a secretion prediction score of 0.996 using TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000). Immunolocalization studies indicated that a SlPRALF signal is punctate, with an increasing gradient of the protein toward the tip of the tube (Fig. 3A). Protein-blotting experiments showed that the pollen tube-localized protein may correspond to the unprocessed form of SlPRALF, based on the size of immune-reactive bands (≥17 kD; Fig. 3B). The diffuse bands observed may represent either posttranslationally modified versions or oligomers that are insensitive to SDS-reducing agent treatment (Leite et al., 2000). A similar diffuse banding pattern was observed when AtRALF23 was overexpressed in Arabidopsis and resolved on denaturing gels (Srivastava et al., 2009). The processed form of SlPRALF (approximately 7 kD) was not detected (even after overloading the gel with more than 100 μg of protein per lane) either in total pollen tube extracts or in subcellular fractions of pollen extracts, indicating that it is either degraded rapidly or immediately secreted to outside the pollen tube after processing. The processed form of SlPRALF was detected only in the medium, and no protein larger than the processed form was found in the medium, suggesting that only the mature peptide is secreted to outside the pollen tube.
Figure 3.
Detection of SlPRALF in pollen and medium. A, Immunolocalization of SlPRALF in tomato pollen tubes. Tomato pollen was germinated for 1 h, fixed and incubated with polyclonal antibodies raised against recombinant SlPRALF, and detected using TRITC-conjugated secondary antibodies (top) or preimmune serum detected using FITC-conjugated secondary antibodies (bottom). Pollen tubes were imaged both in bright-field and fluorescent modes with a confocal microscope. Fluorescent images of representative pollen tubes are presented on the left, bright-field images in the middle, and an overlay of both on the right. Intense signal in the exine is due to autofluorescence. B, Processed SlPRALF is secreted into the medium. The immunoblot was probed by anti-SlPRALF antibody. Lane 1, 20 pmol of synthetic active SlPRALF peptide; lane 2, 70 μg of total protein extracted from germinated pollen; lane 3, 25 μg of proteins secreted into PGM; lane 4, 25 pmol of recombinant preproSlPRALF expressed in Escherichia coli.
SlPRALF Alkalinization Activity
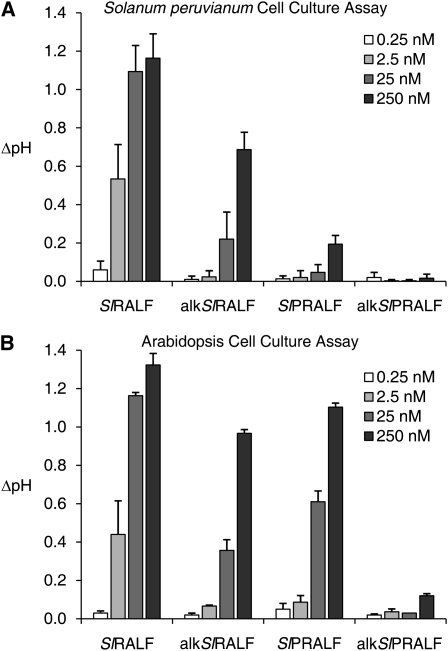
A synthetic form of the predicted mature SlPRALF peptide sequence derived from the SGN unigene U324197 was tested for activity in somatic cell culture assays using Solanum peruvianum and Arabidopsis suspension cultures. Interestingly, S. peruvianum cells, which exhibited a strong alkalinizing response with tomato and tobacco RALFs purified from leaves, responded very weakly to SlPRALF. However, both the SlRALF and the SlPRALF peptides produced a very strong alkalinizing response with Arabidopsis suspension cells, and this assay was utilized for the purification of oxidized SlPRALF (Fig. 4).
Figure 4.
Suspension cell culture alkalinization assay. Change in pH was measured in cell suspension culture 15 min after exposure to unaltered or alkalinized vegetative (SlRALF and alkSlRALF) and pollen (SlPRALF and alkSlPRALF) RALF peptides at 0.25 nm (white bars), 2.5 nm (light gray bars), 25 nm (dark gray bars), and 250 nm (black bars). A, S. peruvianum suspension cell culture assay. B, Arabidopsis suspension cell culture assay.
To test whether a similar alkalinization response could be detected in an in vitro pollen germination system, synthetic SlPRALF was added to a final concentration of 0.1 μm to approximately 3.65 × 105 tomato (cv VF36) pollen grains in 0.5 mL of buffered or unbuffered pollen germination medium (PGM). No significant change in pH was detected (Supplemental Table S1). However, it is important to note that localized changes in pH at the pollen tube plasma membrane would not be detected using this assay.
SlPRALF Does Not Affect Pollen Hydration or Viability
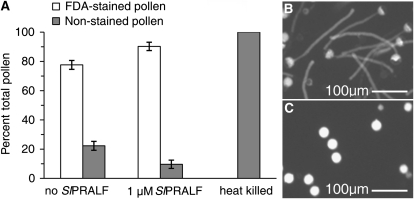
Synthetic SlPRALF was tested for effects on tomato pollen hydration and viability. The peptide did not affect pollen hydration in the range of 0.005 to 1 μm (Table I). Viability of treated pollen was tested using the fluorochromatic response with the stain fluorescein diacetate (Rotman and Papermaster, 1966; Heslop-Harrison and Heslop-Harrison, 1970). As shown in Figure 5, the presence of the SlPRALF peptide at 1 μm, the highest concentration tested, did not significantly affect pollen viability compared with untreated controls (Fig. 5A). The slight increase in viability seen in the presence of SlPRALF is likely due to the lack of elongating pollen tubes, which are vulnerable to breakage (Fig. 5, B and C).
Table I. SlPRALF effects on pollen hydration.
Values are average percentages of NHP from five flowers ± se at each indicated peptide concentration after 1 h in pollen germination medium.
| SlPRALF | NHP |
| μm | % |
| 0.0 | 21.9 ± 1.82 |
| 0.005 | 20.8 ± 3.15 |
| 0.01 | 23.1 ± 4.54 |
| 0.025 | 24.03 ± 2.67 |
| 0.05 | 21.9 ± 4.26 |
| 0.1 | 20.2 ± 4.89 |
| 0.25 | 22.12 ± 4.78 |
| 0.5 | 20.7 ± 3.09 |
| 1.0 | 18.64 ± 0.85 |
Figure 5.
Pollen viability assessed by FDA staining. A, Histogram of pollen fluorescence, with white bars representing positive FDA-stained fluorescent (live) pollen and gray bars representing nonfluorescent (dead) pollen from nontreated (no SlPRALF), treated (1 μm SlPRALF), and heat-killed pollen. FDA staining was done 1 h after the start of pollen to germination, ±sd, n ≥ 100 pollen grains. B, Micrograph of stained untreated pollen. C, Micrograph of stained SlPRALF-treated pollen.
SlPRALF Affects Pollen Tube Elongation
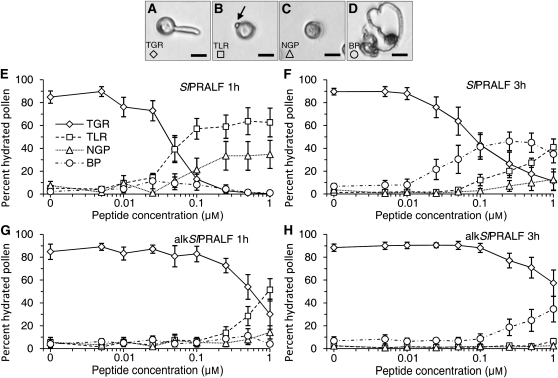
Although SlPRALF had no effect on pollen hydration or viability, this peptide clearly had a dramatic effect on pollen tube elongation (Fig. 5C). To test the effect of the peptide, pollen was placed in PGM with varying concentrations of SlPRALF (Fig. 6). After 1 and 3 h, hydrated pollen was classified as having tubes greater than the radius (TGR; Fig. 6A), tubes less than the radius (TLR; Fig. 6B), no detectable tubes (nongerminated pollen [NGP]; Fig. 6C), or burst pollen (BP; Fig. 6D). In the absence of peptide, approximately 85% of pollen grains had TGR after 1 h. As shown in Figure 6E, at SlPRALF concentrations as low as 0.01 μm, a decrease in the TGR class of pollen was detected, with an almost complete absence at 0.1 μm SlPRALF. Instead, at concentrations higher than 0.05 μm SlPRALF, the predominant class of pollen was TLR, with small incipient pollen tubes (4.58 ± 1.9 μm), which we call “glebula” (“small mound” in Latin; arrow in Fig. 6B). For example, at 0.1 μm SlPRALF, about 57% of the pollen had glebula (TLR). Since early germination events including glebula formation do not appear to be affected by SlPRALF at low concentrations, the primary process affected by SlPRALF seems to be the elongation of pollen tubes.
Figure 6.
Effects of SlPRALF and alkSlPRALF on pollen tube germination and growth. Pollen was exposed to exogenous concentrations of SlPRALF peptide ranging from 0.005 to 1 μm. Peptide effects were classified according to pollen morphology. A, Micrograph of TGR class. B, Micrograph of TLR class. The arrow indicates the glebula. C, Micrograph of NGP class. D, Micrograph of BP class. Bars = 25 μm. E, The average percentage of hydrated pollen in each classification group, ±se, for pollen treated with each concentration of SlPRALF peptide for 1 h. F, The average percentage of hydrated pollen in each classification group, ±se, for pollen treated with SlPRALF peptide for 3 h. G, The average percentage of hydrated pollen in each classification group, ±se, for pollen treated with alkSlPRALF for 1 h. H, The average percentage of hydrated pollen in each classification group, ±se, for pollen treated with alkSlPRALF peptide for 3 h. n ≥ 100 for each treatment.
After prolonged treatment (3 h; Fig. 6F), increases in both the normal pollen (TGR) and in the BP are observed relative to the shorter, 1-h treatment. For example, at 3 h in 0.1 μm SlPRALF, 43.1% of pollen falls into the TGR class and 41.3% into the BP class. This increase in burst or normal pollen with a concomitant decrease in the TLR class suggests that, over time, pollen with arrested tubes can either recover or will tend to burst, possibly due to a buildup of turgor pressure as tube elongation is inhibited.
SlPRALF that has been reduced and alkylated by treatment with iodoacetamide (alkSlPRALF) cannot form disulfide bridges. alkSlPRALF was much less effective in the inhibition of pollen tube elongation (10–20 times higher concentrations are needed to see any effect), as shown in Figure 6, G and H. This result is consistent with the results of Pearce et al. (2001) demonstrating that disulfide bonds are important for SlRALF peptide activity in the cell culture medium alkalinization and root growth inhibition bioassays. At least five times higher concentrations of exogenous vegetatively expressed SlRALF peptide were required to see any affect on pollen tube elongation (Supplemental Fig. S1).
pH Effects on Pollen Germination and Growth
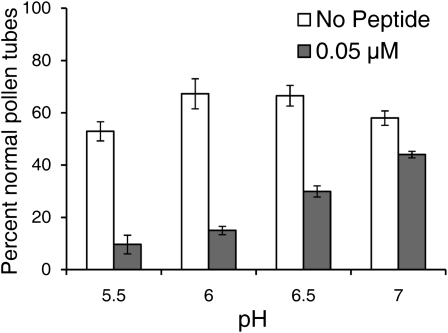
In NaRALF down-regulated plants, a reduced pH of the growth medium partially ameliorated an abnormal root hair growth phenotype (Wu et al., 2007). To test whether the pH of the medium alters SlPRALF effects on pollen tube growth, 0.05 μm peptide was added to PGM at pH 5.5, 6, 6.5, and 7, and the percentage of normal pollen with TGR was calculated (Fig. 7). The highest percentage of normal pollen tubes in the absence of peptide was observed at pH 6 and 6.5, indicating that this range of pH is optimal for pollen germination and tube growth. The greatest effect of the peptide was seen at the lowest pH tested (pH 5.5), and the least effect was seen at the highest pH tested (pH 7). This is not unexpected if exogenous SlPRALF directly or indirectly acts to decrease cytoplasmic pH; a higher external proton concentration would tend to exacerbate an increase of protons inside the pollen tube.
Figure 7.
Effects of peptide on pollen tube growth at different pH environments. Tomato pollen was germinated for 1 h in PGM-MES medium buffered to pH 5.5, 6, 6.5, or 7 containing either no peptide (white bars) or 0.05 μm SlPRALF (gray bars). Average number of normal pollen grains with tubes greater than the radius of the grain is shown as the percentage of normal pollen tubes ± se. n ≥ 100 for each pH.
Reversibility of SlPRALF Effects
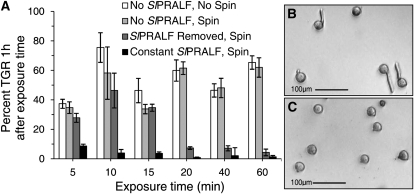
Although seedling root growth is inhibited by SlRALF, seedlings will recover normal root growth when transferred from SlRALF-containing medium to peptide-free medium (Pearce et al., 2001). In order to test whether the inhibitory effects of SlPRALF on pollen tube elongation are reversible, ungerminated pollen was exposed to 0.1 μm SlPRALF for varying time periods and then pelleted and resuspended in peptide-free medium (Fig. 8; Supplemental Fig. S2). Centrifugation had no significant detrimental effect on pollen tube growth. If peptide was removed prior to or at 15 min of exposure, pollen was able to recover and generate normal pollen tubes in peptide-free medium (Fig. 8, A and B). However, pollen exposed to peptide for 20 min or longer was unable to generate normal pollen tubes after peptide was removed (Fig. 8, A and C). Thus, after 20 min of continuous exposure to SlPRALF, pollen tube elongation is irreversibly inhibited.
Figure 8.
Reversibility of SlPRALF inhibition. A, Histogram of percentage of hydrated pollen with TGR ± se 1 h after time-limited treatment with 0.1 μm SlPRALF. Pollen was treated with peptide for designated exposure times, pelleted by centrifugation, and resuspended in medium containing 0.1 μm SlPRALF (constant SlPRALF, spin = black bars) or no peptide (SlPRALF removed, spin = dark gray bars). Samples not treated with peptide were analyzed without centrifugation (no SlPRALF, no spin = white bars) or with centrifugation (no SlPRALF, spin = light gray bars). n ≥ 100. B, Micrograph of pollen in peptide-removed PGM 1 h after 15 min of SlPRALF treatment. C, Micrograph of pollen in peptide-removed PGM 1 h after 20 min of SlPRALF treatment.
SlPRALF Effects on Growth of Elongating Pollen Tubes
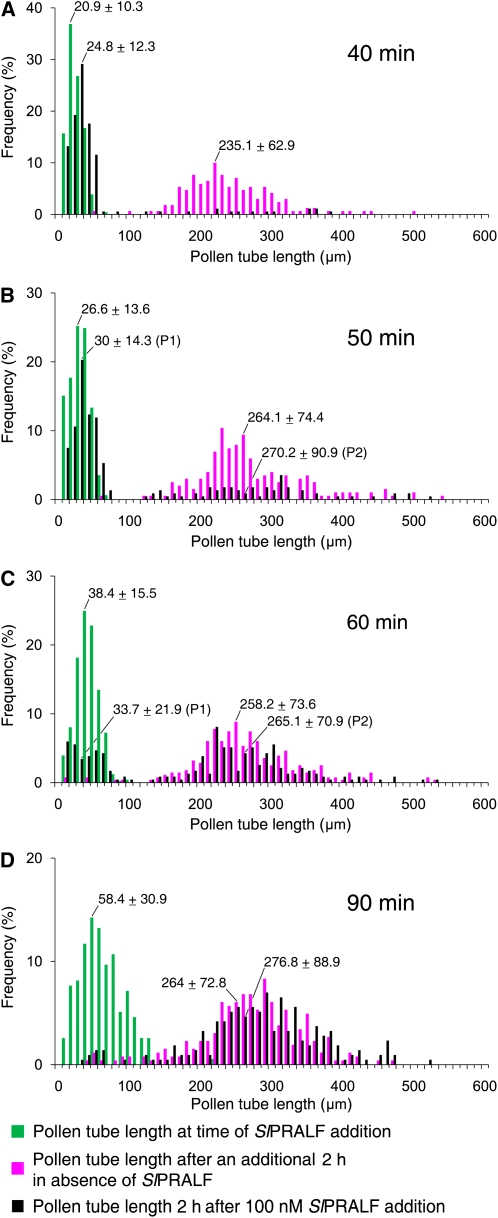
To assess the effect of peptide on actively growing pollen tubes, pollen tubes were allowed to grow prior to the addition of 0.1 μm SlPRALF. As shown in Figure 9A, SlPRALF completely inhibited further pollen tube elongation when added up to 40 min after pollen germination. However, when SlPRALF was added at 50 min, a small population of tubes appeared to be resistant to the peptide (Fig. 9B). When peptide was added at 60 min, two distinct populations of tubes were observed (Fig. 9C, P1 and P2). At this time, about half of the tubes (46.8%) were sensitive to SlPRALF and did not elongate further after peptide addition. However, about half of the tubes (53.2%) were resistant to inhibition by the peptide and grew at the same rate as untreated pollen tubes. The lack of tubes of intermediate lengths suggests that tube growth is either completely inhibited or completely insensitive to inhibition. The bimodal distribution of sensitive and resistant pollen tubes at this time point is most likely due to variation in pollen tube length within the normal distribution of the pollen population, rather than to genetic variation in pollen from the highly inbred cv VF36 (Williams and St. Clair, 1993). When pollen tube lengths average 58 μm at 90 min (Fig. 9D), elongating tubes were no longer sensitive to inhibition by peptide. From these results, we hypothesize that when tubes that have reached a critical threshold length between 38.4 and 58 μm, they become resistant to SlPRALF peptide.
Figure 9.
SlPRALF effects on elongating pollen tubes. Histograms show pollen tube lengths of pollen tubes exposed to 0.1 μm SlPRALF at 40 (A), 50 (B), 60 (C), or 90 (D) min after addition of pollen to PGM. Pollen tube lengths were measured at the time of SlPRALF addition (green bars) and again 2 h later for nontreated (magenta bars) and 0.1 μm SlPRALF-treated (black bars) samples. Pollen tube lengths are grouped into 10-μm bin increments and represented as frequency of pollen tube lengths. The average pollen tube length for each treatment ± se is shown above the corresponding peak. n ≥ 100. The average pollen tube length for populations showing bimodal distributions (B and C) were calculated separately (P1 and P2).
Migration of the Male Germ Unit as a Reference Point for Sensitivity to SlPRALF
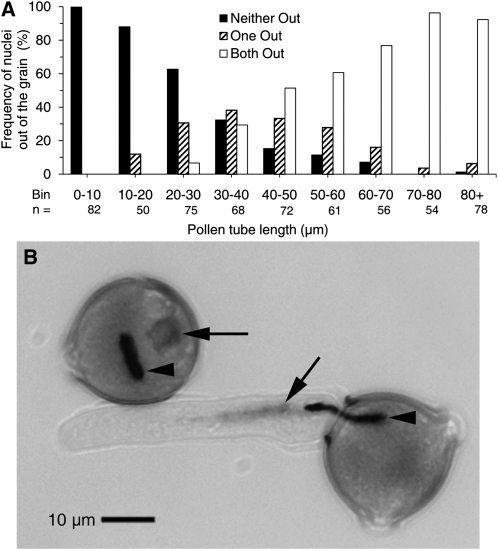
We have demonstrated that pollen tubes are inhibited by exogenous SlPRALF within a defined period; pollen tubes become resistant to SlPRALF once they have reached a length of about 50 μm. Generative cell mitosis and callose plug formation are distinctive cellular events in pollen tube growth, but both of these occur much later than this time frame. The migration of the male germ unit (MGU; comprising the vegetative nucleus and the generative cell) from the pollen grain into the pollen tube is an earlier event that may occur during this period. Migration of the MGU was assessed with regard to tube length by fixing and staining of pollen tubes with 4′,6-diamidino-2-phenylindole (DAPI) after 0.5 to 2 h of growth. As shown in Figure 10, the first migration events are detected when tubes are 10 to 20 μm. The majority of germinated pollen has at least one nucleus/generative cell in the tube by 30 to 40 μm, and the majority have both nuclei in the tube by 50 to 60 μm. MGU migration is essentially complete when tubes are greater than 70 μm in length. Thus, the movement of the MGU into the pollen tube correlates developmentally with the end of the window of pollen tube sensitivity to exogenous SlPRALF. While the mechanism of SlPRALF action is probably not directly related to MGU movement, this event provides a convenient reference point for further investigation into the developmental timing of sensitivity to SlPRALF.
Figure 10.
Migration of the MGU into pollen tubes analyzed by DAPI staining. A, Histogram of MGU components (vegetative nucleus and/or generative cell) in pollen tubes of different lengths. Bars are designated as follows: neither out (black bars) indicates the absence of either the vegetative nucleus or the generative cell in pollen tubes; one out (striped bars) indicates the presence of the leading edge of either (single) component in pollen tubes; and both out (white bars) indicates the presence of the leading edge of both the vegetative nucleus and the generative cell in pollen tubes. Total pollen number (n) analyzed is shown for each pollen tube bin length. B, Micrograph showing the diffusely stained vegetative nucleus (arrows) and intensely stained generative cell (arrowheads) entering tubes in DAPI-stained pollen. The top grain would be scored as neither out, and the bottom grain would be scored as both out.
DISCUSSION
RALFs are recently discovered plant peptides that can increase the pH of suspension cell culture medium more rapidly than systemin (Pearce et al., 2001). However, RALF genes are not induced by wounding, suggesting that RALF peptides do not play a role in defense like the systemin family of peptides. Rather, a role for RALF peptides in development is suggested by the inhibition of root growth by exogenously added peptide (Pearce et al., 2001; Mingossi et al., 2010) and by the tissue-specific regulation of RALF gene expression (Germain et al., 2005; Punwani et al., 2007; Mingossi et al., 2010). Down-regulation of a root-expressed NaRALF gene results in increased root growth and abnormal root hair growth, further supporting this notion (Wu et al., 2007). A growth inhibitory role for RALF is also consistent with the dwarf phenotypes that are observed in transgenic overexpression lines of AtRALF1 and AtRALF23 (Matos et al., 2008; Srivastava et al., 2009). Analogous to its vegetative orthologs, the pollen-specific tomato SlPRALF gene described in this article seems to be a negative regulator of growth, as suggested by the effects of a synthetic SlPRALF peptide on pollen tube growth in vitro.
The finding that SlPRALF inhibits pollen tube growth may at first seem puzzling, since pollen itself produces this peptide. However, pollen tube germination and growth is known to be self-regulating. For example, the peptide phytosulfokine-α is produced by pollen and stimulates germination and pollen tube growth (Chen et al., 2000). The proposed role of the small Cys-rich pollen protein LAT52 is more complex. From antisense studies, LAT52 protein appears to be required for pollen hydration and germination (Muschietti et al., 1994). This positive regulation may be mediated by LAT52 binding to the pollen LePRK2 kinase receptors (Tang et al., 2002). After these early events, the LAT52 protein may be displaced by a stigma factor, possibly LeSTIG1, which binds to the LePRK2 receptor and promotes pollen tube growth (Tang et al., 2004). In this way, LAT52 has an autocrine-like effect on several different aspects of pollen tube growth. SlPRALF-mediated inhibition of growth could have a role in modulating pollen tube growth at its earliest phase in response to an unfavorable environment, for example a nonreceptive immature stigma. Transient inhibition of pollen tube growth by SlPRALF could be abrogated by female factors once pollen tube growth is established within a receptive pistil.
Although the dynamics of endogenous SlPRALF protein production and processing are yet to be determined, our results indicate that the preproprotein appears to enter the endomembrane system, where it may be processed at a dibasic site similarly to other RALFs (Matos et al., 2008; Srivastava et al., 2009) and released into the medium as an active peptide (Fig. 2), indicating that the peptide hormone acts extracellularly. The localization of the SlPRALF preprotein within pollen tubes and the active peptide in the medium is in accordance with the distribution of unprocessed and processed forms of several well-characterized peptide growth regulators (Iida and Shibata, 1994; Shankaran et al., 2008). These results are also consistent with the localization of a Nicotiana benthamiana RALF fused to GFP, which is localized first to the endoplasmic reticulum and later to the cell wall in N. benthamiana leaf cells (Escobar et al., 2003). The extracellular location of SlPRALF also provides the opportunity for this peptide to interact with the cell wall-localized pollen LRX protein.
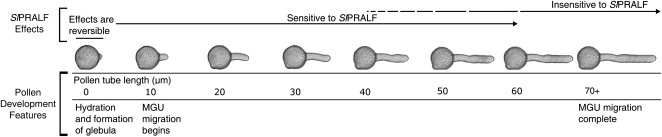
Exogenously added synthetic SlPRALF has a negative effect on pollen tube growth, and the peptide is effective only during a specific window in tube development (Fig. 11). SlPRALF effects are reversible within the first 20 min after pollen has been exposed to the peptide, and actively growing tubes become resistant to exogenously added SlPRALF once they are 40 to 60 μm long. The exit of the MGU from the pollen grain into the pollen tube provides a developmental reference point for the onset of resistance, in that both MGU movement and pollen sensitivity to exogenous SlPRALF characterize an early phase of pollen tube growth. How this window of sensitivity is delimited is not currently understood and will require further experimentation. A recent spatiotemporal study of membrane-related proteins during lily (Lilium longiflorum) pollen tube hydration, early germination, and later tube growth revealed that ion transport-associated proteins were particularly dynamic during early germination (Pertl et al., 2009). The determination of the as yet unknown identity of a putative membrane-localized SlRALF-binding protein could be helpful in elucidating the dynamics of this process (Scheer et al., 2005). In any case, the results presented here are consistent with the idea that SlPRALF can regulate the elongation of pollen tubes within a specific developmental window.
Figure 11.
Summary of exogenous SlPRALF effects on pollen tube growth. Pollen tubes are sensitive to inhibition by SlPRALF peptide after hydration and early germination events, including glebula formation. This inhibition is reversible up to 15 min of exposure to peptide. Growing pollen tubes become resistant to exogenously added SlPRALF once they have reached lengths of 40 to 60 μm, when MGU migration is nearing completion.
A consistent picture of RALF function as a growth inhibitor is emerging from experiments with exogenously added peptide and gene regulation studies. Exogenous RALF peptide inhibits root and root hair growth, sugarcane microcalli development, and Arabidopsis hypocotyl elongation (Pearce et al., 2001; Mingossi et al., 2010). We have shown that exogenous SlPRALF peptide inhibits pollen tube growth. Overexpressed AtRALF1 and AtRALF23 causes stunted vegetative growth (Matos et al., 2008; Srivastava et al., 2009). In contrast, down-regulation of NaRALF promotes root growth and causes abnormal bulbous root hair growth, followed by cell bursting (Wu et al., 2007). Given these observations and our own results, it is possible that SlPRALF plays a role in regulating localized cell expansion in emerging pollen tubes to produce a cylindrical allometrically expanding structure with growth limited to the tip (Lancelle and Hepler, 1992; Geitmann and Dumais, 2009).
The mechanism(s) by which RALFs regulate cell growth will require further study. RALFs affect a number of processes, including proton flux, mitogen-activated protein kinase activity, and cytoplasmic Ca2+ levels (Pearce et al., 2001; Wu et al., 2007; Haruta et al., 2008; Srivastava et al., 2009). A dramatic increase in extracellular pH was the basis for the initial discovery of RALF peptides. Our observations of an increased peptide effect with increased extracellular proton concentration (Fig. 7) are consistent with observations in roots of transgenic down-regulated NaRALF plants (Wu et al., 2007), wherein a decreased pH of the medium partially rescued an abnormal growth phenotype. If proton flux regulation is the primary action of RALFs, these peptides may function to modulate the rigidity of the extracellular landscape of the cells by influencing the activity of pH-sensitive cell wall modification proteins, including pectin methylesterases, exo-α-glucanases, and/or expansins (Kotake et al., 2000; Cosgrove et al., 2002; Holdaway-Clarke and Hepler, 2003; Bosch and Hepler, 2005; Jiang et al., 2005; Sampedro and Cosgrove, 2005; Choi et al., 2006). Alternatively, RALFs could exert their effects intracellularly by modifying the pH of the cytoplasm. In pollen tubes, for example, an internal alkaline band that may be required for normal growth has been observed behind the growing tip (Feijo et al., 1999). It is possible that SlPRALF only acts in a localized fashion in vivo, perhaps increasing pollen tube elongation rates (e.g. by fine-tuning the localization of necessary proton fluxes). However, changes in proton fluxes may be downstream from initial events triggered by RALFs, as suggested by the observation that both down-regulation (Wu et al., 2007) and overexpression (Srivastava et al., 2009) of root-expressed RALFs result in transgenic plants being unable to acidify growth medium. These observations make a simple direct proton-pump activation model for RALFs improbable. Activation of mitogen-activated protein kinases occurs just as rapidly (within 5 min) as alkalinization of cell medium (Pearce et al., 2001). RALFs can cause even more rapid changes in cytoplasmic Ca2+ concentration (Haruta et al., 2008). AtRALF1 was identified as the predominant peptide factor in seedling lysates that could elicit cytoplasmic Ca2+ elevation in Arabidopsis seedlings and roots within 40 s (at nanomolar concentrations equivalent to those used in these studies), suggesting that a Ca2+ signaling cascade may be the primary RALF response. It is well known that highly regulated ion flow is required for normal tube growth (Feijo et al., 1995, 1999; Messerli and Robinson, 1998; Messerli et al., 1999; Robinson and Messerli, 2002; Holdaway-Clarke and Hepler, 2003; Bosch and Hepler, 2005; Lovy-Wheeler et al., 2006). In particular, a steep Ca2+ gradient at the pollen tube tip has been shown to be a critical regulator of pollen tube growth (Pierson et al., 1996; Hepler, 1997; Holdaway-Clarke et al., 2003). Given the well-characterized physiology of growing pollen tubes and their relatively simple cellular structure, pollen may provide the ideal system for probing the molecular mechanisms of RALF action. In any case, RALF peptides are likely to be key regulators of plant growth and development and are richly deserving of intensive investigation.
MATERIALS AND METHODS
Plants
Tomato (Solanum lycopersicum ‘VF36’) plants were grown in a greenhouse in Pro Mix BX soil (Premier Horticulture) and supplemented weekly with 10:20:10 nitrogen:phosphorus:potassium fertilizer. Plants were grown throughout the year, and natural photoperiod was supplemented to a 12-h-light (24°C)/12-h-dark (21°C) photoperiod. Mature flower-producing plants were used for pollen collection.
Pollen Collection
Tomato pollen was collected from stage 15 flowers (Brukhin et al., 2003) using a vibrating hand-held tooth polisher (Dental Concepts) into tubes with PGM. PGM solutions contained 24% (w/v) polyethylene glycol (PEG) 4000, 0.01% (w/v) boric acid, 2% (w/v) Suc, 20 mm MES buffer, pH 6.0, 3 mm Ca(NO3)2·4H2O, 0.02% (w/v) MgSO4·7H2O, and 1 mm KNO3. To prevent pollen from clumping in PGM, the collection tubes were vortexed for 15 s using the tooth polisher. Aliquots of the pollen/PGM mixture were then immediately further diluted for each experiment as described below.
Yeast Two-Hybrid Screen
A yeast two-hybrid library constructed using mature tomato cv VF36 pollen cDNA cloned into pAD-GAL4-2.1 (Stratagene) was a generous gift from Dr. Sheila McCormick (Tang et al., 2002). Sequences encoding the recognition domain (amino acids 22–392, from the mature N terminus to the end of the Cys-rich region adjacent to the extensin domain) of the tomato pollen LRX gene (Stratford et al., 2001) were cloned in-frame with the DNA-binding domain of pBD-GAL4 Cam (Stratagene) and used as bait. A modified lithium acetate method (Gietz et al., 1992) was used to transform PJ69-4A yeast cells (James et al., 1996) with bait plasmid, and positive transformants were transformed with DNA from the pAD-GAL4 pollen library. Selection was on synthetic complete medium lacking His, Leu, and Trp and containing 3 mm 3-amino-1,2,4-triazole.
Sequence Alignment
Sequences were aligned using the MUSCLE algorithm (Edgar, 2004) within Jalview (Clamp et al., 2004). The alignments were then edited by eye to produce the final alignment and colored using Boxshade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html).
RNA Gel-Blot Analysis
Tissue was harvested from tomato plants, frozen in liquid nitrogen, and stored at −80°C until processed. Tissue was either powdered using a mortar and pestle in liquid nitrogen and added to Trizol (Invitrogen) or Trizol was added to the frozen tissue, which was then disrupted using a micropestle attached to an electric motor. All subsequent steps followed the manufacturer's protocol. Five micrograms of total RNA from each tissue was electrophoresed on a 1.2% (w/v) agarose gel containing 2.2 m formaldehyde, transferred to a nylon membrane, and probed for the presence of SlPRALF RNA using standard protocols (Sambrook et al., 1989). The probe, prepared using the RediPrime II kit (Amersham Biosciences) according to the manufacturer's protocol, consisted of the Pol2 cDNA clone (encoding 48 of the 50 amino acid residues in the putative active peptide). The washed blot was exposed to a phosphorimaging screen and read using a Storm model 840 (Molecular Dynamics).
Antibodies
Polyclonal antibodies were raised in a rabbit using a recombinant fusion protein composed of mature SlPRALF sequences fused to glutathione S-transferase (GST) as an antigen at La Trobe University. IgG fractions from immune and preimmune sera were purified using protein A-Sepharose. Anti-SlPRALF antibodies were further purified from anti-GST antibodies before use using recombinant GST protein bound to glutathione-Sepharose.
Immunolocalization
Tomato pollen was germinated in PGM for 1 h and pelleted at 3,000 rpm for 1 min. Pollen tubes were fixed in HistoChoice (Amresco) for 20 min and washed three times in wash buffer (50 mm Tris-0.15 m NaCl [pH 7.5] containing 0.05% Tween 20, 0.02% sodium azide, and 1% bovine serum albumin). The sample was divided into two aliquots and incubated with either immune serum or preimmune serum diluted 1:500 in wash buffer for 2 to 3 h. Pollen tubes were washed four times in wash buffer and incubated in goat anti-rabbit antibodies conjugated with either tetramethyl rhodamine isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC; Sigma-Aldrich) at 1:1,000 dilution in wash buffer. Samples were washed three times in wash buffer and imaged using an Olympus inverted microscope (model IX18) equipped with a CSU22 spinning-disc confocal system. The samples were excited with 473-nm (for FITC) or 561-nm (for TRITC) lasers, and fluorescent and differential interference contrast images were collected using Slide Book version 5 software.
PGM and Pollen Protein Analysis
Since PEG interfered with the analysis of proteins in the medium, pollen was germinated for 1 h in a PEG-free medium. In the modified PGM, PEG was substituted with 12% Suc and 2% raffinose. Pollen proteins were extracted by maceration and heated to 70°C for 10 min in SDS sample buffer containing sample reducing agent as recommended by the manufacturer (Invitrogen). Proteins of all sizes were efficiently retrieved from PGM using a chlorophenol extraction method (Ohyama et al., 2008). Protein was quantified using a Bradford microassay (Bio-Rad). Proteins were separated in a Bis/Tricine gel system (4%–12% acrylamide gradient; Invitrogen) and blotted onto Immobilin-Psq membranes (Millipore). Blots were blocked in 2% gelatin and probed with SlPRALF antiserum (1:5,000 dilution). Signals were detected by chemiluminescence using horseradish peroxidase-conjugated goat anti-rabbit secondary antiserum.
SlPRALF and alkSlPRALF Synthesis
The synthetic peptide sequence was based upon the Solanum pennellii unigene SGN-U324197 from the SGN Web site (http://www.sgn.cornell.edu/). SlPRALF was synthesized by 9-fluorenylmethyl chloroformate solid-phase synthesis with p-methyl benzyhydrylamine resin according to the manufacturer's protocol (model 431A; Applied Biosystems). The synthesized peptide was oxidized as described previously (Pearce et al., 2001). A mixture of oxidized products was obtained, and the active form was identified by fractionation on strong cation-exchange HPLC and utilization of the alkalinization assay (Pearce et al., 2001). Strong cation-exchange HPLC was performed on a poly-SULPHOETHYL aspartamide column (5 μm, 4.6 × 200 mm; Nest Group). The column was equilibrated in 5 mm potassium chloride, pH 3, in 25% (v/v) acetonitrile, and the oxidized SlPRALF (20 mg) was loaded onto the column. After 2 min, a 90-min gradient was applied to 100% (v/v) elution buffer, consisting of 5 mm potassium phosphate and 1 m potassium chloride, pH 3, in 25% (v/v) acetonitrile. A flow rate of 1 mL min−1 was employed, and 1-min fractions were collected and assayed in the suspension cell alkalinization assay as described below. The active fraction (72–74 min) was desalted by C18 HPLC and lyophilized.
Active synthetic SlPRALF was reduced and alkylated using dithiothreitol and iodoacetamide as described previously (Pearce et al., 2001). The resulting alkSlPRALF peptide displayed no activity in the Arabidopsis (Arabidopsis thaliana) suspension cell alkalinization assay.
Oxidized and alkylated SlPRALF were analyzed for correctness and purity by matrix-assisted laser desorption-ionization mass spectrometry using a PerSeptive Biosystems Voyager time of flight mass spectrometer with α-cyano-4-hydroxycinnamic acid as the matrix.
The disulfide linkages of the purified, oxidized SlPRALF were determined by fragmenting the peptide by cyanogen bromide treatment to cleave the peptide at Met-9 and Met-29 and analyzing the products by matrix-assisted laser desorption-ionization mass spectrometry. Major mass peaks found after cyanogen bromide treatment included 3,404 D (amino acids 1–29), 2,274 D (amino acids 10–29), and 2,405 D (amino acids 30–48). These masses indicate that the disulfide bonds of the active SlPRALF were the same as those of the oxidized RALFs from tobacco (Nicotiana tabacum) and tomato: cys1-cys2 and cys3-cys4 (Pearce et al., 2001).
Suspension Cell Alkalinization Assay
Solanum peruvianum and Arabidopsis suspension cells were maintained in Murashige and Skoog medium as described previously (Pearce et al., 2001; Huffaker et al., 2006). Suspension cells were transferred on a weekly basis (2.5 mL per 40 mL of medium) and utilized 3 to 5 d after transfer. One-milliliter aliquots of Arabidopsis suspension cells were transferred onto 24-well culture cluster plates and allowed to equilibrate for 1 h on an orbital shaker at 160 rpm. HPLC fractions of 5-μL aliquots were added, and after 15 min, the pH of the medium was recorded.
Pollen Viability Staining
Pollen exposed to SlPRALF peptide was assessed for viability using fluorescein diacetate (FDA). Pollen was collected as described above and immediately diluted 1:10 into PGM plus or minus 1 μm SlPRALF, and a negative “heat-killed” control aliquot was exposed to 50°C for 10 min. Aliquots were transferred into individual wells of a flat-bottom 96-well microtiter plate and allowed to germinate and grow for 1 h. At 1 h, FDA was added to a final concentration of 0.2 mg mL−1. After a 10-min incubation with FDA at room temperature, images were captured using a Nikon DIAPHOT-TMD inverted microscope, a Photometrics Cool Snap CF camera, and Image Pro Software at 100× magnification with UV excitation and a long-pass GFP emission filter (FDA has a broad range emission, with a peak at 518 nm). A minimum of 100 pollen grains were counted for each treatment.
Assay for Effects of SlPRALF Peptides on Pollen Hydration and Germination
Pollen from five flowers was collected separately into tubes containing PGM, vortexed, and further diluted 1:10 into individual treatment reaction tubes containing peptide at final concentrations of 0, 0.005, 0.01, 0.025, 0.05, 0.1, 0.25, 0.5, and 1 μm. Each treatment tube was vortexed for 10 s after addition of pollen. Aliquots of 100 μL from each reaction were transferred into wells of a flat-bottomed 96-well microtiter plate, which was then enclosed in a humidified container. Pollen in wells was examined using an inverted microscope as described previously. Bright-field images encompassing at least 100 pollen grains were photographed at 100× magnification for each treatment reaction after 1 and 3 h.
Pollen was classified based upon specific morphological characteristics. Nonhydrated pollen grains (NHP) were classified by their oblong morphology, which resembles a deflated football, as opposed to round hydrated pollen. Quantification of NHP was scored as the percentage of the total pollen population for each peptide concentration after 1 h. se was calculated using data with pollen from five flowers for each concentration of peptide after 1 h of germination. Similar results were obtained with SlPRALF treatment for 3 h and for alkSlPRALF treatment for 1 and 3 h (data not shown).
After 1 and 3 h of peptide treatment, hydrated pollen grains were categorized into four classifications. The first class consisted of hydrated pollen with TGR. The average hydrated pollen radius is 15 μm. The second class is pollen with TLR. The third class consists of grains with no apparent tube or protrusion, designated as NGP (but hydrated) grains. The fourth class was BP grains, which exhibit extruded cytoplasm from a grain or tube. Each class of hydrated pollen grain was scored and represented as a percentage of total hydrated pollen ± se for each peptide concentration.
pH Effects on Pollen Germination and Growth
Pollen was collected separately from three flowers into nonbuffered PGM (pH 6.07) and diluted 1:10 into PGM with 20 mm MES buffered to pH 5.5, 6, 6.5, or 7. Each treatment tube was vortexed for 10 s after addition of pollen. After 1 h, samples encompassing at least 100 pollen grains were photographed at 100× magnification for each pH treatment and categorized into four morphological classifications as described previously. The TGR class of hydrated pollen grain was scored and represented as a percentage of total hydrated pollen ± se for each pH treatment.
Reversibility Tests
Pollen was collected separately from three flowers and further diluted 1:10 in PGM. One sample was incubated at room temperature in plain PGM (“no-SlPRALF” control), while the other samples were incubated in PGM plus 0.1 μm SlPRALF. Exposure times were 5, 10, 15, 20, 40, and 60 min. After the designated exposure time, tubes were spun at 13,000 rpm for 30 s and an aspirator was used to carefully remove the supernatant from the pollen pellet. Pollen in the no-SlPRALF control and the “SlPRALF-removed” SlPRALF-treated pellets were resuspended in plain PGM. The “constant-SlPRALF” pellet was resuspended in PGM containing 0.1 μm SlPRALF. Each treatment aliquot was gently vortexed for 10 s at a low setting to resuspend the pollen pellet. Images were collected as described previously 1 h after resuspension. The average percentages of TGR ± se, TLR ± se, NHP ± se, and BP ± se were calculated for three replicates of each treatment at each exposure time. To confirm that centrifugation does not alter pollen tube growth, additional noncentrifuged pollen samples with either 0 or 0.1 μm SlPRALF were compared with centrifuged samples. At least 100 hydrated pollen grains were analyzed for each treatment time.
Assay for Effects of SlPRALF on Pollen Tube Elongation
Pollen from three flowers was collected separately in PGM, further diluted 1:10 into PGM, and allowed to germinate and grow for 40, 50, 60, or 90, min. Two 50-μL aliquots were transferred into separate wells for each time treatment interval. One aliquot received an equal volume of PGM as an untreated control, and the other received an equal volume of 0.2 μm SlPRALF in PGM for a final concentration of 0.1 μm SlPRALF. Images were collected as described previously. Pollen tubes were imaged after 0.5, 1, and 2 h from the time of peptide addition. For simplicity, only the data for 2 h after peptide addition are shown. Pollen tube lengths were measured using ImageJ 1.33u (http://rsb.info.nih.gov/ij/) immediately before peptide addition and 2 h after peptide addition for both untreated and 0.1 μm SlPRALF treatments. Tube lengths for each time treatment were grouped in bins of 10 μm as frequencies of lengths. The mean pollen tube length ± se was calculated for the bell curve of each treatment. Calculation of the mean for data demonstrating a bimodal distribution was determined by manually defining a cutoff value of the two peaks. The peak at shorter pollen tube lengths (P1) was then calculated separately from the longer pollen tube length peak (P2). At least 100 pollen grains were analyzed for each treatment.
Pollen DNA Staining
Pollen was collected separately from three flowers and then diluted 1:5 into PGM. Pollen was germinated for 1 to 3 h. Pollen was then fixed by carefully removing the PGM from the pollen and replacing it with a fixative solution of ethanol:glacial acetic acid (3:1, v/v).
To stain, fixative was removed from the pollen, gently mixed with 10 μL of 0.4 mg mL−1 DAPI in buffer (0.1 m NaH2PO4, pH 7, 1 mm EDTA, and 0.1% [v/v] Triton X-100), and stained for 15 min. Stained pollen was imaged using oil immersion at 400× magnification with UV excitation and a DAPI emission filter (461 nm). Tube length and the distance of the leading edge of the vegetative nucleus (VN) or generative cell (GC) to the edge of the pollen grain were measured as described previously. Tube lengths were grouped in bins of 10 μm. Any tube length greater than 80 μm was placed in one bin, 80+. For each bin length, the frequency of nuclear migration was scored. Tubes that did not show any migration of the leading edge of the VN or GC into the pollen tube were scored as “neither out,” and those that showed at least one, or both, leading edge of the VN or GC migrating into the pollen tube were scored as “one out” or “both out,” respectively.
Sequence data from this article can be found in the SGN (www.sgn.cornell.edu) data library under accession numbers SGN_U316452 (SlRALF), SGN_U324197 (SlPRALF), and SGN_U207478 (PhanthRALF) and in the GenBank/EMBL data library under the Arabidopsis Genome Initiative accession number At1g28270 (AtRALFL4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Vegetative SlRALF has a reduced effect on pollen.
Supplemental Figure S2. SlPRALF inhibition reversibility for abnormal pollen morphology classes.
Supplemental Table S1. Effect of SlPRALF on media pH.
Supplementary Material
Acknowledgments
We thank Ruth Arthur-Asmah, Sandra Duwaik, Tara Schnieder, and Elizabeth Piesman for assistance in data analysis and Irene Day for critical reading of the manuscript.
References
- Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, Simmons MP, Bedinger P, Goff SA, Ringli C, et al. (2003) Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice: a conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. (2005) Peptide signalling in plant development and self/non-self perception. Curr Opin Cell Biol 17: 116–122 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukhin V, Gonzalez HN, Chevalier C, Mouras A. (2003) Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex Plant Reprod 15: 311–320 [Google Scholar]
- Chen YF, Matsubayashi Y, Sakagami Y. (2000) Peptide growth factor phytosulfokine-alpha contributes to the pollen population effect. Planta 211: 752–755 [DOI] [PubMed] [Google Scholar]
- Choi D, Cho HT, Lee Y. (2006) Expansins: expanding importance in plant growth and development. Physiol Plant 126: 511–518 [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. (2004) The Jalview Java Alignment Editor. Bioinformatics 12: 426–427 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. (2002) The growing world of expansins. Plant Cell Physiol 43: 1436–1444 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K. (2003) High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15: 1507–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhi N, Whitelegge JP, Brusslan JA. (2008) Plant peptides and peptidomics. Plant Biotechnol J 6: 105–134 [DOI] [PubMed] [Google Scholar]
- Feijo JA, Malho R, Obermeyer G. (1995) Ion dynamics and its possible role during in vitro pollen germination and tube growth. Protoplasma 187: 155–167 [Google Scholar]
- Feijo JA, Sainhas J, Hackett GR, Kunkel JG, Hepler PK. (1999) Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J Cell Biol 144: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Geitmann A, Dumais J. (2009) Not-so-tip-growth. Plant Signal Behav 4: 136–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain H, Chevalier E, Caron S, Matton DP. (2005) Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 220: 447–454 [DOI] [PubMed] [Google Scholar]
- Germain H, Chevalier E, Matton DP. (2006) Plant bioactive peptides: an expanding class of signaling molecules. Can J Bot 84: 1–19 [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Constabel CP. (2003) Rapid alkalinization factors in poplar cell cultures: peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol 131: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Monshausen G, Gilroy S, Sussman MR. (2008) A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47: 6311–6321 [DOI] [PubMed] [Google Scholar]
- Hepler PK. (1997) Tip growth in pollen tubes: calcium leads the way. Trends Plant Sci 2: 79–80 [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y. (1970) Evaluation of pollen viability by enzymatically induced fluorescence: intracellular hydrolysis of fluorescein diacetate. Stain Technol 45: 115–120 [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Hepler PK. (2003) Control of pollen tube growth: role of ion gradients and fluxes. New Phytol 159: 539–563 [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Weddle NM, Kim S, Robi A, Parris C, Kunkel JG, Hepler PK. (2003) Effect of extracellular calcium, pH and borate on growth oscillations in Lilium formosanum pollen tubes. J Exp Bot 54: 65–72 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H, Shibata Y. (1994) Phasic secretion of newly synthesized atrial natriuretic factor from unstimulated atrial myocytes in culture. Circ Res 74: 659–668 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. (2003) On your mark, get set, GROW! LePRK2-LAT52 interactions regulate pollen tube growth. Trends Plant Sci 8: 97–99 [DOI] [PubMed] [Google Scholar]
- Kotake T, Li YQ, Takahashi M, Sakurai N. (2000) Characterization and function of wall-bound exo-β-glucanases of Lilium longiflorum pollen tubes. Sex Plant Reprod 13: 1–9 [Google Scholar]
- Lancelle SA, Hepler PK. (1992) Ultrastructure of freeze-substituted pollen tubes of Lilium longiflorum. Protoplasma 167: 215–230 [Google Scholar]
- Lay FT, Anderson MA. (2005) Defensins: components of the innate immune system in plants. Curr Protein Pept Sci 6: 85–101 [DOI] [PubMed] [Google Scholar]
- Leite A, Kemper EL, da Silva MJ, Luchessi AD, Siloto RMP, Bonaccorsi ED, El-Dorry HF, Arruda P. (2000) Expression of correctly processed human growth hormone in seeds of transgenic tobacco plants. Mol Breed 6: 47–53 [Google Scholar]
- Lovy-Wheeler A, Kunkel JG, Allwood EG, Hussey PJ, Hepler PK. (2006) Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell 18: 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. (2008) A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett 582: 3343–3347 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. (2006) Peptide hormones in plants. Annu Rev Plant Biol 57: 649–674 [DOI] [PubMed] [Google Scholar]
- Messerli MA, Danuser G, Robinson KR. (1999) Pulsatile influxes of H+, K+ and Ca2+ lag growth pulses of Lilium longiflorum pollen tubes. J Cell Sci 112: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Messerli MA, Robinson KR. (1998) Cytoplasmic acidification and current influx follow growth pulses of Lilium longiflorum pollen tubes. Plant J 16: 87–93 [Google Scholar]
- Mingossi FB, Matos JL, Rizzato AP, Medeiros AH, Falco MC, Silva-Filho MC, Moura DS. (2010) SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S. (1994) LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J 6: 321–338 [DOI] [PubMed] [Google Scholar]
- Nakayama K. (1997) Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J 327: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Mundy J, Skriver K. (2002) Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol 2: 441–451 [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA., Jr (2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertl H, Schulze WX, Obermeyer G. (2009) The pollen organelle membrane proteome reveals highly spatial-temporal dynamics during germination and tube growth of lily pollen. J Proteome Res 8: 5142–5152 [DOI] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, van Aken J, Hackett G, Hepler PK. (1996) Tip-localized calcium entry fluctuates during pollen tube growth. Dev Biol 174: 160–173 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD. (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Rabiger DS, Drews GN. (2007) MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19: 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KR, Messerli MA. (2002) Pulsating ion fluxes and growth at the pollen tube tip. Sci STKE 2002: PE51. [DOI] [PubMed] [Google Scholar]
- Rotman B, Papermaster BW. (1966) Membrane properties of living mammalian cells as studied by enzymatic hydrolysis of fluorogenic esters. Biochemistry 55: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Broadwater AH, Lowrey KB, Bedinger PA. (1995a) Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci USA 92: 3086–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Marquez J, Suarez-Cervera M, Bedinger PA. (1995b) Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell 7: 2211–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA, Pearce G. (2003) Systemins: a functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA (Suppl 2) 100: 14577–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA, Pearce G, Scheer J, Moura DS. (2002) Polypeptide hormones. Plant Cell (Suppl) 14: S251–S264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sampedro J, Cosgrove DJ. (2005) The expansin superfamily. Genome Biol 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Pearce G, Ryan CA. (2005) LeRALF, a plant peptide that regulates root growth and development, specifically binds to 25 and 120 kDa cell surface membrane proteins of Lycopersicon peruvianum. Planta 221: 667–674 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. (1999) The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chrétien M. (1999) Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res 848: 45–62 [DOI] [PubMed] [Google Scholar]
- Shankaran SS, Capell A, Hruscha AT, Fellerer K, Neumann M, Schmid B, Haass C. (2008) Missense mutations in the progranulin gene linked to frontotemporal lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J Biol Chem 283: 1744–1753 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Stratford S, Barnes W, Hohorst D, Sagert J, Golubiewski A, Cotter R, McCormick S, Showalter A, Bedinger P. (2001) A leucine-rich repeat region is conserved in pollen extensin-like (Pex) protein in monocots and dicots. Plant Mol Biol 46: 43–56 [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A. (2000) The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA 97: 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S. (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14: 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S. (2004) LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J 39: 343–353 [DOI] [PubMed] [Google Scholar]
- Williams CE, St Clair DA. (1993) Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random amplified polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome 36: 619–630 [DOI] [PubMed] [Google Scholar]
- Wu J, Kurten EL, Monshausen G, Hummel GM, Gilroy S, Baldwin IT. (2007) NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J 52: 877–890 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.