Abstract
A collection of 130 new plant cell wall glycan-directed monoclonal antibodies (mAbs) was generated with the aim of facilitating in-depth analysis of cell wall glycans. An enzyme-linked immunosorbent assay-based screen against a diverse panel of 54 plant polysaccharides was used to characterize the binding patterns of these new mAbs, together with 50 other previously generated mAbs, against plant cell wall glycans. Hierarchical clustering analysis was used to group these mAbs based on the polysaccharide recognition patterns observed. The mAb groupings in the resulting cladogram were further verified by immunolocalization studies in Arabidopsis (Arabidopsis thaliana) stems. The mAbs could be resolved into 19 clades of antibodies that recognize distinct epitopes present on all major classes of plant cell wall glycans, including arabinogalactans (both protein- and polysaccharide-linked), pectins (homogalacturonan, rhamnogalacturonan I), xyloglucans, xylans, mannans, and glucans. In most cases, multiple subclades of antibodies were observed to bind to each glycan class, suggesting that the mAbs in these subgroups recognize distinct epitopes present on the cell wall glycans. The epitopes recognized by many of the mAbs in the toolkit, particularly those recognizing arabinose- and/or galactose-containing structures, are present on more than one glycan class, consistent with the known structural diversity and complexity of plant cell wall glycans. Thus, these cell wall glycan-directed mAbs should be viewed and utilized as epitope-specific, rather than polymer-specific, probes. The current world-wide toolkit of approximately 180 glycan-directed antibodies from various laboratories provides a large and diverse set of probes for studies of plant cell wall structure, function, dynamics, and biosynthesis.
Cell walls play important roles in the structure, physiology, growth, and development of plants (Carpita and Gibeaut, 1993). Plant cell wall materials are also important sources of human and animal nutrition, natural textile fibers, paper and wood products, and raw materials for biofuel production (Somerville, 2007). Many genes thought to be responsible for plant wall biosynthesis and modification have been identified (Burton et al., 2005; Lerouxel et al., 2006; Mohnen et al., 2008), and 15% of the Arabidopsis (Arabidopsis thaliana) genome is likely devoted to these functions (Carpita et al., 2001). However, phenotypic analysis in plants carrying cell wall-related mutations has proven particularly difficult. First, cell wall-related genes are often expressed differentially and at low levels between cells of different tissues (Sarria et al., 2001). Also, plants have compensatory mechanisms to maintain wall function in the absence of a particular gene (Somerville et al., 2004). Thus, novel tools and approaches are needed to characterize wall structures and the genes responsible for their synthesis and modification.
Monoclonal antibodies (mAbs) developed against cell wall polymers have emerged as an important tool for the study of plant cell wall structure and function (Knox, 2008). Previous studies have utilized mAbs that bind epitopes present on rhamnogalacturonan I (RG-I; Freshour et al., 1996; Jones et al., 1997; Willats et al., 1998; McCartney et al., 2000; Clausen et al., 2004; Altaner et al., 2007), homogalacturonan (Willats et al., 2001; Clausen et al., 2003), xylogalacturonan (Willats et al., 2004), xylans and arabinoxylans (McCartney et al., 2005), xyloglucan (Freshour et al., 1996, 2003; Marcus et al., 2008), arabinogalactan(protein) (Pennell et al., 1991; Puhlmann et al., 1994; Dolan et al., 1995; Smallwood et al., 1996), and extensins (Smallwood et al., 1995) to localize these epitopes in plant cells and tissues. In addition, mAbs have been used to characterize plants carrying mutations in genes thought to be associated with cell wall biosynthesis and metabolism (Orfila et al., 2001; Seifert, 2004; Persson et al., 2007; Cavalier et al., 2008; Zabotina et al., 2008). Despite their utility, the available set of mAbs against carbohydrate structures is relatively small given the structural complexity of wall polymers (Ridley et al., 2001; O'Neill and York, 2003), and knowledge of their epitope specificity is limited. Thus, additional mAbs specific to diverse epitope structures and methods for rapid epitope characterization are needed (Somerville et al., 2004).
Here, we report the generation of 130 new mAbs that bind to diverse epitopes present on a broad spectrum of plant cell wall glycans. In addition, approximately 50 previously reported or generated mAbs were included in the ELISA-based screens used to group the antibodies according to their binding patterns against a diverse panel of 54 polysaccharides. The resulting ELISA data were analyzed by hierarchical clustering to illustrate the relationships between the available mAbs. Nineteen groups of mAbs were identified from the clustering analysis. Some initial information regarding possible epitopes recognized by some of these antibodies could be inferred from the clustering analysis.
RESULTS
Immobilization of Plant Polysaccharides for ELISA
Eight commercially available 96-well plates were tested to determine their suitability for plant cell wall polysaccharide-based ELISAs. These plates were tested simultaneously using a standard ELISA protocol and 12 mAb/polysaccharide pairs: CCRC-M1/sycamore xyloglucan, CCRC-M2/gum karaya, CCRC-M7/sycamore pectic polysaccharides, CCRC-M10/mustard seed mucilage, CCRC-M14/Arabidopsis RG-I, CCRC-M16/soybean RG-I, CCRC-M30/Arabidopsis seed mucilage, CCRC-M58/tamarind xyloglucan, PN16.4B4/gum arabic, JIM5/citrus pectin, JIM13/gum ghatti, and LM10/4-O-methylglucuronoxylan. Of the ELISA plates tested, Costar 3598 gave the highest mean signal across all of the mAb/polysaccharide pairs tested, showing a mean optical density (OD) of 0.551 ± 0.150 (Supplemental Fig. S1). Other plate types, including Immulon 1B (0.340 ± 0.131), Immulon 4HB (0.442 ± 0.173), Immulon 2HB (a plate that we had used previously [Puhlmann et al., 1994]; 0.422 ± 0.161), Nunc 269620 (0.425 ± 0.150), Nunc 439454 (0.454 ± 0.154), Costar 2507 (0.335 ± 0.141), and Costar 3590 (0.435 ± 0.118), gave lower mean signals. Costar 3598 plates also displayed the highest minimum signal (minimum OD = 0.316) for the mAb/polysaccharide pairs tested. Immulon 1B (0.101), Immulon 4HB (0.213), Immulon 2HB (0.195), Nunc 269620 (0.179), Nunc 439454 (0.265), Costar 2507 (0.185), and Costar 3590 (0.250) had lower threshold OD values. Based on these results, Costar 3598 plates were used to carry out all subsequent ELISAs.
Data Correlation Analysis Emphasizes Reproducibility of ELISAs
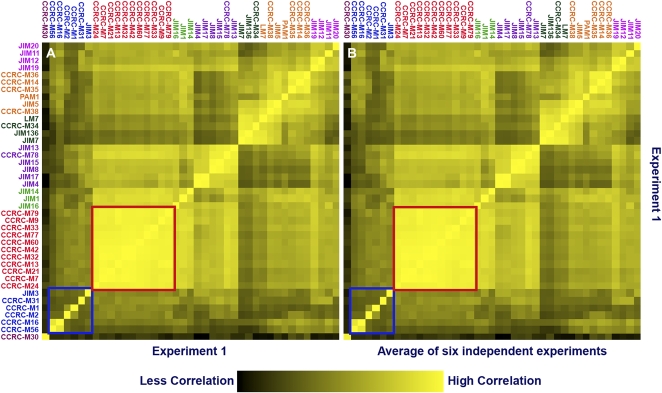
ELISA analyses were used to test mAb-binding specificities against a diverse panel of plant polysaccharide preparations. The reproducibility of the ELISA data pattern for each antibody was examined by generating a correlation heat map (Supplemental Materials and Methods S1). This correlation heat map analysis was done using the data obtained from six replicate experiments involving ELISA screening of 41 antibodies against a panel of diverse polysaccharides. In the correlation heat map, the value (color) of each square corresponds to the correlation of the ELISA response vector for one mAb in one experiment to the ELISA response vector for each mAb in another experiment or group of experiments. Perfect reproducibility corresponds to a heat map with all diagonal elements equal to 1.0 (brightest yellow) and perfect symmetry about the diagonal. (This would be the result if a data set were compared with itself, as shown in Fig. 1A). A second heat map, shown in Figure 1B, depicts the correlation of a randomly selected ELISA panel test replicate with the average of six replicates. In this case, each diagonal heat map element correlates the response pattern of a specific mAb in the selected experiment with the average ELISA response pattern for that mAb. Almost all of the correlation coefficients were greater than 0.98 (Supplemental Table S3). Each off-diagonal heat map element in Figure 1B shows the correlation of the ELISA response pattern of a mAb in the selected experiment to the mean ELISA response pattern of a different mAb. The presence of very few deviations from symmetry about the diagonal in the correlation heat map indicates that the ELISAs are highly reproducible. The reproducibility of ELISAs is also emphasized by the significant resemblance of the experimental correlation heat map (Fig. 1B) to the autocorrelation heat map (Fig. 1A).
Figure 1.
Reproducibility of ELISAs. Data correlation heat maps generated by comparison of one ELISA data set with itself (A) and with the average of six ELISA replicates (B). Within each square in this heat map, increasing yellow color indicates greater correlation between two mAb-binding patterns. The bright yellow diagonal elements depict the high degree of correlation of specific mAb-binding patterns between replicates. Each off-diagonal heat map element shows the correlation of the ELISA response pattern of a mAb in the selected experiment to the mean ELISA response pattern of a different mAb. The high color intensity of the diagonal and high degree of symmetry about the diagonal in B indicate high reproducibility among independently performed ELISA analyses. Well-defined clades correspond to bright correlation squares (e.g. the squares outlined in red). Poorly defined clades correspond to darker correlation squares (e.g. the squares outlined in blue).
The correlation heat maps (Fig. 1) also group antibodies that show similar binding patterns to the panel of polysaccharides. These groups (clades) of antibodies are highlighted by the coloring of the correlation heat map (from black to bright yellow; Fig. 1). For example, the relatively brighter yellow “correlation square” of the CCRC-M79 group (red-highlighted antibodies, red outline; Fig. 1B) indicates that the binding patterns of mAbs in this clade are very similar. In contrast, the relatively dark coloring of the correlation square in the lower left corner of the map (corresponding to the clade containing JIM3 and five other antibodies [blue-highlighted antibodies, blue outline; Fig. 1B]) indicates that the binding patterns of the mAbs in this clade are not closely related; such outliers fall into a single cluster only because they do not fit into any well-defined cluster within the scope of this analysis.
Polysaccharide Panel Screening and Hierarchical Clustering of mAbs
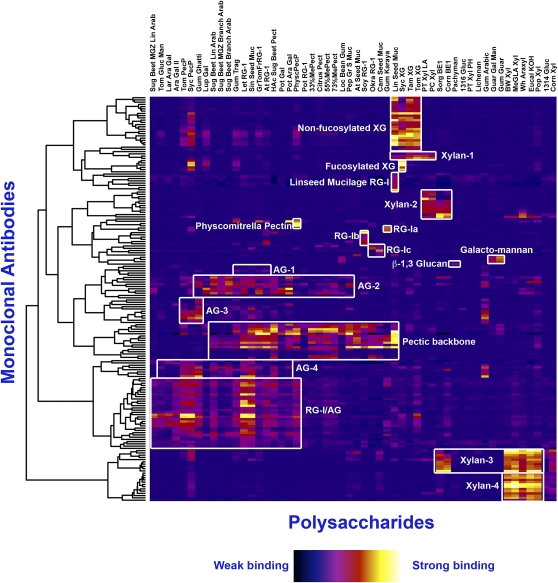
The current toolkit of mAbs studied here, when screened against 54 diverse plant polysaccharide preparations whose detailed chemical compositions are either previously known or determined during this study (Supplemental Table S1), yielded diverse polymer-binding patterns when viewed as a whole. However, subsets of the collection shared similar polymer-binding patterns, suggesting that hierarchical relationships exist among these mAbs. To study these relationships in greater detail and to group mAbs based on their polymer-binding fingerprints, hierarchical clustering analysis of the ELISA data was performed using a modification of previously described methods (Ferguson et al., 1988). The goal of this clustering was to compare the ELISA responses for each mAb when tested against the panel of polysaccharides. The raw ELISA data for each mAb constitutes a vector (i.e. an ordered list of values, one for each polysaccharide). A matrix, in which each row consisted of the ELISA response vector for a particular mAb, was generated. The rows and columns in the matrix were clustered (Supplemental Materials and Methods S1) to generate dendrograms that allowed similarities in the ELISA response patterns of mAbs (rows) and polysaccharides (columns) to be visualized. The clustered ELISA data are represented as a heat map (Fig. 2) along with the dendrograms that were used to order the data. The color of each cell in the heat map represents the ELISA response of a particular mAb when tested against a particular polysaccharide.
Figure 2.
Clustering of plant cell wall glycan-directed mAbs based on their polysaccharide recognition patterns. The heat map represents the hierarchical clustering of data obtained from ELISAs of plant cell wall glycan-directed mAbs against a diverse panel of plant cell wall polysaccharides. Each row in the heat map reflects the binding pattern of a single mAb against the panel of polysaccharides. Each column reflects the binding of all antibodies against a given polysaccharide. The color of each element in the map reflects the strength of the ELISA signal (white = strongest binding, dark blue = no binding). Groups of mAbs that show similar binding patterns to the polysaccharide preparations are identified by white rectangles. The polysaccharide preparations used for the screen and their glycosyl compositions are listed in Supplemental Table S1. The identity of the mAbs and their order within each group are given in Supplemental Table S2.
Dendrograms generated by our initial clustering experiments, performed essentially as described previously (Ferguson et al., 1988), were often in disagreement with groupings obtained by manual comparison of the ELISA responses. We showed that this initial approach, which is based on using the Pearson correlation coefficient as the distance metric for clustering, can lead to dendrograms that imply close associations between dissimilar patterns. Therefore, we used a different approach in which the inverse cosine of the dot product of each pair of ELISA response vectors was used as the distance metric for clustering. This is similar to the use of Pearson correlation coefficients in that it builds dendrograms using response patterns rather than absolute responses. When applied to our ELISA data, this new approach produced dendrograms that were more consistent with the mAb groupings obtained by manual comparison of the ELISA responses.
Encouraged by these results, we used the R language (R Development Core Team, 2006) to develop a software application that uses the alternative approach. Given the ELISA response data for a collection of mAbs against the panel of polysaccharides, the software provides dendrograms for the mAbs and the polysaccharides and a heat map ordered using the dendrograms. The software also allows the subtrees of a selected vertex in either dendrogram to be reversed, which does not formally or materially alter the dendrogram but can provide images that are easier to interpret. This software also can produce a heat map that illustrates the correlation of one data set to another, providing a rapid method of assessing reproducibility between data sets and identifying those data points and ELISA response patterns that differ significantly between the two data sets. We are making this software freely available for use by others (http://glycomics.ccrc.uga.edu/cluster/).
The hierarchical clustering analysis grouped the mAbs into well-resolved clades that are characterized by commonalities in polymer recognition (Fig. 2). Based on this clustering analysis, we identified 19 groups of mAbs that recognize a range of glycostructures covering most major cell wall polysaccharides (outlined in white boxes in Fig. 2). Some examples include a nonfucosylated xyloglucan-directed clade of mAbs, a fucosylated xyloglucan-directed clade, the pectic backbone-directed clade, the RG-I/AG clade, four distinct xylan-directed clades of mAbs (Xylan-1 to -4), and several arabinogalactan-directed clades (AG-1 to -4). The mAbs that are grouped within each clade are identified in Supplemental Table S2. Thus, the clustering analysis yielded important information identifying polysaccharide preparations rich in epitopes recognized by these new mAbs that can be used to focus future, more detailed epitope characterization studies.
Very few of the clades of antibodies showed polymer-specific binding patterns. Those mAbs that show polymer-specific binding patterns include a set of mAbs that bind only to linseed mucilage (Linseed Mucilage RG-I clade), two sets of xylan-directed mAbs (Xylan-2 and -4), a set of mAbs that bind only to galactomannans, the β-1,3-glucan-directed antibody (LAMP), and a set of antibodies that selectively recognize a pectic polysaccharide preparation from Physcomitrella patens. The majority of the antibodies in the toolkit show less specificity with respect to the polysaccharide preparations that they recognize, reflecting a broader distribution of the epitopes recognized by these mAbs among plant cell wall glycans and/or covalent linkages between different glycans.
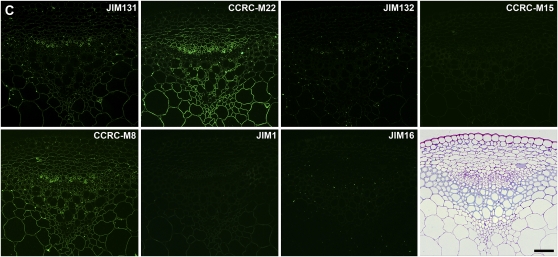
The collection of mAbs screened against the polysaccharide panel included those whose generation and partial characterization had been reported previously (e.g. CCRC-M1 to -M12, PN and MH series, JIM series, MAC series, LM series, PAM1, AX1, LAMP). These previously generated mAbs were broadly distributed among the antibody clades that emerged from the hierarchical clustering analyses of the entire mAb collection. In the case of several JIM and MAC series antibodies, the current clustering analysis led to new groupings of these antibodies relative to groupings that had emerged from previous analyses (Yates and Knox, 1994; Moller et al., 2008). Thus, the mAbs in the former “HRGP” group are now divided among two clades that we have called AG-1 and AG-2. The mAbs in the AG-1 clade (with JIM11, JIM20, JIM93, JIM94, and MAC204) bind to gum tragacanth and to lettuce and green tomato RG-I preparations. The mAbs in the AG-2 clade (with JIM12, JIM14, JIM19, MAC207, LM5, and LM6) bind to linear and branched arabinans and RG-I preparations from diverse plants but do not bind to larch arabinogalactan. The mAbs in the former “AGP” group are distributed among three distinct clades of mAbs: RG-I/AG, AG-3, and AG-4. The mAbs in the RG-I/AG clade (which includes JIM1, JIM16, JIM131, and JIM132) bind to RG-I preparations from a broad range of plants but do not bind to gum arabic. The mAbs in the AG-3 clade (which includes JIM4, JIM17, JIM8, and JIM15) bind strongly to gum ghatti and gum arabic and also to pectic polysaccharide preparations from tomato and sycamore maple. The mAbs in the AG-4 clade (with JIM13 and JIM133) bind to RG-I preparations from a broad range of plants and also bind to gum arabic.
Immunolabeling
Immunolabeling studies were carried out on Arabidopsis inflorescence stems (Figs. 3–5) to obtain independent verification of the clades or subclades resulting from the hierarchical clustering of ELISA data. These studies were done using three sets of mAbs that resulted from the hierarchical clustering analyses (Fig. 2; Supplemental Table S2), two distinct sets of xylan-directed mAbs (Xylan-3 and -4), and another set of mAbs directed against the arabinogalactan side chains of RG-I (RG-I/AG).
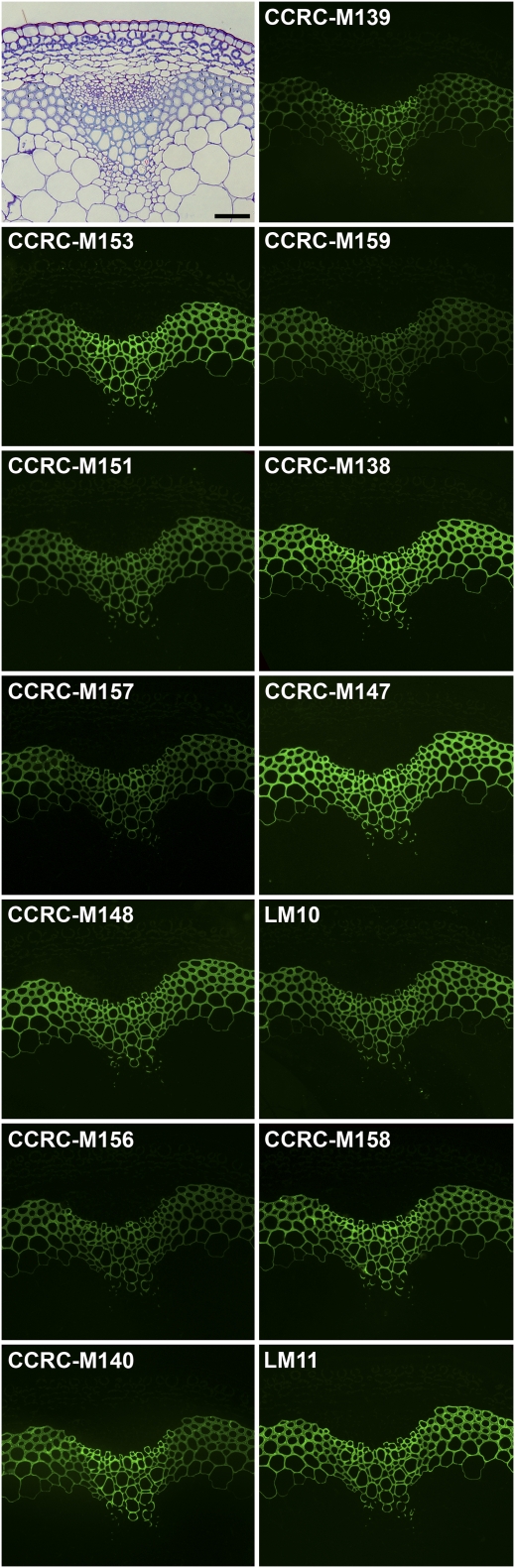
Figure 3.
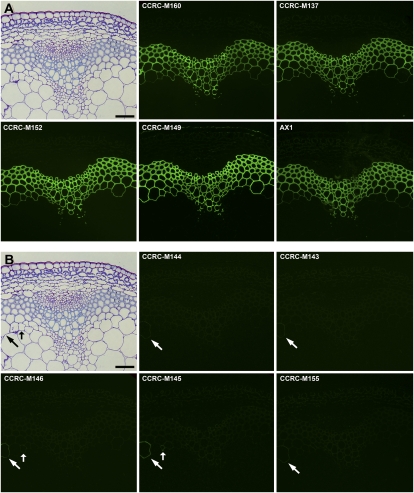
Immunofluorescent labeling of Arabidopsis stems with the xylan-binding mAbs in Xylan-4. Transverse sections (250 nm) were taken from the base of inflorescence stems of 40-d-old Arabidopsis plants. Immunolabeling was carried out using a group of xylan-binding antibodies (Xylan-4) identified by hierarchical clustering of ELISA data obtained from the polysaccharide panel screens (Fig. 2; Supplemental Table S2). Bar = 50 μm.
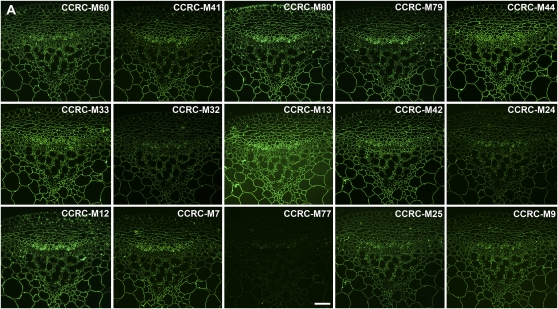
Figure 5.
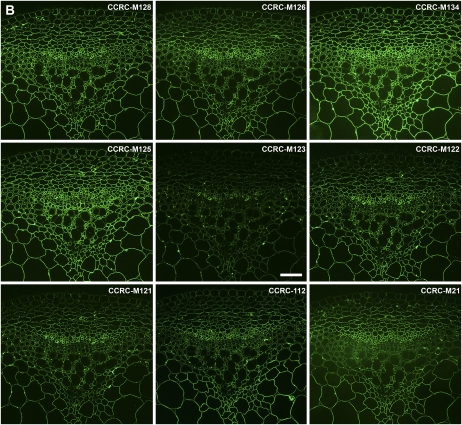
Immunofluorescent labeling of Arabidopsis stems using pectic arabinogalactan-binding mAbs. Transverse sections (250 nm) were taken from inflorescence stems of 40-d-old Arabidopsis plants. Immunolabeling was carried out using a group of pectic arabinogalactan-binding mAbs (RG-I/AG) identified by hierarchical clustering of ELISA data obtained from the polysaccharide panel screens (Fig. 2; Supplemental Table S2). One toluidine blue-stained section is included in C for orientation of all immunofluorescence panels. A, Immunolabeling with the top subclade of RG-I/AG mAbs (CCRC-M9 to CCRC-M60; Supplemental Table S2). B, Immunolabeling with the middle subclade of RG-I/AG mAbs (CCRC-M21 to CCRC-M128; Supplemental Table S2). C, Immunolabeling with the bottom subclade of RG-I/AG mAbs (JIM16 to JIM131; Supplemental Table S2). Bar = 50 μm.
All of the mAbs in the Xylan-4 clade labeled xylem tissues in Arabidopsis stems in a similar fashion, although the labeling intensity differed among some of the mAbs (Fig. 3). These intensity differences likely result from differences in the epitope structures recognized by the mAbs and/or the different epitope density distribution patterns within the xylans synthesized in different cells. The similar localization patterns among these xylan-directed mAbs support the results of the hierarchical clustering that grouped these mAbs into a tight cluster (Fig. 2). Interestingly, another set of xylan-directed mAbs, which form a distinct subset within the Xylan-3 clade, also label xylem tissue in a similar pattern as observed with the Xylan-4 mAbs (Fig. 4A). This suggests that the xylans made in Arabidopsis stem xylem tissues carry at least two distinct epitopes recognized by these two groups of xylan-directed antibodies, which is consistent with the known structural complexity of xylans synthesized by dicots (Ebringerová et al., 2005).
Figure 4.
Immunofluorescent labeling of Arabidopsis stems with the xylan-binding mAbs in Xylan-3. Transverse sections (250 nm) were taken from the base of inflorescence stems of 40-d-old Arabidopsis plants. Immunolabeling was carried out using two subgroups (A and B) of xylan-binding antibodies within the Xylan-3 clade identified by hierarchical clustering of ELISA data obtained from the polysaccharide panel screens (Fig. 2; Supplemental Table S2). The toluidine blue section shown in A and B is identical to that shown in Figure 3 and is included for orientation of the immunofluorescent panels. Arrows identify the labeled cells in B. Bars = 50 μm.
The Xylan-3 clade is subdivided into two distinct subclades based on the hierarchical clustering of ELISA data obtained from the polysaccharide screens (Fig. 2). Immunolabeling data support this subdivision. One subset of mAbs in the Xylan-3 clade (CCRC-M137, CCRC-M149, CCRC-M160, and AX1) all labeled xylem tissues in the stems in a similar fashion (Fig. 4A). The mAbs in the other subset in the Xylan-3 clade (CCRC-M143, CCRC-M144, CCRC-M145, CCRC-M146, and CCRC-M155) all show a labeling pattern that is very distinct from the first subset of Xylan-3 mAbs, labeling only specific cells in Arabidopsis stems (Fig. 4B).
The RG-I/AG clade of mAbs was grouped into three subclades by the hierarchical clustering (Fig. 2; RG-I/AG clade [top group of 15 mAbs, middle nine mAbs, and the bottom seven mAbs]); two of those subclades appeared to be distinguished only by their ability to bind to larch and soybean arabinans/galactans, while the third subclade showed more diverse binding patterns against the pectic arabinogalactans tested here. The first two subclades showed very similar immunolabeling patterns in Arabidopsis stems, although differences in labeling intensities and subtle differences in labeling patterns were observed (Fig. 5, A and B). Antibodies in the third RG-I/AG subclade showed disparate labeling patterns from each other and from the other two subclades (Fig. 5C). Thus, the observed immunolabeling patterns support the subclade structure within the RG-I/AG clade of mAbs that had been defined by the hierarchical clustering of the ELISA data.
DISCUSSION
The complexity of the plant cell wall glycome necessitates the development of a broad diversity of tools in order to gain a deeper understanding of cell wall structure, function, and biology. Here, we describe a comprehensive toolkit of approximately 180 plant cell wall-directed mAbs, of which approximately 130 are newly generated and reported here for the first time. These antibodies are annotated in a database accessible on the Internet (http://www.WallMabdb.net), and the antibodies are available to the cell wall research community from CarboSource (http://www.CarboSource.net) for the CCRC, MH, PN, JIM, and MAC series of antibodies and from PlantProbes (http://www.PlantProbes.net) for LM and JIM antibodies as well as from other individual laboratories that generated the mAbs (Supplemental Table S2). The world-wide collection of cell wall glycan-directed mAbs is now sufficiently large and diverse to constitute a comprehensive resource that will prove invaluable for detailed studies of the structure, dynamics, function, and biosynthesis of plant cell walls.
The ELISA used in this study for determination of antibody-binding specificities is a reliable and a highly reproducible assay for quantifying the binding of mAbs to polysaccharide antigens. One key aspect of this method that is different from many ELISA applications is the drying down of the polysaccharides to the bottom of the ELISA plate wells, rather than allowing adsorption to take place only in the liquid phase, as is commonly done. Adsorption of polysaccharides in the liquid phase yielded greater variability in the ELISA data, probably due to less consistent immobilization of diverse polysaccharides to the ELISA plates (data not shown). The ability of a polysaccharide to bind to the plate under our assay conditions does not depend on its glycosyl composition or charge; both neutral and charged polysaccharides of diverse structures bind to the plates. However, the molecular size of a polysaccharide does affect its ability to adhere to the ELISA plates. For example, unmodified oligosaccharides and low molecular mass polysaccharides, such as RG-II (which has a molecular mass of approximately 5–10 kD; O'Neill et al., 2004) do not adhere to the plastic plates and must be coupled to carriers or modified in other ways in order to immobilize them on the plates. Of the eight different commercially available ELISA plates that were tested, Costar 3598 showed the most uniform binding of diverse polysaccharides under our assay conditions; as a result, these plates were used throughout this study.
ELISA analyses were done to probe the binding specificities of the antibodies against a panel of 54 polysaccharide preparations representing diverse plant cell wall polysaccharides. Hierarchical clustering analysis of ELISA responses of all these antibodies against this panel of polysaccharides resulted in the clustering of the 180 mAbs into 19 well-resolved groups of antibodies (Fig. 2; Supplemental Table S2). These mAb clusters are also supported by immunolocalization studies (Figs. 3–5). Comparisons of the glycosyl compositions of the polysaccharide preparations (Supplemental Table S1) recognized by each group of antibodies reveal compositional commonalities. The polysaccharide preparations used in the panel can each be viewed as a collection of epitopes, each of which typically ranges in size from one to eight glycosyl residues (Kabat, 1966; Reimer et al., 1992; Puhlmann et al., 1994; Steffan et al., 1995; Clausen et al., 2003). Different polysaccharide preparations will vary in both the types of epitopes (structural features) present and the amounts of each epitope. A minor polysaccharide component will contribute a correspondingly low proportion of epitopes to the overall epitope composition of a given polysaccharide preparation. The ELISAs provide both qualitative and quantitative measures of the epitope composition of each of the polysaccharide preparations. Hierarchical clustering as applied to the ELISA data will then group the antibodies based both on which polysaccharide preparations are recognized and on the strength of the ELISA signal for each polysaccharide. Minor contaminants in an individual polysaccharide preparation will not significantly affect the outcome of the clustering. This can be most clearly seen in the heat map (Fig. 2) in the case of the two xyloglucan clusters. These antibodies show some cross-reactivity with a sycamore pectic polysaccharide preparation, but the hierarchical clustering still groups these antibodies on the basis of the strong signals to the xyloglucans and clearly distinguishes these two groups of mAbs from each other and from other groups of mAbs that bind to RG-I epitopes. Thus, the power of the hierarchical clustering approach is that it can identify the commonalities in epitope recognition patterns across the collection of antibodies and polysaccharides and group the antibodies accordingly.
This study included a larger number of mAbs and utilized a greater diversity of polysaccharides in the binding studies used as the foundation for the hierarchical clustering than were used in a previous study that yielded a dendrogram of cell wall-reactive mAbs (Moller et al., 2008), resulting in differences in the clade compositions between the two studies. For example, JIM5 and JIM7, which fell into the same cluster previously (Moller et al., 2008), are clearly resolved into distinct, but related, subclusters in our analysis (Fig. 2; Supplemental Table S2). The separate clustering of JIM5 and JIM7 noted here is consistent with the reported differences in epitopes recognized by these mAbs (Clausen et al., 2003). Likewise, several JIM and MAC mAbs that recognize AGP and/or extensin (HRGP) epitopes and had been grouped largely into two clades in previous analyses (Yates and Knox, 1994; Moller et al., 2008) are now distributed among several clades, each of which shows distinct arabinogalactan glycan-binding patterns. These new clusterings suggest that the set of cell wall glycoprotein-binding JIM and MAC mAbs bind to a greater diversity of glycans, specifically pectic arabinogalactans, than had previously been recognized and cannot be viewed as being specific to a particular class of cell wall glycoproteins.
The observed complexities in the mAb-binding patterns to plant cell wall glycans reflect the known structural complexities of plant cell wall polysaccharides (Ridley et al., 2001; O'Neill and York, 2003). Some of the observed cross-reactivities can be readily explained by the covalent association of different glycans with one another in the wall and in glycan preparations. Thus, antibodies that bind to homogalacturonan epitopes (e.g. CCRC-M38, JIM5, CCRC-M34, JIM7) also bind to a broad range of RG-I preparations due to the covalent association between homogalacturonan and RG-I (Mohnen, 2008). In other cases, the cross-reactivities can be explained by the presence of similar/identical epitopes on a given polysaccharide isolated from different plants. Thus, the xylan-binding antibodies in the Xylan-3 and Xylan-4 clades bind epitopes that are present on both monocot and dicot xylans.
In still other cases, the cross-reactivities can be explained by the fact that some structural features (epitopes) are present on multiple glycans that occur in plant cell walls. This is particularly the case with antibodies that bind to epitopes containing arabinosyl and/or galactosyl residues, which are present in multiple structural contexts within diverse plant cell wall glycans. For example, mAbs that bind to arabinogalactan side chains of RG-I frequently, but not always, also bind to free and/or protein-linked arabinogalactans, which contain similar structural features (Ridley et al., 2001; Seifert and Roberts, 2007). The data presented here emphasize that glycan-directed antibodies should be utilized as epitope-directed reagents and frequently are not polymer selective.
Some observed cross-reactivities are not as readily explained based on current knowledge of cell wall glycan structures. For example, mAbs that bind to fucosylated xyloglucans (e.g. CCRC-M1) also bind strongly to sycamore RG-I (but not to other RG-Is included in this study). This cross-reactivity is not due to contamination of the sycamore RG-I preparation with xyloglucan, since treatment of sycamore RG-I with a xyloglucan-specific endoglucanase did not affect binding of CCRC-M1 (data not shown). The cross-reactivity of the mAbs in the Xylan-1 clade with xyloglucans included in this study is also not readily explained. The epitope(s) recognized by the Xylan-1 mAbs appears not to be present on all xyloglucans, as these mAbs do not label any cells in Arabidopsis tissues (data not shown). Interestingly, carbohydrate-binding modules that recognize xylan have been reported to also bind to xyloglucans (Boraston et al., 2001; Gunnarsson et al., 2006). Xyloglucan and xylan are not known to be covalently linked or to share common structural features (except that both have a β-1,4-linked backbone composed of pyranosyl residues in which all exocyclic oxygens are equatorial). Resolution of these cross-reactivities must await detailed characterizations of the epitopes recognized by these mAbs.
Most of the mAb clades identified through hierarchical clustering are divided further into subclades. Some of these subdivisions are informative with respect to possible epitopes recognized by newly generated mAbs due to tight clustering with previously characterized mAbs. For example, several new mAbs (CCRC-M39, CCRC-M84, CCRC-M102, CCRC-M106) cluster with CCRC-M1, suggesting that the newly generated mAbs bind to the same or similar fucosylated xyloglucan epitope recognized by CCRC-M1 (Puhlmann et al., 1994). Other newly reported mAbs cluster in distinct subclades with previously characterized mAbs directed against homogalacturonans (Fig. 2; Supplemental Table S2). CCRC-M34, CCRC-M130, and JIM136 cluster closely in a subclade with JIM7 and LM7, mAbs that bind to densely methyl-esterified homogalacturonan epitopes (Clausen et al., 2003), suggesting that these three mAbs bind to methylated homogalacturonan epitopes. In contrast, CCRC-M38, CCRC-M131, and CCRC-M132 cluster tightly in a subclade with JIM5, a mAb that binds to a homogalacturonan epitope having a low density of methyl esterification (Clausen et al., 2003), suggesting that these three mAbs bind to a largely or completely deesterified homogalacturonan epitope. Lastly, about a dozen newly generated mAbs cluster tightly in a subclade of the RG-I/AG clade (Fig. 2; Supplemental Table S2) that includes CCRC-M7, suggesting that these mAbs recognize a β-1,6-galactan epitope similar or identical to that recognized by CCRC-M7 (Steffan et al., 1995). Verification of these tentative epitope assignments awaits more detailed studies, which are currently under way in our laboratory.
MATERIALS AND METHODS
Polysaccharides
Polysaccharides from various plant sources were obtained from commercial sources (Megazyme, Sigma, and Sunkist) and various laboratories at the University of Georgia's Complex Carbohydrate Research Center and elsewhere. Detailed information about these polysaccharides, such as glycan class, preparation, source, and sugar composition, are given in Supplemental Table S1. Stock solutions were prepared by dissolving the polysaccharides at 1 mg mL−1 in deionized water and were stored at −20°C.
mAbs
mAbs were obtained as hybridoma cell culture supernatants either from laboratory stocks (CCRC series, MH series, PN series, JIM series, MAC series; available from CarboSource [http://www.carbosource/net]) or from Plant Probes (LM series, PAM1 [http://www.plantprobes.net/]) unless otherwise indicated. A detailed list of all mAbs included in this study showing the immunogens used to develop them, their isotype, and the cell wall polysaccharide class they primarily recognize is provided in Supplemental Table S2.
ELISA
Flat-bottom 96-well plates tested for use in the ELISA were Immulon 1B, Immulon 2HB, Immulon 4HB, Nunc 269620, and Nunc 439454 (Thermo Fisher Scientific) and Costar 2507, Costar 3590, and Costar 3598 (Corning Life Sciences). Initial experiments were carried out with several polysaccharides over a broad concentration range (1 ng well−1 to 10 μg well−1) in order to determine the maximum loading of polysaccharides onto the plates. These studies showed that an amount of 0.5 μg well−1 saturates the ELISA wells with a given polysaccharide antigen (data not shown). Polysaccharides were applied (50 μL of 10 μg mL−1 in deionized water per well) to 96-well plates and were dried to the well surfaces by evaporation overnight at 37°C. Control wells were coated with deionized water. The plates were blocked with 200 μL of 1% (w/v) instant nonfat dry milk (Carnation) in Tris-buffered saline (50 mm Tris-HCl, pH 7.6, containing 100 mm sodium chloride) for 1 h. All subsequent aspiration and wash steps were performed using an ELx405 microplate washer (Bio-Tek Instruments). Blocking agent was removed by aspiration, and 50 μL of undiluted hybridoma supernatant were added to each well and incubated for 1 h at room temperature. Supernatant was removed and wells were washed three times with 300 μL of 0.1% (w/v) instant nonfat dry milk in Tris-buffered saline (wash buffer). Peroxidase-conjugated goat anti-mouse IgG or goat anti-rat IgG antibodies (Sigma-Aldrich), depending on the primary antibody used, was diluted 1:5,000 in wash buffer, and 50 μL were added to each well and incubated for 1 h. Note that the secondary antibodies used in this study are generated against whole immunoglobin molecules and thus bind to several isotypes of primary antibodies, including IgGs, IgMs, and IgAs, according to the manufacturers. Wells were then washed five times with 300 μL of wash buffer. 3,3′,5,5′-Tetramethylbenzidine substrate solution (Vector Laboratories) was freshly prepared according to the manufacturer's instructions, and 50 μL were added to each well. After 20 min, the reaction was stopped by adding 50 μL of 0.5 n sulfuric acid to each well. The OD of each well was read as the difference in A450 and A655 using a model 680 microplate reader (Bio-Rad). The reading from each test well was subtracted from that of a control well on the same plate that contained the same primary and secondary antibodies but no immobilized polysaccharide.
Polysaccharide Panel Screening
Polysaccharide panel screening of mAbs was carried out by ELISA against 54 plant polysaccharides (Supplemental Table S1) immobilized to Costar 3598 96-well plates. A single preparation of each polysaccharide was used for all experiments reported here.
Hierarchical Clustering
Hierarchical clustering of the ELISA results for binding of each mAb to each polysaccharide in the panel was carried out using previously described methods (Ferguson et al., 1988) with modifications (Supplemental Materials and Methods S1). The R language for statistical computing was used for these analyses (R Development Core Team, 2006).
Plant Culture Conditions
Seeds of Arabidopsis (Arabidopsis thaliana ecotype Columbia) were surface sterilized by immersion in 70% (v/v) aqueous ethanol for 2 min followed by a 2-min immersion in 1% sodium hypochlorite solution (20% [v/v] Clorox containing 0.02% [v/v] Triton X-100). Seeds were then rinsed three times with sterile distilled water. Sterilized seeds were germinated and grown in sterile petri dishes on 1% (w/v) agar with Murashige and Skoog basal salt medium (Sigma-Aldrich) supplemented with 1% (w/v) Suc, pH 6.9. The petri dishes were oriented vertically, maintained at 23°C, and received 12 h of fluorescent illumination (100 μE m−2 s−1) daily. Two-week-old seedlings grown on the plates were then transferred to 4-inch pots containing compost with vermiculite and perlite. These plants were grown in a growth chamber at 20°C with a 16-h-light/8-h-dark cycle.
Tissue Fixation
Forty-day-old Arabidopsis inflorescence stems were fixed for 2.5 h in 1.6% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in 25 mm sodium phosphate buffer, pH 7.1. Tissue was then washed with buffer twice for 15 min, washed with water twice for 15 min, and dehydrated through a graded ethanol series (35%, 50%, 75%, 95%, 100%, 100%, and 100% [v/v] ethanol) for 30 min each step. The dehydrated tissue was moved to 4°C and then gradually infiltrated with cold LR White embedding resin (Ted Pella) using 33% (v/v) and 66% (v/v) resin in 100% ethanol for 24 h each, followed by 100% resin for 24 h three times. The infiltrated tissue was transferred to gelatin capsules containing 100% resin for embedding, and resin was polymerized by exposing the capsules to 365-nm UV light at 4°C for 48 h.
Immunolabeling
Semithin sections (250 nm) were cut with a Leica EM UC6 ultramicrotome (Leica Microsystems) and mounted on glass slides (colorfrost/plus; Fisher Scientific). Immunolabeling was performed at room temperature by applying and removing a series of 10-μL droplets of the appropriate reagents to the sections as follows. Sections were blocked with 3% (w/v) nonfat dry milk in KPBS (0.01 m potassium phosphate, pH 7.1, containing 0.5 m NaCl) for 45 min and then were washed with KPBS for 5 min. Undiluted hybridoma supernatant of the mAb under study was applied and incubated for 120 to 150 min. Sections were then washed with KPBS three times for 5 min each, and goat anti-mouse IgG or goat anti-rat IgG conjugated to Alexa-fluor 488 (Invitrogen) diluted 1:100 in KPBS was applied and incubated for 90 to 120 min. Sections were then washed with KPBS for 5 min and distilled water for 5 min. Prior to applying a coverslip, Citifluor antifade mounting medium AF1 (Electron Microscopy Sciences) was applied.
Light Microscopy
Light microscopy was carried out on an Eclipse 80i microscope (Nikon) equipped with epifluorescence optics. Images were captured with a Nikon DS-Ri1 camera head using NIS-Elements Basic Research software, and images were assembled without further processing using Adobe Photoshop (Adobe Systems).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Suitability of 96-well plates for polysaccharide ELISAs.
Supplemental Table S1. Plant polysaccharide preparations used in this study.
Supplemental Table S2. mAbs included in this study.
Supplemental Table S3. Coefficients correlating the ELISA responses obtained in one experiment with the averages of ELISA responses from six experiments.
Supplemental Materials and Methods S1. Hierarchical clustering of mAbs.
Supplementary Material
Acknowledgments
We thank the following scientists for their generous contributions of polysaccharide preparations included in the screening panel of polysaccharides. Members of the Complex Carbohydrate Research Center, University of Georgia, are thanked for their contribution of the following polysaccharide preparations: Chenghua Deng for green tomato fruit RG-I and lettuce RG-I and Christian Heiss for tamarind xyloglucan. We also thank Mark Davis of the National Renewable Energy Laboratory (Golden, Colorado) for the corn xylan and Ian Sims of Industrial Research Limited (Lower Hutt, New Zealand) for the samples of Phormium xylans. We thank Dr. Henk Schols (Laboratory of Food Chemistry, Wageningen, The Netherlands) for providing several different xylan and arabinogalactan preparations and okra RG-I. Lastly, we acknowledge the generosity of Keith Roberts and Nick Brewin (John Innes Institute, Norwich, UK) in providing us with the JIM and MAC hybridoma lines and Dr. Fabienne Guillon (INRA, Nantes, France) for supplying us with the AX1 mAb.
References
- Altaner C, Hapca AI, Knox JP, Jarvis MC. (2007) Detection of β-1-4-galactan in compression wood of Sitka spruce [Picea sitchensis (Bong.) Carriere] by immunofluorescence. Holzforschung 61: 311–316 [Google Scholar]
- Boraston AB, Creagh AL, Alam MM, Kormos JM, Tomme P, Haynes CA, Warren RAJ, Kilburn DG. (2001) Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40: 6240–6247 [DOI] [PubMed] [Google Scholar]
- Burton RA, Farrokhi N, Bacic A, Fincher GB. (2005) Plant cell wall polysaccharide biosynthesis: real progress in the identification of participating genes. Planta 221: 309–312 [DOI] [PubMed] [Google Scholar]
- Carpita N, Tierney M, Campbell M. (2001) Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol Biol 47: 1–5 [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen MH, Ralet MC, Willats WGT, McCartney L, Marcus SE, Thibault JF, Knox JP. (2004) A monoclonal antibody to feruloylated-(1→4)-β-d-galactan. Planta 219: 1036–1041 [DOI] [PubMed] [Google Scholar]
- Clausen MH, Willats WGT, Knox JP. (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Dolan L, Linstead P, Roberts K. (1995) An AGP epitope distinguishes a central metaxylem initial from other vascular initials in the Arabidopsis root. Protoplasma 189: 149–155 [Google Scholar]
- Ebringerová A, Hromádková Z, Heinze T. (2005) Hemicellulose. Adv Polym Sci 186: 1–67 [Google Scholar]
- Ferguson MW, Wycoff KL, Ayers AR. (1988) Use of cluster analysis with monoclonal antibodies for taxonomic differentiation of phytopathogenic fungi and for screening and clustering antibodies. Curr Microbiol 17: 127–132 [Google Scholar]
- Freshour G, Bonin CP, Reiter WD, Albersheim P, Darvill AG, Hahn MG. (2003) Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis thaliana. Plant Physiol 131: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson LC, Zhou Q, Montanier C, Karlsson EN, Brumer H, Ohlin M. (2006) Engineered xyloglucan specificity in a carbohydrate-binding module. Glycobiology 16: 1171–1180 [DOI] [PubMed] [Google Scholar]
- Jones L, Seymour GB, Knox JP. (1997) Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA. (1966) The nature of an antigenic determinant. J Immunol 97: 1–11 [PubMed] [Google Scholar]
- Knox JP. (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11: 308–313 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. (2006) Biosynthesis of plant cell wall polysaccharides: a complex process. Curr Opin Plant Biol 9: 621–630 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Herve C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP. (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- McCartney L, Ormerod AP, Gidley MJ, Knox JP. (2000) Temporal and spatial regulation of pectic (1→4)-β-d-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J 22: 105–113 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Mohnen D, Bar-Peled M, Somerville C. (2008) Cell wall polysaccharide synthesis. Himmel DE, , Biomass Recalcitrance: Deconstructing the Plant Cell Wall for Bioenergy. Blackwell Publishing, Oxford, pp 94–187 [Google Scholar]
- Moller I, Marcus SE, Haeger A, Verhertbruggen Y, Verhoef R, Schols H, Ulvskov P, Mikkelsen JD, Knox JP, Willats W. (2008) High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj J 25: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Ishii T, Albersheim P, Darvill AG. (2004) Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol 55: 109–139 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, York WS. (2003) The composition and structure of primary cell walls. Rose JKC, , The Plant Cell Wall. Blackwell Publishers, Oxford, pp 1–54 [Google Scholar]
- Orfila C, Seymour GB, Willats WGT, Huxham IM, Jarvis MC, Dover CJ, Thompson AJ, Knox JP. (2001) Altered middle lamella homogalacturonan and disrupted deposition of (1→5)-α-l-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol 126: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K. (1991) Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG. (1994) Generation of monoclonal antibodies against plant cell wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2006) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing (http://www.R-project.org)
- Reimer KB, Gidney MAJ, Bundle DR, Pinto BM. (1992) Immunochemical characterization of polyclonal and monoclonal Streptococcus group A antibodies by chemically defined glycoconjugates and synthetic oligosaccharides. Carbohydr Res 232: 131–142 [DOI] [PubMed] [Google Scholar]
- Ridley BL, O'Neill MA, Mohnen D. (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, Raikhel NV. (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127: 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ. (2004) Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol 7: 277–284 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Martin H, Knox JP. (1995) An epitope of rice threonine- and hydroxyproline-rich glycoprotein is common to cell wall and hydrophobic plasma-membrane glycoproteins. Planta 196: 510–522 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Yates EA, Willats WGT, Martin H, Knox JP. (1996) Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 198: 452–459 [Google Scholar]
- Somerville C. (2007) Biofuels. Curr Biol 17: R115–R119 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Steffan W, Kovác P, Albersheim P, Darvill AG, Hahn MG. (1995) Characterization of a monoclonal antibody that recognizes an arabinosylated (1→6)-β-d-galactan epitope in plant complex carbohydrates. Carbohydr Res 275: 295–307 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Marcus SE, Knox JP. (1998) Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydr Res 308: 149–152 [DOI] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Steele-King CG, Marcus SE, Mort A, Huisman M, Van Alebeek GJ, Schols HA, Voragen AGJ, Le Goff A, et al. (2004) A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta 218: 673–681 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Orfila C, Limberg G, Buchholt HC, Van Alebeek GJWM, Voragen AGJ, Marcus SE, Christensen TMIE, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls: implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Yates EA, Knox JP. (1994) Investigations into the occurrence of plant cell surface epitopes in exudate gums. Carbohydr Polym 24: 281–286 [Google Scholar]
- Zabotina OA, van de Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. (2008) Arabidopsis XXT5 gene encodes a putative α1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.