Abstract
Herein, we sought to explore the contribution of cellulose biosynthesis to the shape and morphogenesis of hexagonal seed coat cells in Arabidopsis (Arabidopsis thaliana). Consistent with seed preferential expression of CELLULOSE SYNTHASE9 (CESA9), null mutations in CESA9 caused no change in cellulose content in leaves or stems, but caused a 25% reduction in seeds. Compositional studies of cesa9 seeds uncovered substantial proportional increases in cell wall neutral sugars and in several monomers of cell wall-associated polyesters. Despite these metabolic compensations, cesa9 seeds were permeable to tetrazolium salt, implying that cellulose biosynthesis, via CESA9, is required for correct barrier function of the seed coat. A syndrome of depleted radial wall, altered seed coat cell size, shape, and internal angle uniformity was quantified using scanning electron micrographs in cesa9 epidermal cells. By contrast, morphological defects were absent in cesa9 embryos, visually inspected from torpedo to bent cotyledon, consistent with no reduction in postgermination radical or hypocotyl elongation. These data implied that CESA9 was seed coat specific or functionally redundant in other tissues. Assessment of sections from glutaraldehyde fixed wild-type and cesa9 mature seeds supported results of scanning electron micrographs and quantitatively showed depletion of secondary cell wall synthesis in the radial cell wall. Herein, we show a nonredundant role for CESA9 in secondary cell wall biosynthesis in radial cell walls of epidermal seed coats and document its importance for cell morphogenesis and barrier function of the seed coat.
Perhaps one of the most important reasons for the successful radiation of land plants into the many diverse and extreme environments of our planet can be found in the evolution of seeds (Lidgard and Crane, 1988; Knapp et al., 2005). At the heart of this evolutionary step, from spore-mediated reproduction to seed-mediated reproduction (Holsinger, 2000), is the mechanistic structure of the seed. In a simple model, the seed is categorized into three components, the embryo, the endosperm, and the seed coat (testa; Fahn, 1990). With respect to the angiosperm testa, this portion of the seed consists of several layers of specialized tissues that are maternally inherited and differentiated from cells of the ovule integuments following fertilization (Vaughan and Whitehouse, 1971; Corner, 1976; Sagasser et al., 2002). Comprising the outermost cell layers of the seed, the testa is uniquely positioned at the interface between the embryo and the external environment and thus has evolved as a dynamic and specialized structure capable of protecting the embryo from environmental insults such as desiccation, mechanical stress, pathogen attack, and UV damage (Windsor et al., 2000; Haughn and Chaudhury, 2005). For instance, there are numerous dispersal mechanisms that, whether mediated by animals, wind, or water, all require specific adaptations of the seed coat (Howe and Smallwood, 1982). The testa cells also play a major role in maintaining the dehydrated dormant state of the embryo until appropriate conditions exist (Windsor et al., 2000). A good example of the highly specialized role of testa cells is found in the epidermal seed coat layer of cotton (Gossypium hirsutum) from which the economically important cotton trichome or fiber is produced (Kim and Triplett, 2001). The visual appearance of the testa is also frequently used for taxonomic descriptions and to distinguish between closely related plant species (Rodin and Kapil, 1969; Chuang and Heckard, 1972).
Seed development in Arabidopsis (Arabidopsis thaliana; fruiting body are dehiscent siliques) has received substantial scientific scrutiny, and seed coats have been cytologically (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000) and genetically (Reiser and Fischer, 1993; Klucher et al., 1996; Western et al., 2001, 2004; Dean et al., 2007; Arsovski et al., 2009a, 2009b) studied. The epidermal cells of the testa in Arabidopsis form a hexameric matrix. Each epidermal testa cell also has a central columella (volcano shaped) separated from the radial wall by a mucilage pocket (Haughn and Chaudhury, 2005). Beneath the surface view of the hexagon cells, histological studies have revealed an elegant developmental strategy whereby mucilage is deposited in the apoplast at the junction of the outer tangential and radial walls gradually forming the mucilage pocket (Western et al., 2000, 2001). Subsequently, a thick secondary cell wall is deposited along the radial boundary wall and the outer tangential wall forming the volcano-shaped columella. Upon imbibition, the pectinaceuous mucilage swells, ruptures the primary radial wall and extrudes from the seed coat, and germination begins (Gutterman and Shemtov, 1996; Western et al., 2000, 2001). The formation of the secondary cell wall includes reinforcement of the radial wall that assists in forming the unique hexagonal shapes of testa epidermal cells. This radial wall must inherently possess substantial biomechanical strength. The major load-bearing constituent of higher plant cell walls is cellulose, a polymer of 1,4-β-d-Glc residues (Brown et al., 1996). However, the molecular underpinning of seed coat reinforcement remains poorly characterized, particularly with respect to the contribution of cellulose biosynthesis.
The overarching goal of this study sought to explore cellulose deposition in the uniquely hexagonally shaped epidermal seed coat (testa) cells. In this study, a reverse genetic approach in Arabidopsis was used to show that CELLULOSE SYNTHASE9 (CESA9), which was previously proposed to be a redundant component of cellulose biosynthesis, is central to the formation of the secondary wall in this cell type. With no change observed in embryogenesis, these results inferred intriguing cell type specificity for cellulose biosynthesis in the seed.
RESULTS
Gene Expression Analysis for CESA9
CESA9 encodes a 1,088-amino acid protein and comprises 12 introns and 13 exons (Richmond, 2000). Gene expression (mRNA transcript abundance) of CESA9 was interrogated using GENEINVESTIGATOR expression profiling tool (Zimmermann et al., 2004). CESA9 gene expression was highest during fruit development, specifically, after stage 3 of seed development. CESA9 expression increased and peaked between stage 5 and stage 9 of seed development (data not shown; see GENEINVESTIGATOR output). Expression was low in rapidly elongating tissue such as hypocotyls or roots. Consistent with these data, coexpression analysis (www.atted.bio.titech.ac.jp; Obayashi et al., 2009) using CESA9 as bait did not reveal coexpression with any other primary or secondary cell wall genes (Supplemental Fig. S1). Contrastingly, genes associated with both primary and secondary cell wall cellulose biosynthesis have previously been shown to cluster tightly together (Brown et al., 2005; Persson et al., 2005). For example, coexpression analysis performed using CESA3 as bait identified CESA1, CESA2, CESA6, COBRA, and KORRIGAN all following a tight transcriptional coexpression pattern (Supplemental Fig. S1), consistent with Persson et al. (2005). Alternatively, transcripts that are coexpressed with CESA9 included an endoplasmic reticulum lumen protein-retaining receptor family protein (At3g25160), ATOEP16-S protein (At4g16160), a hydrophobic protein responsive to low temperature and salt (At2g38905), two independent Gly-rich proteins/oleosins (At3g18570 and At2g25890), thioredoxin-like2 (At3g14950), Gln synthase (At1g48470), and Suc phosphate synthase (At1g04920). These transcripts have no published association with cellulose biosynthesis. The presence in this cluster of oleosins, which are known to be seed-specific oil-body proteins, show that gene coexpression may be due only to seed-specific transcripts and thus be unrelated to cell wall biosynthesis.
Isolation of T-DNA Mutants for CESA9
Gene expression analyses showed that CESA9 was expressed during fruit development. However, whether CESA9 was expressed in the embryo (Beeckman et al., 2002) or the seed coat was unclear. To address this and explore the role of CESA9 in seed physiology, a reverse genetic approach was taken and two alleles for CESA9 were identified and named cesa9-1 (Persson et al., 2007) and cesa9-2 (Supplemental Fig. S2). Phenotypes described herein were consistent among these alleles. Persson et al. (2007) and Harris et al. (2009) previously reported the isolation of homozygous T-DNA insertion alleles for CESA9 (cesa9-1; At2g21770) and in neither case was the phenotype of the seed examined and reported. Plants homozygous for cesa9-1 and cesa9-2 alleles were viable and displayed no growth feature differences in the mature plant, consistent with previous reports (Persson et al., 2007; Fig. 1A). More specifically, when grown at 22°C, the growth habit of the cesa9 seedlings and mature plants showed no substantial radial swelling or dwarfing phenotypes as was expected for a cellulose-deficient mutant (Sugimoto et al., 2001).
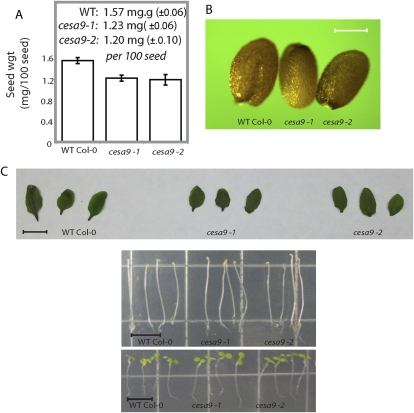
Figure 1.
Examination of cesa9-1 and cesa9-2 plants imply that phenotypes are restricted to the seed. A, Analysis of seed weight was performed on 100 individual seed. Error bars are se of three replicates for two alleles (P > 0.01 ANOVA). B, Light microscopy was used to image wild-type (WT), cesa9-1, and cesa9-2 seed (scale bar = 100 μm). C, Phenotypic examination of rosette leaves (scale bar = 1 cm), dark-, and light-grown seedlings (scale bar = 7 mm) comparing cesa9-1 and cesa9-2 with wild type. Col-0, Columbia-0. [See online article for color version of this figure.]
Phenotypic Analysis of cesa9 Plants
Quantitative (Fig. 1A) and visual inspection (Fig. 1B) revealed that cesa9 seeds were smaller in size than wild-type seed. Specifically, seed weights were 1.57 mg 100 seed−1 (±6) in the wild type relative to 1.23 mg 100 seed−1 (±6) cesa9-1 and 1.20 mg 100 seed−1 (±10) cesa9-2 (Fig. 1A). Since cesa9 displayed a dramatic seed size phenotype, we asked whether morphogenesis was affected during embryogenesis. Embryos were dissected from cesa9 and wild-type ovules, and the embryonic stages of development were compared. The early stages of cesa9 mutant development from globular to heart stage (Supplemental Fig. S3; Supplemental Materials and Methods S1) exhibited no visible phenotypes relative to wild type, which was consistent to mature embryo stage (data not shown). No differences were observed in elongating seedlings (light or dark grown) and rosette leaf size between wild-type and the cesa9-1 and cesa9-2 alleles. Altered seed coat cell morphology was observed when performing differential interference contrast microscopy (Nomarski optics; Fig. 2A) and incomplete clearing of testa pigment by the Hoyer's solution revealed that the ultrastructure of cesa9 was different to wild-type cells. In particular, the cell shape of cesa9 appeared less uniform than that of wild-type seed coats (Fig. 2A).
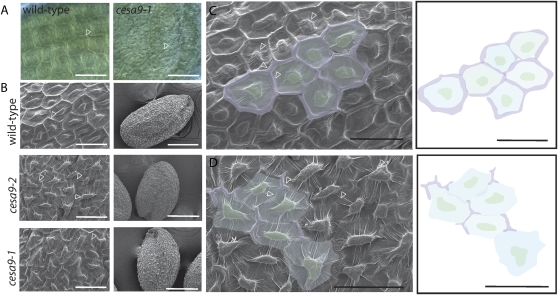
Figure 2.
Mutations in CESA9 cause distorted cell shape and morphogenesis in seed coat epidermal cells. A, Nomarski optics were used to visualize the epidermal cells of seed excised from siliques at the 14th silique posterior to gynoecium protrusion (scale bar = 50 μm). B, SEMs of seed coat epidermal cells in mature wild-type, cesa9-1, and cesa9-2 seed (scale bar = 50 μm for high magnification [left sections] and 250 μm for image of entire seed [right sections]). C, SEM of wild-type epidermal seed coat (scale bar = 50 μm). Adjacent section is a schematic illustration of the cell boundary (purple), trough (blue), and columella (green; scale bar = 50 μm). D, SEM of cesa9 epidermal seed coat (scale bar = 50 μm). Adjacent section is a schematic illustration of the cell boundary (purple), trough (blue), and columella (green; scale bar = 50 μm). Arrowheads in C and D indicate radial cell wall. [See online article for color version of this figure.]
To gain further insight into the defective cell shape phenotype, examination by scanning electron microscopy (SEM) was performed on cesa9-1 and cesa9-2 (Fig. 2B). Epidermal cell morphology was severely distorted among null cesa9 seeds relative to the uniform cell shape in wild-type seeds (Fig. 2B). In mature cesa9 seed imaged by SEM the radial cell wall was either thinner or not observable at all beneath the outer tangential cell wall draping over the columella (Fig. 2, C and D), suggesting that collapse in radial cell walls led to an indistinguishable border between neighboring cells and the appearance of cell fusion. Highly irregular shape was also observable in cesa9 by SEM relative to uniform cell hexagonal shapes in wild type. Hence, loss of uniform cell shape in the mature seed was established in cesa9.
Quantitative SEM Study of Ablated Epidermal Cell Shape and Morphogenesis in cesa9 Seed
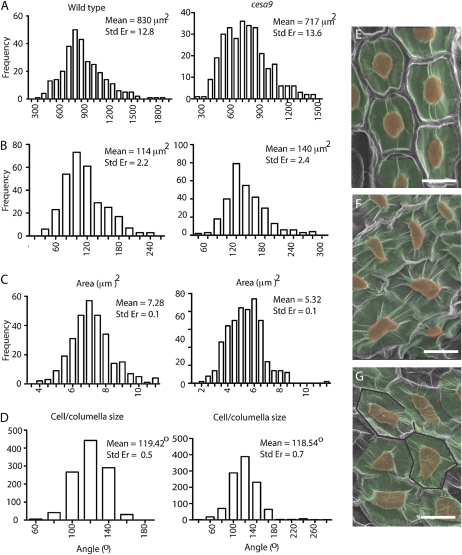
To further distinguish altered shape parameters in the cesa9 seed coat epidermal cells, we quantitatively analyzed cell area (Fig. 3A), the area of columella (Fig. 3B), cell area/columella area (Fig. 3C), and internal angles between borders (Fig. 3D) using frequency distribution comparisons between wild-type (Fig. 3E) and cesa9 (Fig. 3, F and G) cells. Comparisons between cesa9 and wild-type cell area used a minimum population of 10 different seeds from each genetic background and 30 cells from each seed were selected for measurement (n = 300; Fig. 3A). A region of cells, in the middle of a seed that did not extend to the border regions, was selected to promote comparison with planar uniformity. Results showed that the mean cell area in wild-type cells was 830 μm2 (se of 12.8 μm2) relative to 717 μm2 (se of 13.6 μm2) in cesa9 (P > 0.05, Wilcoxan ranked signed test; Fig. 3), demonstrating a smaller overall cell size among epidermal testa cells in cesa9. In contrast to total cell area, columella area was measurably greater in the cesa9 (n = 300; 140 μm2; se of 2.4 μm2) relative to 114 μm2 in wild type (n = 300; se of 2.2 μm2; Fig. 3B). Cell area/columella area was therefore greater in the wild-type testa cells and had a ratio of 7.28 whereas the ratio for cesa9 was 5.32 (Fig. 3C). However, the number did not provide a sufficient measure of the obvious nonuniformity seen in the cesa9 cells relative to wild type. In an effort to obtain such an assessment, we determined the internal angle between the sides of the hexagonally shaped wild-type cells with those of cesa9 (Fig. 3D). In wild-type cells, the average internal angle was 119.4°C measured over 30 different seeds and composed of 1,086 independent angle measurements. Not surprisingly, given the geometric constraints, the internal angles in cesa9 cells also averaged out to 118.5°C (n = 1,080); however, the range of angles observed in the cesa9 was substantially greater than wild type. For instance, the minimum internal angle was 43°C and 64°C, respectively, and the maximum internal angle was 257°C and 162°C, respectively (Fig. 3D). Internal angles ranging from 43°C to 257°C are consistent with careful visual inspection that showed highly irregular and inconsistent shapes occurring between neighboring cells in cesa9 (Fig. 3, E and G).
Figure 3.
Quantitative analysis of cell morphogenesis using SEM. A, Epidermal cell area (μm2) in seeds from cesa9-1 and wild type shows a greater variation in cell area in cesa9 relative to wild-type seed. Area measurements on 300 cells in 10 different seeds displaying significantly smaller cell size. B, Columella area measurements of the cesa9 mutant columella relative to wild type (n = 300). C, Analysis of columella area relative to the cell size. Columella area was approximately 14% of the wild-type cell area, and 20% of the cesa9 mutant. D, Analysis of the internal angles between the cell sides (n = 1,080). Statistical significance of distribution shifts was calculated by using the Wilcoxon rank sum test for cesa9 relative to wild type (P < 0.01). Wild-type and cesa9 cell area (P > 0.001), wild-type and cesa9 columella area (P > 0.008), wild-type and cesa9 cell area/columella area (P > 0.001), and wild-type and cesa9 internal angle (P > 0.5). E and F, Provides an example of morphological variation between wild-type and cesa9 epidermal testa cells (scale bars = 30 μm). Std Er, Standard error. [See online article for color version of this figure.]
Analysis of the Structure and Development of Wild-Type and cesa9 Seed Coats
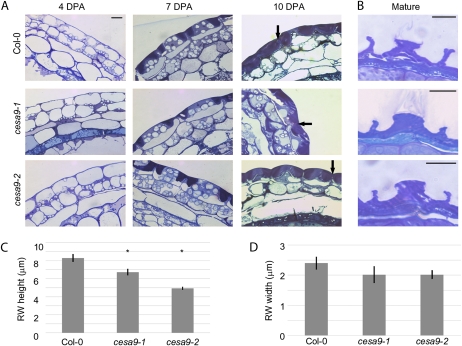
The surface SEM data suggest that the cell wall structure of the epidermal cells are altered in the cesa9 mutant. To investigate this hypothesis further, developmental analysis was performed on toluidine blue-stained sections of wild-type and cesa9 seed coat cells cryofixed at 4, 7, and 10 DPA (Fig. 4A). No difference was observed between wild-type and mutant genotypes prior to secondary cell wall deposition, at 4 and 7 DPA. At 10 DPA, a secondary cell wall deposited along the membrane of the radial and outer tangential sides of the cell membrane can be observed. Careful visual examination suggested that less secondary cell wall material is present below the mucilage pocket and along the radial wall of cesa9 mutants compared to wild type (Fig. 4A, see arrows). To better examine this aspect of the phenotype, mature and wild-type cesa9 seeds were fixed under aqueous conditions (3% glutaraldehyde), causing the mucilage pocket to burst and allowing a clear picture of the remaining epidermal secondary wall material in mature seeds. The height and width of the mutant radial wall, although variable, appeared smaller than in wild type (Fig. 4B). To quantify these changes, the radial wall height and width were measured from a total of 10 cells for each genotype. The results (Fig. 4C) indicate significant differences in the height of the radial walls when compared to wild-type cells. Although a decrease in radial wall width was also observed the differences were not significant (Fig. 4D).
Figure 4.
Analysis of the structure and development of wild-type and cesa9 seed coats. A, Epidermal cell morphology of wild-type and cesa9 toluidine blue-stained sections of cryrofixed 4 DPA, 7 DPA, and 10 DPA seeds. Arrows on 10 DPA images indicate the location of the radial wall where secondary cell wall synthesis is occurring (scale bar = 10 mm, all images same magnification). B, Epidermal cell morphology of wild-type and cesa9 toluidine blue-stained sections of aqueous (3% v/v) glutaraldehyde fixed mature seeds (scale bar = 10 mm). C, Average height of the radial wall (RW) of wild-type and cesa9 seed coat cells (mm). D, Average width of the radial wall of wild-type and cesa9 seed coat cells (mm): error bars are se from the mean. The asterisk (*) in C indicated significant difference from wild type based on one-way ANOVA at P > 0.05 (Student's t test). Col-0, Columbia-0. [See online article for color version of this figure.]
Histological and Compositional Studies of cesa9 Seed
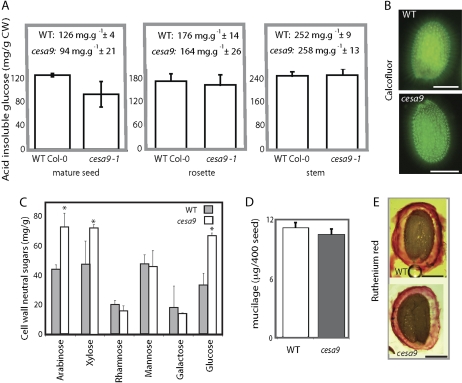
The relative amounts of acid-insoluble Glc (relative estimate of crystalline cellulose) were examined in seed, stems, and leaves of cesa9-1 and wild type (Fig. 5A). Total acid-insoluble Glc of the cesa9 mutant was on average 94 mg g−1 (±21) versus 126 mg g−1 (±4) in the wild-type seed, representing a 25.4% reduction. Further measurements indicated no change in acid-insoluble Glc content in stems or leaves, consistent with no difference in radical and hypocotyl emergence and rosette leaf morphology (Fig. 1C). Staining with Calcofluor, a fluorescent stain for β-glycans including cellulose (Windsor et al., 2000; Willats et al., 2001; Macquet et al., 2007), allowed the observation of the mucilage strands extending laterally from the seed coat epidermis (Fig. 5B), consistent with previous reports (Macquet et al., 2007). The main difference evident from Calcofluor staining was that cesa9-1 mucilage strands arose from the imbibed seed at slightly irregular angles and were more diffuse in its distribution relative to wild type (Fig. 5B). To further explore biosynthetic feedback into other cell wall polysaccharides, such as that identified by Burton et al. (2000), we examined the content of Ara, Xyl, Man, Glc, Rha, and Gal in mature seed. Starch was removed from the seed by washing with acetone and then treating the raw seed biomass with amylase enzyme (Sigma Aldrich) prior to derivatization of the neutral sugars and analysis by gas chromatography (GC). Results showed that cesa9 seed contained significantly more Ara (44 mg g−1 wild type versus 74 mg g−1 cesa9), Glc (36 mg g−1 wild type versus 67 mg g−1 cesa9), and Xyl (48 mg g−1 wild type versus 72 mg g−1 cesa9) relative to wild type. Man (49 mg g−1 wild type versus 46 mg g−1 cesa9), Rha (21 mg g−1 wild type versus 17 mg g−1 cesa9), and Gal (18 mg g−1 wild type versus 15 mg g−1 cesa9) were not different in the mutant background relative to wild type (Fig. 5C). Comparing cell wall neutral sugars as a total of cell wall biomass documented that cesa9 had increased neutral sugar content (48.8% of cell wall biomass) relative to wild type (34.8%; Fig. 5C). An important component of epidermal seed coat cells is mucilage (Western et al., 2000). Therefore we quantitatively determined mucilage content by isolating and weighing the mucilage from 40-mg batches of seed (n = 3) and found that cesa9 seed contained 1.16 mg mucilage 40 mg seed−1 whereas wild-type seed contained 1.08 mg mucilage 40 mg seed−1 (Fig. 5D). Histological staining of mucilage using ruthenium red further confirmed mucilage synthesis and extrusion was unaffected in cesa9 (Fig. 5E).
Figure 5.
Composition of wild-type (WT) and cesa9 seed cell walls. A, Acid-insoluble crystalline cellulose content of various tissues was determined colorometrically for mature seed, leaves, and stems. Error bars are ses of three technical replicates from three independent batches of seed as biological replicates. CW, Cell wall. B, Calcofluor staining and subsequent illumination with UV light (scale bar = 250 μm). C, GC analysis of cell wall neutral sugars. Error bars are se of three replicates. D, Mucilage weights determined for wild-type and cesa9 seed, error bars are se of three replicates. E, Ruthenium red-stained seed visualized by light microscopy (scale bar = 200 μm). Col-0, Columbia-0. [See online article for color version of this figure.]
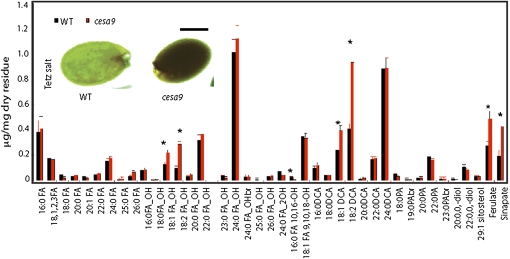
When seeds were incubated in a solution of tetrazolium salts (Debeaujon et al., 2001), cesa9 seeds were found to be more sensitive to salt uptake than wild-type seed (Fig. 6, inset). To determine whether the defect in cellulose synthesis resulted in cell wall-associated polyesters in the seed coat, we analyzed the composition of lipid polyester monomers arising from whole seeds of the cesa9 mutant compared with wild-type seed. The data show that several polyester monomers such as 16:0 fatty acid (FA), 10:16-diOH FA, 25:0 ω-OH FA, 24:0 2-OH FA, and 23:0 ω-OH FA were reduced (Fig. 6). However, a net increase in total polyester monomers was observed due to an increase in some aliphatics (18:1 dicarboxylic acid [DCA], 18:2 DCA, 18:1 and 18:2 ω-hydroxy FAs) as well as the two ester-bound aromatics (ferulate and sinapate). These data suggest that ablated cellulose, via the action of CESA9, resulted in altered polyester monomer composition and organization in the seed coat.
Figure 6.
Polyester and aliphatic monomer composition for wild-type (WT) and cesa9 seed. GC and GC-MS analysis of mature cesa9 seed assessed lipid polyester monomers from seeds of wild-type and cesa9 plants. The insoluble dry residue obtained after grinding and delipidation of tissues with organic solvents was depolymerized by acid-catalyzed methanolysis and aliphatic and aromatic monomers released were analyzed by GC-MS. Error bars are the se of four replicates. Black bar is wild type and red bar is cesa9. PAs, Primary alcohols; br., branched. Inset documents tetrazolium salt uptake into the cesa9 seed relative to wild type (scale bar = 250 μm). [See online article for color version of this figure.]
DISCUSSION
The process of testa cell development in Arabidopsis has been elegantly dissected microscopically (Beeckman et al., 2000; Western et al., 2000, 2001; Haughn and Chaudhury, 2005), revealing a complex process whereby mucilage is secreted and sequestered in the apoplast between the primary cell wall and the plasma membrane at the junction of the radial and outer tangential cell walls. Concurrent to mucilage production, the cytoplasm and starch granules of testa cells are shaped into a column in the center of the cell (Western et al., 2000). Following mucilage secretion, an elaborate secondary cell wall is produced around the cytoplasmic column forming the columella, and along the radial cell wall (Western et al., 2000; Windsor et al., 2000). During the final step of seed maturation, “the mucilage and outer wall dehydrate leaving the columella and radial walls visible as the epidermal plateau and reticulations visible on the mature seed coat” (Windsor et al., 2000, p. 483). Analysis of developing seed by both SEM and sectioning showed that the radial wall was present during early developmental stages in cesa9 (Fig. 4) but failed to show secondary cell wall thickening relative to wild type during the latter stages of development (Fig. 4). For instance, careful examination of cross-sectioned seed coat cells stained with toluidine blue showed that the secondary cell wall of wild-type cells extended up the entire radial wall whereas in cesa9 the secondary radial wall was less developed. Quantitative analysis of the height of radial walls in cross-sectioned tissue clearly demonstrated a significant reduction in radial wall height. Therefore, it is likely that after cesa9 seed matures and dehydrates, the unreinforced radial cell walls partially collapse due to a lack of cesa9-dependent cellulose biosynthesis, resulting in the appearance that neighboring cells lack radial walls visualized by SEM (Fig. 2). A series of surface defects in cell shape and morphology were quantifiable in the mature cesa9 seed coat cells, such as cell area, columella area, and internal angle uniformity (Fig. 3). Given that the function of cellulose biosynthesis is to provide rigidity to the cell wall, one plausible explanation for the changes in cell morphogenesis is that the constraint of internal turgor pressure is disturbed. This may explain the collapsed cell boundary phenotype, irregular internal angles, and altered cell and columella areas (Figs. 2 and 3). Such a scenario has previously been documented for other tissue types (Burton et al., 2000; Desprez et al., 2007).
Previous studies have shown that where cellulose biosynthesis is inhibited in either primary cell wall or secondary cell wall xylem thickening, compensation by other cell wall polymers, such as hemicellulose, attempt to overcome structural weakness (Turner and Somerville, 1997; Burton et al., 2000; Cano-Delgado et al., 2003; Bosca et al., 2006; Taylor, 2008). Evidence for proportional increase in cell wall polymers other than cellulose were also observed in cesa9 seed on a total cell wall basis (Fig. 5C). Noncellulosic cell wall polysaccharides (neutral sugars) and aliphatic monomers and some polyesters (Fig. 6) such as 18:1 DCA, 18:2 DCA, 18:1 and 18:2 FAs, ferulate, and sinapate were proportionally more abundant in cesa9 relative to wild type. Despite increased biosynthesis of noncellulosic cell wall polymers, the uptake of tetrazolium salts could not be prevented (Fig. 6, inset), suggesting that the role of the seed coat in osmoprotection (boundary function) was compromised. Furthermore, the up-regulation in noncellulosic cell wall polymers could not rescue cell shape and morphogenesis, supporting a nonredundant role for regulated cellulose biosynthesis, via CESA9, in these tissues.
Given the paucity of cell type specificity for CESA subunits, results presented herein for CESA9 provide important evidence for specific requirements of secondary cell wall biosynthesis in epidermal seed coat radial cell walls that are distinct from secondary xylem thickening and primary cell wall cellulose synthesis. A nonredundant role for CESA9 in secondary cell wall thickening in seed coat cells (Figs. 4–6) documents a previously uncharacterized complexity in cellulose biosynthesis. Prior studies conclude that at least three distinct protein-protein interactions constitute the planar CESA rosette formation (Taylor et al., 2003; Desprez et al., 2007; Persson et al., 2007), which was recently demonstrated for the secondary cell wall using yeast (Saccharomyces cerevisiae) two-hybrid interactions in vitro and bimolecular fluorescence complementation in planta (Timmers et al., 2009). Conceptually, if the hexameric model holds true for seed coat secondary cell wall synthesis, CESA9 must occupy one stoichiometric location in the CESA complex during radial cell wall thickening. Because seed coat cell shape and morphogenesis phenotypes are distorted in cesa9 alleles, there is no evidence to suggest that a different subunit can fulfill its role. A reason for this scenario may be based on the specific cellular requirements of the testa cell layer (Haughn and Chaudhury, 2005), which may not offer the same cellular cues required to compensate for the loss of cesa9, such as up-regulation of CESA2, CESA5, CESA6, or CESA10 as may occur in elongating tissues (Persson et al., 2007). Alternatively, encoded within CESA9 may be subtle differences in amino acid composition that facilitate some of the unique features of cellulose deposition in this hexagonally shaped cell type. Indeed, the contribution of structural macromolecules to highly specialized cell types remains a poorly understood area of plant cell biology and beckons further exploration.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were of the Columbia-0 ecotype. Seeds were surface sterilized using 30% bleach solution and stratified for 3 d in 0.15% agar at 4°C. For phenotypic analysis and growth assays, plants were exposed to light for 1 h and grown in either continuous light (200 mmol m2 s−1) or complete darkness at 22°C on plates containing 0.5× Murashige and Skoog mineral salts (Sigma) and 1% agar.
Identification of T-DNA Insertions in CESA9
PCR-confirmed homozygous alleles carrying exonal T-DNA insertion were as follows: cesa9-1 (SALK_107750C; Persson et al., 2007; Harris et al., 2009) and cesa9-2 (SALK_046455; genotyping presented in Supplemental Fig. S3). These alleles were sourced from the Arabidopsis Biological Resource Center as a fee for service product and identified through The Arabidopsis Information Resource, and genotyped homozygous lines redeposited as a public resource.
Staging of Floral Age
For developmental studies performed using light microscopy, pollination was defined as the time in days when the flower was beginning to open (DPA). At this stage the stamen are beginning to grow over the gynoecium and the pollen is released (anthesis). Petals can be seen extending beyond the tops of the sepals. Flowers at anthesis were marked using water-soluble paint, and a different color was used for each day of marking. By contrast, SEM and Nomarski optics used a different staging regime. Gyneoecium protrusion was defined as the point when the flower was fully open and the elongating gynoecium was clearly elongated beyond the petals and stamen. At this stage, the flower was tagged and each silique posterior to this event counted.
Resin Embedding for Bright-Field Microscopy
Siliques staged at 4, 7, and 10 DPA were removed from the plant and dissected using a razor blade. The seeds were removed and the seed coat was punctured by either an insect pin, or a razor blade. High-pressure freezing, freeze substitution, and resin embedding were performed according to Western et al. (2000). Samples were loaded onto copper hats (Ted Pella) containing 1-hexadecene and frozen under high pressure using a Bal-Tec HPM 010 high-pressure freezer (RMC Products). Copper hats were then transferred to frozen cryovials containing freeze-substitution medium consisting of 2% (w/v) osmium tetroxide in acetone with 8% (v/v) dimethoxypropane. Freeze substitution was performed for 6 d at −80°C by incubation in an electron microscope automatic freeze substitution chamber (Leica), followed by an incubation at −20°C over 20 h to allow for reaction of the fixatives. Samples were then removed from the copper hats and rinsed in anhydrous acetone several times and slowly infiltrated and embedded in Spurr's epoxy resin (Canemco; Spurr, 1969). Alternatively for the analysis of mature seeds, samples were fixed in 3% glutaraldehyde in 0.1 m KH2PO4 buffer pH 7.0 or FAA (4% [v/v] paraformaldehyde [Canemco], 15% [v/v] acetic acid, and 50% [v/v] ethanol), and directly dehydrated without post fixation. All samples were then transferred to a propylene oxide solution and slowly infiltrated with Spurr's epoxy resin (Canemco). Dehydration, embedding, and sectioning were performed as described by Western et al. (2000). Sections were photographed using an Axioskop 2 microscope (Carl Zeiss) and Q Capture pro imaging software (Q imaging). Resin-embedded samples were then thick sectioned (0.5 mm) and stained with 1% (w/v) toluidine blue O in 1× (w/v) sodium borate (pH 11). Sections were examined by light microscopy and evaluated for intact seed coat cells and proper developmental stage based on the morphological criteria described by Western et al. (2000). Intact seed coat cells with a complete columella and radial wall were quantitatively assessed for height and width of the wall. The height of the wall was measured from the bottom of the mucilage pocket to the top of the wall. Width was measured for each radial wall. A total of 10 walls were measured for each genotype. These measurements were made on the 3% glutaraldehyde-fixed samples, because it enabled visual clarity of where the radial wall began and ended. Significance of comparisons was established based on a Student's t test with a two-tailed analysis.
Histochemical Analyses
Tetrazolium Salt Uptake
Assaying the ability of seeds to uptake salt was achieved by placing whole seeds in an aqueous solution of 1% (w/v) tetrazolium violet (2,3,5-triphenyltetrazolium) at 30°C for 4 to 24 h (DeBolt et al., 2009).
Ruthenium Red
A total of 0.03% (w/v) ruthenium red (Sigma) in water for 30 min at 25°C was used to test for aberrant mucilage production according to Beeckman et al. (2000).
Cellulose Staining Using Calcofluor
Seeds were imbibed in 25 μg mL−1 Calcofluor (Sigma-Aldrich) for 15 to 30 min and then observed with an Olympus compound microscope (Olympus Microscopes) equipped with UV illumination (Willats et al., 2001).
Analysis of Seed Lipid Polyesters
Experimental procedures to extract soluble lipids and analyze polyesters of the cell wall residue were previously described in Molina et al. (2008). In brief, for each replicate, 50 to 100 mg mature seeds of wild type and cesa9 were grinded, delipidated, and the dried residue was depolymerized using methanolysis. Released monomers were acetylated or sylilated and then separated, identified, and quantified by GC-mass spectrometry (MS) using a splitless injection. MS was performed using electron impact ionization, and peak quantification achieved on the basis of their total ion current. For additional details on GC-MS analysis see Molina et al. (2008).
Seed Cell Wall Preparation and Analysis
Cellulose content was measured colorimetrically and neutral sugar composition was determined by GC by using 500 mg (dry weight) of ball-milled material as described (Blakeney et al., 1983; Harris et al., 2009). Sugars from mature seed samples were prepared by sequential washing (five times) with 70% ethanol for 45 min at 70°C followed by five sequential acetone washes at room temperature for 2 min each. The main goal was to remove starch from the seed. Samples were then dried under vacuum and 500 mg were then redissolved in 1 mL of α-amylase solution (Sigma Aldrich no. A8220) according to manufacturer's instruction and incubated for 15 min at 37°C followed by being washed and vacuum dried. Exactly 20 mg of the air-dry cell wall material was measured into glass tubes containing 250 μL of 1 m sulfuric acid, vortexed, and autoclaved for 40 min at 120°C (Bioclave). Depolymerized sugars were then neutralized by adding 100 μL of 9 m NH4OH. Samples were dried and redissolved in pyridine with 100 μL 25 mg mL−1 hydroxylamine and incubated at 75°C for 30 min. This was followed by the addition of 100 μL BIS-trimethyl-silyltrifluoroacetamine (Supelco) directly to the glass vial, which was then incubated for 30 min at 75°C. For standards 5 mg of neutral sugars Ara, Rha, Man, Gal, Glc, and myoinositol were measured into a glass vial and the above-mentioned procedure repeated without depolymerization. Neutral sugars were analyzed using GC using a DB-5 column from Agilent (Agilent Technologies) with a length of 30 m, I.D. 0.250, film 0.25 μm (Phenyl Methyl Siloxane). The flow rate used was 1.6 mL/min He. Thermal profile 170°C, ramp 4°C min−1 to 230°C, hold for 10 min with a total run time of 25 min per sample.
Mucilage Content Determination
To determine the ammonium-oxalate soluble mucilage content, methods defined in Arsovski et al. (2009b) were followed whereby 40 mg (dry weight) of Arabidopsis seed were incubated in 0.2% (w/v) of aqueous ammonium-oxalate solution at 37°C for 8 h. To facilitate extraction of mucilage, test tubes were vortexed for 1 min every hour. After incubation the seeds were removed by centrifugation, and 5 volumes of ethanol were added to the aqueous phase. Polysaccharides were allowed to precipitate for 30 min on ice, and then centrifuged in a benchtop centrifuge for 30 min at 21,000g in 2-mL Eppendorf tubes. Precipitated mucilage was washed with 70% ethanol, air dried, and weighed. Averages and ses were calculated based on six mucilage weight replicates for cesa9 and wild-type seeds.
SEM and Image Analysis
Arabidopsis seeds were attached to standard electron microscope stubs, and sputter coated with gold-palladium alloy using Hummer VI sputtering system (Anatech). Specimens were visualized using Hitachi model S-800 scanning electron microscope (Hitachi), images were captured using Evex Nano Analysis digital imaging system. Image analysis was performed using the ImageJ (W. Rasband, National Institute of Health) software. Area measurement for each cell and columella area used area measurement output after tracing the polygon via the freehand selection tool (ImageJ). The angle tool (ImageJ) was used to calculate the internal angles in the hexagonal-shaped seed cells. Data was organized in the spreadsheet program Microsoft Excel and transferred to GraphPad Prism (Prism-4, GraphPad) for generating histograms and performing statistical comparisons.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AC007019.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene expression analysis.
Supplemental Figure S2. PCR genotyping of cesa9-2.
Supplemental Figure S3. Embryo morphology for cesa9 and the wild type.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
The authors acknowledge the access and assistance from the University of Kentucky and the University of British Columbia Electron Microscopy Facilities, and Staffan Persson (Max Planck Institute of Molecular Plant Physiology) for technical assistance. The authors are grateful to the National Science Foundation-funded projects, the Arabidopsis Biological Resource Center, and The Arabidopsis Information Resource community resource infrastructure.
References
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. (2009a) AtBXL1 encodes a bifunctional β-d-xylosidase/alpha-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Villota MM, Rowland O, Subramaniam R, Western TL. (2009b) MUM enhancers are important for seed coat mucilage production and mucilage secretory cell differentiation in Arabidopsis thaliana. J Exp Bot 60: 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inze D. (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inze D, Berleth T. (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeney AB, Harris PJ, Henry RJ, Stone BA. (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299 [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM, Saxena IM, Kudlicka K. (1996) Cellulose biosynthesis in higher plants. Trends Plant Sci 1: 149–156 [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, Baulcombe DC, Fincher GB. (2000) Virus-induced gene silencing of a plant cellulose synthase gene. Plant Cell 12: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M. (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Chuang TI, Heckard LR. (1972) Seed coat morphology in Cordylanthus (Scrophulariaceae) and its taxonomic significance. Am J Bot 59: 258–265 [Google Scholar]
- Corner EJHT. (1976) The Seeds of Dicotyledons. Vols 1 and 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Koornneef M. (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Scheible WR, Schrick K, Schaller H, Beisson F, Bischoff V, Bouvier-Nave P, Li Y, Nair M, Carroll A, et al. (2009) Mutations in UDP glucose:sterol-glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol 151: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn A. (1990) Plant Anatomy, Ed 4 Pergamon Press, Oxford [Google Scholar]
- Gutterman Y, Shemtov S. (1996) Structure and function of the mucilaginous seed coats of Plantago coronopus inhabiting the Negev Desert of Israel. Isr J Plant Sci 44: 125–133 [Google Scholar]
- Harris D, Stork J, DeBolt S. (2009) Genetic modification in cellulose-synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. GCB Bioenergy 1: 51–61 [Google Scholar]
- Haughn G, Chaudhury A. (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Holsinger KE. (2000) Reproductive systems and evolution in vascular plants. Proc Natl Acad Sci USA 97: 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe HF, Smallwood J. (1982) Ecology of seed dispersal. Annu Rev Ecol Syst 13: 201–228 [Google Scholar]
- Kim HJ, Triplett BA. (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M, Stockler K, Havell D, Delsuc F, Sebastiani F, Lockhart PJ. (2005) Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (Southern Beech). PLoS Biol 3: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidgard S, Crane PR. (1988) Quantitative-analyses of the early angiosperm radiation. Nature 331: 344–346 [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM. (2007) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M. (2008) Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J 53: 437–449 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei HR, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Fischer RL. (1993) The ovule and the embryo sac. Plant Cell 5: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. (2000) Higher plant cellulose synthases. Genome Biol 4: 30011–30016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin RJ, Kapil RN. (1969) Comparative anatomy of the seed coats of gnetum and their probable evolution. Am J Bot 56: 420–431 [Google Scholar]
- Sagasser M, Lu GH, Hahlbrock K, Weisshaar B. (2002) A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev 16: 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. (2001) Wall architecture in the cellulose-deficient alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215: 172–183 [DOI] [PubMed] [Google Scholar]
- Taylor NG. (2008) Cellulose biosynthesis and deposition in higher plants. New Phytol 178: 239–252 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RGF, Trindade LM. (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583: 978–982 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan J, Whitehouse J. (1971) Seed structure and the taxonomy of the Cruciferae. Bot J Linn Soc 64: 383–409 [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW. (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP. (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM. (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.