The cell walls of plants are the most abundant source of organic carbon on the planet. This photosynthetically fixed carbon is recycled by microbial enzymes that convert cell wall polysaccharides to monosaccharides and oligosaccharides, a process that is of biological and industrial importance (Sticklen, 2008; Himmel and Bayer, 2009). Plant cell walls are recalcitrant to biological depolymerization, as the extensive interactions between polysaccharides, and between polysaccharides and lignin, restrict access to the battery of microbial glycoside hydrolases, pectate lyases, and esterases that break down these composite structures (for review, see Mohnen, 2008). Since the early 1990s, there has been an explosion of structural information on both the catalytic and noncatalytic components of these enzymes. This review will provide an overview/update of the structure-function relationships of the enzymes that catalyze plant cell wall deconstruction.
THE PLANT CELL WALL
Plant cell walls are composed predominantly of the polysaccharides cellulose, hemicellulose, and pectin, although secondary walls are often rigidified by the impregnation of lignin, a heterogenous aromatic polymer. The structure of the plant cell has been extensively reviewed previously and will be described briefly here (for an overview of plant cell wall structure, see Harris and Stone, 2008; Mohnen, 2008; Mohnen et al., 2008).
Cellulose is a β-1,4-linked Glc molecule that is substantially crystalline. All hemicellulosic polysaccharides contain a β-linked sugar backbone. In xylans, mannans, and xyloglucans, the backbone sugars are β-1,4-d-Xyl, β-1,4-d-Man, and β-1,4-d-Glc, respectively, while in glucomannan, the backbone consists of randomly dispersed β-1,4-Glc and β-1,4-Man sugars. The backbones of hemicellulosic polysaccharides are decorated with a variety of sugars and acetyl groups, explaining why these polymers are not crystalline. There are three major forms of pectin: homogalacturonan, rhamnogalacturonan I, and rhamnogalacturonan II (for review, see Mohnen, 2008). Homogalacturonan consists of a polygalacturonic acid backbone (Mohnen, 2008). Rhamnogalacturonan I displays a backbone composed of an alternating disaccharide, [(α-1,4)-d-GalA→(α-1,2)-l-Rha]n, that contains extensive decorations at the O4 of the Rha residues (Mohnen, 2008). Rhamnogalacturonan II is the most structurally complex of the three pectic polysaccharides, consisting of 13 different sugars and over 20 different linkages (for an extensive review, see O'Neill et al., 2004).
CAZY
Enzymes that modify complex carbohydrates, together with their accessory noncatalytic carbohydrate-binding modules (CBMs), have been grouped into sequence-based families on the continuously updated Carbohydrate-Active EnZymes (CAZy) database (Cantarel et al., 2009; http://www.cazy.org/). Members of the same enzyme family display a common fold, while the catalytic apparatus and mechanism are similarly conserved. Currently, 44 of the 115 glycoside hydrolase families (GHs) contain enzymes that contribute to plant cell wall deconstruction. Crystal structures of relevant enzymes in 41 of these 44 GHs have been reported. With respect to polysaccharide lyase families (PLs) and carbohydrate esterase families (CEs), six out of 21 PLs and 11 out of 16 CEs contain enzymes that play a role in plant cell wall metabolism. Of the 59 CBM families, around half of these modules bind to components of the plant cell wall, and structural information is available for all but three of these families.
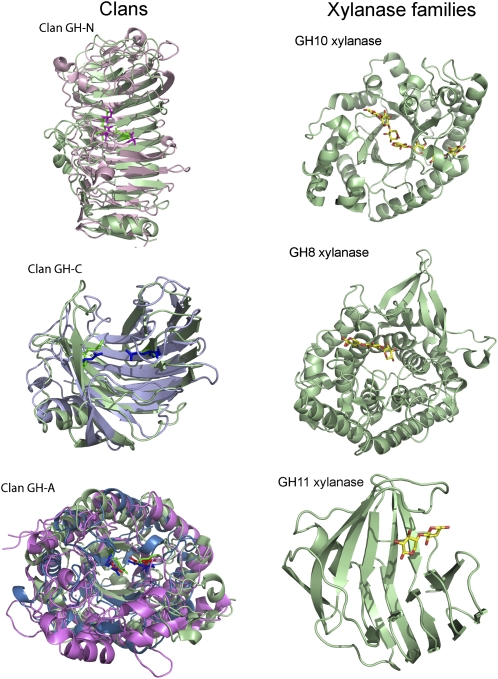
While the structures of CEs, PLs, and CBMs are dominated by the α/β-hydrolase (Correia et al., 2008), parallel β-helix (Pickersgill et al., 1994), and jelly roll (or β-sandwich; Czjzek et al., 2001) folds, respectively, there are a large number of different folds within the GHs, which are discussed below. Indeed, the same criteria used to include enzymes in the same GH have now been used to cluster a proportion of the GHs into 14 different clans (Cantarel et al., 2009). The structural biology of plant cell wall-degrading systems provides elegant examples of both convergent and divergent evolution (Fig. 1).
Figure 1.
Structural convergence and divergence in plant cell wall hydrolases. In the examples shown, a GH28 polygalacturonase (green; Protein Data Bank [PDB] no. 1BHE) and GH49 dextranase (pink; PDB 1OGM) are the clan GH-N representative enzymes. The clan GH-C enzymes are a GH11 xylanase (green; PDB 1BCX) and a GH12 endoglucanase (light blue; PDB 1OA4). The clan GH-A enzymes are a GH5 endoglucanase (magenta; PDB 1A3H), GH26 mannanase (blue; PDB 2BVT), and a GH53 endo-β-1,4-galactanase (green; PDB 1R8L). The catalytic residues are shown in stick format in a darker form of the respective color of the protein fold. Convergent evolution is evident by the observation that xylanases are found in three glycoside hydrolase families that display very different folds.
MECHANISM OF PLANT CELL WALL DECONSTRUCTION
The vast majority of glycoside hydrolases cleave glycosidic bonds by either a single or double displacement mechanism, which leads to inversion or retention of anomeric configuration, respectively (for review, see Rye and Withers, 2000). Polysaccharide lyases cleave their scissile bond through a β-elimination mechanism (Herron et al., 2000). While carbohydrate esterases generally hydrolyze ester linkages through a double displacement mechanism in which Ser (Schubot et al., 2001) or Asp in CE8 (Fries et al., 2007) functions as the catalytic nucleophile, exceptions to this mode of action are apparent in CE4, where catalysis is metal dependent (Taylor et al., 2006).
CATALYTIC MODULES OF GLYCOSIDE HYDROLASES
Currently, the crystal structure of the catalytic modules of representatives of nearly all the relevant GHs, PLs, CEs, and CBM families, which contribute to plant cell wall deconstruction, have been reported (Cantarel et al., 2009). Some structural folds have given rise to a myriad of enzymes that display significant differences in specificity, exemplified by the GHs located in clan GH-A. Members of this clan display a (β/α)8-fold in which the catalytic residues are presented at the C terminus of β-strands 4 and 7 (Henrissat et al., 1995; Jenkins et al., 1995). While the enzymes all hydrolyze an equatorial glycosidic bond, their mode of action (exo and endo), specificity for the sugar at the catalytic −1 subsite and more distal regions of the substrate-binding region (Xyl, Man, Glc, Araf, Gal), and the linkage cleaved (e.g. β-1,4, β-1,3) vary between enzymes (Fig. 1). The same enzyme activity can often be found in multiple GHs, located in distinct clans, as a consequence of convergent evolution (Fig. 1). For example, cellulases are located in 11 GHs, with seven of these families distributed across four different clans, while four of these GHs currently are not linked to a clan. There have been several reviews on the three-dimensional structure of the catalytic modules of glycoside hydrolases, including plant cell wall-degrading enzymes (Davies and Henrissat, 1995; Henrissat and Davies, 1997, 2000; Davies et al., 2005; Gilbert et al., 2008). Therefore, this Update will provide a brief overview of the structures of these enzymes and a more detailed description of recent structural information.
Cellulases
Cellulose utilization is believed to be mediated by endo-β-1,4-glucanases, cellobiohydrolases (also called exo-β-1,4-glucanases), and β-glucosidases. Classically, cellulose hydrolysis, of which the Hypocrea jecorina (formerly Trichoderma reesei) system is the archetype, is viewed as a synergistic process; endo-acting cellulases create new ends from which the exo-acting cellobiohydrolases can release cellobiose from either the reducing (GH7 and GH48) or nonreducing (GH6) end of the cellulose chains (for review, see Kleywegt et al., 1997; Teeri, 1997). This model, however, is inconsistent with several features of cellulose degradative systems. Thus, biochemical and structural data indicate that GH6 cellobiohydrolases are not, exclusively, exo acting (Amano et al., 1996; Armand et al., 1997; Varrot et al., 1999). Furthermore, some highly active cellulase systems lack a classic pair of cellobiohydrolases that act from the reducing and nonreducing ends of cellulose chains, respectively (Xie et al., 2007; Weiner et al., 2008). Indeed, one of the most distinctive features of the cellulose-degrading bacterium, Cytophaga hutchinsonii, is the absence of GH6, GH48, or GH7 cellobiohydrolases (Xie et al., 2007), although it is possible that the bacterium contains novel cellobiohydrolases. An intriguing report by Tolonen et al. (2009) showed that a single endo-processive GH9 cellulase was essential for cellulose degradation in Clostridium phytofermentans. Given the redundancy in cellulase systems, demonstration that a single enzyme is essential for a functional degradative system is rare and questions the classical synergy model. While there now does not appear to be a single unifying model for cellulose hydrolysis, recent studies, deploying atomic force microscopy to visualize the movement of cellulase molecules on its crystalline substrate, will likely provide novel insights into the mechanism by which these enzymes function (Igarashi et al., 2009).
Plant cellulases are restricted to a very small number of families exemplified by Arabidopsis (Arabidopsis thaliana), whose genome encodes 25 cellulases (endoglucanases) all in GH9. Phylogenetic analysis of the Arabidopsis GH9s points to three distinct subfamilies: α, β, and γ. Biochemical studies on γ-endoglucanases show that they are approximately 100-fold less active than the corresponding microbial enzymes, reflecting the loss of a critical aromatic residue at the −2 subsite (Master et al., 2004). This reinforces a remodeling role, rather than a degradative role, for these membrane-associated cellulases. A cohort of the α-endoglucanases contains a C-terminal module that binds to cellulose (Urbanowicz et al., 2007), which has functional implications discussed below.
Xyloglucan
The β-1,4-glucan backbone of xyloglucan is hydrolyzed by specific endoglucanases (i.e. endo-xyloglucanase or xyloglucan endo-hydrolases) from GH5, GH7, GH12, GH16, GH44, and GH74. GH12 enzymes can tolerate the side chains in xyloglucan. Indeed, GH5 and GH74 endoxyloglucanases can make productive interactions with the α-1,6-Xyl decorations and, in the case of the GH5 enzymes, Gal pendants of the Xyl residues (Martinez-Fleites et al., 2006; Gloster et al., 2007). Maybe the most interesting aspect of xyloglucan modification is found in GH16, where enzymes may display endoxyloglucanase activity or, in the case of XETs, remodel the structure of the polysaccharide through transglycosylation reactions (Baumann et al., 2007). This article will not discuss these GH16 enzymes, which are covered in detail in the review by Eklöf and Brumer (2010; this issue).
β-Mannanases
β-Mannanases display a (β/α)8 barrel fold and are located within GH5 and GH26, while β-mannosidases are GH2 enzymes; all three GHs are within clan GH-A. The crystal structures of β-mannanases generally reveal an open active-site cleft with at least four subsites. An unusual feature of β-mannanases is that substrate specificity is not conferred by the recognition of Man in its relaxed chair conformation (4C1) at the critical −1 subsite (glycosidic bond cleavage occurs between the sugars bound at the −1 and +1 subsites; Davies et al., 1997) but through the B2,5 topology displayed by the oxocarbonium transition state (Ducros et al., 2002; Tailford et al., 2007; Cartmell et al., 2008).
In addition to mannan, β-mannanases hydrolyze glucomannan, a heterogenous β-1,4-linked polymer of Glc and Man. β-Mannanases are defined by their capacity to hydrolyze mannosidic bonds, which requires that Man is positioned in the −1 subsite. Recognition of Man and Glc at subsites distal to −1 is highly variable, although some general trends are emerging that point to a divergence in specificity between GH5 and GH26 mannanases. GH5 mannanases are able to accommodate Glc at the −2 and +1 subsites (Tailford et al., 2009) and are thus able to hydrolyze mannosidic linkages flanked by Man or Glc. Indeed, one of these enzymes, BaMan5A, does not recognize O2 as a specificity determinant at any subsite distal to −1. Thus, while BaMan5A hydrolyzes only mannosidic bonds, the topographical features of the substrate-binding cleft of this enzyme are optimized to utilize glucomannan as its preferred substrate (Tailford et al., 2009). The relaxed specificity for Glc or Man, apart from the critical −1 subsite, is a feature shared with the other GH5 mannanases, where structural information is available.
In contrast, the GH26 mannanases characterized to date generally display tight specificity for Man at both the −2 and −1 subsites. Indeed, a cohort of GH26 mannanases contain an Arg at the −2 subsite that makes extensive interactions with the substrate and appears to confer unusually high activity against small mannooligosaccharides (Ducros et al., 2002; Cartmell et al., 2008). Screening genomic databases for other GH26 enzymes that retain this Arg may facilitate the identification of novel mannooligosaccharidases. Currently, the two Cellvibrio enzymes that contain a high-affinity −2 subsite do not possess additional negative binding subsites, which may explain why the high activity displayed against mannotriose and mannotetraose is not translated to the hydrolysis of polysaccharides (Hogg et al., 2001; Cartmell et al., 2008).
Xylan Degradation
The xylan backbone is hydrolyzed primarily by GH10 and GH11 xylanases, while the Araf side chains are removed by arabinofuranosidases from GH43, GH51, GH54, and GH62 (for review of xylan degradation, see Gilbert et al., 2008). The uronic side chains are released from the nonreducing end of xylooligosaccharides by GH67 α-glucuronidases (Nurizzo et al., 2002), although recent data showed that GH115 α-glucuronidases remove the uronic acid decorations from the internal regions of xylan (Ryabova et al., 2009). Each of these families contains at least one structural representative, with the exception of GH62 and GH115 (http://www.cazy.org). GH43 enzymes may display the highest level of substrate diversity, exemplified by the activity of two arabinofuranosidases from this family that remove the O3 side chain from Xyl residues that are decorated at both O2 and O3 with Araf (van den Broek et al., 2005; Sorensen et al., 2006). The crystal structure of this enzyme (H.J. Gilbert, unpublished data) reveals an extended substrate-binding pocket that interacts with both O2- and O3-linked Araf. By contrast, an arabinoxylan-specific GH43 arabinofuranosidase, which removes O2- or O3-linked Araf side chains from singularly substituted Xyl residues, contains a small substrate-binding pocket embedded in a shallow cleft that is optimized to bind the 3-fold helical structure of the xylan backbone (Vandermarliere et al., 2009). Recent protein crystallographic studies have shown that xylan side chains can be accommodated and can actually be exploited as specificity determinants (Pell et al., 2004; Vardakou et al., 2005), while a GH5 xylanase displays an absolute requirement for 4-O-methyl-d-GlcUA appended to the Xyl positioned at the −2 subsite (Vrsanska et al., 2007). There are two structures of this enzyme (Larson et al., 2003; St John et al., 2009); however, the mechanism by which the enzyme recognizes the uronic acid side chain remains unclear.
Pectin Degradation
The structures of pectinases (polygalacturonases), pectate lyases, and pectin methylesterases have been extensively described and reviewed previously (Jenkins and Pickersgill, 2001). In general, these enzymes display a right-handed parallel β-helix topology. Exceptions include PL10 pectate lyases, which adopt an (α/α)6 toroid conformation (Charnock et al., 2002b), and PL2 lyases, which display a (α/α)7 barrel and utilize manganese rather than calcium in the active site (Abbott and Boraston, 2007). The catalytic apparatus in PL10, and those displaying a β-helix fold, is conserved, providing an example of convergent evolution (Charnock et al., 2002b). An Arg is the most likely candidate catalytic base in these PLs. The basic residue abstracts the C5 proton, which, in several PL families (PL2, PL9, and PL10), results in the formation of an enolate-enolate intermediate in which the two negatively charged oxygens are stabilized by calcium and hydrogen bonds. The collapse of the intermediate results in the cleavage of the scissile bond, although the mechanism by which the leaving group (glycosidic oxygen) is protonated remains unclear. An interesting variation of this catalytic mechanism has been proposed for PL1 lyases. It was suggested that the PL1 lyase generates an enol-enolate through donation of a proton by a nearby Lys to one of the oxygen atoms of the carboxylate. The authors suggest that through this intermediate, PL1 lyases are more active than PL10 and PL9 enzymes that can only generate the enolate-enolate intermediate (Seyedarabi et al., 2010).
Recent advances have also been made in understanding the processive mechanism displayed by pectin methyl esterases, which yield blocks of nonmethylated GalUA (GalA). Structural and biochemical data show that the enzyme demethylates the sugar at the +1 subsite and uses the negative charge of the carboxylate as a specificity determinant at the −1 subsite and to some extent at −2, while +3 makes hydrophobic contact with the methyl group of the esterified uronic acid (Fries et al., 2007). Thus, after removing the methyl group, the GalA generated then slides along the substrate-binding cleft to occupy the −1 site; thus, a new methylated GalA is presented in the crucial +1 subsite. This progressive sliding of pectin along the substrate-binding cleft is encouraged further by the specificity displayed by the +3 and −2 subsites.
STRUCTURAL CHANGES THAT MODULATE THE MODE OF ENZYME ACTION
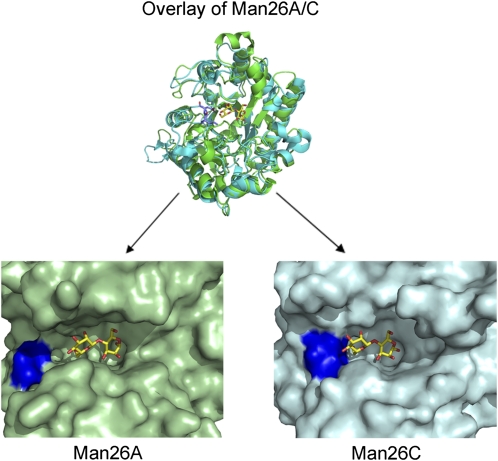
The structural basis for the GH6 and GH7 cellobiohydrolases and endoglucanases is well established and has been extensively reviewed (Kleywegt et al., 1997; Teeri, 1997; Varrot et al., 1999). Recent structural data have also provided insight into how subtle structural changes can convert endo-acting glycoside hydrolases and polysaccharide lyases into exo-acting enzymes. Thus, small loop extensions surrounding the distal subsite that accommodates the nonreducing end of the substrate create steric constraints that prevent extension of the substrate beyond this subsite. Variants of these enzymes, in which the loop extensions have been removed, display an endo mode of action (Proctor et al., 2005; Cartmell et al., 2008; Ochiai et al., 2009; Fig. 2).
Figure 2.
An overlay of an endo (CjMan26A; green) and an exo (CjMan26C; cyan) β-mannanase from Cellvibrio japonicas. A small extension of the loop at the distal −2 subsite presents two residues, Asp-130 and Leu-129, shown in stick format in purple. These residues present a steric block that prevents extension of substrate distal to the sugar bound at the −2 subsite. The residues shown in dark blue, in the surface representations of the two enzymes, are Asp-130 in CjMan26C and the equivalent amino acid (Glu-121) in CjMan26A.
PLANT CELL WALL GLYCOSYLTRANSFERASES
The crystal structures of several glycosyltransferases in numerous GT families have been reported in the last decade. The data have revealed only two major folds for these enzymes, while also providing insights into the likely catalytic mechanisms displayed by inverting and retaining glycosyltransferases (for review, see Lairson et al., 2008). These studies, however, have focused, almost exclusively, on enzymes that are not membrane associated, and currently, there is no high-resolution crystal structural information on glycosyltransferases that contribute to plant cell wall synthesis. Cellulose synthase, however, can be visualized by freeze-fracture techniques, in conjunction with immunological methods. The data revealed six globular complexes approximately 25 nm in diameter. Each of the six subunits (each subunit contains multiple cellulose synthase molecules) of these rosettes synthesize multiple β-1,4-glucan chains, which cocrystallize to form microfibrils (for review, see Somerville, 2006). Although issues remain concerning the nature of the primer and the direction of chain growth elongation (although elongation from the nonreducing end is the preferred model), the lack of detailed structural information on these enzymes precludes further discussion of this enzyme system here. It is evident, however, that using genetic approaches, Arabidopsis plant cell wall glycosyltransferases have been identified and, in some instances, predicted activities have been verified by detecting appropriate transfer reactions in heterologous hosts (for review, see Liepman et al., 2010). It is evident that in the next few years there will be rapid advances in the identification of glycosyltransferase genes that encode plant cell wall-synthesizing enzymes. It is highly likely that the resultant data will underpin the much needed detailed structural and biochemical information of these plant cell wall glcosyltransferases.
CBMS
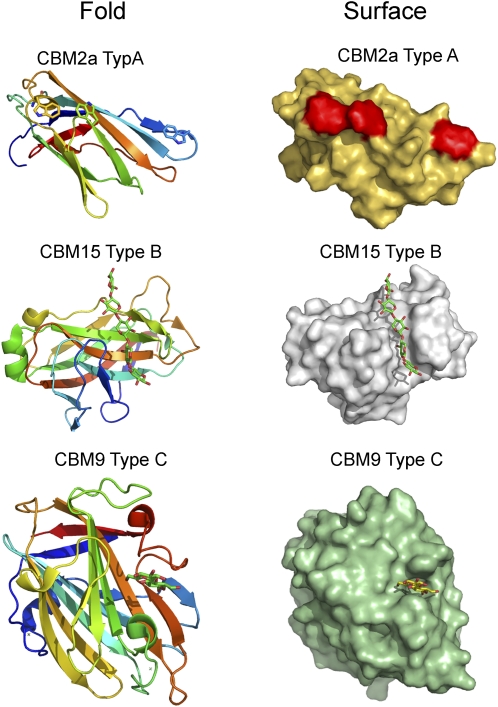
Microbial plant cell wall hydrolases display complex molecular architectures in which the catalytic module is appended, by flexible linker sequences, to one or more CBMs (for review, see Boraston et al., 2004). In some of the 59 CBM families, exemplified by CBM1, CBM10, and CBM20, ligand specificity is invariant (Linder and Teeri, 1997; Southall et al., 1999; Raghothama et al., 2000), while in some families, such as CBM6 (Czjzek et al., 2001; Pires et al., 2004), CBM4 (Boraston et al., 2002b), and CBM35 (Tunnicliffe et al., 2005; Montanier et al., 2009b), carbohydrate recognition is highly variable. In addition to defining a phylogenetic relationship between CBMs by clustering these modules into sequence-based families, they have also been classified into three categories (types A, B, and C) based on the topology of their ligand-binding sites and their mode of ligand recognition (for review, see Boraston et al., 2004; Fig. 3).
Figure 3.
Examples of type A, type B, and type C CBMs. CBM2a is derived from the Cellulomonas fimi xylanase Xyn10A (Protein Data Bank [PDB] 1XG), CBM15 is a component of the C. japonicas xylanase Xyn10C (PDB 1GNY), and CBM9 is from a Thermotoga maritima GH10 xylanase (PDB 1I82). The folds are ramped from blue (N terminus) to red (C terminus). The three aromatic residues that form a ligand-binding apolar surface in the CBM2a module are colored red and are shown in stick format in the respective surface and fold depictions of the protein.
CBMs have now been described that bind to the major polysaccharides found in the plant cell wall (for review, see Boraston et al., 2004), while modules that recognize the side chains of these polymers, and the products released through their deconstruction, have also been identified (Notenboom et al., 2001; Miyanaga et al., 2004; Montanier et al., 2009b). In general, the ligand specificity of CBMs reflects the substrate cleaved by the cognate enzyme (discussed further below). Many of these enzymes, however, also contain a CBM that binds to crystalline cellulose (Kellett et al., 1990; McKie et al., 2001; Hogg et al., 2003). It has been suggested that once bound, these type A modules are able to slide across the surface of cellulose (Jervis et al., 1997), enabling the substrate-specific type B and type C CBMs to lock onto its ligand and thus direct the enzyme to its target glycosidic bonds (Kellett et al., 1990).
In general, the affinity of CBMs for their target plant-derived ligands is low (Kd of approximately 100 μm; Boraston et al., 2004). Some enzymes, however, contain multiple copies of CBMs that display the same specificity, and in these proteins, avidity effects between these modules have led to increased affinity for polysaccharides (Bolam et al., 2001; Freelove et al., 2001; Boraston et al., 2002a). It is interesting that nature has deployed CBM duplication as a mechanism for increased affinity rather than increasing the interactions between ligand and a single CBM module. It is possible that as CBMs generally bind to ligands that are in intimate contact with other components of the plant cell wall, steric constraints prevent extensive interactions between the protein and target carbohydrate.
Plant CBMs
CBMs are less prevalent among plant glycoside hydrolases that cleave structural polysaccharides; however, several CBM49 and CBM22 modules are present in plant cellulases and xylanases, respectively. The CBM49 modules are located in a subfamily of GH9 plant endoglucanases, and one of these modules was shown to bind tightly to crystalline cellulose (Urbanowicz et al., 2007). It is possible that the GH9 CBM49-containing endoglucanases play a role in modulating the structure of crystalline cellulose.
HOW DO CBMS POTENTIATE CATALYSIS?
The mechanism by which CBMs potentiate catalysis remains unclear. It has been hypothesized that cellulose-specific CBMs may play a key role in disrupting the ordered hydrogen-bonding network in crystalline cellulose, making the surface chains accessible to the appended cellulase (Knowles et al., 1987; Teeri, 1997). There is biochemical, biophysical, and microscopic data (Din et al., 1994; Wang et al., 2008) indicating that CBMs mediate changes to the surface structure of cellulose. Furthermore, the addition of CBMs in trans to the cognate catalytic module has led to a modest potentiation (0.2- to 1.5-fold) in catalytic activity against insoluble substrates (Din et al., 1994; Moser et al., 2008). Cellulases, typically endoglucanases, however, are often 3 orders of magnitude more active against soluble forms of cellulose than the crystalline polysaccharide (Durrant et al., 1991; Irwin et al., 1993). Thus, CBMs acting in trans have only a minor influence on the access problem, although this might reflect the dissociation of the targeting and (possible) disrupting function of these modules. Of potential significance is the location of crystalline cellulose-specific CBMs in many enzymes that display no cellulase activity (Kellett et al., 1990; McKie et al., 2001; Hogg et al., 2003; Vincent et al., 2010), which argues against these modules having a specialized function in cellulose degradation. A more likely explanation for the capacity of CBMs to increase the activity of glycoside hydrolases against insoluble substrates is that they reduce the “accessibility problem” by bringing the appended catalytic modules into intimate and prolonged association with their target substrate, thereby enhancing catalytic efficiency.
In several organisms, however, there are populations of CBMs that are not components of enzymes, and these modules may destabilize the crystalline structure of some polysaccharides. Thus, CBM33, which is highly expressed in chitin-degrading bacteria such as Serratia marcescens, potentiates the chitinases from this organism, particularly during the latter stages of the degradative process when the glycoside hydrolases are attacking highly crystalline forms of the polysaccharide (Vaaje-Kolstad et al., 2005a). More modest potentiation of cellulases by “noncatalytic” bacterial CBMs has also been reported (Moser et al., 2008), while it has been suggested that several noncatalytic fungal proteins may play a role in plant cell wall disruption. It is believed that the primary function of GH61s (now established as fungal noncatalytic carbohydrate-binding proteins) is to disrupt plant cell wall structure and thus increase the access of degradative enzymes to their substrates (Rosgaard et al., 2006; Karkehabadi et al., 2008; Harris et al., 2010). Indeed, fungi often contain multiple copies of this protein, and they are coexpressed with a range of cellulases (Vanden Wymelenberg et al., 2009). Another potential fungal CBM33 analog is swollenin from Trichoderma reesei, which appears to have disruptive effects on cellulose, although recent studies suggest that the protein may display endoglycanase activities (Yao et al., 2008). In addition to microorganisms, plants (and plant cell wall-degrading nematodes) also produce proteins, referred to as expansins, that mediate a relaxation in the structure of the cell wall (for review, see Cosgrove, 2000). Expansins mechanically weaken plant cell walls (McQueen-Mason and Cosgrove, 1994), and their use in improving cellulase efficiency has been reported (Han and Chen, 2007). Currently, GH61s and expansins appear to disrupt the cellulose-hemicellulose interface, while the functional importance of swollenin remains opaque.
The structure of a CBM33 reveals a binding surface that contains several conserved polar residues that are pivotal to the synergistic effects of this protein with chitinases (Vaaje-Kolstad et al., 2005b). Significantly, mutations that prevented the CBM33 from potentiating chitinase activity had little effect on the affinity of the protein for chitin. This led to the proposal that the specific polar interactions between chitin and the protein disrupt the hydrogen-binding network between individual polysaccharide chains (Vaaje-Kolstad et al., 2005b). This could also explain the specificities displayed by CBM33 modules. The crystal structures of GH61s from Hypocrea jecorina (Karkehabadi et al., 2008) and Thielavia terrestris (Harris et al., 2010) reveal a similar surface to CBM33, again pointing to a disruptive function for these proteins.
While the effects of noncatalytic proteins on cellulose hydrolysis, to date, have been disappointing, continued efforts at identifying the functional significance of these molecules is merited. For example, it is possible that specific combinations of these proteins are required to disrupt the structure of cellulose, while, currently, the influence of these accessory proteins has been explored only in isolation.
THE STRUCTURAL BASIS FOR CBM SPECIFICITY
The three-dimensional structures of representatives of 21 of the 24 CBM families that target the plant cell wall have been determined (for review, see Boraston et al., 2004). The vast majority of these modules display a jelly roll fold comprising two antiparallel β-sheets that form the two surfaces of these proteins. Ligand binding occurs on the concave surface presented by one of the β-sheets (Boraston et al., 2002b) or in the loops that connect the two β-sheets (defined hereafter as site 1; Czjzek et al., 2001; Montanier et al., 2009b).
Crystalline Cellulose Recognition
CBMs that bind crystalline cellulose contain three aromatic amino acids that adopt a planar topology with respect to each other (Kraulis et al., 1989; Xu et al., 1995; Tormo et al., 1996; Raghothama et al., 2000). These residues make extensive hydrophobic contacts with fully exposed sugar rings presented at the 110 face of cellulose crystals (Lehtio et al., 2003). Ligand recognition by these CBMs is driven primarily through an increase in entropy, resulting in the desolvation of the interacting macromolecules (Creagh et al., 1996). By contrast, enthalpy drives the binding of CBMs to discreet polysaccharide chains, where both polar and apolar interactions occur, while entropy has a negative impact on overall affinity (Charnock et al., 2000, 2002a; Bolam et al., 2001; Boraston et al., 2002a, 2002b). The negative entropy may reflect conformational restriction of the ligand bound to the protein, which is not entirely offset by the release of tightly bound water molecules (for review, see Boraston et al., 2004). It should be recognized, however, that the energetic freedom of the solvating molecules of the protein is a controversial issue; thus, the molecular basis for the thermodynamic forces that drive ligand recognition in type B CBMs remains unclear.
Xylan versus Cellulose Recognition
Subtle changes in structure can lead to significant changes in ligand specificity, exemplified by xylan and cellulose specificity within CBM2, which is defined by the conformation adopted by the surface aromatic residues, which are perpendicular in xylan-binding modules and planar in cellulose-binding modules (Simpson et al., 1999, 2000), consistent with the conformation adopted by the two polysaccharides. The perpendicular arrangement of surface Trps in xylan-binding CBM2s is mediated by an Arg, while in cellulose-binding CBM2s, the basic residue is replaced by a Gly, enabling the Trp to collapse onto the surface of the protein and adopt a planar orientation with respect to the other aromatic residues (Simpson et al., 2000). While the perpendicular arrangement of aromatic residues in xylan-binding CBMs is a common feature, modules that recognize the hemicellulosic polymer can adopt different ligand-binding strategies. Thus, in CBM4, CBM6, and CBM22, xylan recognition is dominated by a single Xyl residue that is sandwiched between a pair of planar aromatic residues within a deep ligand-binding cleft (Czjzek et al., 2001; Charnock et al., 2002a; Simpson et al., 2002). While this binding mode confers higher affinity for isolated xylan chains, CBMs that recognize xylan through the asymmetric distribution of aromatic residues display more versatile ligand recognition; they are able to bind to the hemicellulose within in a variety of terrestrial plant cell walls, a specificity that is not displayed by the modules from CBM4, CBM6, and CBM22 (McCartney et al., 2006).
The Topology of the Ligand-Binding Cleft Influences Specificity
While CBMs that bind to internal regions of polysaccharides display an open cleft topology, the shape of the cleft influences specificity. This is exemplified in CBM4, where structurally related modules bind to linear β-1,4-polysaccharides, such as cellulose, or highly curved structures, such as β-1,3-glucan (Boraston et al., 2002a). In CfCBM4-1, both ends of the cleft are open, enabling the protein to bind linear glucan chains such as cellulose. However, insertions in two loops confer a U-shape topology on the longitudinal axis of the binding cleft of TmCBM4-2, which is complementary to the curved conformation adopted by its ligand, β-1,3-glucan. A more extreme example of how topological changes can cause a dramatic change in ligand specificity is evident in site 1 in CBM6 modules. This site may adopt a pocket-like topology and thus recognize the termini of polysaccharide chains (Pires et al., 2004) or display an open cleft and bind to the internal regions of xylan (Czjzek et al., 2001). From the discussion above, it is apparent that CBMs, in common with lectins, display preformed carbohydrate-recognition sites that mirror the solution conformations of their target ligands, thereby minimizing the energetic penalty paid upon binding.
Recognition of Heterogenous Polymers
Glucomannan (contains a random distribution of β-1,4-linked d-Man and d-Glc residues) presents a significant challenge with respect to CBM specificity. While mannan-specific CBMs recognize the regions of glucomannan containing successive Man residues (Tunnicliffe et al., 2005), two CBM families, CBM29 and CBM16, contain proteins that display specificity for the heterogenous polymer in addition to cellulose and mannan. In these modules, the aromatic residues in the binding cleft make planar contacts with sugars at n and n + 2 and thus avoid steric clashes with the axial O2 in the Man residues. In addition, several polar residues are capable of making hydrogen bonds with the axial or equatorial O2 of Man or Glc, respectively; while at other sugar-binding subsites, O2 is not a specificity determinant (Charnock et al., 2002a; Bae et al., 2008). CBMs have also been shown to harness both the backbone and side chain of decorated glucans such as xyloglucan (Najmudin et al., 2006). Specificity for this polymer has also been engineered into a xylan-specific module (Gunnarsson et al., 2006), while recent structural information on this protein provides insight into how the observed change in specificity was achieved (Gullfot et al., 2010).
Calcium
CBMs, which display a jelly roll fold, contain a highly conserved structural calcium (for review, see Boraston et al., 2004). Furthermore, there are increasing examples of CBMs where calcium plays a direct or indirect role in ligand recognition in site 1. Thus, in Aga16B-CBM6-2, calcium orientates a Tyr such that it can interact with the neoagarose ligand (Henshaw et al., 2006), while in CBM36 (and a second xylan-specific CBM; H.J. Gilbert, unpublished data), the metal ion coordinates with the O2 and O3 of Xyl residues within xylan (Jamal-Talabani et al., 2004). A cohort of six CBM35s (three of these modules are identical but are located in different xylan-degrading enzymes) were recently shown to bind to uronic acids, where calcium makes critical electrostatic interactions with the C6 carboxylate (Montanier et al., 2009b). Subtle differences in the ligand-binding site in this cohort of CBM35s confer differences in ligand specificity. The modules derived from the pectin-metabolizing enzymes bind only to Δ4,5-anhydrogalacturonic acid, while the other CBM35s recognize both the pectin degradation product and GlcUA (Montanier et al., 2009b).
DUAL CATALYTIC AND NONCATALYTIC BINDING FUNCTIONS FOR AN ESTERASE
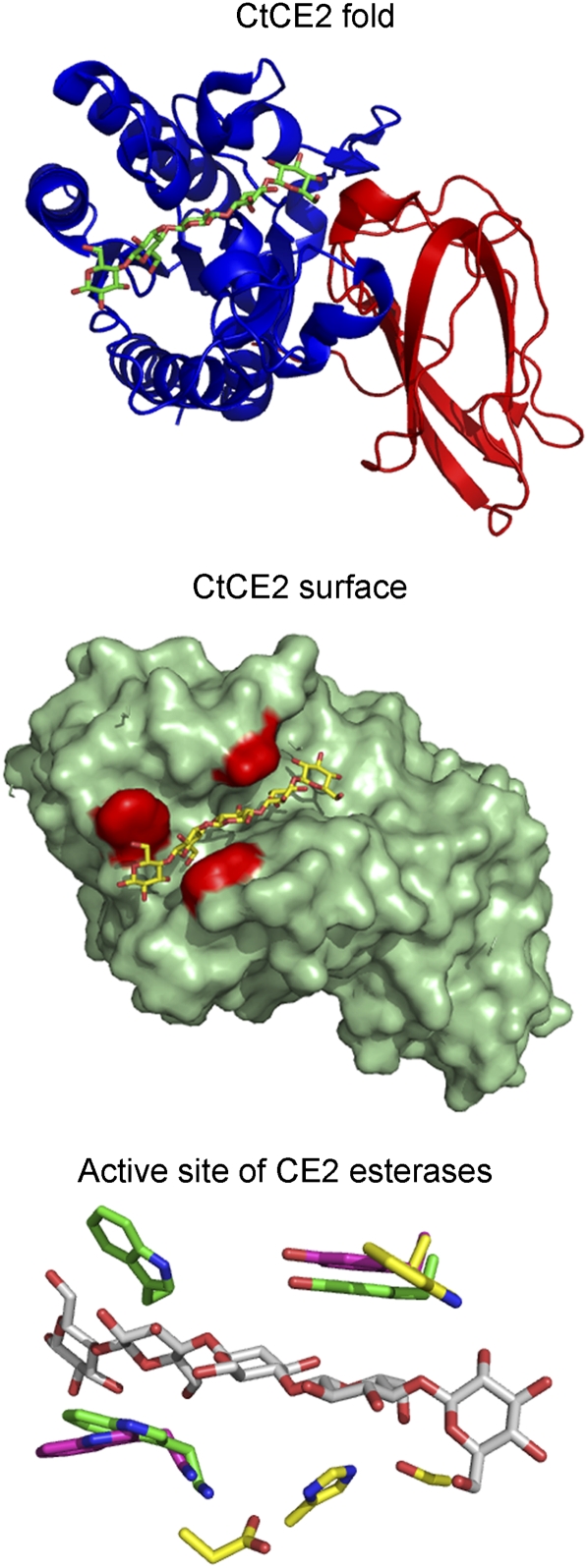
Within the context of plant cell wall degradation, the catalytic and CBM functions are conferred by discrete regions of the multimodular enzymes that catalyze this process. Recently, however, a CE2 esterase, CtCE2, which is appended to a GH5 endoglucanase, was shown to have a noncatalytic cellulose-binding function in addition to displaying esterase activity; other modules in this esterase family (which are not appended to other catalytic modules) do not recognize cellulose. The crystal structure of CtCE2 shows that cellulose binds to the active site of the esterase through hydrophobic interactions with three aromatic residues and by hydrogen bonds with components of the catalytic apparatus. The crystal structures of other CE2 esterases showed that these enzymes lack one or more of the three aromatic residues, explaining why they do not recognize cellulose. The CE2 family encapsulates the requirement for multiple activities by biocatalysts that attack challenging macromolecular substrates such as the plant cell wall, including the grafting of a second, powerful, and discrete noncatalytic binding functionality into the active site of an enzyme. This report provides a rare example of “gene sharing” (Montanier et al., 2009a), where the introduction of a second functionality into the active site of an enzyme does not compromise the original activity of the biocatalyst (Fig. 4).
Figure 4.
The structural features of the active site of CtCE2 that displays esterase and a CBM function. The catalytic module of CtCE2 contains two discrete domains that display a typical α/β-hydrolase fold (blue), evident in Ser esterases, and a jelly roll fold (red), respectively. The surface representation of CtCE2 reveals a cleft that accommodates cellopentaose (shown in yellow) and houses the active site of the esterase. The three aromatic residues that play a key role in binding cellulose are shown in red. The active site of three CE2 esterases show cellopentaose (gray) and the three aromatic residues (green) from CtCE2, the two aromatic residues (magenta) in CjCE2B, and the single aromatic amino acid (yellow) in CjCE2A. All three enzymes are Ser esterases, and the catalytic triad (Ser-160, His-335, Asp-333) of CE2A is displayed in yellow.
FUNCTIONAL SIGNIFICANCE FOR CBM LIGAND RECOGNITION
As discussed above, type B CBMs generally bind to substrates of the catalytic modules. Exceptions to this rule include a xylanase-derived CBM9, which binds the reducing end of xylan or cellulose (Notenboom et al., 2001), while CBM35s, located in pectin-metabolizing enzymes, bind to the reaction products generated by pectate lyases (Montanier et al., 2009b). It would appear, therefore, that these CBMs are recruiting enzymes to regions of the cell wall where the target substrates for the appended enzymes are undergoing degradation; thus, in a sense, the modules are directing the catalytic apparatus to areas of the wall that are susceptible to degradation. The CBM35 appended to three xylan-degrading enzymes binds to both GlcUA (GlcA) and the unsaturated product released by pectate lyases but not to 4-O-methyl-d-GlcUA, the more common uronic acid found in xylans. The biological rationale for this dual specificity is unclear. It has been proposed that in model plants, such as Arabidopsis, in rapidly dividing cells there are significant quantities of unmethylated GlcA (Pena et al., 2007). This has led to the hypothesis that, by targeting unmethylated GlcA, the CBM is directing enzymes to more open structures that are particularly susceptible to enzyme degradation. It is possible that this cohort of CBM35s initially direct the xylan-degrading apparatus to regions of cell walls that are being actively degraded, for which anhydrogalacturonic is a marker, but, as xylan structures are revealed, the enzyme is shuttled onto the hemicellulosic polysaccharide, affording the enzyme access to its target substrate (Montanier et al., 2009b).
Recent studies have also shown that CBMs can display a bacterial anchoring function. Thus, the CBM35 from the Amycolatopsis orientalis exo-β-d-glucosaminidase (Chi-CBM35) tethers the enzyme to the cell wall of the bacterium (Montanier et al., 2009b). Similarly, a family of CBMs unique to Ruminococcus albus, which bind to a wide spectrum of β-linked plant structural polysaccharides (CBM37), anchor their cognate enzymes to the surface of the bacterium (Ezer et al., 2008). The biological rationale for this CBM function appears to be to keep the enzymes in close proximity to the bacterium. However, Ezer et al. (2008) also proposed a model in which the CBM37 acts as a shuttle that transfers the appended enzymes from the bacterial surface to the plant cell wall. In any event, these recent reports of the cell adhesion role of CBMs, which was previously unconsidered, may prove to factor prominently in the function of these protein modules in the future.
FUTURE PERSPECTIVES
In the last 15 years, there have been significant advances in the three-dimensional structural analysis of plant cell wall-degrading enzymes. The data have informed our understanding of the mechanism of both catalysis and substrate recognition, which has led to the identification of numerous “specificity motifs,” some of which are described in this article. It is evident, however, that the explosion of genomic and metagenomic information is resulting in an exponential increase in the identification of CAZy enzymes. This has resulted in a significant imbalance between the number of enzymes in CAZy families and the biochemical/structural analysis of these proteins. Indeed, only around 3% of the proteins in CAZy have a characterized biochemical activity, while three-dimensional structural information is only available for 0.3% of these enzymes (Cantarel et al., 2009). It is estimated that we can safely predict the activities of no more than 20% of the proteins within CAZy. The situation is compounded further by the difficulties in determining the biochemical properties of plant cell wall-degrading enzymes, where the chemical complexity and requirement for a hierarchical degradative process create significant functional barriers. Notwithstanding these problems, continued biochemical and structural information is urgently required if we are to fully integrate information obtained from the “omics” technologies to understand the biology of plant cell wall deconstruction. Indeed, integrating structure, function, and phylogenetics to develop predictive models for ligand/substrate specificity is an important goal for structural biologists working on plant cell wall-modifying enzymes. An example of such an analysis was developed recently by Abbott et al. (2009). Deploying CBM6 as a model system, they were able to identify two regions that appear to be “hot spots” of primary and tertiary structure variation, which confer functional specificity in these modules, a view supported by the recent characterization of a Xyl-specific CBM6. A more general phylogenetic analysis of endoglucanases belonging to several GHs was also insightful in providing a predictive platform for glycoside hydrolase activities (Vlasenko et al., 2010). As discussed above, the characterization of glycosyltransferases that catalyze the synthesis of plant structural polysaccharides represents the biggest challenge in the cell wall field. It is evident that a significant investment is required to develop our understanding of the structure-function relationships of these enzymes, which is essential if we are to fully understand the mechanism for the biogenesis of the plant cell wall.
References
- Abbott DW, Boraston AB. (2007) A family 2 pectate lyase displays a rare fold and transition metal-assisted beta-elimination. J Biol Chem 282: 35328–35336 [DOI] [PubMed] [Google Scholar]
- Abbott DW, Ficko-Blean E, van Bueren AL, Rogowski A, Cartmell A, Coutinho PM, Henrissat B, Gilbert HJ, Boraston AB. (2009) Analysis of the structural and functional diversity of plant cell wall specific family 6 carbohydrate binding modules. Biochemistry 48: 10395–10404 [DOI] [PubMed] [Google Scholar]
- Amano Y, Shiroishi M, Nisizawa K, Hoshino E, Kanda T. (1996) Fine substrate specificities of four exo-type cellulases produced by Aspergillus niger, Trichoderma reesei, and Irpex lacteus on (1→3), (1→4)-beta-D-glucans and xyloglucan. J Biochem 120: 1123–1129 [DOI] [PubMed] [Google Scholar]
- Armand S, Drouillard S, Schulein M, Henrissat B, Driguez H. (1997) A bifunctionalized fluorogenic tetrasaccharide as a substrate to study cellulases. J Biol Chem 272: 2709–2713 [DOI] [PubMed] [Google Scholar]
- Bae B, Ohene-Adjei S, Kocherginskaya S, Mackie RI, Spies MA, Cann IKO, Nair SK. (2008) Molecular basis for the selectivity and specificity of ligand recognition by the family 16 carbohydrate-binding modules from Thermoanaerobacterium polysaccharolyticum ManA. J Biol Chem 283: 12415–12425 [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklof JM, Michel G, Kallas AM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolam DN, Xie H, White P, Simpson PJ, Hancock SM, Williamson MP, Gilbert HJ. (2001) Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry 40: 2468–2477 [DOI] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston AB, McLean BW, Chen G, Li A, Warren RA, Kilburn DG. (2002a) Co-operative binding of triplicate carbohydrate-binding modules from a thermophilic xylanase. Mol Microbiol 43: 187–194 [DOI] [PubMed] [Google Scholar]
- Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, Davies GJ. (2002b) Differential oligosaccharide recognition by evolutionarily-related beta-1,4 and beta-1,3 glucan-binding modules. J Mol Biol 319: 1143–1156 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell A, Topakas E, Ducros VM, Suits MD, Davies GJ, Gilbert HJ. (2008) The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J Biol Chem 283: 34403–34413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Bolam DN, Nurizzo D, Szabo L, McKie VA, Gilbert HJ, Davies GJ. (2002a) Promiscuity in ligand-binding: the three-dimensional structure of a Piromyces carbohydrate-binding module, CBM29-2, in complex with cello- and mannohexaose. Proc Natl Acad Sci USA 99: 14077–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Bolam DN, Turkenburg JP, Gilbert HJ, Ferreira LM, Davies GJ, Fontes CM. (2000) The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39: 5013–5021 [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Brown IE, Turkenburg JP, Black GW, Davies GJ. (2002b) Convergent evolution sheds light on the anti-beta-elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc Natl Acad Sci USA 99: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia MA, Prates JA, Bras J, Fontes CM, Newman JA, Lewis RJ, Gilbert HJ, Flint JE. (2008) Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J Mol Biol 379: 64–72 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Creagh AL, Ong E, Jervis E, Kilburn DG, Haynes CA. (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc Natl Acad Sci USA 93: 12229–12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czjzek M, Bolam DN, Mosbah A, Allouch J, Fontes CM, Ferreira LM, Bornet O, Zamboni V, Darbon H, Smith NL, et al. (2001) The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J Biol Chem 276: 48580–48587 [DOI] [PubMed] [Google Scholar]
- Davies G, Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3: 853–859 [DOI] [PubMed] [Google Scholar]
- Davies GJ, Gloster TM, Henrissat B. (2005) Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struct Biol 15: 637–645 [DOI] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J 321: 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N, Damude HG, Gilkes NR, Miller RC, Warren RAJ, Kilburn DG. (1994) C-1-C-X revisited: intramolecular synergism in a cellulase. Proc Natl Acad Sci USA 91: 11383–11387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducros VM, Zechel DL, Murshudov GN, Gilbert HJ, Szabo L, Stoll D, Withers SG, Davies GJ. (2002) Substrate distortion by a beta-mannanase: snapshots of the Michaelis and covalent-intermediate complexes suggest a B(2,5) conformation for the transition state. Angew Chem Int Ed Engl 41: 2824–2827 [DOI] [PubMed] [Google Scholar]
- Durrant AJ, Hall J, Hazlewood GP, Gilbert HJ. (1991) The non-catalytic C-terminal region of endoglucanase E from Clostridium thermocellum contains a cellulose-binding domain. Biochem J 273: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer A, Matalon E, Jindou S, Borovok I, Atamna N, Yu Z, Morrison M, Bayer EA, Lamed R. (2008) Cell surface enzyme attachment is mediated by family 37 carbohydrate-binding modules, unique to Ruminococcus albus. J Bacteriol 190: 8220–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freelove AC, Bolam DN, White P, Hazlewood GP, Gilbert HJ. (2001) A novel carbohydrate-binding protein is a component of the plant cell wall-degrading complex of Piromyces equi. J Biol Chem 276: 43010–43017 [DOI] [PubMed] [Google Scholar]
- Fries M, Ihrig J, Brocklehurst K, Shevchik VE, Pickersgill RW. (2007) Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J 26: 3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HJ, Stalbrand H, Brumer H. (2008) How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol 11: 338–348 [DOI] [PubMed] [Google Scholar]
- Gloster TM, Ibatullin FM, Macauley K, Eklof JM, Roberts S, Turkenburg JP, Bjornvad ME, Jorgensen PL, Danielsen S, Johansen KS, et al. (2007) Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem 282: 19177–19189 [DOI] [PubMed] [Google Scholar]
- Gullfot F, Tan TC, von Schantz L, Karlsson EN, Ohlin M, Brumer H, Divne C. (2010) The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module. Proteins 78: 785–789 [DOI] [PubMed] [Google Scholar]
- Gunnarsson LC, Zhou Q, Montanier C, Karlsson EN, Brumer H, III, Ohlin M. (2006) Engineered xyloglucan specificity in a carbohydrate-binding module. Glycobiology 16: 1171–1180 [DOI] [PubMed] [Google Scholar]
- Han YJ, Chen HZ. (2007) Synergism between corn stover protein and cellulase. Enzyme Microb Technol 41: 638–645 [Google Scholar]
- Harris PJ, Stone BA. (2008) Chemistry and molecular organization of plant cell walls. Himmel ME, , Biomass Recalcitrance. Blackwell, Oxford, pp 60–93 [Google Scholar]
- Harris PV, Welner D, McFarland KC, Re E, Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, et al. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49: 3305–3316 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA 92: 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7: 637–644 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Davies GJ. (2000) Glycoside hydrolases and glycosyltransferases: families, modules, and implications for genomics. Plant Physiol 124: 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshaw J, Horne-Bitschy A, van Bueren AL, Money VA, Bolam DN, Czjzek M, Ekborg NA, Weiner RM, Hutcheson SW, Davies GJ, et al. (2006) Family 6 carbohydrate binding modules in beta-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J Biol Chem 281: 17099–17107 [DOI] [PubMed] [Google Scholar]
- Herron SR, Benen JA, Scavetta RD, Visser J, Jurnak F. (2000) Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc Natl Acad Sci USA 97: 8762–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Bayer EA. (2009) Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr Opin Biotechnol 20: 316–317 [DOI] [PubMed] [Google Scholar]
- Hogg D, Pell G, Dupree P, Goubet F, Martin-Orue SM, Armand S, Gilbert HJ. (2003) The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem J 371: 1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg D, Woo EJ, Bolam DN, McKie VA, Gilbert HJ, Pickersgill RW. (2001) Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J Biol Chem 276: 31186–31192 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Koivula A, Wada M, Kimura S, Penttila M, Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J Biol Chem 284: 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DC, Spezio M, Walker LP, Wilson DB. (1993) Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol Bioeng 42: 1002–1013 [DOI] [PubMed] [Google Scholar]
- Jamal-Talabani S, Boraston AB, Turkenburg JP, Tarbouriech N, Ducros VM, Davies GJ. (2004) Ab initio structure determination and functional characterization of CBM36: a new family of calcium-dependent carbohydrate binding modules. Structure 12: 1177–1187 [DOI] [PubMed] [Google Scholar]
- Jenkins J, Lo Leggio L, Harris G, Pickersgill R. (1995) Beta-glucosidase, beta-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold beta/alpha architecture and with two conserved glutamates near the carboxy-terminal ends of beta-strands four and seven. FEBS Lett 362: 281–285 [DOI] [PubMed] [Google Scholar]
- Jenkins J, Pickersgill R. (2001) The architecture of parallel beta-helices and related folds. Prog Biophys Mol Biol 77: 111–175 [DOI] [PubMed] [Google Scholar]
- Jervis EJ, Haynes CA, Kilburn DG. (1997) Surface diffusion of cellulases and their isolated binding domains on cellulose. J Biol Chem 272: 24016–24023 [DOI] [PubMed] [Google Scholar]
- Karkehabadi S, Hansson H, Kim S, Piens K, Mitchinson C, Sandgren M. (2008) The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J Mol Biol 383: 144–154 [DOI] [PubMed] [Google Scholar]
- Kellett LE, Poole DM, Ferreira LM, Durrant AJ, Hazlewood GP, Gilbert HJ. (1990) Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J 272: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Zou JY, Divne C, Davies GJ, Sinning I, Stahlberg J, Reinikainen T, Srisodsuk M, Teeri TT, Jones TA. (1997) The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes. J Mol Biol 272: 383–397 [DOI] [PubMed] [Google Scholar]
- Knowles J, Lehtovaara P, Teeri T. (1987) Cellulase families and their genes. Trends Biotechnol 5: 255–261 [Google Scholar]
- Kraulis J, Clore GM, Nilges M, Jones TA, Pettersson G, Knowles J, Gronenborn AM. (1989) Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei: a study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 28: 7241–7257 [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77: 521–555 [DOI] [PubMed] [Google Scholar]
- Larson SB, Day J, Barba de la Rosa AP, Keen NT, McPherson A. (2003) First crystallographic structure of a xylanase from glycoside hydrolase family 5: implications for catalysis. Biochemistry 42: 8411–8422 [DOI] [PubMed] [Google Scholar]
- Lehtio J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. (2003) The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci USA 100: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wightman R, Geshi N, Turner SR, Sheller HV. (2010) Arabidopsis: a powerful model system for plant cell wall research. Plant J 61: 1107–1121 [DOI] [PubMed] [Google Scholar]
- Linder M, Teeri TT. (1997) The roles and function of cellulose-binding domains. J Biotechnol 57: 15–28 [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Guerreiro CI, Baumann MJ, Taylor EJ, Prates JA, Ferreira LM, Fontes CM, Brumer H, Davies GJ. (2006) Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem 281: 24922–24933 [DOI] [PubMed] [Google Scholar]
- Master ER, Rudsander UJ, Zhou W, Henriksson H, Divne C, Denman S, Wilson DB, Teeri TT. (2004) Recombinant expression and enzymatic characterization of PttCel9A, a KOR homologue from Populus tremula x tremuloides. Biochemistry 43: 10080–10089 [DOI] [PubMed] [Google Scholar]
- McCartney L, Blake AW, Flint J, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. (2006) Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc Natl Acad Sci USA 103: 4765–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie VA, Vincken JP, Voragen AG, van den Broek LA, Stimson E, Gilbert HJ. (2001) A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochem J 355: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. (2004) Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J Biol Chem 279: 44907–44914 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Mohnen D, Bar-Peled M, Somerville C. (2008) Biosynthesis of plant cell walls. Himmel ME, , Biomass Recalcitrance. Blackwell Publishing, Oxford, pp 94–187 [Google Scholar]
- Montanier C, Money VA, Pires VM, Flint JE, Pinheiro BA, Goyal A, Prates JA, Izumi A, Stalbrand H, Morland C, et al. (2009a) The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol 7: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanier C, van Bueren AL, Dumon C, Flint JE, Correia MA, Prates JA, Firbank SJ, Lewis RJ, Grondin GG, Ghinet MG, et al. (2009b) Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci USA 106: 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser F, Irwin D, Chen SL, Wilson DB. (2008) Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol Bioeng 100: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Najmudin S, Guerreiro CI, Carvalho AL, Prates JA, Correia MA, Alves VD, Ferreira LM, Romao MJ, Gilbert HJ, Bolam DN, et al. (2006) Xyloglucan is recognized by carbohydrate-binding modules that interact with beta-glucan chains. J Biol Chem 281: 8815–8828 [DOI] [PubMed] [Google Scholar]
- Notenboom V, Boraston AB, Kilburn DG, Rose DR. (2001) Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 40: 6248–6256 [DOI] [PubMed] [Google Scholar]
- Nurizzo D, Nagy T, Gilbert HJ, Davies GJ. (2002) The structural basis for catalysis and specificity of the Pseudomonas cellulosa alpha-glucuronidase, GlcA67A. Structure 10: 547–556 [DOI] [PubMed] [Google Scholar]
- Ochiai A, Itoh T, Mikami B, Hashimoto W, Murata K. (2009) Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases. J Biol Chem 284: 10181–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Ishii T, Albersheim P, Darvill AG. (2004) Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol 55: 109–139 [DOI] [PubMed] [Google Scholar]
- Pell G, Taylor EJ, Gloster TM, Turkenburg JP, Fontes CM, Ferreira LM, Nagy T, Clark SJ, Davies GJ, Gilbert HJ. (2004) The mechanisms by which family 10 glycoside hydrolases bind decorated substrates. J Biol Chem 279: 9597–9605 [DOI] [PubMed] [Google Scholar]
- Pena MJ, Zhong R, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill R, Jenkins J, Harris G, Nasser W, Robert-Baudouy J. (1994) The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Biol 1: 717–723 [DOI] [PubMed] [Google Scholar]
- Pires VM, Henshaw JL, Prates JA, Bolam DN, Ferreira LM, Fontes CM, Henrissat B, Planas A, Gilbert HJ, Czjzek M. (2004) The crystal structure of the family 6 carbohydrate binding module from Cellvibrio mixtus endoglucanase 5a in complex with oligosaccharides reveals two distinct binding sites with different ligand specificities. J Biol Chem 279: 21560–21568 [DOI] [PubMed] [Google Scholar]
- Proctor MR, Taylor EJ, Nurizzo D, Turkenburg JP, Lloyd RM, Vardakou M, Davies GJ, Gilbert HJ. (2005) Tailored catalysts for plant cell-wall degradation: redesigning the exo/endo preference of Cellvibrio japonicus arabinanase 43A. Proc Natl Acad Sci USA 102: 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama S, Simpson PJ, Szabo L, Nagy T, Gilbert HJ, Williamson MP. (2000) Solution structure of the CBM10 cellulose binding module from Pseudomonas xylanase A. Biochemistry 39: 978–984 [DOI] [PubMed] [Google Scholar]
- Rosgaard L, Pedersen S, Cherry JR, Harris P, Meyer AS. (2006) Efficiency of new fungal cellulase systems in boosting enzymatic degradation of barley straw lignocellulose. Biotechnol Prog 22: 493–498 [DOI] [PubMed] [Google Scholar]
- Ryabova O, Vrsanska M, Kaneko S, van Zyl WH, Biely P. (2009) A novel family of hemicellulolytic alpha-glucuronidase. FEBS Lett 583: 1457–1462 [DOI] [PubMed] [Google Scholar]
- Rye CS, Withers SG. (2000) Glycosidase mechanisms. Curr Opin Chem Biol 4: 573–580 [DOI] [PubMed] [Google Scholar]
- Schubot FD, Kataeva IA, Blum DL, Shah AK, Ljungdahl LG, Rose JP, Wang BC. (2001) Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum. Biochemistry 40: 12524–12532 [DOI] [PubMed] [Google Scholar]
- Seyedarabi A, To TT, Ali S, Hussain S, Fries M, Madsen R, Clausen MH, Teixteira S, Brocklehurst K, Pickersgill RW. (2010) Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase. Biochemistry 49: 539–546 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Bolam DN, Cooper A, Ciruela A, Hazlewood GP, Gilbert HJ, Williamson MP. (1999) A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure 7: 853–864 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Jamieson SJ, Abou-Hachem M, Karlsson EN, Gilbert HJ, Holst O, Williamson MP. (2002) The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 41: 5712–5719 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Xie H, Bolam DN, Gilbert HJ, Williamson MP. (2000) The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J Biol Chem 275: 41137–41142 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Sorensen HR, Jorgensen CT, Hansen CH, Jorgensen CI, Pedersen S, Meyer AS. (2006) A novel GH43 alpha-L-arabinofuranosidase from Humicola insolens: mode of action and synergy with GH51 alpha-L-arabinofuranosidases on wheat arabinoxylan. Appl Microbiol Biotechnol 73: 850–861 [DOI] [PubMed] [Google Scholar]
- Southall SM, Simpson PJ, Gilbert HJ, Williamson G, Williamson MP. (1999) The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett 447: 58–60 [DOI] [PubMed] [Google Scholar]
- Sticklen MB. (2008) Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet 9: 433–443 [DOI] [PubMed] [Google Scholar]
- St John FJ, Godwin DK, Preston JF, Pozharski E, Hurlbert JC. (2009) Crystallization and crystallographic analysis of Bacillus subtilis xylanase C. Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Ducros VM, Flint JE, Roberts SM, Morland C, Zechel DL, Smith N, Bjornvad ME, Borchert TV, Wilson KS, et al. (2009) Understanding how diverse beta-mannanases recognize heterogeneous substrates. Biochemistry 48: 7009–7018 [DOI] [PubMed] [Google Scholar]
- Tailford LE, Money VA, Smith NL, Dumon C, Davies GJ, Gilbert HJ. (2007) Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the beta-mannosidase, BtMan2A. J Biol Chem 282: 11291–11299 [DOI] [PubMed] [Google Scholar]
- Taylor EJ, Gloster TM, Turkenburg JP, Vincent F, Brzozowski AM, Dupont C, Shareck F, Centeno MS, Prates JA, Puchart V, et al. (2006) Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J Biol Chem 281: 10968–10975 [DOI] [PubMed] [Google Scholar]
- Teeri TT. (1997) Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol 15: 160–167 [Google Scholar]
- Tolonen AC, Chilaka AC, Church GM. (2009) Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Mol Microbiol 74: 1300–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, Shoham Y, Steitz TA. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J 15: 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe RB, Bolam DN, Pell G, Gilbert HJ, Williamson MP. (2005) Structure of a mannan-specific family 35 carbohydrate-binding module: evidence for significant conformational changes upon ligand binding. J Mol Biol 347: 287–296 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Catala C, Irwin D, Wilson DB, Ripoll DR, Rose JK. (2007) A tomato endo-beta-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). J Biol Chem 282: 12066–12074 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G, Horn SJ, van Aalten DM, Synstad B, Eijsink VG. (2005a) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem 280: 28492–28497 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G, Houston DR, Riemen AH, Eijsink VG, van Aalten DM. (2005b) Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J Biol Chem 280: 11313–11319 [DOI] [PubMed] [Google Scholar]
- van den Broek LA, Lloyd RM, Beldman G, Verdoes JC, McCleary BV, Voragen AG. (2005) Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol 67: 641–647 [DOI] [PubMed] [Google Scholar]
- Vanden Wymelenberg A, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D, Cullen D. (2009) Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol 75: 4058–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermarliere E, Bourgois TM, Winn MD, van Campenhout S, Volckaert G, Delcour JA, Strelkov SV, Rabijns A, Courtin CM. (2009) Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family. Biochem J 418: 39–47 [DOI] [PubMed] [Google Scholar]
- Vardakou M, Flint J, Christakopoulos P, Lewis RJ, Gilbert HJ, Murray JW. (2005) A family 10 Thermoascus aurantiacus xylanase utilizes arabinose decorations of xylan as significant substrate specificity determinants. J Mol Biol 352: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Varrot A, Schulein M, Davies GJ. (1999) Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry 38: 8884–8891 [DOI] [PubMed] [Google Scholar]
- Vincent F, Molin DD, Weiner RM, Bourne Y, Henrissat B. (2010) Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown. FEBS Lett 584: 1577–1584 [DOI] [PubMed] [Google Scholar]
- Vlasenko E, Schulein M, Cherry JR, Xu F. (2010) Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour Technol 101: 2405–2411 [DOI] [PubMed] [Google Scholar]
- Vrsanska M, Kolenova K, Puchart V, Biely P. (2007) Mode of action of glycoside hydrolase family 5 glucuronoxylan xylanohydrolase from Erwinia chrysanthemi. FEBS J 274: 1666–1677 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang Y, Gao P. (2008) A novel function for the cellulose binding module of cellobiohydrolase I. Sci China C Life Sci 51: 620–629 [DOI] [PubMed] [Google Scholar]
- Weiner RM, Taylor LE, II, Henrissat B, Hauser L, Land M, Coutinho PM, Rancurel C, Saunders EH, Longmire AG, Zhang H, et al. (2008) Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T. PLoS Genet 4: e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, et al. (2007) Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol 73: 3536–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GY, Ong E, Gilkes NR, Kilburn DG, Muhandiram DR, Harris-Brandts M, Carver JP, Kay LE, Harvey TS. (1995) Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 34: 6993–7009 [PubMed] [Google Scholar]
- Yao Q, Sun TT, Liu WF, Chen GJ. (2008) Gene cloning and heterologous expression of a novel endoglucanase, swollenin, from Trichoderma pseudokoningii S38. Biosci Biotechnol Biochem 72: 2799–2805 [DOI] [PubMed] [Google Scholar]