Abstract
Three genes encoding flavonoid 3′-hydroxylase (F3′H) in apple (Malus × domestica), designated MdF3′HI, MdF3′HIIa, and MdF3′HIIb, have been identified. MdF3′HIIa and MdF3′HIIb are almost identical in amino acid sequences, and they are allelic, whereas MdF3′HI has 91% nucleotide sequence identity in the coding region to both MdF3′HIIa and MdF3′HIIb. MdF3′HI and MdF3′HII genes are mapped onto linkage groups 14 and 6, respectively, of the apple genome. Throughout the development of apple fruit, transcriptional levels of MdF3′H genes along with other anthocyanin biosynthesis genes are higher in the red-skinned cv Red Delicious than that in the yellow-skinned cv Golden Delicious. Moreover, patterns of MdF3′H gene expression correspond to accumulation patterns of flavonoids in apple fruit. These findings suggest that MdF3′H genes are coordinately expressed with other genes in the anthocyanin biosynthetic pathway in apple. The functionality of these apple F3′H genes has been demonstrated via their ectopic expression in both the Arabidopsis (Arabidopsis thaliana) transparent testa7-1 (tt7) mutant and tobacco (Nicotiana tabacum). When grown under nitrogen-deficient conditions, transgenic Arabidopsis tt7 seedlings expressing apple F3′H regained red color pigmentation and significantly accumulated both 4′-hydrylated pelargonidin and 3′,4′-hydrylated cyanidin. When compared with wild-type plants, flowers of transgenic tobacco lines overexpressing apple F3′H genes exhibited enhanced red color pigmentation. This suggests that the F3′H enzyme may coordinately interact with other flavonoid enzymes in the anthocyanin biosynthesis pathway.
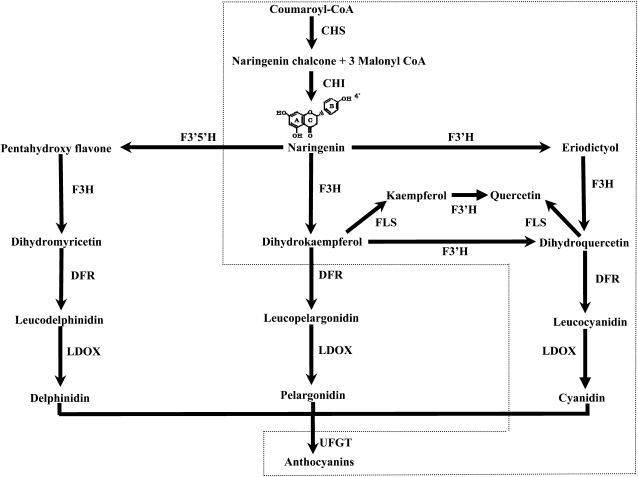
Flavonoids are ubiquitous secondary metabolites in higher plants and play important roles in myriad activities, such as protecting plants from UV radiation and pathogen infection, providing flowers and seeds with pigmentation to attract pollinators and seed dispersers, and reducing the risk of oxidative damage to human health (Regan et al., 2001; Schaefer et al., 2004; Veeriah et al., 2006). Flavonoids consist mainly of anthocyanins, chalcone, flavone, flavonol, flavanone, and isoflavonoids. Of these flavonoid molecules, anthocyanins are broadly distributed in flowering plants and predominantly contribute to both flower and fruit colors. The biosynthetic pathway of anthocyanins has been well established in different plants, including petunia (Petunia hybrida), snapdragon (Antirrhinum majus), and maize (Zea mays; Holton and Cornish, 1995; Winkel-Shirley, 2001; Grotewold, 2006). Briefly, the biosynthesis of anthocyanins begins with the condensation of malonyl-CoA with 4-coumaroyl-CoA, leading to the formation of naringenin chalcone (Fig. 1), and this reaction is catalyzed by the enzyme chalcone synthase (CHS). The chalcone is subsequently converted into naringenin by chalcone flavanone isomerase (CHI). Naringenin is then hydroxylated, at the 3′ position of the central ring, by flavanone 3-hydroxylase (F3H) to produce dihydrokaempferol (DHK). DHK can be further hydroxylated at the 3′ position or at both 3′ and 5′ positions of the B-ring to produce dihydroquercetin and dihydromyricetin, respectively. DHK, dihydroquercetin, and dihydromyricetin generally lead to the production of the brick-red/orange pelargonidin-, red/pink cyanidin-, and blue/violet delphinidin-based pigments, respectively (Grotewold, 2006). Therefore, the hydroxylation pattern plays an important role in coloration. Moreover, the hydroxylation pattern is also an important determinant of the flavonoid stability and antioxidant capacity (Rice-Evans et al., 1996; Croft, 1998).
Figure 1.
The biosynthesis pathways of the most abundant anthocyanin pigments. The proposed pathway in apple is marked with dotted lines.
The hydroxylation pattern of the B-ring is controlled by two members of the vast and versatile cytochrome P450 family, flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H). Both F3′H and F3′5′H are microsomal cytochrome P450-dependent monooxygenases that require NADPH as a cofactor. F3′H and F3′5′H introduce hydroxyl groups at the 3′ or both 3′ and 5′ positions of the B-ring of the flavonoid molecule, respectively, leading to the formation of 3′,4′- and 3′,4,′5′-hydroxylated flavonoids, respectively. Some plants such as Arabidopsis (Arabidopsis thaliana), apple (Malus × domestica), and rose (Rosa species) do not have functional F3′5′H enzymes (Forkmann, 1991). To date, flavonoid hydroxylases have been investigated in plants, as they highly influence flower coloration. Genes encoding F3′H and F3′5′H have been isolated in myriad plant species, including petunia (Holton et al., 1993; Menting et al., 1994; Brugliera et al., 1999), Arabidopsis (Schoenbohm et al., 2000), lisianthus (Eustoma grandiflorum; Noda et al., 2004), and grape (Vitis vinifera; Bogs et al., 2006), among others. Manipulation of F3′H and F3′5′H genes has been effective in genetic engineering of floral crops to develop new genotypes with novel flower colors for ornamental purposes (Shimada et al., 1999; Mori et al., 2004; Nakatsuka et al., 2007).
Apples are among the most important fruit trees grown around the world and are reported to possess high levels of antioxidants when compared with other groups of fruits, vegetables, and even tea (Camellia sinensis; Chinnici et al., 2004). The domesticated apple belongs to the family Rosaceae. It is self-incompatible and a highly heterozygous diploid with a haploid chromosome number of 17 (2n = 34; Tatum et al., 2005; Korban et al., 2009). Fruit color is one of the most important commercial traits, as it strongly influences consumer purchase and consumption of apple fruit. Generally, red-skinned apples are preferred over other colors of apples, as consumers tend to associate these with better taste, ripeness, and flavor (King and Cliff, 2002).
The molecular mechanism underlying color development in apple fruits has not been well investigated. To date, cDNA clones encoding secondary metabolic enzymes such as dihydroflavonol reductase (DFR) and anthocyanidin synthase have been isolated from apple (Kim et al., 2003). Transcription factors that coordinately regulate genes involved in the anthocyanin biosynthesis pathway in apple have also been identified (Espley et al., 2006, 2009; Takos et al., 2006). However, genes encoding F3′H have not yet been reported in apple, although they play important roles in both flower and fruit coloration. Recently, a large number of EST sequences from apple have been developed in our laboratory and deposited in the GenBank/EMBL/DDBJ databases (Gasic et al., 2009). These EST sequences together with our previously constructed bacterial artificial chromosome (BAC) libraries (Xu et al., 2001, 2002) provide us with a unique opportunity to investigate genes involved in flavonoid biosynthesis in apple.
In this study, we report on the isolation of a gene family encoding F3′H in apple and investigate the functionality of these F3′H genes via their ectopic expression in both Arabidopsis and tobacco (Nicotiana tabacum). This knowledge elucidates the mechanism responsible for the hydroxylation of flavonoids in both apple and other higher plants. Moreover, this will aid in future efforts to modify anthocyanin biosynthesis in apple as well as other plants.
RESULTS
Isolation and Sequence Analysis of Three Gene Copies Encoding F3′H in Apple
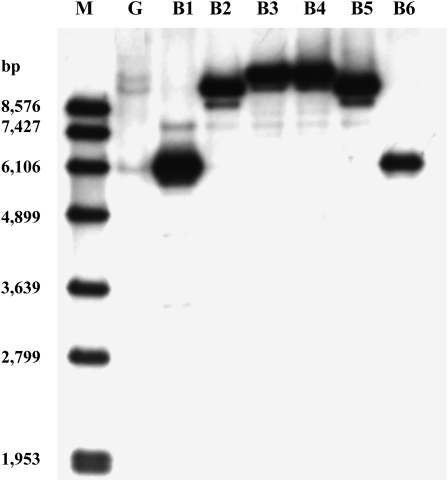
A total of six positive apple BAC clones, designated B1 to B6, were identified. BAC DNA of these six clones together with genomic DNA of apple cv GoldRush were subjected to DNA-blot analysis, and three different sizes of bands were generated (Fig. 2). This indicated that three copies of genes encoding F3′H were detected in apple. Moreover, three pairs of BAC clones, B1/B6, B2/B5, and B3/B4, yielded low-, middle-, and high-Mr bands, respectively, suggesting that each pair of BAC clones contained a different copy of genes encoding F3′H. Therefore, BAC clones B1, B2, and B3 were selected and subjected to subcloning.
Figure 2.
Southern-blot analysis of apple genomic DNA and BAC DNAs. The cDNA probe corresponds to a partial region of the last exon of MdF3′HI. M, Standard DNA marker; G, apple genomic DNA. B1 to B6 correspond to six positive BAC clones. Each BAC DNA yields a strong band corresponding to one of the three bands from the genomic DNA.
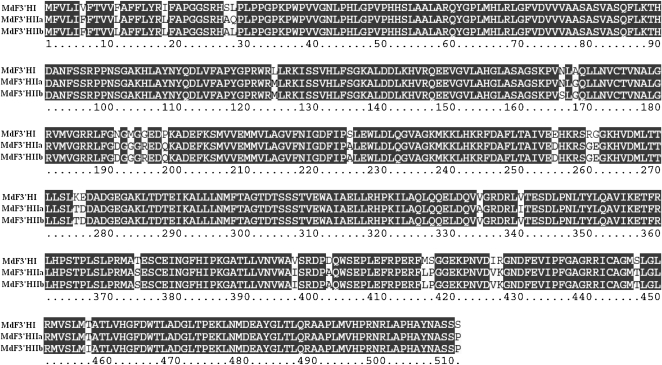
Three F3′H genes, designated MdF3′HI (GenBank accession no. FJ919631), MdF3′HIIa (GenBank accession no. FJ919632), and MdF3′HIIb (GenBank accession no. 919633), have been isolated and sequenced. All MdF3′H genes are composed of three exons with an open reading frame of 1,536 bp encoding a putative protein of 511 amino acids. Exons of MdF3′HI, MdF3′HIIa, and MdF3′HIIb span 3,651, 3,272, and 3,884 bp of genomic DNA fragments, respectively. MdF3′HI shows approximately 90% and approximately 65% nucleotide sequence identities, in coding and genomic regions, respectively, with either MdF3′HIIa or MdF3′HIIb. MdF3′HIIa and MdF3′HIIb share 99% and 97% nucleotide sequence identities in coding and genomic regions, respectively. MdF3′HI shows 95% amino acid sequence identity with both MdF3′HIIa and MdF3′HIIb. The deduced amino acid sequences of MdF3′HIIa and MdF3′HIIb are almost identical with only four different sequences (Fig. 3).
Figure 3.
Comparisons of the deduced amino acid sequences of the three apple F3′H genes. Alignment was carried out using ClustalX. Sequence mismatches are noted with lowercase letters.
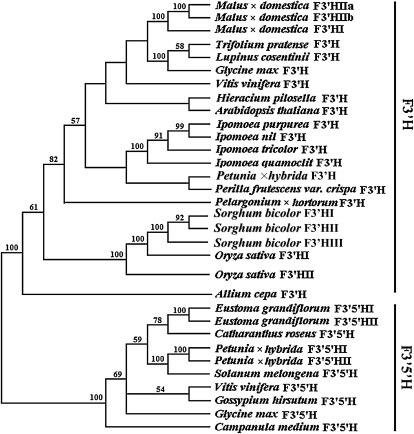
A phylogenetic analysis was performed using deduced amino acid sequences of genes encoding flavonoid hydroxylase from apple and from other plants, and two clades, designated F3′H and F3′5′H clades, were generated (Fig. 4). These two clades were highly supported with 100% bootstrap values. The three apple F3′H genes, MdF3′HI, MdF3′HIIa, and MdF3′HIIb, were grouped into the F3′H clade, indicating that they were all genes encoding the F3′H.
Figure 4.
Phylogenetic tree derived from amino acid sequences of genes encoding flavonoid hydroxylase in plants. The phylogenetic analysis was performed using the maximum parsimony method. Numbers on branches correspond to bootstrap estimates for 100 replicate analyses using 500× stepwise addition of taxa; values less than 50% are not indicated.
Physical Relationships between MdF3′HIIa and MdF3′HIIb Genes
MdF3′HI, MdF3′HIIa, and MdF3′HIIb were isolated from BAC clones B1, B2, and B3, respectively. To identify the physical relationships among these BAC clones, a BAC-based physical map of the whole apple genome was used (Han et al., 2007). It was found that B2 and B3 overlapped and were located on the same BAC contig 2917 (Supplemental Fig. S1). This indicated that MdF3′HIIa and MdF3′HIIb might either be allelic or clustered. To further clarify the physical relationships between B2 and B3, the following was pursued. First, genomic DNA fragments, approximately 7 kb in size, downstream of 3′ untranslated regions of both MdF3′HIIa and MdF3′HIIb were sequenced. Sequence alignment revealed that these two fragments were highly similar, with more than 99% identity in nucleotide sequences, and suggesting that genomic fragments of MdF3′HIIa and MdF3′HIIb overlapped at the same locus. Second, a BAC library of apple cv GoldRush, constructed using HindIII and representing approximately 5× haploid apple genome equivalents, was screened, and a total of eight BAC clones were identified to contain F3′H genes. A DNA-blotting analysis indicated that all eight BAC clones, similar to those six BAC clones, B1 to B6, from a BamHI-constructed BAC library of apple cv GoldRush, contained only a single copy of F3′H. This suggested that F3′H genes were not clustered within the apple genome. Altogether, these results strongly demonstrated that MdF3′HIIa and MdF3′HIIb were allelic.
Tagging and Mapping of MdF3′H Genes
Analysis of genomic DNA sequences indicated that the second intron of all three MdF3′H genes contained a (CT)n repeat. Therefore, two pairs of primers (5′-CGCCAAGCTCACAGACACTG-3′/5′-CGGGTTGATTGTTGCACATC-3′ and 5′-GGATGATGCTGACGGTGAG-3′/5′-TCACTTCTTGAGCGTTCATCTT-3′) flanking the (CT)n repeat were designed. Two gene-tagged simple sequence repeat (SSR) markers, designated as F3′HI-SSR and F3′HII-SSR, were successfully developed for MdF3′HI and MdF3′HII, respectively. Genomic DNA sequence comparisons between MdF3′HIIa and MdF3′HIIb revealed the presence of an approximately 540-bp insertion/deletion (indel) in the first intron. A pair of primers flanking the indel (forward, 5′-CCACGATGGCGGATGTTACG-3′; reverse, 5′-CGGTCAAGAAGGCATCGAAC-3′) were then designed and successfully used to develop a gene-tagged sequence-tagged-site marker, designated F3′HII-Indel for the MdF3′HII gene.
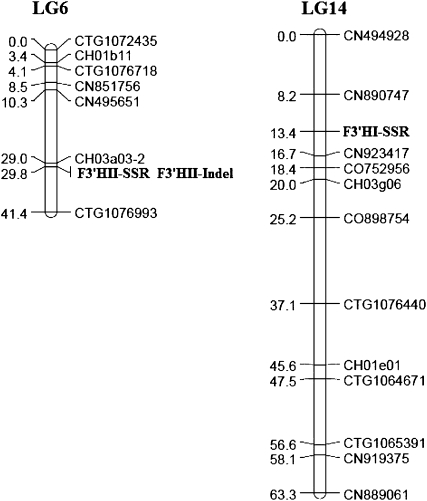
Recently, we developed an EST-SSR-based genetic linkage map for the apple genome using an apple segregating mapping population derived from a cross between Co-op 17 and Co-op 16 (Y. Han, D. Zheng, S. Vimolmangkang, J.E. Beever, and S.S. Korban, unpublished data). To genetically map these F3′H genes in apple, the three gene-tagged markers, F3′HI-SSR, F3′HII-SSR, and F3′HII-Indel, were used to screen this segregating population. The results revealed that MdF3′HI and MdF3′HII genes mapped onto linkage groups 14 and 6, respectively (Fig. 5).
Figure 5.
Genetic mapping of F3′H genes in apple. Markers linked to F3′H genes are marked in boldface. The MdF3′HI gene is tagged with F3′HI-SSR, while the MdF3′HII gene is anchored by F3′HII-SSR or F3′HII-Indel. LG6 and LG14 represent linkage groups 6 and 14, respectively.
Expression Profiles of MdF3′H Genes and Other Anthocyanin Biosynthetic Genes in Apple
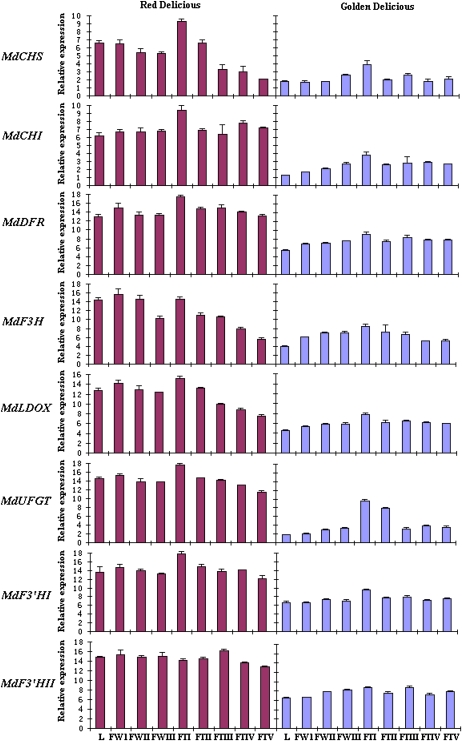
Expression profiles of MdF3′HI and MdF3′HII genes in a red-colored fruiting apple, cv Red Delicious, and a yellow-colored fruiting apple, cv Golden Delicious, were investigated using real-time PCR. Both MdF3′HI and MdF3′HII genes were expressed in all analyzed tissues, including leaves, flowers, and fruits (Fig. 6). Transcriptional levels of both MdF3′HI and MdF3′HII in all tissues in Red Delicious were higher than those in Golden Delicious. Accumulation of MdF3′HI transcripts reached a peak in fruits of both Red Delicious and Golden Delicious at the early developmental stage, 2 weeks after pollination, and subsequently showed a slight decline throughout fruit development. Transcript accumulation of MdF3′HII in both Red Delicious and Golden Delicious was slightly enhanced throughout fruit development, with a peak at the mid stage of development (15 weeks after pollination). Transcriptional levels of MdF3′HI and MdF3′HII were relatively higher in developing flowers than those in young leaves of both Red Delicious and Golden Delicious.
Figure 6.
Analysis of expression profiles of anthocyanin genes in apple cv Red Delicious (red skin) and cv Golden Delicious (yellow skin) using real-time PCR. The cDNA templates are listed as follows: L, young leaves; FWI, flower buds at the pink stage; FWII, flower buds at the balloon stage; FWIII, flowers at full bloom; FTI, 2 weeks after pollination (WAP); FTII, 6 WAP; FTIII, 15 WAP; FTIV, 20 WAP; FTV, 24 WAP. Transcriptional levels were normalized to expression of an apple actin gene. All data correspond to mean values of three biological replicates. [See online article for color version of this figure.]
HPLC analysis demonstrated that Red Delicious had higher levels of flavonols, proanthocyanidins (PAs), and anthocyanidins than Golden Delicious (Table I). To monitor flavonoid pathway activity, expression profiles of six other anthocyanin biosynthetic genes, MdCHS, MdCHI, MdDFR, MdF3H, MdLDOX, and MdUFGT, were also measured in Red Delicious and Golden Delicious by real-time PCR (Fig. 6). Similar to MdF3′H genes, these genes showed higher levels of transcripts in Red Delicious than in Golden Delicious in almost all tissues analyzed. The accumulation of these gene transcripts in fruits from both Red Delicious and Golden Delicious reached a peak at the early developmental stage (2 weeks after pollination) and declined thereafter until fruit maturity. Transcript levels of MdUFGT, involved in the last step of anthocyanin synthesis, were significantly lower in fruits of Golden Delicious than in Red Delicious. Thus, expression of anthocyanin biosynthetic genes was consistent with the accumulation of flavonoids in apple fruits.
Table I. HPLC analysis of flavonoid in fruits of apple cv Red Delicious and cv Golden Delicious.
Fruit stage was the same as indicated in Figure 6 as follows: FTI, 2 weeks after pollination (WAP); FTII, 6 WAP; FTIII, 15 WAP; FTIV, 20 WAP; FTV, 24 WAP. N/D indicates that a peak was not detected, and Tr indicates that the peak was present but its level was too low. All data correspond to mean values of three biological replicates.
| Cultivar | Skin Color | Stage | Flavonol |
Proanthocyanidin |
Anthocyanidin |
|||
| Kaempferol | Quercetin | Catechin | Epicatechin | Pelargonidin | Cyanidin | |||
| μg g−1 | ||||||||
| Red Delicious | Red | FTI | 24.8 | 958.5 | 0.8 | 0.5 | N/D | 6.5 |
| FTII | 10.7 | 705.0 | 0.8 | 1.0 | N/D | 8.3 | ||
| FTIII | 1.0 | 127.5 | 6.6 | 24.0 | N/D | 5.6 | ||
| FTIV | Tr | 69.3 | 1.6 | 10.2 | N/D | 7.5 | ||
| FTV | Tr | 41.9 | 1.0 | 6.5 | N/D | 9.5 | ||
| Golden Delicious | Yellow-green | FTI | 28.5 | 781.2 | 0.5 | 0.1 | N/D | 4.9 |
| FTII | 9.5 | 428.4 | 0.4 | 0.3 | N/D | 5.8 | ||
| FTIII | 0.9 | 113.1 | 1.2 | 14.9 | N/D | 2.9 | ||
| FTIV | Tr | 115.7 | 0.7 | 6.2 | N/D | 2.2 | ||
| FTV | 0.8 | 86.0 | 0.3 | 4.8 | N/D | 2.8 | ||
Functional Analysis of MdF3′H Genes in an Arabidopsis Mutant and in Tobacco
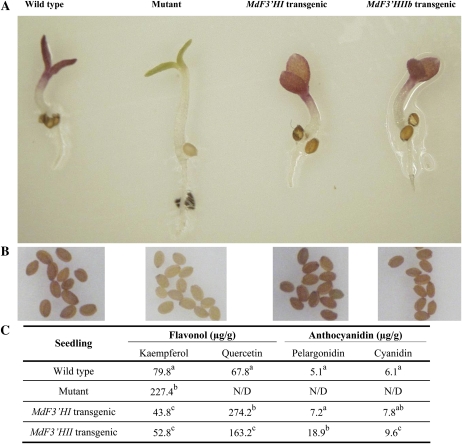
Of the three apple F3′H genes, MdF3′HIIa and MdF3′HIIb were allelic and almost identical in amino acid sequences. Therefore, only two genes, MdF3′HI and MdF3′HIIb, were subjected to functional analysis. The Arabidopsis transparent testa7-1 (tt7-1) mutant, lacking a flavonoid 3′-hydroxylase, was selected to investigate the functionality of MdF3′H genes. Coding region sequences encoding MdF3′HI and MdF3′HIIb were separately transferred into the Arabidopsis tt7-1 mutant under the control of the cauliflower mosaic virus 35S promoter, and several transgenic lines were generated for each construct. Seeds of the Arabidopsis tt7-1 mutant, T2 transgenic lines, and wild-type Arabidopsis were germinated and grown on half-strength Murashige and Skoog (MS) medium but without nitrogen. Germinating seedlings of wild-type plants and transgenic lines expressing either MdF3′HI or MdF3′HIIb had red cotyledons, whereas cotyledons of the Arabidopsis tt7-1 mutant were green (Fig. 7A). Moreover, seeds collected from kanamycin-resistant T2 plants showed pigmentation characteristic of the wild-type Arabidopsis, while seeds of the Arabidopsis tt7-1 mutant were pale brown in color (Fig. 7B).
Figure 7.
Complementation of the pigmentation of Arabidopsis tt7 mutant seedlings of the ecotype Landsberg erecta with apple F3′H genes. A, Phenotypes of wild-type, mutant, and transgenic Arabidopsis seedlings grown in nitrogen-deficient MS medium. B, Phenotypes of wild-type, mutant, and transgenic Arabidopsis seeds. C, Contents of anthocyanidins and flavonols in Arabidopsis seedlings grown in nitrogen-deficient MS medium. Data correspond to means of three biological replicates. Means with different letters within the same column are significantly different at the 0.01 level of probability. Two additional transgenic lines each of MdF3′HI and MdF3′HII were analyzed, and these exhibited similar phenotypes and HPLC profiles as shown. N/D, Not determined.
HPLC analysis of seedlings grown on half-strength MS medium without nitrogen revealed that transgenic Arabidopsis lines contained higher levels of quercetin, pelargonidin, and cyanidin but lower levels of kaempferol than wild-type Arabidopsis (Fig. 7C). These results clearly demonstrated that both MdF3′HI and MdF3′HIIb were functional. In addition, anthocyanidins, including pelargonidin and cyanidin, were identified in transgenic and wild-type Arabidopsis seedlings grown under nitrogen-deficient conditions, but these were not detectable in Arabidopsis tt7-1 mutant seedlings. These findings strongly suggested that F3′H genes might also play important roles in the synthesis of both 3′,4′-hydroxylated cyanidin and 4′-hydroxylated pelargonidin.
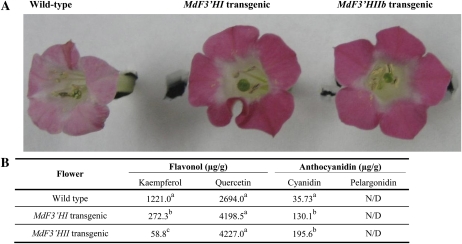
Coding region sequences of MdF3′HI and MdF3′HIIb were also separately transferred into tobacco under the control of the cauliflower mosaic virus 35S promoter. T2 transgenic tobacco lines expressing MdF3′HI or MdF3′HIIb showed markedly enhanced intensity of flower color when compared with wild-type tobacco plants (Fig. 8A). Transgenic lines had red flowers, whereas wild-type plants produced pale pink flowers. HPLC analysis of these tissues indicated that transgenic flowers produced higher levels of cyanidin than wild-type flowers (Fig. 8B). Flowers of those transgenic lines expressing either MdF3′HI or MdF3′HIIb also showed significantly higher amounts of quercetin but lower levels of kaempferol than those of nontransgenic control tobacco. However, pelargonidin was not detected in wild-type and transgenic lines.
Figure 8.
Functional characterization of MdF3′H genes following their overexpression in transgenic tobacco lines. A, Differences in color between transgenic and wild-type tobacco flowers. B, Contents of anthocyanidins and flavonols in transgenic and wild-type tobacco flowers. All data correspond to mean values of three biological replicates. Values with different letters within the same column are significantly different at the 0.01 level of probability. Two transgenic lines each of MdF3′HI and MdF3′HII were analyzed and showed similar phenotypes and HPLC profiles as shown. N/D, Not determined.
DISCUSSION
Genes encoding F3′H and F3′5′H have been well investigated in several ornamental plants such as petunia, rose, and carnation (Dianthus caryophyllus). However, there are few reports on genes encoding flavonoid hydroxylase from fruit trees. Recently, Bogs et al. (2006) has reported on the identification of VvF3′H and VvF3′5′H genes in grapevine. In this study, we report on the isolation and functional analysis of F3′H genes from apple. It is important to note that apple does not have functional F3′5′H enzymes, while grapevine has both F3′H and F3′5′H enzymes. Thus, patterns of anthocyanin accumulation must be different between apple and grapevine. Thereby, findings reported in this study will aid in a comprehensive understanding of F3′H genes in different fruit crops.
Duplication of F3′H Genes in Plants
Gene duplication is assumed to be a major driving force for recruitment of genes for secondary metabolism (Pichersky and Gang, 2000). This reported gene duplication in plants may arise from polyploidy (genome duplication) and/or segmental duplication. These two patterns of gene duplications have also been detected during evolutionary development of F3′H genes in plants. For example, there are two copies of F3′H genes in the rice (Oryza sativa) genome (http://rgp.dna.affrc.go.jp/E/toppage.html), and these are clustered on chromosome 10, suggesting that they were derived following segmental duplication. In this study, two F3′H genes, MdF3′HI and MdF3′HII, sharing 91% nucleotide sequence identity in the coding region, have been identified in the apple genome. Genetic mapping results have indicated that MdF3′HI and MdF3′HII genes are located on linkage groups 14 and 6, respectively. These results together with the reported allopolyploid origin of the apple genome suggest that duplication of F3′H genes in apple is likely derived from whole-genome duplication during the process of speciation (Xu and Korban, 2004). Moreover, polyploidization is a significant evolutionary process in higher organisms, and genomes of flowering plants are reported to have incurred one or more polyploidization events during evolution (Masterson, 1994). Whole-genome duplication has also occurred during the evolutionary process of speciation of Arabidopsis (Ermolaev et al., 2003). However, database analysis of the whole-genome sequence of Arabidopsis indicates that there is only a single copy of the F3′H gene in the Arabidopsis genome (http://www.arabidopsis.org/). Thus, the evolutionary processes of Arabidopsis and rice F3′H genes are consistent with previously reported findings that gene copies involved in secondary metabolism are likely to be retained when derived from segmental duplication, whereas duplicated gene copies following whole-genome duplications are rapidly lost (Maere et al., 2005). If it is true that F3′H genes derived following whole-genome duplication are likely to be lost, then further studies must be conducted to clarify whether or not the two apple F3′H genes are likely derived from a segmental duplication followed by translocation.
MdF3′H Genes Coordinately Expressed with Other Anthocyanin Biosynthetic Genes
Expression profiles of genes involved in anthocyanin synthesis have been investigated in both red- and non-red-colored peel and flesh apple genotypes (Espley et al., 2006; Takos et al., 2006). For all genes assayed, including MdCHS, MdCHI, MdF3H, MdDFR, MdFLS, MdLAR, MdANR, MdLDOX, and MdUFGT, transcriptional levels in fruit tissues of either red skin and/or flesh apples have been significantly higher than those of non-red skin or flesh genotypes. The F3′H gene is associated with accumulation of cyanidin pigments, and it is demonstrated to play an important role in plant coloration (Hoshino et al., 2003; Todaa et al., 2005). However, there are no reports on apple MdF3′H genes. In this study, transcriptional levels of apple MdF3′H and other anthocyanin biosynthetic genes in red- and yellow-colored skin genotypes have been investigated (Fig. 6). Similar to other anthocyanin biosynthesis genes, transcriptional levels of both MdF3′HI and MdF3′HII in all tested tissues, including leaves, flowers, and fruits, are found to be higher in red-colored cv Red Delicious than in the yellow-colored cv Golden Delicious. This finding further suggests that MdF3′H genes are coordinately expressed with other anthocyanin biosynthetic genes in apple. Moreover, expression profiles of MdF3′H genes are quite similar to those of the VvF3′H gene in grapevine. Transcriptional levels of the VvF3′H gene in grape are also reported to be higher in red than in white berries (Bogs et al., 2006). Thus, it seems that the F3′H gene family is also one of the important determinant factors that influence the accumulation of anthocyanin in fruit. In addition, Red Delicious has accumulated higher levels of PAs and anthocyanidins than Golden Delicious (Table I). Taken together, our results demonstrate that transcriptional levels of anthocyanin biosynthesis genes are responsible for fruit color differences among different apple genotypes.
Based on the flavonoid biosynthesis pathway, CHS is the first enzyme in this pathway, while F3′H provides a branching point for the biosynthesis of flavonols (Grotewold, 2006). In this study, the structural genes MdCHS and MdF3′H exhibit slight down-regulated expression during fruit development in the red-skinned Red Delicious (Fig. 6). This finding is consistent with the observed decline in accumulation of flavonols in apple fruit throughout the duration of fruit development. Likewise, LDOX competes with leucoanthocyanidin reductase for the same substrate leucocyanidin and catalyzes the synthesis of cyanidin, which is one of the precursors of PAs (Grotewold, 2006). In this study, levels of PAs decrease during the late stages of fruit development, and this is consistent with down-regulation of MdLDOX expression in apple fruit. On the other hand, other structural genes, such as MdCHI and MdDFR, display relatively stable levels of expression throughout fruit development in Red Delicious. Expression of these genes is consistent with the observed stable levels of anthocyanins detected in apple fruit.
In Arabidopsis, specific regulators of the anthocyanin biosynthesis pathway have been identified. For example, MYB11, MYB12, and MYB111 show a high degree of functional similarity and display very similar target gene specificities for several flavonoid biosynthesis genes, including AtCHS, AtCHI, AtF3H, and AtFLS (Stracke et al., 2007). The transcription factor AtTT2 encodes an R2R3-MYB domain protein and regulates AtDFR, AtLDOX, and AtANR (Nesi et al., 2001). In apple, genes involved in anthocyanin synthesis show higher levels of expression in red skin/flesh than in non-red skin/flesh, suggesting that the expression of flavonoid genes is likely to be controlled by a common regulator. More recently, two transcription factors, MdMYB1 and MdMYB10, have been reported in apple (Bogs et al., 2006; Espley et al., 2006, 2009). Transcriptional levels of MdMYB1 and MdMYB10 in fruits are significantly different between red and non-red skin or flesh apples. Thus, it is reasonable to speculate that the two transcription factors coordinately regulate genes in the anthocyanin biosynthesis pathway. As there are several gene families involved in the anthocyanin biosynthesis pathway, and each gene family may consist of several members, further studies must be conducted in the future to clarify how transcriptional factors influence several different genes involved in this pathway.
Functional Conservation of F3′H Genes in Plants
Complementation of petunia and Arabidopsis flavonoid mutants with maize genes has revealed that genes encoding CHS, CHI, and dihydroflavonol 4-reductase are functionally conserved (Meyer et al., 1987; Dong et al., 2001). Recently, Bogs et al. (2006) have isolated the VvF3′H gene from grapevine and demonstrated the functionality of this gene by ectopic expression in petunia ht1 mutant line Skr4 × Sw63. Transgenic petunia lines have shown changes in both flower color and flavonoid composition. In this study, MdF3′H genes have been separately transferred into the Arabidopsis tt7 mutant, and transgenic seedlings grown under nitrogen stress conditions have shown similar patterns of anthocyanin accumulation as those of wild-type Arabidopsis. Moreover, flavonoid accumulation in transgenic seedlings grown without nitrogen stress treatment has also been determined (Supplemental Table S2). Similar to wild-type Arabidopsis, transgenic seedlings accumulate quercetin-type flavonols and cyanidin-based pigments. Therefore, it is evident that F3′H genes are also functionally exchangeable among different plant species. In addition, transgenic tobacco seedlings expressing these MdF3′H genes have also shown higher levels of accumulation of cyanidin pigments than those of wild-type tobacco. This has suggested that manipulation of the F3′H gene family will contribute to modification of coloration in plants and, therefore, is a viable approach for engineering plants to modify coloration.
In this study, two F3′H loci, MdF3′HI and MdF3′HII, have been identified in the apple genome. MdF3′HI and MdF3′HII have 91% and 95% nucleotide and amino acid sequence identities, respectively. Ectopic expression of MdF3′H genes has demonstrated that MdF3′HI transgenic Arabidopsis lines accumulate significantly higher levels of quercetin but significantly lower levels of both pelargonidin and cyanidin than MdF3′HII transgenic Arabidopsis lines (Fig. 7C). MdF3′HI transgenic tobacco lines accumulate lower levels of cyanidin but significantly higher levels of kaempferol than MdF3′HII transgenic tobacco lines (Fig. 8B). It is not clear whether these observed differences in flavonoid accumulation among transgenic Arabidopsis and tobacco lines carrying different F3′H genes suggest a divergence of MdF3′HI from MdF3′HII. To address this question, substrate specificities of these two genes will be investigated in future experiments.
The two alleles MdF3′HIIa and MdF3′HIIb were identified and mapped, based on two gene-tagged markers in the first and second introns, onto linkage group 6. Likewise, a gene-tagged SSR marker was also developed, based on a (CT)n repeat in the second intron, for the MdF3′HI gene. The SSR marker was used to screen a segregating population derived from a cross between Co-op 17 and Co-op 16, and three alleles were identified (Supplemental Fig. S2). It has been reported that the substitution of a single amino acid could lead to changes in substrate specificities of enzymes involved in anthocyanin biosynthesis (Johnson et al., 2001). Therefore, it would be useful to determine whether there is any functional divergence between/among alleles of MdF3′HI and MdF3′HII genes. In addition, those gene-tagged markers developed in this study provide a molecular tool for functional studies and for marker-assisted selection of F3′H genes in apple.
Characterization of Flavonoid Biosynthesis in Apple
Several plant species such as petunia, tobacco, and grapevine do not produce pelargonidin-based anthocyanins, as their DFR cannot utilize DHK as a substrate (Meyer et al., 1987; Aharoni et al., 2001; Bogs et al., 2006). In this study, components of anthocyanidin have been determined in apple fruits, and no pelargonidin was detected in both red-colored and yellow-colored genotypes (Table I). This finding demonstrates that the apple DFR cannot efficiently reduce DHK. Previously, it has been reported that there is no functional F3′5′H in apple, an enzyme involved in the biosynthesis of delphinidin-based anthocyanin (Forkmann, 1991). Thus, apple only produces cyanidin-based anthocyanin, and the proposed pathway is presented in Figure 1. The substrate specificity of DFR enzymes is noteworthy. Johnson et al. (2001) have compared DFRs of petunia with those in several other plants such as maize, gerbera (Gerbera hybrida), and rose that can catalyze the reduction of DHK and have found a presumed substrate-binding region (Supplemental Fig. S3). Furthermore, a change of a single amino acid in the region has further demonstrated that a residue at the 134th position plays an important role in determining substrate specificity. A DFR that cannot accept DHK as a substrate contains an Asp residue at the 134th position, whereas a DFR that can reduce DHK contains an Asn residue (Johnson et al., 2001). An apple DFR has an Asn residue at the 134th position, but it cannot utilize DHK as a substrate (Supplemental Fig. S3). Therefore, the substrate specificity of DFR is probably determined not only by either Asn or Asp at the 134th position but also by other residue(s) in the substrate-binding region.
Previous studies have demonstrated that non-red-colored skin apple cultivars are capable of synthesizing PAs and flavonols but do not synthesize anthocyanin (Lata et al., 2005; Vanzani et al., 2005; Takos et al., 2006). In this study, when the yellow-skinned cv Golden Delicious is subjected to flavonoid analysis, PAs, flavonols, and cyanidin are identified throughout fruit development (Table I). It has been widely documented that ripe Golden Delicious fruit lacks anthocyanin (Vrhovsek et al., 2004; Takos et al., 2006; Lata, 2007; Stracke et al., 2009). Thus, it is clear that the deficiency of anthocyanin in Golden Delicious is attributed to a block in the last step in the anthocyanin biosynthesis pathway that is catalyzed by the UFGT enzyme. The level of expression of MdUFGT is lower in Golden Delicious than that in the red-skinned Red Delicious (Fig. 6). Currently, an effort is under way to determine whether or not a functional MdUFGT gene(s) is present in Golden Delicious. The results will elucidate the mechanism underlying the deficiency of anthocyanin in non-red skin colored apple cultivars.
Levels of the flavonols kaempferol and quercetin are high during early fruit development in apple, and these decline throughout subsequent stages of fruit development. Those upstream genes in the flavonoid biosynthesis pathway, including MdCHS, MdCHI, MdF3H, and MdF3′H, exhibit higher levels of expression during the early stages of fruit development (Fig. 6). Thus, accumulation of flavonols is consistent with expression of those upstream genes in the flavonoid biosynthesis pathway. Levels of kaempferol dramatically decline to almost zero during later stages of fruit development. However, expression of MdFLS is slightly down-regulated during late stages of fruit development (Supplemental Fig. S4). This inconsistency may be attributed to relatively high levels of expression of MdF3′H genes that convert kaempferol to quercetin and to the competition of MdF3′H and MdFLS for the same substrate, DHK. Similarly, the content of quercetin declines significantly during apple fruit development. This may be due to the competition of MdDFR with MdFLS for the DHK substrate and to relatively stable levels of expression of MdDFR. Therefore, it is clear that MdF3′H influences the biosynthesis of flavonols in apple.
Leucocyanidin can be converted into the monomeric PA units catechin and epicatechin through two pathways, either via a single-step reaction catalyzed by LAR or a two-step reaction catalyzed by LDOX and BANYULS. In this study, accumulation of both catechin and epicatechin is low in young fruits, but these reach peak values at the mid stage of fruit development and then gradually drop as the fruit reaches maturity. The expression of MdF3′HII shows a peak at the mid stage of fruit development, and both MdF3′HI and MdF3′HII gradually display down-regulated expression during late fruit development. The observed consistency between the expression of MdF3′H genes and PA accumulation during late fruit development suggests that MdF3′H may affect the biosynthesis of PAs in apple. Finally, cyanindin content is relatively stable, while levels of MdF3′H gene expression are high throughout fruit development. Moreover, expression patterns of MdF3′H genes correspond to accumulation patterns of 3′,4′-hydroxylated flavonoids. Altogether, expression of MdF3′H genes is consistent with the biosynthesis of flavonols, PAs, and anthocyanins in apple fruit.
Effects of Nitrogen Stress on Flavonoid Biosynthesis in Arabidopsis
It has been reported that Arabidopsis DFR enzymes can utilize DHK as a substrate, but they fail to do so in Arabidopsis plants when a functional F3′H is present (Dong et al., 2001). Thus, an Arabidopsis DFR preferentially reduces dihydroquercetin in wild-type seedlings grown under low nitrogen stress, resulting in high levels of cyanidin-based anthocyanins and no detectable pelargonidin-based pigments (Dong et al., 2001). Interestingly, in this study, wild-type and transgenic Arabidopsis seedlings grown on a nitrogen-deficient medium have accumulated both pelargonidin and cyanidin and produced red pigments in cotyledons (Fig. 7). Moreover, wild-type and transgenic Arabidopsis seedlings grown without nitrogen stress have accumulated high levels of pelargonidin but only small amounts of cyanidin (Supplemental Table S2). Olsen et al. (2009) have recently reported on the effects of nitrogen on regulators and those products of the flavonoid biosynthesis pathway. In this study, Arabidopsis seedlings were grown on either nitrogen-deficient medium or standard MS medium, whereas Dong et al. (2001) grew their seedlings only under low nitrogen stress. Thus, nitrogen stress may significantly affect the accumulation patterns of anthocyanin in Arabidopsis.
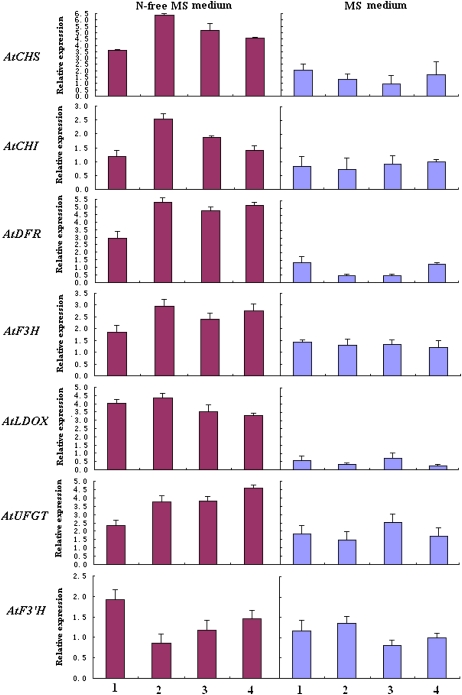
To gain insights into the mechanism underlying the effects of nitrogen stress on anthocyanin in Arabidopsis, we have analyzed the expression of anthocyanin pathway genes in both wild-type and transgenic seedlings grown with or without nitrogen stress (Fig. 9). Overall, these genes, including AtCHS, AtCHI, AtDFR, AtF3H, AtLDOX, AtF3′H, and AtUFGT, demonstrate higher levels of expression in seedlings grown under nitrogen-deficient stress compared with seedlings grown without nitrogen stress. It is worth noting that expression levels of the two genes AtDFR and AtLDOX are significantly enhanced in seedlings grown under nitrogen-deficient stress. Further studies are needed to clarify whether these observed enhanced levels of expression of anthocyanin biosynthesis genes, particularly AtDFR and AtLDOX, are responsible for changes in accumulation patterns of anthocyanin in Arabidopsis seedlings grown under nitrogen-deficient stress.
Figure 9.
Analysis of expression profiles of anthocyanin genes in Arabidopsis seedlings grown in both MS medium and nitrogen-free MS medium using real-time PCR. The cDNA templates are listed as follows: 1, the wild type; 2, tt7 mutant; 3, MdF3′HI transgenic line; 4, MdF3′HIIb transgenic line. Transcriptional levels were normalized to expression of an Arabidopsis actin gene. All data correspond to mean values of three biological replicates. [See online article for color version of this figure.]
The Arabidopsis tt7-1 mutant carries an internal stop codon in the putative AtF3′H gene (Schoenbohm et al., 2000). Previous studies have reported that the Arabidopsis tt7-1 mutant accumulates very low levels of pelargonidin, and as a result either no or barely visible anthocyanin pigments are detected in both seeds and seedlings (Koorneef et al., 1982; Dong et al., 2001). In this study, accumulation of both cyanidin and pelargonidin in Arabidopsis tt7-1 mutant seedlings grown under either nitrogen-deficient or nonstress conditions is not detectable (Supplemental Table S2). This is consistent with the findings that when seedlings of this tt7-1 mutant are grown with or without nitrogen-deficient stress, they reveal no differences in color pigmentation, as both have green coloration. Therefore, it seems that F3′H genes are indispensable for the accumulation of pelargonidin in Arabidopsis seedlings grown under nitrogen-deficient stress conditions. However, it is not clear how F3′H genes induce the biosynthesis of pelargonidin in seedlings grown under nitrogen-deficient conditions. Several studies have indicated that flavonoid biosynthetic enzymes could form large macromolecular complexes through specific protein-protein interactions (Burbulis and Winkel-Shirley, 1999; Winkel-Shirley, 1999). Thus, accumulation of both pelargonidin and cyanidin in Arabidopsis seedlings grown under nitrogen-deficient conditions may be due to interactions between the F3′H enzyme and DFR or to other flavonoid biosynthetic enzymes, thus leading to changes in DFR substrate specificity.
Arabidopsis synthesizes PAs in seed coats, and these PAs contain only epicatechin and no detectable catechin (Abrahams et al., 2002). In this study, we have found that there is no detectable epicatechin and catechin in wild-type plants and in transgenic Arabidopsis seedlings grown under both nitrogen-deficient and nonstress conditions (Supplemental Table S2). These findings indicate that nitrogen stress may have no effect on the synthesis of PAs in Arabidopsis seedlings. In addition, accumulation of flavonols is higher in Arabidopsis seedlings grown under nitrogen-deficient stress compared with seedlings grown without nitrogen stress.
CONCLUSION
Flavonoid biosynthesis is spatially and temporally regulated in apple fruit. In this study, we have identified two apple MdF3′H gene families that have higher levels of expression in the red-skinned cv Red Delicious than in the yellow-skinned cv Golden Delicious. These two gene families are coordinately expressed with other structural genes in the anthocyanin biosynthetic pathway in apple fruit. Expression of MdF3′H genes corresponds to the biosynthesis of flavonoid in apple fruit. Gene expression studies and biochemical analysis reveal that the deficiency of anthocyanin in the fruit of Golden Delicious is due to a block in the final step in the anthocyanin biosynthesis pathway. Ectopic expression studies of MdF3′H genes clearly demonstrate that these play important roles in the biosynthesis of flavonoid and that nitrogen stress has a strong influence on the expression of anthocyanin biosynthetic genes in Arabidopsis.
MATERIALS AND METHODS
Plant Material
Wild-type, tt7 mutant, and T2 transgenic seeds of Arabidopsis (Arabidopsis thaliana) were germinated on half-strength MS medium with or without nitrogen. After 10 d of growth, seedlings were collected and stored at −80°C until needed. Wild-type and T2 transgenic plants of tobacco (Nicotiana tabacum) were grown in the greenhouse, and flowers were harvested at the full-bloom stage. Apple (Malus × domestica) fruits at different stages of development (early to maturity) were collected and stored at −80°C until needed, and the whole fruit was used for gene expression and flavonoid biosynthesis analyses.
Identification of BAC Clones Containing Apple F3′H Genes
The deduced amino acid sequence of an EST contig of accession Apple_0223.261.C2.Contig645 in our apple EST database (http://titan.biotec.uiuc.edu/cgi-bin/ESTWebsite/estima_start?seqSet=apple) is blasted against the GenBank database (http://www.ncbi.nlm.nih.gov). This apple EST contig is highly homologous to F3′H genes from other plants such as grape (Vitis vinifera), soybean (Glycine max), sorghum (Sorghum bicolor), Arabidopsis, and petunia (Petunia hybrida; greater than 70% in amino acid sequences).
This apple EST contig was then used to design a pair of primers (forward, 5′-CGTCATCAAGCACGGTGGAATG-3′; reverse, 5′-CTCAAGCGGTTCGGACCATTG-3′) to screen an apple BAC library according to a previously described PCR-based screening protocol (Xu et al., 2002). The BAC library was developed from apple cv GoldRush using BamHI and corresponded to 5× haploid genome equivalents.
Southern Blotting of Genomic and BAC DNA
A total of 5 μg of genomic DNA from leaves of cv GoldRush and 25 μg of BAC DNA, per positive clone, were digested with BamHI, separated on 0.8% agarose gels, and transferred onto Hybond-N nylon membranes (Amersham Biosciences) using the capillary transfer method. Hybridization was carried out using the DIG Easy Hyb kit (Roche). DNA probes were prepared using the PCR DIG Probe Synthesis Kit (Roche) according to the manufacturer's instructions. Blots were washed once with a low-stringency buffer (2× SSC containing 0.1% SDS) for 10 min at room temperature and twice with a high-stringency buffer (0.5× SSC containing 0.1% SDS) for 15 min at 65°C. Then, they were exposed to a Lumi-Film x-ray film (Hyperfilm; Amersham Biosciences) at room temperature for 25 min.
Subcloning of BAC DNAs into the Plasmid Vector pBluescript SK+
A total of 5 μg of purified BAC DNA was partially digested with Sau3Al. Digested fragments of approximately 8 kb were collected from a 1% agarose gel using a QIAEX II gel extraction kit (Qiagen) and then ligated into a BamHI-digested pBluescript SK+ vector. Ligation products were transformed into Escherichia coli competent cells by electroporation using a Bio-Rad gene pulser.
Recovery of Full-Length cDNA of Apple F3′H Genes
The full-length cDNA fragments of apple F3′H genes were recovered using both 5′- and 3′-RACE. Based on genomic DNA sequences of apple F3′H genes, two pairs of gene-specific primers, 5′-CCGGATCGCGAGATACGGCCCATAC-3′/5′-GGCCCATACGTTGACCAGAAGAGTG-3′ and 5′-GACCCTTGGGCTGCGTATGGTGTCTC-3′/5′-GACCCTTGGGCTGCGTATGGTGTCTC-3′, were designed for 5′- and 3′-RACE, respectively. The 5′- and 3′-RACEs were performed using the BD SMART RACE cDNA Amplification Kit according to the protocol recommended by the manufacturer (BD Biosciences). cDNA templates were synthesized from young fruit tissues of apple cv GoldRush.
Expression Profiles of MdF3′H Genes in Apple Using Real-Time PCR
Total RNA from fruit tissues was extracted according to the protocol described by Gasic et al. (2004). For leaf and flower tissues, total RNA was extracted using the RNAqueous Kit (Ambion/Applied Biosystems) according to the manufacturer's instructions. Approximately 3 μg of total RNA per sample was treated with DNase I (Invitrogen Life Science) and then used for cDNA synthesis.
A SYBR Green-based real-time PCR assay was carried out in a total volume of 25 μL of reaction mixture containing 12.5 μL of 2× SYBR Green I Master Mix (Applied Biosystems), 0.2 μm of each primer, and 100 ng of template cDNA. An actin gene was used as a constitutive control along with the following primer sequences: 5′-CTACAAAGTCATCGTCCAGACAT-3′ and 5′-TGGGATGACATGGAGAAGATT-3′. Reaction mixtures without cDNA templates were also run as negative controls to evaluate the specificity of the real-time PCR. Amplifications were performed using a 7300 Real-Time PCR System (Applied Biosystems). The amplification program consisted of one cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescent product was detected at the last step of each cycle. Melting curve analysis was performed at the end of 40 cycles to ensure the proper amplification of target fragments. Fluorescence readings were consecutively collected during the melting process from 60.0°C to 90.0°C at the heating rate of 0.5°C s−1. A negative control without cDNA template was run with each analysis to evaluate the overall specificity. All analyses were repeated three times using biological replicates. Differences in cycle thresholds between target and actin genes corresponded to levels of gene expression. All primer sequences used for real-time PCR are listed in Supplemental Table S1.
Expression Vector Construction and Plant Transformation
Two pairs of primers, 5′-CCATGGATCCGATGTTTGTTCTCATAGTCTTCACCG-3′/5′-CACGTGAGCTCTCAAGATGATGATGCATTGT-3′ and 5′-CCATGGATCCGATGTTTGTTCTCATATTCTTCACCG-3′/5′-CACGTGAGCTCTCAAGGTGATGACGCATTAT-3′, were designed to amplify whole coding regions of MdF3′HI and MdF3′HII genes, respectively, using cDNA extracted from leaves of cv GoldRush as templates. The forward and reverse primers contained NcoI/BamHI and PmlI/SacI sites at the 5′ end, respectively. PCR products were digested with BamHI and SacI and then inserted into BamHI/SacI-digested pBI121. As a result, two constructs containing coding regions of MdF3′HI and MdF3′HIIb were generated.
The two constructs of MdF3′H genes were separately transformed into the Arabidopsis tt7-1 mutant and tobacco (cv Petite Havana SR1). The Arabidopsis tt7-1 mutant (CS88) with the Landsberg erecta genetic background was obtained from the Arabidopsis Biological Resource Center (Ohio State University). Arabidopsis transformation was carried out according to the floral dip method (Clough and Bent, 1998). For transgenic plant selection, T0 seeds were sterilized and germinated on half-strength MS modified basal salt mixture without nitrogen (PhytoTechnology Laboratories), 0.5% (w/v) agar, and 1% (w/v) Suc. The medium was adjusted to pH 5.7 with potassium hydroxide and containing 12 μg mL−1 kanamycin. Following 1 week of selection, kanamycin-resistant plants with red cotyledons were transplanted to soil and placed in a growth chamber at 22°C and 50% to 80% relative humidity. Tobacco transformation was carried out using an Agrobacterium tumefaciens-mediated leaf transformation protocol as described previously (Horsch et al., 1985).
Flavonoid Analysis
Anthocyanins and flavonols were extracted from 50 mg of finely ground tissues in 250 μL of 80% (v/v) methanol at room temperature and centrifuged at 13,000 rpm for 10 min. Approximately 100 μL of the supernatant was transferred to a fresh tube and acid hydrolyzed by adding 30 μL of 3 n HCl, incubated at 70°C for 1 h, and then mixed with 50 μL of 100% methanol. PAs were extracted using 0.1 g of finely ground plant tissue in 1 mL of 70% (v/v) acetone containing 0.1% (w/v) ascorbate. The extract was incubated at room temperature for 24 h in the dark and then centrifuged at 13,000 rpm for 15 min at room temperature. Approximately 200 μL of supernatant was transferred to a new tube, evaporated at 35°C, and then resuspended in 100 μL of 1% HCl-methanol (v/v) and 100 μL of 200 mm sodium acetate (pH 7.5). Flavonoid contents were determined using HPLC. Flavonol extracts (20 μL) were injected into the Phenomenex Gemini 3u C6-phenyl 110A column. Solvent A consisted of 0.1% formic acid in water, and solvent B consisted of acetonitrile. The flow rate was 250 μL min−1. The gradient conditions were as follows: 0 min, 10% B; 10 min, 50% B; 10.5 to 15 min, 100% B; and 15.5 to 21 min, 10% B. The flavonol compounds were identified with reference to commercial standards of kaempferol, cyanidin, pelargonidin, quercetin, catechin, and epicatechin (Sigma). Analyses of each sample were repeated three times using three biological replicates.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ919631 (MdF3′HI), FJ919632 (MdF3′HIIa), FJ919633 (MdF3′HIIb), BAE71221 (Trifolium pratense F3′H), AAX53074 (Lupinus cosentinii F3′H), AAO47847 (soybean F3′H), CAI54287 (grape F3′H), ABC47161 (Hieracium pilosella F3′H), AAG16746 (Arabidopsis F3′H), BAD00191 (Ipomoea purpurea F3′H), BAC00190 (Ipomoea nil F3′H), BAC00192 (Ipomoea tricolor F3′H), AAS46257 (Ipomoea quamoclit F3′H), AAD56282 (petunia F3′H), BAB59005 (Perilla frutescens var crispa F3′H), AAG49315 (Pelargonium × hortorum F3′H), ABG54319 (sorghum F3′HI), ABG54321 (sorghum F3′HII), ABG54320 (sorghum F3′HIII), AAM00948 (rice F3′HI), NP_001064333 (rice F3′HII), AAS48419 (Allium cepa F3′H), AAB17562 (lisianthus F3′5′HI), BAD34460 (lisianthus F3′5′HII), CAA09850 (Catharanthus roseus F3′5′H), CAA80266 (petunia F3′5′H), CAA80265 (petunia F3′5′H), CAA50155 (Solanum melongena F3′5′H), CAI54277 (grape F3′5′H), AAP31058 (Gossypium hirsutum F3′5′H), AAM51564 (soybean F3′5′H), and BAA03440 (Campanula medium F3′5′H).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A physical contig spanning the MdF3′HII gene in apple.
Supplemental Figure S2. A gene tag for the MdF3′HI gene in apple.
Supplemental Figure S3. Sequence alignment of DFR fragments from different plants.
Supplemental Figure S4. Expression profiles of MdFLS genes in apple fruit.
Supplemental Table S1. Primers for real-time PCR analysis of anthocyanin gene expression in apple.
Supplemental Table S2. HPLC analysis of flavonoids in Arabidopsis seedlings.
Supplementary Material
References
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR. (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O'Connell AP. (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Bogs J, Ebadi A, McDavid D, Robinson SP. (2006) Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol 140: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugliera F, Barri-Rewell G, Holton TA, Mason JG. (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19: 441–451 [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B. (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96: 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnici F, Bendini A, Gaiani A, Riponi C. (2004) Radical scavenging activities of peels and pulps from cv. Golden Delicious apples as related to their phenolic composition. J Agric Food Chem 52: 4684–4689 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Croft KD. (1998) The chemistry and biological effects of flavonoids and phenolic acids: towards prolongation of the healthy life span. Ann N Y Acad Sci 854: 435–442 [DOI] [PubMed] [Google Scholar]
- Dong X, Braun EL, Grotewold E. (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaev MD, Wu M, Eisen JA, Salzberg SL. (2003) The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol 51: 859–866 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volzd R, Putterill J, Schoutene HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Puterill J, Kutty-Amma S, Allan AC. (2006) Red colouration in apple fruit is due to the activity of a MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G. (1991) Flavonoids as flower pigments: the formation of the natural spectrum and its extension by genetic engineering. Plant Breed 106: 1–26 [Google Scholar]
- Gasic K, Gonzalez DO, Thimmapuram J, Liu L, Malnoy M, Gong G, Han Y, Vodkin LO, Aldwinckle HS, Carroll NJ, et al. (2009) Comparative analysis and functional annotation of a large expressed sequence tag collection of apple. Plant Genome 2: 23–38 [Google Scholar]
- Gasic K, Hernandez A, Korban SS. (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22: 437–438 [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Han Y, Gasic K, Marron B, Beever JE, Korban SS. (2007) A BAC-based physical map of the apple genome. Genomics 89: 630–637 [DOI] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JG, Lu CY, Farcy E, Stevenson TW, Cornish EC. (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366: 276–279 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Frayley RT. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hoshino A, Morita Y, Choi J, Saito N, Toki K, Tanaka Y, Iida S. (2003) Spontaneous mutations of the flavonoid 3-hydroxylase gene conferring reddish flowers in the three morning glory species. Plant Cell Physiol 44: 990–1001 [DOI] [PubMed] [Google Scholar]
- Johnson E, Ryu S, Yi HK, Shin B, Cheong H, Choi G. (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25: 325–333 [DOI] [PubMed] [Google Scholar]
- Kim S, Lee J, Hong S, Yoo Y, An G, Kim S. (2003) Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 65: 403–413 [Google Scholar]
- King MC, Cliff MA. (2002) Development of a model for prediction of consumer liking from visual attributes of new and established apple cultivars. J Am Pom Soc 56: 223–229 [Google Scholar]
- Koorneef M, Luiten W, de Vlaming P, Schram AW. (1982) A gene controlling flavonoid 3′-hydroxylation in Arabidopsis. Arabidopsis Inf Serv 19: 113–115 [Google Scholar]
- Korban SS, Wannarat W, Rayburn CM, Tatum T, Rayburn AL. (2009) Genome size and nucleotypic variation in Malus germplasm. Genome 52: 148–155 [DOI] [PubMed] [Google Scholar]
- Lata B. (2007) Relationship between apple peel and the whole fruit antioxidant content: year and cultivar variation. J Agric Food Chem 55: 663–671 [DOI] [PubMed] [Google Scholar]
- Lata B, Przeradzka M, Binkowska M. (2005) Great differences in antioxidant properties exist between 56 apple cultivars and vegetation seasons. J Agric Food Chem 53: 8970–8978 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–424 [DOI] [PubMed] [Google Scholar]
- Menting JGT, Scopes RK, Stevenson TW. (1994) Characterization of flavonoid 3′,5′-hydroxylase in microsomal membrane faction of Petunia hybrida flowers. Plant Physiol 106: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Heidmann I, Forkmann G, Saedler H. (1987) A new petunia flower color generated by transformation of a mutant with a maize gene. Nature 330: 677–678 [DOI] [PubMed] [Google Scholar]
- Mori S, Kobayashi H, Hoshi Y, Kondo M, Nakano M. (2004) Heterologous expression of the flavonoid 3′,5′-hydroxylase gene of Vinca major alters flower color in transgenic Petunia hybrida. Plant Cell Rep 22: 415–421 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M. (2007) Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep 26: 1951–1959 [DOI] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N, Kanno Y, Kato N, Kazuma K, Suzuki M. (2004) Regulation of gene expression involved in flavonol and anthocyanin biosynthesis during petal development in lisianthus (Eustoma grandiflorum). Physiol Plant 122: 305–313 [Google Scholar]
- Olsen KM, Slimestad R, Lea US, Brede C, Løvdal T, Ruoff P, Verheul M, Lillo C. (2009) Temperature and nitrogen effects on regulators and products of the flavonoid pathway: experimental and kinetic model studies. Plant Cell Environ 32: 286–299 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Regan BC, Julliot C, Simmen B, Vienot F, Charles-Dominique P, Mollon JD. (2001) Fruits, foliage and the evolution of primate colour vision. Philos Trans R Soc Lond B Biol Sci 356: 229–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice-Evans CA, Miller NJ, Paganga G. (1996) Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic Biol Med 20: 933–956 [DOI] [PubMed] [Google Scholar]
- Schaefer HM, Schaefer V, Levey DJ. (2004) How plant-animal interactions signal new insights in communication. Trends Ecol Evol 19: 577–584 [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381: 749–753 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Nakano-Shimada R, Ohbayashi M, Okinaka Y, Kiyokawa S, Kikuchi Y. (1999) Expression of chimeric P450 genes encoding flavonoid-3′,5′-hydroxylase in transgenic tobacco and petunia plants. FEBS Lett 461: 241–245 [DOI] [PubMed] [Google Scholar]
- Stracke BA, Rufer CE, Weibel FP, Bub A, Watzl B. (2009) Three-year comparison of the polyphenol contents and antioxidant capacities in organically and conventionally produced apples (Malus domestica Bork. cultivar ‘Golden Delicious’). J Agric Food Chem 57: 4598–4605 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR. (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum T, Stepanovic S, Biradar DP, Rayburn AL, Korban SS. (2005) Variation in nuclear DNA content in Malus species and cultivated apples. Genome 48: 924–930 [DOI] [PubMed] [Google Scholar]
- Todaa K, Akasakab M, Dubouzeta EG, Kawasakic S, Takahashia R. (2005) Structure of flavonoid 3′-hydroxylase gene for pubescence color in soybean. Crop Sci 45: 2212–2217 [Google Scholar]
- Vanzani P, Rossetto M, Rigo A, Vrhovsek U, Mattivi F, D'Amato E, Scarpa M. (2005) Major phytochemicals in apple cultivars: contribution to peroxyl radical trapping efficiency. J Agric Food Chem 53: 3377–3382 [DOI] [PubMed] [Google Scholar]
- Veeriah S, Kautenburger T, Habermann N, Sauer J, Dietrich H, Will F, Pool-Zobel BL. (2006) Apple flavonoids inhibit growth of HT29 human colon cancer cells and modulate expression of genes involved in the biotransformation of xenobiotics. Mol Carcinog 45: 164–174 [DOI] [PubMed] [Google Scholar]
- Vrhovsek U, Rigo A, Tonon D, Mattivi F. (2004) Quantitation of polyphenols in different apple varieties. J Agric Food Chem 52: 6532–6538 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (1999) Evidence of enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107: 142–149 [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Korban SS. (2004) Somatic variation plays a key role in the evolution of the Vf gene family residing in the Vf locus that confers resistance to apple scab disease. Mol Phylogenet Evol 32: 57–65 [DOI] [PubMed] [Google Scholar]
- Xu M, Korban SS, Song J, Jiang J. (2002) Constructing a bacterial artificial chromosome library of the apple cultivar GoldRush. Acta Hortic 595: 103–112 [Google Scholar]
- Xu M, Song J, Cheng Z, Jiang J, Korban SS. (2001) A bacterial artificial chromosome (BAC) library of Malus floribunda 821 and contig construction for positional cloning of the apple scab resistance gene Vf. Genome 44: 1104–1113 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.