Abstract
It is not yet known whether dephosphorylation of proteins catalyzed by phosphatases occurs in the apoplastic space. In this study, we found that tobacco (Nicotiana tabacum) purple acid phosphatase could dephosphorylate the phosphoryl residues of three apoplastic proteins, two of which were identified as α-xylosidase and β-glucosidase. The dephosphorylation and phosphorylation of recombinant α-xylosidase resulted in a decrease and an increase in its activity, respectively, when xyloglucan heptasaccharide was used as a substrate. Attempted overexpression of the tobacco purple acid phosphatase NtPAP12 in tobacco cells not only decreased the activity levels of the glycosidases but also increased levels of xyloglucan oligosaccharides and cello-oligosaccharides in the apoplast during the exponential phase. We suggest that purple acid phosphatase controls the activity of α-xylosidase and β-glucosidase, which are responsible for the degradation of xyloglucan oligosaccharides and cello-oligosaccharides in the cell walls.
Purple acid phosphatase (PAP) belongs to a large family of dinuclear metalloenzymes (LeBansky et al., 1992; Klabunde et al., 1996) and catalyzes the hydrolysis of a wide range of phosphate esters. It is distinguished from other acid phosphatases by its purple color, which is due to a Tyr-to-iron (III) charge transfer transition (Antanaitis et al., 1983). Arabidopsis (Arabidopsis thaliana) contains a large family of PAPs composed of 29 genes, 28 of which have signal peptides that potentially transfer to the wall and/or vacuole. Only a few functions have been suggested for these phosphatases: AtPAP15 seems to modulate ascorbic acid biosynthesis (Zhang et al., 2008), and AtPAP17 may play a role in the metabolism of reactive oxygen species (del Pozo et al., 1999). In other plant species, soybean (Glycine max) GmPAP3 is induced by NaCl stress but not by phosphorus deficiency (Liao et al., 2003), tomato (Solanum lycopersicum) PAP may release phosphate from extracellular phosphate ester under phosphate starvation (Bozzo et al., 2002), and tobacco (Nicotiana tabacum) NtPAP12 could be involved in the deposition of β-glucan (Kaida et al., 2003, 2009; Sano et al., 2003). Mammalian PAPs, which are secretory enzymes, may be involved in iron transport (Nuttleman and Roberts, 1990), generation of reactive oxygen species (Sibille et al., 1987), and bone resorption (Ek-Rylander et al., 1994).
We previously demonstrated that the activities of cellulose and callose synthases are enhanced by overexpression of NtPAP12 in tobacco cells (Kaida et al., 2009). The phosphorylation/dephosphorylation process in those synthases may occur directly on the catalytic subunit itself, which has been predicted to be located on the cytoplasmic side of the plasma membrane (Nühse et al., 2004; Taylor, 2007). This is not compatible with the cell wall localization of NtPAP12. The data also indicate that phosphorylation may play a role in regulating the turnover of cellulose synthase by proteolysis through a proteasome-dependent pathway (Taylor, 2007), which again implies a cytoplasmic phosphorylation event. Thus, we suggested that NtPAP12 could be involved in the regulation of cellulose synthase activity, either by acting on an unidentified membrane protein or by enhancing its activity with an effector, which can lead to the promotion of cellulose synthesis. Nevertheless, this phosphatase may be involved in the activation of synthases indirectly by acting on either apoplastic proteins or unidentified membrane proteins, since the level of activation for glucan synthases was only a 2- to 3-fold increase in the transgenic tobacco cells overexpressing NtPAP12 compared with wild-type cells.
The extracellular phosphorylation network has been proposed by proteomic analysis of Arabidopsis cells due to the identification of phosphorylated Tyr residues in xyloglucanase, putative lectin receptor-like kinase, and putative chitinase (Ndimba et al., 2003). The change in phosphorylation status was also identified in the extracellular peroxidase in maize (Zea mays) cells (Chivasa et al., 2005b). Another analysis has indicated that some potential phosphorylated proteins might be present in the apoplastic space during wall regeneration (Kwon et al., 2005). We previously showed that tobacco PAP had a higher catalytic efficiency for Tyr phosphopeptides (kcat/Km = 1,093–1,335) than for ATP (kcat/Km = 333) and p-nitrophenyl-phosphate (kcat/Km = 379), suggesting that the enzyme could dephosphorylate the phosphoryl residues of proteins in vivo (Kaida et al., 2008). There is still much to be learned, however, including the role that phosphorylation plays in the functions of these proteins. It is possible, for example, that extracellular PAPs might modify the functions of the phosphoproteins by dephosphorylating those proteins in the apoplasts, but to date no evidence has been reported demonstrating this activity. In this study, we searched for substrates of PAP using phosphoproteomic analyses of apoplastic proteins in tobacco cells.
RESULTS
Phosphoproteomic Screen in Apoplast of Tobacco Cells
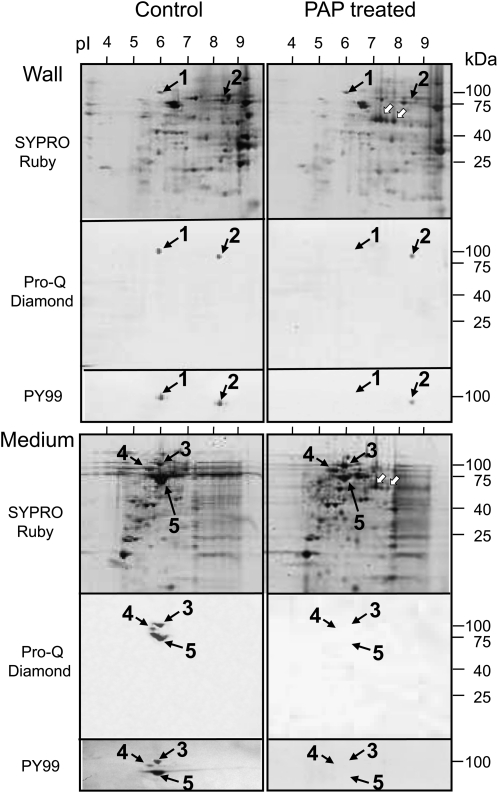
Phosphoproteomic analysis was carried out for extracellular proteins in suspension-cultured tobacco cells using two-dimensional gel electrophoresis (Fig. 1), in which the gel was stained with Pro-Q Diamond for phosphoproteins followed by SYPRO Ruby for total proteins, according to the method described by Steinberg et al. (2003). Two phosphoproteins, spot 1 (100 kD, pI 6.0) and spot 2 (90 kD, pI 8.5), were detected in the walls, and three phosphoproteins, spot 3 (100 kD, pI 6.0), spot 4 (90 kD, pI 5.6), and spot 5 (75 kD, pI 6.0), were detected in the culture medium. Western blotting with the mouse monoclonal antibody PY99, which specifically recognizes phospho-Tyr residues (Abel et al., 2001), showed that all the proteins (spots 1–5) were positively stained (Fig. 1, left panels). To make clear biological repeats, the proteins were prepared several times from different culture flasks at 1- to 2-month intervals. These phosphoproteins were always detected in cells between 2 and 10 d of culture, and the intensity of their expression levels did not change for at least 3 years.
Figure 1.
Images from phosphoproteomic two-dimensional analyses of proteins in the cell walls and culture media. The left panels show the patterns of proteins extracted from wild-type cells, and the right panels show the patterns of proteins extracted from wild-type cells that were digested with purified PAP after their extraction. Proteins were separated according to pH range (pH 3–10) for first-dimensional electrophoresis and by means of SDS-PAGE for second-dimensional electrophoresis. The proteins were stained with SYPRO Ruby, the phosphates with Pro-Q Diamond, and the phospho-Tyrs with PY99 antibody. Spots 1 to 5 are further characterized in Table I and Supplemental Table S1. White arrows indicate native PAP added to the apoplastic protein samples. The analyses by two-dimensional electrophoresis and staining with Pro-Q Diamond and PY99 antibody were performed three times.
After protein mixtures from the walls and culture medium of the wild-type cells had been treated with 10 units of purified PAP (Kaida et al., 2008) for 3 min at 25°C, the phosphate signals of spots 1, 3, 4, and 5 could no longer be detected by means of both Pro-Q Diamond staining and western blotting with PY99; this indicates that those phosphoproteins were dephosphorylated in vitro by the phosphatase treatment (Fig. 1, right panels). Nevertheless, those were detected at a certain level by means of SYPRO Ruby staining. This quick and clear-cut dephosphorylation by 10 units of the enzyme is in agreement with our previous finding that the phosphatase has a higher catalytic efficiency for phosphopeptides (Kaida et al., 2008). It is also likely that the proteins for spots 1, 3, 4, and 5 are not highly phosphorylated at multiple sites, because the dephosphorylation of their proteins did not cause a marked pH shift to basic pI on the two-dimensional gels. The removal of only one phosphate from a phosphoprotein resulted in a basic shift of nearly 0.05 pH units (Yamagata et al., 2002). Two signals of tobacco PAP were detected on the two-dimensional gel of wall proteins in the position corresponding to a molecular mass of 60 kD at pI values of 7.3 and 7.8 (Fig. 1).
Potential Substrates for PAP
Each of the phosphoproteins visualized in spots 1 to 5 was digested with trypsin and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using Mascot search software (Table I; identified peptides are listed in Supplemental Table S1). Besides the National Center for Biotechnology Information nonredundant (NCBInr) database, the original database of tobacco (including N. tabacum, Nicotiana benthamiana, and Nicotiana sylvestris) ESTs was used for research. Peptides from spot 1 matched the ESTs gi|52834489 and gi|39866468. Four peptide sequences detected from these ESTs with P < 0.05 were 93% identical to nasturtium (Tropaeolum majus) α-xylosidase (Crombie et al., 2002). Peptide fragments from spot 1 also matched potato (Solanum tuberosum) α-glucosidase and Arabidopsis α-xylosidase when the NCBInr database was used. Peptides from spot 2 matched Arabidopsis copper ion-binding protein when the NCBInr database was used. Peptides from spot 3 (in the medium) matched the ESTs gi|52834489, gi|76867473, and gi|39866468. Seven peptide fragments detected from these ESTs were 90% identical to nasturtium α-xylosidase (Crombie et al., 2002). Among these seven fragments, four peptides were identical to those from spot 1 (in the walls). Peptides from spot 5 matched the ESTs gi|33667347 and gi|37359708. Two peptide sequences detected from these ESTs were 79% identical to β-glucosidase from Arabidopsis and 88% identical to β-xylosidase from tomato. No sequence similarity was observed for the peptide sequences from spot 4.
Table I. Identification of phosphoproteins in the apoplast of plant cells by LC-MS/MS analysis.
Spot number corresponds to that shown in Figure 1. A Mascot search was performed (P < 0.05).
| Experimental Value |
Matched Sequence |
Predicted Protein |
|||||
| Spot | Molecular Mass | pI | GenBank gi No. | Score | Protein Name | Species | GenBank Accession No. |
| kD | |||||||
| 1 | 100 | 6.0 | gi|52834489 | 208 | α-Xylosidase | Nasturtium | AJ131520 |
| gi|39866468 | 101 | ||||||
| gi|8919178 | 317 | α-Glucosidase | Potato | CAB96077 | |||
| gi|4163997 | 181 | α-Xylosidase | Arabidopsis | NM_105527 | |||
| 2 | 90 | 8.5 | gi|15242108 | 155 | Copper ion binding | Arabidopsis | NM_124527 |
| 3 | 100 | 6.0 | gi|52834489 | 376 | α-Xylosidase | Nasturtium | AJ131520 |
| gi|76867473 | 190 | ||||||
| gi|39866468 | 110 | ||||||
| 4 | 90 | 5.6 | Not identified | ||||
| 5 | 75 | 6.0 | gi|33667347 | 113 | β-Glucosidase | Arabidopsis | Q9LXD3 |
| gi|37359708 | 212 | β-Xylosidase | Tomato | Q76MS4 | |||
Effects of Dephosphorylation and Phosphorylation on Recombinant AtXYL1
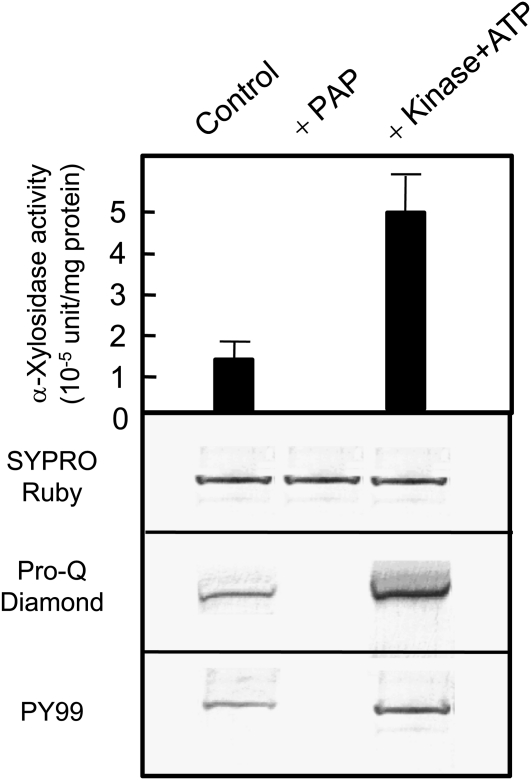
We examined whether the dephosphorylation of potential α-xylosidase protein could affect its activity. When a recombinant protein of Arabidopsis α-xylosidase (AtXYL1; Sampedro et al., 2001) was treated with purified tobacco PAP, the recombinant protein lost a phosphate signal by both Pro-Q Diamond staining and immunostaining with PY99 (Fig. 2). When xyloglucan oligosaccharide was used as a substrate, the dephosphorylation of the protein resulted in a decrease in α-xylosidase activity. A treatment of the recombinant protein with Abl protein Tyr kinase and ATP showed an increase in phosphate signal by both Pro-Q Diamond staining and immunostaining with PY99 and a concomitant 5-fold increase in α-xylosidase activity. The results showed that the dephosphorylation and phosphorylation of recombinant α-xylosidase resulted in a decrease and an increase in its activity, respectively, when xyloglucan heptasaccharide (XXXG) was used as a substrate.
Figure 2.
Dephosphorylation and phosphorylation of recombinant protein (AtXYL1) from S. cerevisiae. The top panel shows the levels of α-xylosidase activity in the recombinant protein treated with purified PAP or Abl protein Tyr kinase. The activity levels were measured in triplicate, and average and sd values are shown. The other panels show SDS-PAGE of the recombinant protein stained with SYPRO Ruby (second panel), Pro-Q Diamond (third panel), and western blot with PY99 antibody (bottom panel).
Attempted Overexpression of NtPAP12 in Tobacco Cells
Transgenic cells overexpressing NtPAP12 under the control of a constitutive promoter were generated (Kaida et al., 2009). The activity of the phosphatase was greater in the walls of the transgenic cells than in the wild-type cells by a factor of 20 (Table II). Based on two-dimensional electrophoresis in the transgenic cells overexpressing NtPAP12, the phosphate signals of spots 1, 3, and 5 corresponding to α-xylosidase and β-glucosidase were detected by staining with SYPRO Ruby but not by staining with Pro-Q Diamond (Supplemental Data S1; Supplemental Fig. S1), showing that the glycosidases might be less phosphorylated in the transgenic cells. In fact, the activity levels of α-xylosidase and β-glucosidase in the transgenic cells were lower than those in the wild-type cells due to their potential substrates as XXXG and cellobiose (Table II). It has been expected that activities of α-xylosidase and β-glucosidase decrease in the transgenic cells, which results in an increase in the accumulation of xyloglucan oligosaccharides. Therefore, we analyzed the amount and type of oligosaccharides present. It should be noted that levels of xyloglucanase and cellulase activities in the apoplastic spaces were almost similar between the transgenic and wild-type cells (data not shown).
Table II. Enzyme activity in the apoplast of the tobacco cells at 5 d after onset of culture.
Data represent means ± sd of three independent experiments. n.d., Not detected.
| Fraction | Tobacco Cells | α-Xylosidase | β-Glucosidase | β-Xylosidase | PAP | Protein |
| 10−3 units per flask | μg per flask | |||||
| Wall | Wild type | 73.9 ± 4.7 | 123 ± 11.7 | n.d. | 0.8 ± 0.1 | 56 |
| Transgenic | 8.5 ± 1.3 | 82 ± 5.1 | n.d. | 16.5 ± 1.9 | 55 | |
| Medium | Wild type | 39.9 ± 6.3 | 41 ± 3.8 | 29.9 ± 1.9 | n.d. | 48 |
| Transgenic | 4.8 ± 0.5 | 8 ± 0.7 | 9.2 ± 1.0 | n.d. | 43 | |
We estimated the amount of xyloglucan oligosaccharides in the culture medium of the transgenic cells at more than 11 mg L−1 as a 4,6-linked Glc and more than 20 mg L−1 as a xylosyl-Glc oligosaccharide. As shown in Table III, the medium in which cells had been cultured for 5 d contained oligosaccharides and polysaccharides for both wild-type cells (2.4 mg per 20 mL) and transgenic cells (2.8 mg per 20 mL). The difference in the quantities of the oligosaccharides in the transgenic cells could be calculated as more than 0.40 mg per 20 mL of culture medium at 5 d.
Table III. Xyloglucan oligosaccharide content at 5 d after onset of culture.
Data represent means ± sd of three independent experiments.
| Medium | Total Carbohydrate | Xyloglucan |
Xyloglucan Oligosaccharides |
||
| Iodine Complex | 4,6-Glc | as 4,6-Glc | as Xylosyl-Glc | ||
| mg per 20 mL | |||||
| Wild type | 2.44 ± 0.25 | 1.25 ± 0.11 | 0.30 ± 0.05 | 0.001 | 0.002 |
| Transgenic | 2.83 ± 0.41 | 1.30 ± 0.12 | 0.52 ± 0.06 | 0.22 | >0.40 |
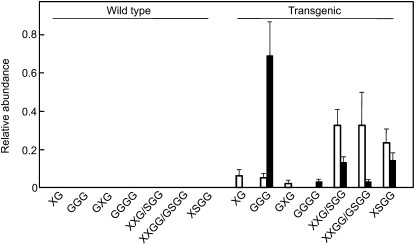
Five-day-old transgenic lines gave seven apparent ions detected by matrix-assisted laser-desorption ionization time of flight/MS, which were consistent with the structures of XG, GXG, XXG/SGG, XXGG/GSGG, and XSGG for xyloglucan oligosaccharides and GGG and GGGG for cello-oligosaccharides in tobacco (Fig. 3). In the wild type, however, no ions were detected at this level. This is in agreement with the content of xyloglucan oligosaccharides (Table III), that the oligosaccharides appeared in the culture medium of transgenic cells at 5 d. Five of the oligosaccharides, XXG/SGG, XXGG/GSGG, XSGG, GGG, and GGGG, also occurred in the cultured medium of the transgenic cells at 7 d, but in different proportions (Fig. 3). This remarkable accumulation of oligosaccharides could be explained by the reduction of α-xylosidase and β-glucosidase activities in the apoplast of the transgenic cells (Table II), since a certain level of xyloglucanase and cellulase activities should cause the degradation of xyloglucan and amorphous cellulose to form their fragment oligosaccharides.
Figure 3.
The relative abundance of each oligosaccharide released into the culture medium from the wild-type and transgenic cells. The white bars indicate samples taken after 5 d of cell culture, and the black bars indicate samples taken after 7 d of culture. Each culture medium was analyzed in triplicate, and average and sd values are shown. The relative abundance of each oligosaccharide (XG, GGG, GXG, GGGG, XXG/SGG, XXGG/GSGG, and XSGG; for nomenclature, see Fry et al., 1993) was calculated from the MS peaks with 496, 527, 659, 689, 791, 953, and 1,085 mass-to-charge ratio values, respectively (Supplemental Fig. S2).
DISCUSSION
Here, we have proposed a potential role for extracellular PAP in tobacco cells: that of a protein phosphatase that appears to catalyze the dephosphorylation of α-xylosidase and β-glucosidase and to modulate their activity in cell walls. There is the possibility that phosphorylation occurs in the walls, because ATP is released from plant cells into the extracellular space (Chivasa et al., 2005a). However, Jamet et al. (2008) reported that neither adenylate kinases nor kinase domains have yet been found in wall proteins through plant cell wall proteomics studies or through bioinformatic searches. Thus, the dephosphorylation of the glycosidases might not be involved in reversible protein phosphorylation, which has been proposed to be a central aspect of cell function and which uses the phosphorylation and dephosphorylation of proteins catalyzed by protein kinases and protein phosphatases (Cohen, 2002). Nothing is known of the phosphorylation mechanism in terms of whether some proteins might be phosphorylated in the simplast and secreted to the apoplast or whether proteins in the ER and Golgi are phosphorylated during the sorting pathway. Nevertheless, there is evidence that phosphoproteins occur in the apoplastic spaces, the walls, and the medium of suspension-cultured tobacco cells (Fig. 1).
We have demonstrated that tobacco PAP can catalyze the dephosphorylation of phosphorylated tobacco wall proteins. One potential α-xylosidase, not only its wall-bound protein but also its recombinant protein, could be neither longer stained with Pro-Q Diamond nor recognized by PY99 antibody against phosphotyrosyl residues (Abel et al., 2001) after treatment with the phosphatase. The treatment with the phosphatase also resulted in a drastic decrease in α-xylosidase activity in the wall protein preparation as well as the recombinant protein (Fig. 2). When the protein could be stained with both Pro-Q Diamond and PY99 antibody (Figs. 1 and 2), they exhibited a certain level of α-xylosidase activity. The phosphorylation with Abl protein Tyr kinase enhanced the level of staining with Pro-Q Diamond and the activity of α-xylosidase (Fig. 2). We propose that the phosphatase acts in vivo on several phosphoproteins, such as potential α-xylosidase and β-glucosidase, because they all occur closely together in the same apoplastic space during growth. In addition, the phosphatase might not be specific for its substrate for phosphoryl residues (Sarmiento et al., 2000; Zhang, 2002).
Overexpression of NtPAP12 decreased the activity levels of α-xylosidase and β-glucosidase and increased the levels of xyloglucan oligosaccharides and cello-oligosaccharides in the apoplast. This occurred not only because these glycosidases degrade xyloglucan oligosaccharides by alternately removing single Xyl and Glc residues from the nonreducing termini (Koyama et al., 1983; Iglesias et al., 2006) but also because β-glucosidase acts on cello-oligosaccharides to release Glc residues from the nonreducing termini. Cello-oligosaccharides are known to act as effectors or primers for cellulose and callose synthases (Hayashi et al., 1987). Treatment of pea (Pisum sativum) hypocotyl segments with auxin increased the solubilization of both xyloglucan oligosaccharides and cello-oligosaccharides in the apoplast of pea stems (Labavitch and Ray, 1974; Tominaga et al., 1999). Taken together, we suggest that the cellulose and callose synthase activities are promoted by the increase in the oligosaccharides caused by the action of PAP. Nevertheless, the phosphatase could have several functions in the phenomena of plant cells due to the various substrate preferences. Studies on the knockout lines of its homolog (AtPAP10) will be included in a subsequent report.
MATERIALS AND METHODS
Culture of Tobacco Cells
The tobacco cell line XD-6, derived from Nicotiana tabacum ‘Xanthi’, was cultured in 40 mL of Murashige and Skoog medium containing 3% Suc and 4.5 μm 2,4-dichlorophenoxyacetic acid in a 100-mL flask. The flask was shaken on a reciprocal shaker at 110 rpm at 25°C for 10 d. Three milliliters of cultured cells was transferred to fresh medium.
Preparation of Wall-Bound Enzymes
Five-day-old suspension-cultured tobacco cells (200 g) in the logarithmic phase of growth were homogenized in 50 mm HEPES-KOH (pH 7.0) buffer using a Teflon-glass homogenizer at 1,500 rpm for 20 min. Insoluble wall materials were collected on nylon mesh (pore size, 50 μm) and washed five times with 10 mm HEPES-KOH buffer (pH 7.0) by centrifugation at 5,000g for 20 min. Enzymes bound to the cell walls were extracted with the same buffer containing 1 m NaCl. The extracts were concentrated by means of precipitation with ammonium sulfate at 65% saturation. Following centrifugation at 14,000g for 30 min, the pellet was dissolved with 10 mm HEPES-KOH (pH 7.0) and dialyzed against the same buffer. We used this fraction as wall enzymes.
Purified PAP was obtained according to the method described in the previous paper (Kaida et al., 2008). The insoluble wall material was washed with 10 mm HEPES-KOH buffer (pH 7.0) followed by the same buffer containing 0.2 m NaCl. The wall-bound phosphatase was extracted with the buffer containing 0.7 m NaCl. After the proteins were concentrated by precipitation with ammonium sulfate and dialyzed against the same buffer, the phosphatase was purified through several columns of hydroxyapatite (Seikagaku), Toyopearl-butyl (Tosoh), and a Superdex 200 HR 10/30 column (10 × 300 mm; GE Healthcare). The purified enzyme was precipitated with ammonium sulfate and stored at −80°C.
Phosphoproteomic Analysis
After 5 d of culture, tobacco cells were homogenized with 50 mm HEPES-KOH buffer (pH 7.0) containing 100 μm vanadate and 1 mm dithiothreitol, and the cell walls were washed with buffer six times. Wall proteins were extracted from the wall residues with the same buffer containing 0.7 m NaCl. Proteins in the medium were precipitated with 70% ammonium sulfate in the presence of 2 mm EGTA and dialyzed against distilled water containing 100 μm vanadate.
For first-dimensional electrophoresis, 50 μg of protein solubilized in rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% dithiothreitol, and 0.5% IPG buffer, pH 3–10 [GE Healthcare]) was loaded onto an Immobiline Dry Strip (70 × 3 × 0.5 mm; GE Healthcare) with a linear pH 3 to 10 gradient and focused using a MultiphorII electrophoresis system (GE Healthcare) at 20°C with a gradient program that increased the voltage (from 200 V for 30 min, to 3,500 V for 90 min, and finally to 3,500 V for 60 min). For second-dimensional SDS-PAGE, the protein was reduced for 15 min in reducing equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and 1% dithiothreitol) and then alkylated for 15 min in an alkylating equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and 2.5% iodoacetamide). After electrophoresis, the gel was stained with Pro-Q Diamond followed by SYPRO Ruby (Invitrogen) and then imaged with an FX Molecular Imager laser scanner (Bio-Rad). PY99 (Santa Cruz Biotechnology; Abel et al., 2001), a mouse monoclonal antibody designed to specifically detect phosphorylated Tyr residues, was used for western-blot analysis.
Protein Identification by MS
After detecting proteins of wild-type tobacco cells on a two-dimensional gel with Pro-Q Diamond and SYPRO Ruby with an image scanner, the gel was washed and the proteins were again stained with silver. Protein spots were excised from the gels and subjected to trypsin (Promega) digestion. The digests were separated using a Magic 2002 capillary HPLC system (Michrom BioResources) with a C-18 RP column (150 mm × 200 μm; GL Sciences) and a linear gradient of 5% to 65% solvent B to solvent A (solvent A, 0.1% formic acid in acetonitrile:distilled water [2:98, v/v]; solvent B, 0.1% formic acid in acetonitrile:distilled water [95:5, v/v]). The column was directly interfaced to an LCQ IT mass spectrometer (ThermoElectron) equipped with a nanoelectrospray ion source. The MS/MS spectra were searched against a NCBInr database and an original tobacco EST database using Mascot search software (version 2.1.03; Matrix Science). The following parameters were used for the search: enzyme, trypsin; missed cleavage, one; variable modification, oxidation of Met; fixed modification, carbamidomethylation of Cys and monoisotopic peptide masses. The peptide sequence was subjected to multiple sequence alignments using dynamic programming.
Recombinant AtXYL (α-Xylosidase)
The AtXYL1 (Sampedro et al., 2001) coding region, excluding the potential signal peptide, was amplified by PCR using cDNAs derived from Arabidopsis (Arabidopsis thaliana) mRNAs as a template, a forward primer containing an EcoRI site (5′-GAATTCTACAAAACCATCGGCAAAGGCTA-3′), and a reverse primer containing an internal NotI site (5′-GCGGCCGCTTAATTGATACCCATTTTCCAGGA-3′). A PCR product was ligated to the pYES2/CT expression vector (Invitrogen), and recombinant protein was overexpressed in yeast (Saccharomyces cerevisiae) transformant (strain INVSc1). Protein expression was induced in minimal medium with 2% Gal at 30°C overnight with continuous shaking. Yeast cells were disrupted in breaking buffer containing 50 mm Tris-HCl (pH 8.5), 5 mm EDTA, 10 μm pyridoxal-5′-phosphate, and 30% glycerol by vortexing with glass beads at 4°C. Cell fragments and nuclei were pelleted at 4,500g for 15 min at 4°C. The recombinant His-tagged protein in yeast was isolated using a Ni2+-charged HisTrap column.
In Vitro Dephosphorylation and Phosphorylation of AtXYL
Some of the protein was dephosphorylated at 30°C for 3 min in a reaction mixture containing 10 units of the purified PAP and 50 mm sodium acetate, pH 5.8, with a total volume of 200 μL. The remainder was phosphorylated at 30°C for 30 min in a reaction mixture containing 10 mm MgCl2, 0.1 mm EDTA, 1 mm dithiothreitol, 0.015% Brij 35, 100 μg mL−1 bovine serum albumin, 0.1 mm ATP, 200 units of Abl protein Tyr kinase (Calbiochem), and 10 mm Tris-HCl, pH 7.5, with a total volume of 200 μL.
Enzyme Assays
The activity levels of various wall-bound enzymes were assayed using the wall-bound preparation as described above.
Phosphatase activity was assayed in a reaction mixture with a total volume of 0.1 mL that included the enzyme, 1 mm phospho-Tyr, and 50 mm sodium acetate buffer (pH 5.8). The amount of phosphate released over the course of 3 min at 30°C was assayed according to the method described by Van Veldhoven and Mannaerts (1987). One unit of activity was defined as the amount of enzyme necessary to produce 1 μmol of phosphate per min at 30°C.
α-Xylosidase was assayed in a reaction mixture with a total volume of 0.6 mL that included the enzyme and 5 mm XXXG in 100 mm sodium acetate buffer (pH 4.5). After incubation for 30 min at 30°C, the reaction mixtures were applied to a column (5 mm × 20 mm) of Dowex H+ and OH− (Dow Chemical) and the eluate was collected. The eluted sugars were converted to their alditol acetates according to the method described by Blakeney et al. (1983). Three to 60 μg of 2-deoxyglucose was used as an internal standard. The sugars were analyzed through gas chromatography (Agilent Technologies) using a DB-225 glass capillary column (0.25 mm × 15 m; Agilent Technologies). One unit of activity was defined as the amount of enzyme required to catalyze 1 μmol of Xyl per min at 30°C. It should be noted that p-nitrophenyl α-xylopyranoside does not serve as a substrate for plant α-xylosidase (Koyama et al., 1983; Sampedro et al., 2001).
β-Glucosidase activity was assayed in a reaction mixture with a total volume of 0.1 mL that included the enzyme preparation, 2 mm reduced cellobiose, and 50 mm sodium acetate buffer (pH 5.0). After incubation at 30°C for 10 min, Glc released was estimated as reducing sugar by the Nelson-Somogyi method (Somogyi, 1952). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of reducing sugar per min at 30°C.
β-Xylosidase activity was assayed in a reaction mixture with a total volume of 0.1 mL that included the enzyme preparation, 2 mm reduced xylobioase, and 50 mm sodium acetate buffer (pH 5.0). After incubation at 30°C for 10 min, the amount of Xyl released was estimated based on the amount of reducing sugar according to the Nelson-Somogyi method (Somogyi, 1952). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of reducing sugar per min at 30°C.
Transformation of Tobacco Cells
NtPAP12 was amplified from tobacco cDNA by PCR using a forward primer (5′-CTGGATCCTGGTGTGCAAAAATGGGTGTG-3′) and a reverse primer (5′-TTCACGATTTGGTTACCATGGACTC-3′). Next, the cDNA was digested with BamHI and SacI and then introduced into the binary vector under the control of an enhanced cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996), and the plasmid was transformed into Agrobacterium tumefaciens LBA4404.
The transformation of tobacco cells was performed according to a previously described method (Kaida et al., 2009). Tobacco cells (cell line XD-6) were cocultivated with the bacteria for 4 d at 26°C. Then the cells were cultured in 40 mL of Murashige and Skoog medium supplemented with 50 μg mL−1 kanamycin and 500 μg mL−1 cefotaxime. When the culture reached the stationary phase, 3 mL of culture was transferred to fresh medium supplemented with 200 μg mL−1 kanamycin and 500 μg mL−1 cefotaxime. After an additional 10 culture cycles in the selective medium, we had established a transgenic cell line that had been subcultured more than 200 times in the presence of 50 μg mL−1 kanamycin. During inoculation, the phosphatase activity was always determined using phospho-Tyr as a substrate. All subsequent experiments, described below, were performed using medium without antibiotics.
Analysis of Xyloglucan and Its Oligosaccharides
The culture medium of tobacco cells was boiled for 5 min, dialyzed against water using a Spectra/Por MW 100 membrane (Spectrum), and freeze dried. Xyloglucan was quantified using the iodine-sodium sulfate method (Kooiman, 1960). Total xyloglucans, including their fragment oligosaccharides, were measured by methylation analysis, in which their quantity was set at 4,6-linked Glc per total methylated sugars in the presence of laminarin (3-linked Glc) as an internal standard (Hayashi, 1989).
Xyloglucan oligosaccharides were analyzed by oligosaccharide mass profiling (Obel et al., 2006) with a Voyager DE-Pro matrix-assisted laser-desorption ionization time of flight instrument using an acceleration voltage of 20,000 V with a delay time of 350 ns. Mass spectra were obtained in reflectron mode using 2,5-dihydroxybenzoic acid (10 mg mL−1) as the matrix mixed 1:1 (v/v) with solubilized sugars (Lerouxel et al., 2002). A PERL-based program was used for downstream processing of the oligosaccharide mass profiling data (Supplemental Fig. S1). The relative abundance of each oligosaccharide was shown by the mean of the data in three experiments.
The sequences of NtPAP12, NtPAP21, and AtXYL1 can be found in the GenBank/EMBL data libraries under accession numbers AB017967 AB084124, and NM_105527, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Images of phosphoproteomic two-dimensional analyses of proteins in apoplast of the transgenic tobacco cells overexpressing NtPAP12.
Supplemental Figure S2. Matrix-assisted laser-desorption ionization time of flight mass spectra of oligosaccharides released into the culture medium.
Supplemental Figure S3. Images of phosphoproteomic two-dimensional analyses of plasma membrane proteins prepared from the transgenic tobacco cells overexpressing NtPAP12.
Supplemental Table S1. Peptide sequences identified by LC-MS/MS analysis.
Supplemental Data S1. Phosphoproteomic screen in apoplast of the transgenic tobacco cells overexpressing NtPAP12.
Supplemental Data S2. Phosphoproteomic screen in plasma membrane of the transgenic tobacco cells overexpressing NtPAP12.
Supplemental Materials and Methods S1. Preparation of plasma membrane proteins.
Supplementary Material
Acknowledgments
We thank Ali Masoudi-Nejad and Susumu Goto (Bioinformatics Center, Institute for Chemical Research, Kyoto University) for producing the assembly of tobacco EST.
References
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. (2001) Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409: 729–733 [DOI] [PubMed] [Google Scholar]
- Antanaitis BC, Aisen P, Lilienthal HR. (1983) Physical characterization of two-iron uteroferrin. J Biol Chem 258: 3166–3172 [PubMed] [Google Scholar]
- Blakeney AB, Harris PJ, Henry RJ, Stone BA. (1983) Simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299 [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Lindsey K, Slabas AR. (2005a) Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. Plant Cell 17: 3019–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Simon WJ, Yu XL, Yalpani N, Slabas AR. (2005b) Pathogen elicitor-induced changes in the maize extracellular matrix proteome. Proteomics 5: 4894–4904 [DOI] [PubMed] [Google Scholar]
- Cohen P. (2002) The origins of protein phosphorylation. Nat Cell Biol 4: E127–E130 [DOI] [PubMed] [Google Scholar]
- Crombie HJ, Chengappa S, Jarman C, Sidebottom C, Reid JS. (2002) Molecular characterisation of a xyloglucan oligosaccharide-acting alpha-D-xylosidase from nasturtium (Tropaeolum majus L.) cotyledons that resembles plant ‘apoplastic’ alpha-D-glucosidases. Planta 214: 406–413 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, del la Pena A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress condition. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Ek-Rylander B, Flores M, Wendel M, Heinegards D, Andersson G. (1994) Dephosphorylation of osteopontin and bone sialoprotein by osteoclastic tartrate-resistant acid phosphatase. J Biol Chem 269: 14853–14856 [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Hayashi T. (1989) Measuring β-glucan deposition in plant cell walls. Linskens HF, Jackson JF, , Modern Methods of Plant Analysis: Plant Fibers, 10. Springer-Verlag, Berlin, pp 138–160 [Google Scholar]
- Hayashi T, Marsden MPF, Delmer DP. (1987) Pea xyloglucan and cellulose. V. Xyloglucan-cellulose interactions in vitro and in vivo. Plant Physiol 83: 384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Abelenda JA, Rodino M, Sampedro J, Revilla G, Zarra I. (2006) Apoplastic glycosidases active against xyloglucan oligosaccharides of Arabidopsis thaliana. Plant Cell Physiol 47: 55–63 [DOI] [PubMed] [Google Scholar]
- Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R. (2008) Recent advances in plant cell wall proteomics. Proteomics 8: 893–908 [DOI] [PubMed] [Google Scholar]
- Kaida R, Hayashi T, Kaneko T. (2008) Purple acid phosphatase in the walls of tobacco cells. Phytochemistry 69: 2546–2551 [DOI] [PubMed] [Google Scholar]
- Kaida R, Sage-Ono K, Kamada H, Okuyama H, Syono K, Kaneko TS. (2003) Isolation and characterization of four cell wall purple acid phosphatase genes from tobacco cells. Biochim Biophys Acta 1625: 134–140 [DOI] [PubMed] [Google Scholar]
- Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS. (2009) Activation of β-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol 150: 1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Sträter N, Fröhlich R, Witzel H, Kreb B. (1996) Mechanism of Fe(III)-Fe(II) purple acid phosphatase based on crystal structure. J Mol Biol 259: 737–748 [DOI] [PubMed] [Google Scholar]
- Kooiman P. (1960) A method for determination of amyloid in plant seeds. Recl Trav Chim Pays Bas 79: 675–678 [Google Scholar]
- Koyama T, Hayashi T, Kato Y, Matsuda K. (1983) Degradation of xyloglucan by wall-bound enzymes from soybean tissue. II. Degradation of the fragment heptasaccharide from xyloglucan and the characteristic action pattern of the β-D-xylosidase in the enzyme system. Plant Cell Physiol 24: 155–162 [Google Scholar]
- Kwon H-K, Yokoyama R, Nishitani K. (2005) A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol 46: 843–857 [DOI] [PubMed] [Google Scholar]
- Labavitch JM, Ray PM. (1974) Relationship between promotion of xyloglucan metabolism and induction of elongation by indoleacetic acid. Plant Physiol 54: 499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBansky BB, McKnight TD, Griffing LR. (1992) Purification and characterization of a secreted purple phosphatase from soybean suspension cultures. Plant Physiol 99: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M. (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Wong FL, Phang TH, Cheung MY, Li WYF, Shao G, Yan X, Lam HM. (2003) GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318: 103–111 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Ndimba BK, Chivasa S, Hamilton JM, Simon WJ, Slabas AR. (2003) Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttleman PR, Roberts RM. (1990) Transfer of iron from uteroferrin (purple acid phosphatase) to transferrin related to acid phosphatase activity. J Biol Chem 265: 12192–12199 [PubMed] [Google Scholar]
- Obel N, Erben V, Pauly M. (2006) Functional wall glycomics through oligosaccharide mass profiling. Hayashi T, , The Science and Lore of the Plant Cell Wall: Biosynthesis, Structure and Function. BrownWalker, Boca Raton, FL, pp 258–266 [Google Scholar]
- Sampedro J, Sieiro C, Revilla G, Gonzalez-Villa T, Zarra I. (2001) Cloning and expression pattern of a gene encoding an α-xylosidase activity against xyloglucan oligosaccharides from Arabidopsis. Plant Physiol 126: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano A, Kaida R, Maki H, Kaneko TS. (2003) Involvement of an acid phosphatase on cell wall regeneration of tobacco protoplast. Physiol Plant 119: 121–125 [Google Scholar]
- Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y, Lawrence DS, Almo SC, Zhang ZY. (2000) Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry 39: 8171–8179 [DOI] [PubMed] [Google Scholar]
- Sibille JC, Doi K, Aisen P. (1987) Hydroxyl radical formation and iron-binding proteins. J Biol Chem 262: 59–62 [PubMed] [Google Scholar]
- Somogyi M. (1952) Notes on sugar determination. J Biol Chem 195: 19–23 [PubMed] [Google Scholar]
- Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, Hendrickson J, Beechem JM, Haugland RP, Patton WF. (2003) Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics 3: 1128–1144 [DOI] [PubMed] [Google Scholar]
- Taylor NG. (2007) Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites: a potential role in regulating protein degradation. Plant Mol Biol 64: 161–171 [DOI] [PubMed] [Google Scholar]
- Tominaga R, Samejima M, Sakai F, Hayashi T. (1999) Occurrence of cello-oligosaccharides in the apoplast of auxin-treated pea stems. Plant Physiol 119: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven PP, Mannaerts GP. (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem 161: 45–48 [DOI] [PubMed] [Google Scholar]
- Yamagata A, Kristensen DB, Takeda Y, Miyamoto Y, Okada K, Inamatsu M, Yoshizato K. (2002) Mapping of phosphorylated proteins on two-dimensional polyacrylamide gels using protein phosphatase. Proteomics 2: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Zhang W, Gruszewski HA, Chevone BI, Nessler CL. (2008) An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY. (2002) Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol 42: 209–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.