Arabinogalactan-proteins (AGPs) are undoubtedly one of the most complex families of macromolecules found in plants, perhaps matched only by the polyphenolics (lignins/cutins/suberins) and pectins. Their complexity arises from the incredible diversity of the glycans decorating the protein backbone, the array of peripheral sugars decorating the large arabinogalactan (AG) chains, the microheterogeneity of protein backbone glycosylation, and the diversity of protein backbones containing AG glycomodules. It has been postulated that up to 40% of the Arabidopsis (Arabidopsis thaliana) proteins predicted to be glycosylphosphatidylinositol anchored have the potential to be substituted with AG chains; whether these are all AGPs remains to be established. This complexity is reminiscent of the mammalian extracellular glycoproteins/proteoglycans that are known to be critically important as both structural and functional determinants. These parallels have provided the impetus for plant scientists to establish roles for AGPs in plant growth and development as biological regulatory molecules, with some tantalizing possibilities emerging. They have also been widely used by society for industrial and food applications due to their general emulsifying, adhesive, and water-holding properties. Some AGs/AGPs, such as larch AG and gum arabic, have more recently been investigated as potential immunomodulators of the human immune system.
AGPs belong to a large family of wall glycoproteins/proteoglycans, the Pro/Hyp-rich glycoproteins, originally classified into three separate classes: Pro-rich proteins (PRPs), extensins, and AGPs (Showalter, 1993; Nothnagel, 1997). From various experimental and genomic analyses, it is clear that this family comprises a continuum of molecules from the nonglycosylated/minimally glycosylated PRPs, the moderately glycosylated extensins, to the highly glycosylated AGPs (for review, see Clarke et al., 1979; Fincher et al., 1983; Kieliszewski and Lamport, 1994; Nothnagel, 1997; Cassab, 1998; Serpe and Nothnagel, 1999; Gaspar et al., 2001; Showalter, 2001; Johnson et al., 2003b; Seifert and Roberts, 2007). Despite this continuum and the identification of chimeric and hybrid molecules, there are a large number of molecules that reflect the original classification of extensins, PRPs, and AGPs. This complexity is reminiscent of animal extracellular matrix glycoproteins/proteoglycans and reflects their varied roles as both key structural and regulatory molecules (Filmus et al., 2008; Schaefer and Schaefer, 2010).
AGPs are found on the plasma membrane, in the wall, in the apoplastic space, and in secretions (e.g. stigma surface and wound exudates). They have been found in detergent-resistant membranes in Arabidopsis (Arabidopsis thaliana), suggesting their presence in lipid rafts (Borner et al., 2005). Key distinguishing features (with notable exceptions) of AGPs appear in (1) their carbohydrate, primarily O-linked to the Hyp residues of the protein backbone, that constitutes 90% to 98% (w/w), usually as branched type II arabino-3,6-galactans (AGs; 5–25 kD), although short oligoarabinosides are found (Fig. 1); (2) their protein that typically constitutes 1% to 10% (w/w) and is rich in Hyp/Pro, Ala, Ser, and Thr, with the dipeptide motifs Ala-Hyp, Ser-Hyp, Thr-Hyp, Val-Pro, Gly-Pro and Thr-Pro as distinguishing features, although Ser-(Hyp)2-3 (extensin) motifs can also be present; (3) the presence of a glycosylphosphatidylinositol (GPI) membrane anchor, predicted on most, but not all, AGP protein backbones based on the presence of a hydrophobic, C-terminal domain in the proteins encoded by gene sequences (Fig. 1A); and (4) the ability of most, but not all, AGPs to bind a class of synthetic chemical dyes, the Yariv reagents, in particular the β-glucosyl Yariv reagent (Yariv et al., 1967), that has proven extremely useful in their detection, quantification, and precipitation from solution, a useful early step in their purification. Although the precise mechanism of Yariv binding is not known, it requires the presence of both carbohydrate and protein moieties and is highly variable in strength (Pettolino et al., 2006).
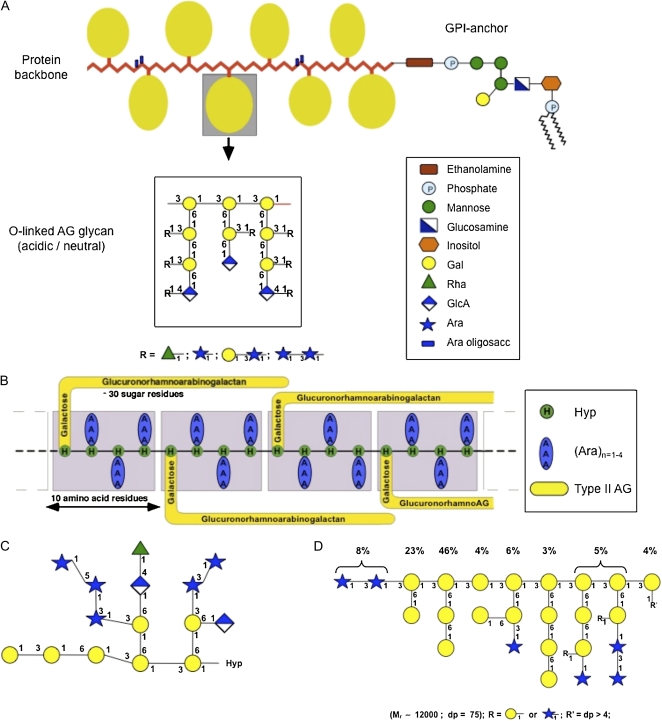
Figure 1.
A, The wattle blossom model of the structure of AGPs with a GPI membrane anchor attached. In this model, there are approximately 25 Hyp residues. Most Hyp residues are noncontiguous and are predicted to bear an AG chain. Each AG chain may contain 15 or more repeats of a β-(1-3)-linked Gal oligosaccharide. There may be a few contiguous Hyp residues bearing short arabino-oligosaccharides. The molecule as a whole is spheroidal. The structure of the GPI anchor shows an ethanolamine-phosphate (P) between the anchor and the C terminus of the protein backbone, which is common to all GPI anchors. The core oligosaccharide of the GPI shown is based on PcAGP1 from pear (Pyrus communis; Oxley and Bacic, 1999), which comprises 2- and 6-linked Manp residues, a 4-linked GlcNH2 residue, and a monosubstituted inositol with a partial Galp residue substitution to C(O)4 of the 6-linked Manp residue. The lipid moiety is a ceramide composed primarily of a phytosphingosine base and tetracosanoic acid. This model is modified from Fincher et al. (1983). B, The twisted hairy rope model of the structure of the GAGP. A hypothetical block size of 7 kD contains 10 amino acid residues (1 kD), 30 sugar residues (4.4 kD), and three Hyp-triarabinosides (1.32 kD). The glucuronorhamnoarabinogalactan has a galactan backbone with GlcpA, Rhap, and Araf side chains similar to that shown in the AG schematic in A. This model is from Qi et al. (1991). C, Primary structure of a representative Hyp-AG polysaccharide (AHP-1) released by base hydrolysis from a synthetic AGP (Ala-Hyp)51 from tobacco BY2 cells. This model is modified from Tan et al. (2004). D, Larch AG structure. R′ = dp > 4 is an undefined AG oligosaccharide. This model is modified from Ponder and Richards (1997).
The high degree of heterogeneity displayed by AGPs is a property of the complexity of both the protein backbone gene families, gleaned largely from the genome sequencing programs, and their carbohydrate sequence and composition, revealed by a combination of chemical studies and the use of antibodies (Knox, 1997). These antibody studies and more recently molecular biology approaches using synthetic Hyp-rich motifs (Tan et al., 2004; Estevez et al., 2006) have also revealed this heterogeneity within and between tissue/cell types, sustaining the interest of researchers regarding their potential as biological regulatory molecules. Mounting evidence indicates that AGPs have a role in controlling plant growth and development. Although much of the evidence is circumstantial (for review, see Seifert and Roberts, 2007), GPI-anchored proteins in mammalian and yeast cells are involved in cell-cell signaling by interacting with other proteins, and the GPI anchor determines their location in membrane microdomains or membrane rafts enriched in sphingolipids and sterols (Orlean and Menon, 2007; Kinoshita et al., 2008). Thus, GPI anchors on AGPs provide a plausible mechanism by which AGPs may be involved in signaling pathways (Schultz et al., 1998; Johnson et al., 2003b). Additionally, it is clear that carbohydrate epitopes on mammalian, Drosophila melanogaster, and protozoan parasite proteoglycans/glycoproteins are key determinants of biological specificity/function (Haltiwanger and Lowe, 2004; Filmus et al., 2008; Schaefer and Schaefer, 2010). However, the key question remains as to whether the function of AGPs resides in the protein backbones, the exquisite microheterogeneity of the glycan epitopes, or both. In this review, we summarize the key structural features of AGPs and overview their putative functions. More exhaustive reviews on each of these topics are cited above.
STRUCTURE
Carbohydrate
The carbohydrate is usually in the form of polysaccharide chains, type II AGs that are O-glycosidically linked to Hyp residues on the protein backbone. These type II AGs have (1-3)-β-d-linked Galp residues that form a backbone substituted at C(O)6 by galactosyl side chains, usually terminating in Araf, Rhap, and Galp residues, giving rise to a neutral glycan chain (Fig. 1). Some AGs, such as those found in gum arabic, are also rich in GlcpA (with or without 4-O-methyl ether) residues and are primarily terminal, giving the molecules an overall negative charge. Native AG chains can range in size from 5 to 25 kD (i.e. approximately 30–120 sugar residues; Fincher et al., 1983; Gane et al., 1995). Chemical analyses of some AG chains suggests that the (1-3)-β-galactan backbone contains repeat blocks of approximately seven Galp residues interrupted by a periodate-sensitive linkage, possibly either (1-6)-Galp or (1-5)-Araf (Churms et al., 1981, 1983; Bacic et al., 1987). Short oligoarabinosides linked O-glycosidically to Hyp residues (Fig. 1) and single Gal residues O-glycosidically linked to Ser/Thr residues are also found on some AGPs (Goodrum et al., 2000).
Characterization of the carbohydrate structures of AGPs is complicated by the difficulties in isolating single AGP molecules and the microheterogeneity of the constituent glycan chains. The application of molecular biology techniques to isolate both heterologously expressed AGP protein backbones and synthetic peptides as GFP fusion proteins by the Kieliszewski/Showalter and Matsuoka laboratories (Shpak et al., 1999; Zhao et al., 2002; Shimizu et al., 2005) was an ingenious innovation and allowed direct testing of the Hyp contiguity hypothesis (Kieliszewski and Lamport, 1994) that predicts glycosylation patterns. Synthetic AGPs with single dipeptide repeats [e.g. (Ala-Hyp)n] reduced the microheterogeneity to a level permitting NMR-based sequencing of AG chains on the synthetic peptides (Fig. 1C), thereby providing major advances in our knowledge. The expression of a major tomato (Solanum lycopersicum) AGP (LeAGP1) as a fusion protein with GFP in tobacco (Nicotiana tabacum) BY-2 suspension culture cells facilitated its purification, and the Hyp glycoside profiles showed that 54% of the total Hyp had polysaccharide substituents with a medium size of 20 residues (approximately 3 kD) but as large as 52 residues (approximately 8 kD; Zhao et al., 2002). The remaining Hyp residues either had oligoarabinosides (28%) or were nonglycosylated (12%–14%). This provided further support for the Hyp contiguity hypothesis, which predicts that blocks of contiguous Hyp residues, such as those occurring in extensins (Ser-Pro3-5), are arabinosylated with oligoarabinosides, whereas noncontiguous Hyp residues, such as those occurring in AGPs (Ala-Hyp, Ser-Hyp), are exclusively substituted with AG polysaccharide chains. Where AGP protein backbones contain both contiguous and noncontiguous Hyp residues such as gum arabic AGP (GAGP; Fig. 1B) and the Gal-rich stylar glycoprotein (GaRSGP) from Nicotiana alata stylar transmitting tract tissue (Sommer-Knudsen et al., 1996), then both type II AG polysaccharide chains and short Ara oligosaccharides are found. Heterologous expression of AGPs in tobacco BY2 cells also provided the first complete sequence of an AG chain on a synthetic peptide. Alkaline hydrolysis of the purified AGP-GFP fusion protein was used to generate a population of Hyp-AGs ranging in size from 13 to 26 sugar residues, with a median size of 15 to 17 residues (Tan et al., 2004). AG-Hyp was purified and the complete primary sequence of the 15-residue glycan chain was established (Fig. 1C), confirming the generally held view of AG structure previously proposed (Fig. 1, A and B). Interestingly, the size of the AG chains elaborated on these synthetic peptides is low compared with even the smallest predicted chain length of native AGs (Fincher et al., 1983; Gane et al., 1995), and the periodate-resistant (1-3)-β-galacto-oligosaccharide “repeats” are much shorter than that estimated from studies on native AGPs (Churms et al., 1981, 1983; Bacic et al., 1987).
The type II AG chains, however, are not restricted to classical AGPs and are also found (1) on chimeric AGPs (e.g. GaRSGP; Sommer-Knudsen et al., 1996) and the chimeric, hybrid AGP 120-kD glycoprotein also from the styles of N. alata (Lind et al., 1994; Schultz et al., 1997; Lee et al., 2008); (2) covalently linked to pectins in Angelica acutiloba root walls (Yamada, 1994), in spent hops (Humulus lupulus) extracts (Oosterveld et al., 2002), and in carrot (Daucus carota) roots (Immerzeel et al., 2006); and (3) as pure polysaccharides with no detectable protein in the secretions from western larch (Larix occidentalis; Ponder and Richards, 1997; Fig. 1D).
In contrast to the lack of well-characterized amino acid sequence motifs defining O-glycosylation, N-glycosylation, when present, only occurs when the protein backbone contains the predicted conserved sequence Asn-Xaa-Ser/Thr, where Xaa can be any amino acid except Pro (Lerouge et al., 1998). Classical AGP protein backbones do not contain this consensus sequence, but many chimeric AGPs, including fasciclin-like AGPs (FLAs), GaRSGP, and an early nodulin-like protein (Johnson et al., 2003a), contain the consensus sequence, and N-glycosylation has been experimentally demonstrated for both GaRSGP (Sommer-Knudsen et al., 1996) and AtFLA7 (Johnson et al., 2003a).
Protein
Cloning of genes encoding AGP backbones has significantly advanced our understanding of AGPs. The first protein backbone genes identified by traditional biochemical approaches required the purification of native AGPs, their subsequent deglycosylation, and Edman degradation N-terminal sequencing (Chen et al., 1994; Du et al., 1994). More recently, whole genomes have been sequenced, revealing the incredible diversity of AGP protein backbones. For example, Arabidopsis has more than 100 genes encoding proteins predicted to have AGP glycomodules. The first thorough bioinformatic analysis of the Arabidopsis genome identified 47 different genes in four classes: 13 classical AGPs, three Lys-rich AGPs, 10 AG peptides, and 21 FLAs (Schultz et al., 2002). A complementary approach, looking for genes encoding GPI-anchored proteins, identified approximately 40 more genes that encoded chimeric proteins containing small AG glycomodules, including proteins such as lipid transfer proteins and selected receptor-like kinases (Borner et al., 2002).
Classical AGPs and AG peptides can be considered the basal form of AGPs in that they have no other domains that might confer functions; as such, the entire protein backbone acts as a glycosylation scaffold. Most classical AGPs have protein backbones that are approximately 100 amino acid residues, whereas most AG peptides have protein backbones of 10 to 13 amino acid residues (Schultz et al., 2002). Purification and protein sequencing of eight of 12 AG peptides from Arabidopsis confirmed prolyl hydroxylation and GPI anchoring of AG peptides and indirectly confirmed AG glycosylation, as the AG peptides could be precipitated with Yariv reagent (Schultz et al., 2004).
FLAs are a large class of chimeric AGPs in Arabidopsis and contain one or two fasciclin domains, thought to be important for protein-protein interactions, and one or two AGP domains (Johnson et al., 2003a). Large gene families of FLAs exist in other dicots, such as cotton (Gossypium hirsutum; Huang et al. 2008), monocots (rice [Oryza sativa] and wheat [Triticum aestivum]; Faik et al., 2006), and gymnosperms (pine [Pinus species]; Li et al., 2009). At least two FLAs (each with only a single fasciclin domain) are found in the moss Physcomitrella patens (Lee et al., 2005). In trees such as eucalypts (Eucalyptus species) and poplars (Populus species), selected FLAs are preferentially expressed in tension wood, suggesting that they contribute to branch strength (Andersson-Gunnerås et al., 2006; Qiu et al., 2008).
The finding that AG glycomodules are found in combinations with other functional domains leads to the question: what is the function of the glycomodule in chimeric AGPs? The range of AG peptides in a few species illustrates the diversity of chimeric AGPs that is being revealed as more cDNA and gene sequences become available. In Arabidopsis, there is an AG peptide, AtAGP24, that has a putative metal-binding domain at the N terminus followed by a short AG glycomodule (HEGHHHHAOAOAOGOAS-GPI anchor, where O is Hyp; Schultz et al., 2004). A search for orthologs identified an EST in barley (Hordeum vulgare; TC147437) that has 12 HXXXH motifs interspersed with short AGP glycomodules and no other domains (W.M.A. Wankamaruddin, J.M. Preuss, M.A. Tester, A.A.T. Johnson, and C.J. Schultz, unpublished data). A similar contig is found in closely related wheat (TC252449), but the apparent rice ortholog (OJ990528_30.2) has the same alternating domain structure, although the sequences between AG glycomodules contain fewer His residues. A gene for a different chimeric AG peptide was recently discovered in Medicago truncatula and seems to be restricted to legumes. The predicted mature protein backbone has an N-terminal domain of unknown function of 21 amino acid residues, followed by an AG glycosylation motif APAPTP (Schultz and Harrison, 2008). This AG peptide, MtAMA1, is only expressed in root tissue colonized with symbiotic arbuscular mycorrhizal fungi, specifically in arbuscule-containing cortical cells, which are the major site of nutrient transfer in this widespread symbiosis. Differences among species highlight the evolutionary complexity of the AG peptide subfamily of AGPs and make functional studies of chimeric AGPs an even greater challenge.
GPI Anchor
A bioinformatic search of the Arabidopsis genome reveals that approximately 248 proteins, belonging to diverse classes of cell surface (glyco)proteins, including enzymes, contain the signal sequence for the addition of a GPI anchor, and approximately 40% of these contain AG glycomodules (Borner et al., 2002, 2003). Most AGP protein backbones contain the expected signals (an N-terminal secretion signal and a C-terminal hydrophobic signal sequence) specifying the cotranslational addition of a GPI anchor (Eisenhaber et al., 2003b; Johnson et al., 2003b). The C-terminal signal sequence contains small aliphatic amino acids at the ω (cleavage) and ω+2 sites, followed by a short spacer, and terminates in a stretch of small hydrophobic amino acids. The structure of the lipid moiety of the GPI anchor of two AGPs has been experimentally determined and is a phosphoceramide (Fig. 1A; Oxley and Bacic, 1999; Svetek et al., 1999). The glycan moiety of one AGP GPI anchor has been fully sequenced and contains the conserved minimal structure found in all eukaryotic GPI anchors [α-d-Manp(1-2)α-d-Manp(1-6)α-d-Manp(1-4)α-d-GlcNH2-inositol] with a plant-specific substitution of a β-d-Galp(1-4) residue on the third Manp (Fig. 1A; Oxley and Bacic, 1999). This remains the only plant GPI anchor sequenced to date.
Tertiary Structure
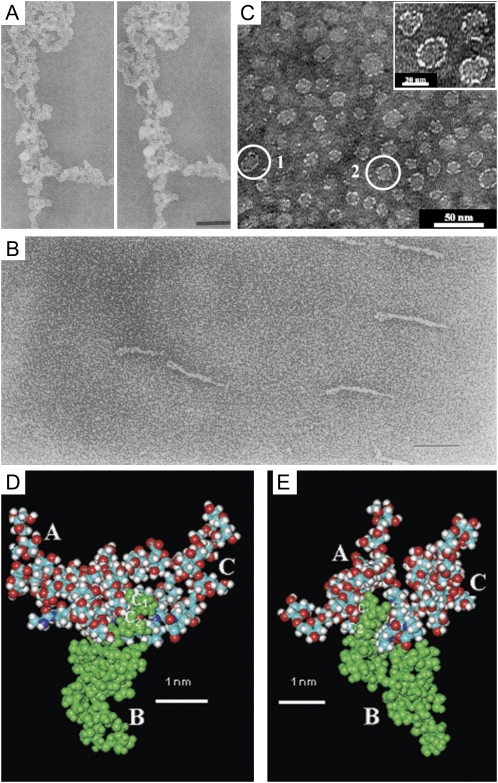
An understanding of the tertiary structure of AGPs will ultimately contribute to an elucidation of their molecular mechanism(s) of action in planta and explain their unique physicochemical properties of having extremely low viscosity at high concentration and a capacity to act as emulsifiers. Until recently, two models have been proposed, the “wattle blossom” model (Fincher et al., 1983), based upon their biophysical properties (Fig. 1A), and the “twisted hairy rope” model (Fig. 1B; Qi et al., 1991), for a GAGP based upon a combination of transmission electron microscopy (TEM) imaging (Fig. 2B) and chemical properties. The twisted hairy rope model envisages an alignment (and stabilization via hydrogen bonding) of the AG chains along the long axis of the protein backbone, forming a molecule of approximately 5 nm diameter, and provides an explanation of how some AGPs can permeate the pores (4–5 nm) of primary walls by reptation. Using direct TEM imaging (Fig. 2A) of an AGP from carrot suspension cultures revealed ellipsoidal rather than rod-like structures that were more consistent with the wattle blossom model (Baldwin et al., 1993). Computer-based molecular modeling of NMR data and compositional studies of a synthetic AGP from tobacco BY2 cells, of three AG chains linked to a (A-O/P)6, revealed that the bulky AG side chains do indeed hydrogen bond with the polypeptide, but in a more complex manner than originally proposed (Fig. 2, D and E). Gum arabic is generally considered to comprise three fractions: two major ones comprising an AG peptide (88.4%, generally referred to as AG and F1 [for fraction 1]), an AGP fraction (10.4%), and a third minor glycoprotein (GAGP) fraction (1.2%; Sanchez et al., 2008). The twisted hairy rope model was derived from studies of this latter GAGP fraction (Figs. 1B and 2B). In 2008, independent studies of the other (major) gum arabic fractions concluded that the AGP fraction was consistent with the wattle blossom model (Mahendran et al., 2008), whereas the AG peptide F1 was a thin oblate ellipsoid that led to the formulation of a “disc-like model” (Fig. 2C), which was difficult to reconcile with the wattle blossom model (Sanchez et al., 2008). The latter study used a combination of small angle neutron scattering coupled to ab initio calculations and various microscopic techniques (TEM and atomic force microscopy). Therefore, it appears that the three fractions from gum arabic have different tertiary structures that undoubtedly reflect the enormous primary structural diversity of both the AG chains and the protein backbones of these AGPs.
Figure 2.
A composite of images of AGPs purified from various plants. A, Stereopair of TEM micrographs showing the three-dimensional structure of a large aggregate of AGP from carrot suspension cultures imaged by the fast-freeze, deep-etch, rotary-shadowed replica technique. Bar = 200 nm. This image is from Baldwin et al. (1993). B, TEM micrograph of Superose-purified GAGP from gum arabic after rotary shadowing. Bar = 100 nm. This image is from Qi et al. (1991). C, TEM micrograph of the AG peptide F1 from acacia gum. The inset shows a magnification of part of the micrograph. This image is modified from Sanchez et al. (2008). D and E, Space-filling CPK models of three AHP-1 AGs (15 glycan residues per polysaccharide chain) from tobacco BY2 cells glycosidically linked to C-4 of each Hyp (O) residue in the 12-residue peptide (A-P-A-O-A-O-A-O-A-P-A-P), in which each O has an AHP-1 substitution (underlined). Nitrogen atoms are shown in dark blue; the oxygen atoms are red; hydrogen atoms are gray; and carbon atoms are turquoise blue. These images are from Tan et al. (2004). D, Glycosylated (A-O/P)6 (see above). Side view of a polysaccharide cluster. Three AHP-1 glycans labeled A to C are O-linked to the Hyp residues of the glycosylated (A-O/P)6 model (residues 4, 6, and 8 of the peptide). The protein backbone lies across the figure, with the N terminus at the far left. Note the close proximity of polysaccharide B (green) to the polypeptide backbone, where the Ara disaccharide residues C1 and C2 form three H bonds as follows: the hydroxymethyl (C-5) of Ara residue C1 to both the carbonyl of Hyp residue 4 and the peptide N of Ala residue 5; and the C-2 hydroxyl of Ara residue C2 to the NH of Ala residue 6. In contrast, the Ara trisaccharide residues at the tip of each polysaccharide form peripheral hook-like projections; these may result in multiple weak interactions (“molecular Velcro”) with the Yariv reagent, which specifically interacts with AGPs. E, Glycopeptide (A-O/P)6. End-on view of a polysaccharide cluster. Reorienting the polypeptide so that it is perpendicular to the plane of the paper shows a syndiotactic propeller-like arrangement of the AG polysaccharides around the polypeptide, providing surfaces for interactions and interdigitation with other matrix molecules.
BIOSYNTHESIS
Pro Hydroxylation
In animals, prolyl 4-hydroxylase (P4H; EC 1.14.11.2) is an α2β2 tetramer that is responsible for hydroxylation of Pro residues in proteins such as collagen (Hill et al., 2000), although Hyp is not glycosylated in animals. The β-subunit is a multifunctional enzyme, protein disulfide isomerase. The α-subunit is the catalytic subunit and is referred to as P4H. In plants, early biochemical analysis suggested that only the catalytic α-subunit was required for modification of Pro to Hyp (Chrispeels, 1984; Bolwell et al., 1985). Sequencing of the Arabidopsis genome and subsequent testing of heterologously expressed candidate genes using synthetic peptides confirmed the activity of two encoded proteins, P4H-1 (Hieta and Myllyharju, 2002) and P4H-2 (Tiainen et al., 2005). The synthetic peptides used in these early studies were more like mammalian collagen and extensin-type motifs ([Ser/Thr]-Pro3-5) rather than the scattered ([Ala/Ser/Thr]-Pro) motifs of AGPs, so while the current knowledge of the protein code for Pro hydroxylation has provided general rules, there are still some ambiguities. For example, why is the Pro in Gly-Pro hydroxylated in the AG peptide, AtAGP24 (Schultz et al., 2004), but not in gum arabic, where Gly-Pro is frequently followed by His (Goodrum et al., 2000) rather than Ala, as in AtAGP24? The first rules suggested by Kieliszewski and Lamport (1994), namely that Lys-Pro, Tyr-Pro, and Phe-Pro are never hydroxylated, whereas Pro-Val is always hydroxylated, remain valid. The importance of the residues on either side of the Pro residue is supported by the recent solving of the crystal structure of the algal P4H from Chlamydomonas rheinhardtii (Koski et al., 2009). The active site of CrPH4-1 contains two loops that bind intimately the Ser-Pro-Ser of peptide (Ser-Pro)5.
The code for hydroxylation was further extended using recombinant sporamin expressed in tobacco BY2 cells. Sporamin is a storage protein from sweet potato (Ipomoea batatas) and contains a single Pro residue that is hydroxylated and glycosylated. Mutation and deletion studies revealed a motif of seven residues ([not basic]-[not T]-[AVSG]-Pro-[AVST]-[GAVPSTC]-[APS]), suggesting a much larger active site (Shimizu et al., 2005). A rather unusual motif, KSPKKS, is hydroxylated in the 120-kD glycoprotein from N. alata (Schultz et al., 1997), highlighting the need to sequence a broad range of native and artificial AGP and extensin substrates before the code can be fully elucidated. The current approach of using plant cell suspension cultures is extremely informative but time-consuming, and downstream analysis of hydroxylation and glycosylation is difficult. It should be possible to increase the throughput of investigating Pro hydroxylation of plant AGP protein backbones using yeast-based systems that have been successful for studying mammalian P4H specificity (Vuorela et al., 1997).
The in vivo location of P4H in plants remains unclear. A recent report suggests that the enzymes are located in both the endoplasmic reticulum (ER) and the Golgi (Yuasa et al., 2005). However, the predicted Golgi localization signals are in the N-terminal ER signal sequence that would be cleaved as the proteins enter the ER cotranslationally and, therefore, would not be available for Golgi targeting from the ER. It is also possible that Golgi localization was an artifact of the BY2 GFP fusion protein expression system. N-terminal sequencing of P4H native proteins is needed to determine if the Golgi signal is retained in any of the mature P4H enzymes.
The Plant GPI Anchor Pathway
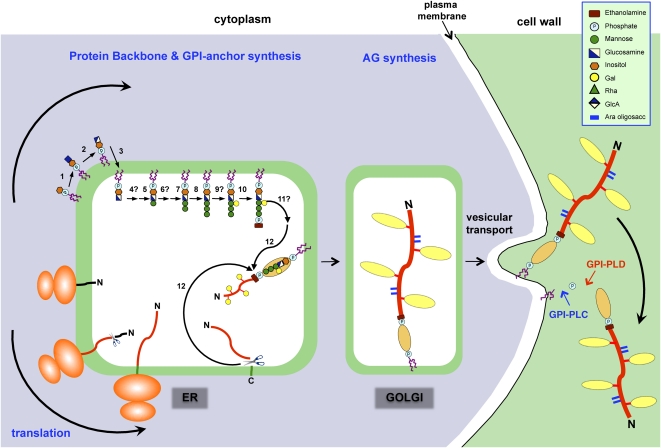
Most of our knowledge of the assembly of the GPI anchor is gleaned from the exhaustive analysis performed in mammalian, yeast, and protozoan systems (for review, see Orlean and Menon, 2007; Fujita and Jigami, 2008; Kinoshita et al., 2008). Given the importance of GPI anchors in eukaryotic systems, it is surprising that there has been little progress in elucidating this pathway in plants. Nonetheless, it is highly likely that the process is conserved, as orthologs of key GPI biosynthetic genes are found in plant genomes (Fig. 3; Table I; Schultz et al., 1998). Lalanne et al. (2004) demonstrated that disruptions in the SETH1 and SETH2 genes that encode orthologs of PIG-C and PIG-A, respectively, two components of the GPI-GlcNH2 complex, specifically block male transmission and pollen function in Arabidopsis. To our knowledge, this is the only study of GPI assembly genes in plants.
Figure 3.
Proposed mechanism for the synthesis and addition of a GPI anchor to an AGP. The addition of a GPI anchor to an AGP likely occurs in several phases, beginning with the synthesis of the GPI moiety on the cytoplasmic surface of the ER. The protein backbone is inserted into the ER cotranslationally, and eventually the two processes converge. The GPI anchor synthesis pathway shown is a composite of that established for mammals and yeast (Orlean and Menon, 2007; Kinoshita et al., 2008) based on the structure of the pear GPI anchor (Fig. 1A). A similar pathway likely exists in plants, since orthologs of the mammalian and yeast genes have been found in plants (Table I). Proteins involved in the biosynthesis pathway (numbered) are summarized in Table I. In both mammals and yeast, any branching glycosylation of the trimannosyl core occurs between steps 8 and 10, and although galactosylation is plant specific, it is presumed to occur at the same stage (step 9). Since substitution of inositol (step 6) and the trimannosyl core (step 11) with acyl and ethanolamine phosphate residues, respectively, has not been demonstrated in plant GPIs, these steps are not shown here, although, interestingly, BLAST searches have identified putative acyl transferases (Table I). Following concomitant removal of the C-terminal GPI signal sequence and addition of the GPI anchor (step 12), numerous Pro residues present within the AGP backbone are hydroxylated to Hyp. Substitution of these Hyp residues with AG chains probably begins within the ER and is completed within the Golgi network. AGPs are transported to the plasma membrane via vesicular transport, where they are either temporarily anchored to the plasma membrane before being released by phospholipases (GPI-PLC and GPI-PLD) or endocytosed. This model is modified from Schultz et al. (1998).
Table I. GPI biosynthesis proteins and their putative plant orthologs (after Orlean and Menon, 2007).
Steps refer to the individual processes involved in GPI anchor biosynthesis and are identical to those used in Figure 3. Putative plant orthologs were identified by tBLASTn or BLASTp using the human and yeast protein sequences to search the Arabidopsis and rice genomes. Searches were performed at the National Center for Biotechnology Information or www.gramene.org using default settings (except to find PIG-Y, where BLASTp word size was changed from n = 3 to n = 2, with no filtering). For Arabidopsis sequences, putative orthologs were cross-referenced, where available, against genes previously identified by Eisenhaber et al. (2003a). n/a, Not applicable.
| Step | Enzyme | Mammalian Protein | Yeast Protein | Putative Plant Orthologs |
|
| Arabidopsis | Rice | ||||
| 1 | GPI-GlcNAc transferase | PIG-A | Gpi3p | SETH2, At3g45100 | Os07g17960 |
| PIG-C | Gpi2p | SETH1, At2g34980 | Os02g0622200 | ||
| PIG-H | Gpi15p | At4g35530 | Os05g07260 | ||
| PIG-P | Gpi19p | At1g61280 | Os03g43900 | ||
| PIG-Q | Gpi1p | At3g57170 | Os05g0290300 | ||
| PIG-Y | Eri1p | AL592312 (83696…83338)a | Os01g0939000 | ||
| DPM2 | At1g74340 | Os02g0140101 | |||
| 2 | GlcNAc-PI de-N-acetylase | PIG-L | Gpi12p | At2g27340 | Os04g0678800 |
| 3 | Flippase | Not identified | |||
| 4 | Inositol acyltransferase | PIG-W | Gwt1p | At4g17910 | Os03g0378200 |
| 5 | α-(1-4)-Mannosyltransferase | PIG-M | Gpi14p | At5g22130 | Os03g0670200 |
| PIG-X | Pbn1p? | At5g46850 | Os12g0596900 | ||
| 6 | EtNP transferase I | PIG-N | Mcd4p | At3g01380 | Os02g0581000 |
| 7 | α-(1-6)-Mannosyltransferase II | PIG-V | Gpi18p | At1g11880 | Os12g0498700 |
| 8 | α-(1-2)-Mannosyltransferase III | PIG-B | Gpi10p | At5g14850 | Os02g0111400, Os01g0580100 |
| 9 | Putative plant-specific GalT | n/a | n/a | Not identified | Not identified |
| 10 | EtNP transferase III | PIG-O | Gpi13p | At5g17250 | Os10g0170300 |
| PIG-F | Gpi11p? | At1g16040 | Os10g0200800 | ||
| 11 | EtNP transferase II | PIG-G | Gpi7p | At2g22530 | Os02g0781500–Os02g0781600 |
| PIG-F | Gpi11p? | At1g16040 | Os10g0200800 | ||
| 12 | GPI transamidase | PIG-K | Gpi8 | At1g08750 | Os02g0219400 |
| GAA1 | Gaa1p | At5g19130 | Os01g0682200 | ||
| PIG-S | Gpi17p | At3g07810 | Os01g0907300 | ||
| PIG-T | Gpi16p | At3g07140 | Os11g28980 | ||
| PIG-U | Gab1p | At1g63110 | Os02g0688900 | ||
The Arabidopsis sequence is not annotated but does contain significant sequence similarity to the rice Os01g0939000 (63% identity, 80% similarity) putative ortholog if the ER signal sequence is removed.
In brief, the addition of a GPI anchor to a protein occurs in several phases, beginning with the early steps in the synthesis of the GPI moiety on the cytoplasmic surface of the ER and the subsequent steps occurring in the ER lumen after the flipping of the anchor from one face to the other. Proteins to be GPI anchored are cotranslationally inserted into the ER, and eventually the two processes converge, when a transamidase complex concomitantly cleaves the C-terminal peptide and transfers the GPI anchor onto the C terminus of the mature protein by a transamidation reaction (Fig. 3). The GPI-anchored protein then undergoes prolyl hydroxylation and galactosylation (see below) before transit to the Golgi, where the AG and possibly the oligoarabinoside chains are elaborated. Within the Golgi, extensive lipid remodeling occurs that is critical to the targeting of GPI-anchored proteins to membrane microdomains or rafts enriched in sphingolipids and sterols (Orlean and Menon, 2007; Fujita and Jigami, 2008; Kinoshita et al., 2008). It is thought that association with membrane rafts is important for GPI-anchored protein function. Lipid rafts appear to exist in plants (Borner et al., 2005), but their composition and function are mostly undefined for plants, although lipid rafts have been implicated in stabilizing cellulose synthase complexes in the plasma membrane (V. Bulone, personal communication).
Biosynthesis of the AG and Arabino-Oligosaccharide Chains
The biosynthesis of glycoconjugates has been extensively studied in mammals, yeast, and bacteria, where many glycosyltransferases (GTs) have been cloned and biochemically characterized (for review, see Breton et al., 2002; Coutinho et al., 2003). However, only a few GTs have been purified and then cloned from plants, and the majority are involved in the assembly of cell wall polysaccharides (for review, see Doblin et al., 2010). Based upon the assembly of mammalian proteoglycans and recent molecular and biochemical studies in Arabidopsis by Strasser et al. (2007) and Qu et al. (2008), it is proposed that the AGs on AGPs are synthesized by type II ER/Golgi-located GTs, including members of the CAZy GT-family-31 GTs that possess (1,3)-β-GalT activity (for review, see Egelund et al., 2010). The O-galactosyltransferase, which is the initial enzyme in the AG biosynthetic pathway (adding the first Gal residue to a Hyp residue in the protein backbone), is predominantly located in ER fractions after Suc density-gradient centrifugation of extracts from Arabidopsis T87 cells (Oka et al., 2010). A model has been proposed in which the addition of the first Gal onto Hyp occurs in the ER (Oka et al., 2010). Early studies by Fincher and colleagues (Mascara and Fincher, 1982; Schibeci et al., 1984) showed that the enzymes responsible for the synthesis of polymers containing (1,6)-β-Gal linkages in Italian ryegrass (Lolium multiflorum) were associated exclusively with the subcellular fractions enriched in Golgi-derived membranes and therefore concluded that the Golgi apparatus plays an important part in the synthesis of the carbohydrate component of AGPs. Although 30 years have passed since these early studies, none of the enzymes responsible for the synthesis of the Gal chains have been cloned and characterized, nor have their substrate specificities been determined biochemically. It is uncertain, therefore, whether the addition of the remaining sugars that make up the AG chains occurs one residue at a time or blockwise, as is the case for N-linked glycans (Kelly et al., 2006), although recent results (Qu et al., 2008; Oka et al., 2010) support the former mechanism. Therefore, it is reasonable to assume that for the assembly of AG chains, several GalTs will be required, such as O-galactosyltransferases, (1,3)-β-GalTs, and (1,6)-β-GalTs, and that these enzymes will work coordinately to regulate the density, length, and sequence of AG chains.
Several GTs are predicted to be responsible for decorating the termini of AG chains (AraTs/GlcATs/RhaTs). Fucf residues are present in AGPs in several dicot plants as terminal residues linked via an α-linkage to the C-2 position of an adjacent Araf residue (Tsumauraya et al., 1988). Two members of CAZy GT-family-37, AtFUT4 and AtFUT6, were recently characterized as α(1,2)-fucosyltransferases (FUTs) that are specific for AGPs (Wu et al., 2010). These are the first enzymes to be characterized that are specific for AGP glycosylation. The authors demonstrated that these two enzymes differentially fucosylate AGPs at different positions on the AG side chains, indicating that the two FUT enzymes may have different physiological roles. An AtFUT6-GFP chimera was localized to the Golgi apparatus, supporting the evidence that much of the biosynthesis of the AG side chains occurs within this organelle (Wu et al., 2010).
The assembly of the arabino-oligosaccharides remains largely unstudied. Two putative Arabidopsis AraT insertional mutants, rra1 and rra2, are presumed to be involved in extensin arabino-oligosaccharide assembly (Egelund et al., 2007) based on reduced residual Ara content in a polymer tightly associated with the cellulosic wall residue. These AraTs possess a type II GT topology and are likely Golgi located (J. Egelund, unpublished data), but whether this reflects the location and activity of the AraTs involved in the synthesis of AGPs remains to be demonstrated. An interesting new approach from the Pauly laboratory (Gille et al., 2009) used glycosyl hydrolases as a forward chemical genetic screen to identify cell wall mutant lines where extensin arabinosylation was altered. It will be of interest to see if any of the other 22 genetic loci identified in this screen uncover AGP-specific GTs or whether it will be necessary to use AGP-specific hydrolases in a future screen.
FUNCTIONS
Biological
The function of AGPs was recently summarized in an excellent review by Seifert and Roberts (2007). Thus, this section will outline key points about AGP function with regard to the GPI anchor, AG side chains, and the fully glycosylated mature protein, highlighting recent studies and identifying future strategies to further clarify AGP function(s).
The function of AGPs has been hard to ascertain for many reasons. The heterogenous nature of the AGP family suggests that AGPs do not have one specific role, as would be anticipated from studies of mammalian glycoproteins/proteoglycans (Filmus et al., 2008; Schaefer and Schaefer, 2010). The complex nature of the carbohydrate side chains buries the protein backbone, preventing simple isolation of individual AGPs, and in many cases prevents detection by protein-specific antibodies. The complex glycosylation and high degree of hydroxylation of AGP protein backbones does not allow expression of correctly glycosylated recombinant proteins in microorganisms, which would allow in vitro examination of function. The large size of the AGP gene family likely leads to functional redundancy, and the production of double mutants of homologous AGPs is often required to produce phenotypes (Motose et al., 2004; Coimbra et al., 2009). For other subclasses of AGPs, such as the three Lys-rich AGPs of Arabidopsis, AtAGP17, AtAGP18, and AtAGP19, each family member may have a unique function based on different phenotypes. It is not known if these differences are due solely to their different expression patterns or whether it is also due to different interacting partners or downstream signaling events (Acosta-Garcia and Vielle-Calzada, 2004; Gaspar et al., 2004; Yang et al., 2007).
Despite these limitations AGPs have been implicated in many processes involved in plant growth and development, including somatic embryogenesis (van Hengel et al., 2002), root growth and development (van Hengel and Roberts, 2003), hormone responses (Park et al., 2003), signaling (Schultz et al., 1998), xylem differentiation (Motose et al., 2004), resistance to Agrobacterium tumefaciens-mediated infection (Gaspar et al., 2004), initiation of female gametogenesis (Acosta-Garcia and Vielle-Calzada, 2004), salt tolerance (Shi et al., 2003; Lamport et al., 2006), cell wall plasticizer (Lamport et al., 2006), cell expansion (Lee et al., 2005; Yang et al., 2007), secretion (Xu et al., 2008a), promotion of pollen tube growth and guidance (Cheung et al., 1995; Wu et al., 1995, 2000; Mollet et al., 2002; Lee et al., 2008), programmed cell death (Gao and Showalter 1999; Chaves et al., 2002), pollen grain development (Pereira et al., 2006; Levitin et al., 2008; Coimbra et al., 2009), and self incompatibility in pollen (Lind et al., 1996; Cruz-Garcia et al., 2005; Hancock et al., 2005; Lee et al., 2008; Table II).
Table II. Summary of recent functional studies of AGPs.
Only studies using forward or reverse genetics providing plant lines with definable phenotypes are listed.
| Name | AGP Class | GPI | Locationa | Genetic Analysis | Phenotype/Observed Result | Proposed Function | Reference |
| PpAGP1 | Classical | Yes | Filament apices | agp1 knockout line | Reduced colony size due to decreased cell length | Apical cell expansion | Lee et al. (2005) |
| AtAGP6 | Classical | Yes | Pollen and pollen tubes | agp6 agp11 double T-DNA insertion mutants, double RNAi knockdowns, AGP6 point mutant | Double insertion mutants show collapsed pollen grains; RNAi double knockdowns have reduced pollen tube length, shorter siliques, and fewer seeds | Pollen grain development and pollen tube growth | Levitin et al. (2008); Coimbra et al. (2009) |
| AtAGP11 | Classical | Yes | Pollen grain and pollen tube | ||||
| CsAGP1 | Lys-rich | Yes | Shoot apices, roots, hypocotyls, and cotyledons | Overexpression in tobacco plants | Tobacco plants overexpressing CsAGP1 are taller with longer internodes | Stem elongation, hormone response | Park et al. (2003) |
| LeAGP1 | Lys-rich | Yes | Maturing metaxylem elements of young stems and petioles, transmitting tissue of cells of the style, inner and outer phloem, cambial zone of young stems and petioles | Overexpression of GFP-LeAGP1 fusion protein in tomato plants | Reduced stem elongation, increased lateral branches, reduced seed size and less seed produced; transgenic plants that produce a non-GPI-anchored form of GFP-LeAGP1 or are missing the Lys-rich domain have wild-type phenotype | Plant growth and development | Gao et al. (1999); Sun et al. (2004) |
| AtAGP17 | Lys-rich | Yes | Flower, leaves, and roots | rat1 T-DNA insertion in promoter (specific inhibition of transcription in the roots) | Reduced Agrobacterium binding to the root surface | Resistance to infection | Nam et al. (1999); Gaspar et al. (2004) |
| AtAGP19 | Lys-rich | Yes | mRNA abundant in stems, moderate levels in flowers and roots, lower levels in leaves | Null T-DNA insertion line | Smaller, rounder rosette leaves, lighter green leaves containing less chlorophyll; delayed growth; shorter hypocotyls and inflorescence stems, fewer siliques, and less seed | Plant growth and development, including cell division and expansion, leaf development, and reproduction | Yang et al. (2007) |
| AtAGP18 | Lys-rich | Yes | Reproductive tissue | RNAi lines | Reduced seed set, and the defect is female specific; no female gametophyte is produced because the megaspore fails to enlarge and divide by mitosis | Initiation of female gametogenesis | Acosta-Garcia and Vielle-Calzada (2004) |
| AtAGP24 | AG peptide | Yes | Abscission zones of seed pods | Overexpression of peptide IDA (for inflorescence deficient in abscission) | Visible deposition of Yariv-positive AGPs at abscission zones; AtAGP24 is the only AGP gene that is overexpressed based on microarray analysis | Metal binding, nonenzymic cleavage of cell walls | Stenvik et al. (2006) |
| GhAGP4 | FLA (one fasciclin domain, two AGP domains) | Yes | Cotton fibers | RNAi line | Suppression of GhAGP4 and partial suppression of three other related FLAs; inhibition of fiber initiation and elongation, significantly shorter fiber length | Initiation and elongation of cotton fibers | Li et al. (2010) |
| SOS5/AtFLA4 | FLA (two fasciclin domains, two AGP domains) | Yes | Flowers, leaves, roots, stems, and siliques | Single-base-pair substitution in conserved fasciclin domain | Swollen root tips and reduced root length under increasing salt concentrations | Salt tolerance and normal root expansion; possible ligand or involved in the production or presentation of a ligand for FEI receptor kinases | Shi et al. (2003); Xu et al. (2008b) |
| AtFLA11, AtFLA12 | FLA (one fasciclin domain, two AGP domains) | Yes | Inflorescence stems | Atfla11/12 double T-DNA insertion mutants | Decreased tensile strength and stiffness in stems; reduced cellulose, Ara, and Gal contents and increased lignin content in stem cell walls | Contributors to plant stem strength | MacMillan et al. (2010) |
| ZeXyp | Chimeric AGPb (contains nonspecific lipid transfer protein domain) | Yes | Immature xylem cells in stems | Overexpression in tobacco suspension cultures | Xylem differentiation (induces tracheary element differentiation) | Soluble signal mediating cell-to-cell interactions in vascular development | Motose et al. (2004) |
| AtXYP1, AtXYP2 | Chimeric AGP (contains nonspecific lipid transfer protein domain) | Yes | Immature xylem cells in stems | Double T-DNA insertion mutants | Discontinuous and thicker veins, incorrect interconnections of tracheary element | Soluble signal mediating cell-to-cell interactions in vascular development | Motose et al. (2004) |
| AtAGP30 | Chimeric AGP (contains Cys-rich domain) | No | Root, root tip, embryos immediately prior to germination | T-DNA insertion line | Mutants germinate more rapidly in the presence of the germination inhibitor abscisic acid; suspension cultures of cells derived from agp30 proliferate successfully but never initiate root formation | Root development and growth; modulation of abscisic acid perception during seed germination | van Hengel and Roberts (2003) |
| NtTTS/NaTTS | Chimeric AGP (contains Cys-rich C-terminal domainb | No | Produced in style, but mature protein taken up by pollen tubes | Antisense line | Reduced fertility and reduced rate of pollen tube growth when style TTS is reduced; antisense pollen grows normally and leads to normal seed set in antisense pollen | Promoting pollen tube growth and guidance | Cheung et al. (1995) |
| Na120 kD | Chimeric and hybridc AGP [contains contiguous and noncontiguous Pro(Hyp)-rich regions and a Cys-rich C-terminal domain] | No | Pistil, and cell wall and mature protein taken up by cytoplasm/vacuoles of pollen tubes | RNAi in self-compatible × self-incompatible hybrid cross | Plants with reduced 120-kD expression unable to perform pollen rejection | Role in S (self incompatibility)-specific pollen rejection | Hancock et al. (2005); Lee et al. (2008) |
Location refers to the expression of mRNA and/or protein as determined by a variety of methods. For details, see references cited within papers describing the genetic analysis.
Chimeric AGPs have one or more non-Pro(Hyp)-rich domains in addition to an AGP domain with noncontiguous Pro(Hyp) residues. Several chimeric AGPs have a conserved C-terminal domain of approximately 140 amino acid residues that contains six conserved Cys residues.
Hybrid AGPs contain, for example, both an AGP domain (noncontiguous Hyp) and an extensin-like domain (contiguous Hyp) and may also be chimeric [containing a non-Pro(Hyp)-rich domain].
Studies of AGPs often use methods that provide a global approach for determining AGP function. Biochemical analysis of AGPs utilizes carbohydrate-specific monoclonal antibodies for which precise epitope specificities are often unknown (for review, see Knox, 1997). β-Glc Yariv, the pink/orange dye that specifically binds AGPs, is used in many studies to bind AGPs, interfering with the activity of all AGPs and thus indirectly elucidating function. Both β-Yariv and the carbohydrate-directed monoclonal antibodies are estimated to bind 50 to 100 different AGP proteins and thus provide a global view of AGP distribution, localization, and function.
The use of molecular techniques has greatly expanded our ability to study individual AGPs directly. These include transcript analysis and the production of mutant plant lines, such as T-DNA insertion lines, RNA interference (RNAi) lines, and lines overexpressing individual genes. These studies are summarized in Table II. This approach is not without difficulties, as, in some cases, a phenotype is only apparent after the production of double knockout lines (Motose et al., 2004), and in others, conditional stress, such as salt, is required to induce a phenotype (Shi et al., 2003). Studies of the same gene by different methods highlight the complex nature of AGP expression. Tissue- and growth stage-specific transcript analysis often does not concur with localization using AGP promoter-driven constructs (Acosta-Garcia and Vielle-Calzada, 2004; Yang and Showalter, 2007), suggesting that multiple approaches are required. For example, a T-DNA insertion in the promoter region of the rat1 mutant line specifically inhibits AtAGP17 expression in roots but not leaves (Gaspar et al., 2004), indicating the importance of the promoter and untranslated regions in controlling AGP expression.
The analysis of RNA transcripts allows us some insights into AGP distribution within plants. In many cases, a clear expression pattern both in tissue location and developmental stage is evident for many individual AGPs (summarized in Table II). For many individual AGPs, transcription occurs at a higher level in specific plant tissues such as roots and root tips (van Hengel and Roberts, 2003), flowers (Acosta-Garcia and Vielle-Calzada, 2004), pollen tubes and pollen (Pereira et al., 2006), young stems (Gilson et al., 2001), differentiating tissue (Motose et al., 2004; Dahiya et al., 2006), shoot apices (Park et al., 2003), and filament apices (Lee et al., 2005). In other cases, specific AGPs are found in several tissue types within the plant; for example, SOS5/FLA4 is expressed ubiquitously throughout the plant but mutant plants only show aberrant root morphology during salt stress (Shi et al., 2003). AGPs have a key function in young or rapidly expanding or elongating tissues and during times of stress.
The presence of a GPI anchor is important when considering AGP function. GPI anchors do not traverse the entire plasma membrane, and in mammalian systems GPI-anchored proteins have been implicated in mediating cell-cell interactions by interacting with other proteins that possess transmembrane domains (Schultz et al., 1998; Johnson et al., 2003b; Filmus et al., 2008; Lim et al., 2008). It is unlikely that direct cell-to-cell interactions occur between GPI-anchored proteins in adjacent plant cells as occurs in mammalian cells, due to the thickness of the wall, estimated to be about 100 nm for a primary wall (Lamport et al., 2006). However, GPI-anchored AGPs and/or their soluble forms could interact with plasma membrane-bound receptor kinases or soluble forms of AGPs could interact with receptors in neighboring cells. The FLAs are expected to be involved in such interactions, as they contain fasciclin domains (Johnson et al., 2003a), which in other eukaryotes are involved in cell adhesion as a result of protein-protein interactions (Elkins et al., 1990; Kawamoto et al., 1998). Recently, it was proposed that SOS5/FLA4 is a ligand of two members of the Leu-rich repeat receptor-like kinase family, FEI1 and FEI2, based on genetic analyses indicating that FEI1 and FEI2 act within the same pathway as SOS5/FLA4 (Xu et al., 2008b).
Localization of GPI-anchored LeAGP1-GFP to both the plasma membrane and Hechtian strands in tobacco suspension cultures demonstrates a link between AGPs, the plasma membrane, and the cytoskeleton (Sun et al., 2004; Sardar and Showalter, 2007). Classical GPI-anchored AGPs have been implicated in controlling cell shape via a connection with the cytoskeleton, due to their involvement in orienting cortical microtubules and influencing F-actin polymerization (Andème-Onzighi et al., 2002; Sardar et al., 2006; Nguema-Ona et al., 2007). It has also been proposed that GPI-anchored AGPs may act as a plasticizer to loosen the pectin network or as a stabilizer of the wall during times of cell stress or expansion (Lamport et al., 2006).
Several studies suggest that the glycan chains are important for AGP function. Addition of a single AG glycosylation site to human growth hormone increased its secretion 100-fold from tobacco suspension cells (Xu et al., 2008a). Evidence for the importance of the carbohydrate chains in determining biological function comes from the mur-1 and reb1-1 mutants. The mur1 mutants are defective in the biosynthesis of Fuc and have aberrant AGP structures and altered root morphology, which is not attributed to defects in other wall components (van Hengel and Roberts, 2002). The reb1-1 mutant plants, defective in Gal biosynthesis (Seifert et al., 2002), have abnormal root extension, bulging of root epidermal hairs, and lack AGPs in their trichoblast cells (Andème-Onzighi et al., 2002). However, reb1-1 mutants also have defects in the xyloglucan content of their walls (Nguema-Ona et al., 2006), highlighting the difficulty of attributing the phenotype from a mutation in a major sugar pathway to a single class of molecules. The production of mutants where the AG glycans are specifically modified will add more insights into precisely how the structure of the carbohydrate effects function (Egelund et al., 2010).
Several recent mutational studies have examined the role of AGP proteins in pollen and pollen tubes. Sexual reproduction in flowering plants involves pollen grains landing on the stigma surface, germination, and growth of the pollen tube via a process of rapid tip elongation through the pistil extracellular matrix. The extracellular matrix is rich in AGP-like molecules, such as the 120-kD and Transmitting Tissue-Specific (TTS) proteins, some of which are internalized by the pollen tube (Lind et al., 1996; Lee et al., 2009). In N. alata, fully glycosylated TTS is implicated in promoting pollen tube growth and affecting pollen tube guidance, whereas the deglycosylated form of TTS is not (Cheung et al., 1995; Wu et al., 1995, 2000). TTS and 120-kD proteins also bind to three interacting partners, a putative Cys protease, a pollen-specific C2 domain-containing protein, and a S-RNase-binding protein (Cruz-Garcia et al., 2005; Hancock et al., 2005). It is proposed that binding of these proteins to pistil AGPs may contribute to signaling and trafficking inside pollen tubes (Lee et al., 2008).
In Arabidopsis, AGP6 and AGP11 were shown to be specific to both pollen tubes and pollen grains (Pereira et al., 2006). Double knockout mutants show aberrant pollen development, including pollen grain collapse and retraction of the plasma membrane from pollen walls (Coimbra et al., 2009). Arabidopsis lines expressing an agp6 agp11 double knockout (RNAi) construct reduce pollen tube growth, and fewer and shorter siliques are produced (Levitin et al., 2008). This implies that AGPs are required for the integrity of the pollen grain and pollen tube growth.
The importance of a subclass of AGPs, the FLAs, is highlighted by recent studies of cotton fiber development (Liu et al., 2008). RNAi lines suppressing GhAGP4 (a FLA) and three other FLAs resulted in inhibition of cotton fiber initiation and reduced fiber elongation (Li et al., 2010).
The essential function of AGPs in varied roles from cell expansion, growth, and differentiation to signaling, secretion, and linking the plasma membrane with the cytoskeleton and wall is clear. However, despite numerous studies, the precise mode of action of AGPs is unknown. The heterogeneous nature of AGPs and the diversity of functions suggest that a more targeted approach is required. It seems likely that the protein backbone, GPI anchor, and AG component all play roles in AGP function, and each of these areas needs to be addressed. With regard to the protein component, a targeted approach on each AGP subfamily is needed, taking into account cell type-specific expression patterns of individual family members. For example, targeted knockout of whole classes or subclasses of AGPs (e.g. by RNAi or using artificial microRNAs; Ossowski et al., 2008) may aid in defining a specific function to a specific group but may only be applicable to recently duplicated family members such as AtAGP6 and AtAGP11. Domain-swapping experiments and the identification of interacting partners are needed to identify regions of the protein backbone that are of biological significance and to indicate pathways in which the AGPs are intrinsically involved.
Commercial
AGPs are important for industrial purposes, primarily due to their emulsifying, adhesive, and water-holding properties. Larch AG and gum arabic (EU code E414; harvested from wounded Acacia senegal trees) are two of the most commercially important gums (for review, see Showalter, 2001). Larch AG is used in the food and pharmaceutical industries as a binder, emulsifier, and stabilizer and in the mining industry to float mineral ores. Gum arabic is considered the premier oil-in-water emulsifier and is used to encapsulate flavor in the manufacture of bakery products, beverages, and desserts (for review, see Pettolino et al., 2006).
AGPs and type II AGs also have potential applications in medicines and have been shown to stimulate animal immune systems by activation of the complement system (for review, see Showalter, 2001; Pettolino et al., 2006). The commercial importance of AGPs is reflected in the vast number of AGP-related patents (http://dk.espacenet.com). Such patents reveal the potential pharmaceutical applications of AGPs, for example, as fat absorbers, in the treatment of diabetes, and for the coating of medical tablets (Ozaki and Kitamura, 2005; Fang and Li, 2007; Malainine, 2007).
CONCLUSION
This is clearly an exciting period for researchers to enter the field of AGPs. Modern molecular and genetic techniques coupled with cell biology and biochemical methods offer the opportunity to rapidly fill in several major gaps in our knowledge of this complex family of molecules, namely unequivocal biological function, the assembly of the glycan chains, and their postassembly processing, both at the cell surface (plasma membrane release/cell wall cross-linking) and following endocytosis (turnover).
Acknowledgments
We are grateful to Ms. Joanne Noble for administrative and editing assistance and figure designs.
References
- Acosta-Garcia G, Vielle-Calzada JP. (2004) A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andème-Onzighi C, Sivaguru M, Judy-March J, Baskin TI, Driouich A. (2002) The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215: 949–958 [DOI] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B. (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45: 144–165 [DOI] [PubMed] [Google Scholar]
- Bacic A, Churms SC, Stephen AM, Cohen PB, Fincher GB. (1987) Fine structure of the arabinogalactan-protein from Lolium multiflorum. Carbohydr Res 162: 85–93 [Google Scholar]
- Baldwin TC, McCann MC, Roberts K. (1993) A novel hydroxyproline-deficient arabinogalactan protein secreted by suspension-cultured cells of Daucus carota. Plant Physiol 103: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Robbins MP, Dixon RA. (1985) Elicitor-induced prolyl hydroxylase from French bean (Phaseolus vulgaris). Biochem J 229: 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P. (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, MacAskill A, Napier JA, Beale MH, Lilley KS, Dupree P. (2005) Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Heissigerova H, Jeanneau C, Moravcova J, Imberty A. (2002) Comparative aspects of glycosyltransferases. Biochem Soc Symp 69: 23–32 [DOI] [PubMed] [Google Scholar]
- Cassab GI. (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281–309 [DOI] [PubMed] [Google Scholar]
- Chaves I, Regalado AP, Chen M, Ricardo CP, Showalter AM. (2002) Programmed cell death induced by (β-D-galactosyl)3 Yariv reagent in Nicotiana tabacum BY-2 suspension-cultured cells. Physiol Plant 116: 548–553 [Google Scholar]
- Chen CG, Pu ZY, Moritz RL, Simpson RJ, Bacic A, Clarke AE, Mau SL. (1994) Molecular cloning of a gene encoding an arabinogalactan-protein from pear (Pyrus communis) cell suspension culture. Proc Natl Acad Sci USA 91: 10305–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM. (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ. (1984) Prolyl hydroxylase in plants. Methods Enzymol 107: 361–369 [Google Scholar]
- Churms SC, Merrifield EH, Stephen AM. (1983) Some new aspects of the molecular-structure of Acacia senegal gum (gum arabic). Carbohydr Res 123: 267–279 [Google Scholar]
- Churms SC, Stephen AM, Siddiqui IR. (1981) Evidence for repeating sub-units in the molecular structure of the acidic arabinogalactan from rapeseed (Brassica campestris). Carbohydr Res 94: 119–122 [Google Scholar]
- Clarke AE, Anderson RL, Stone BA. (1979) Form and function of arabinogalactans and arabinogalactan-proteins. Phytochemistry 18: 521–540 [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. (2009) Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J Exp Bot 60: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Starn M, Blanc E, Henrissat B. (2003) Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci 8: 563–565 [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia F, Hancock CN, Kim D, McClure B. (2005) Stylar glycoproteins bind to S-RNase in vitro. Plant J 42: 295–304 [DOI] [PubMed] [Google Scholar]
- Dahiya P, Findlay K, Roberts K, McCann MC. (2006) A fasciclin-domain containing gene, ZeFLA11, is expressed exclusively in xylem elements that have reticulate wall thickenings in the stem vascular system of Zinnia elegans cv Envy. Planta 223: 1281–1291 [DOI] [PubMed] [Google Scholar]
- Doblin M, Pettolino F, Bacic A. (2010) The skeleton of the plant world and more: role in plant growth, development and biotechnology. Funct Plant Biol (in press) [Google Scholar]
- Du H, Simpson RJ, Moritz RL, Clarke AE, Bacic A. (1994) Isolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell 6: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Ellis MA, Doblin MS, Bacic A. (2010) The genes and enzymes of CAZy GT-family-31: towards unrevealing the function(s) of the novel plant glycosyltransferase family members. Annual Plant Reviews. Vol 41 Plant Polysaccharides. Blackwell Publishing, Oxford: (in press) [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Maurer-Stroh S, Novatchkova M, Schneider G, Eisenhaber F. (2003a) Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays 25: 367–385 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F. (2003b) Glycosylphosphatidylinositol lipid anchoring of plant proteins: sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins T, Hortsch M, Bieber AJ, Snow PM, Goodman CS. (1990) Drosophila fasciclin 1 is a novel homophilic adhesion molecule that along with fasciclin III can mediate cell sorting. J Cell Biol 110: 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez JM, Kieliszewski MJ, Khitrov N, Somerville C. (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol 142: 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Abouzouhair J, Sarhan F. (2006) Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analyses. Mol Genet Genomics 276: 478–494 [DOI] [PubMed] [Google Scholar]
- Fang G, Li J, inventors. October 17, 2007. Preparation method for compound arabinogalactan film coated tablets. China Forestry Intellectual Property Information Center Publication No. CN101053588 (A) [Google Scholar]
- Filmus J, Capurro M, Rast J. (2008) Glypicans. Genome Biol 9: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB, Stone BA, Clarke AE. (1983) Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol 34: 47–70 [Google Scholar]
- Fujita M, Jigami Y. (2008) Lipid remodeling of GPI-anchored proteins and its function. Biochim Biophys Acta 1780: 410–420 [DOI] [PubMed] [Google Scholar]
- Gane AM, Craik D, Munro SLA, Howlett GJ, Clarke AE, Bacic A. (1995) Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata. Carbohydr Res 277: 67–85 [DOI] [PubMed] [Google Scholar]
- Gao M, Kieliszewski MJ, Lamport DTA, Showalter AM. (1999) Isolation, characterization and immunolocalization of a novel, modular tomato arabinogalactan-protein corresponding to the LeAGP-1 gene. Plant J 18: 43–55 [DOI] [PubMed] [Google Scholar]
- Gao M, Showalter AM. (1999) Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan-protein involvement. Plant J 19: 321–331 [DOI] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol 47: 161–176 [PubMed] [Google Scholar]
- Gaspar YM, Nam J, Schultz CJ, Lee LY, Gilson PR, Gelvin SB, Bacic A. (2004) Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiol 135: 2162–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson P, Gaspar YM, Oxley D, Youl JJ, Bacic A. (2001) NaAGP4 is an arabinogalactan protein whose expression is suppressed by wounding and fungal infection in Nicotiana alata. Protoplasma 215: 128–139 [DOI] [PubMed] [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ. (2000) Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54: 99–106 [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. (2004) Role of glycosylation in development. Annu Rev Biochem 73: 491–537 [DOI] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure BA. (2005) The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J 43: 716–723 [DOI] [PubMed] [Google Scholar]
- Hieta R, Myllyharju J. (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. J Biol Chem 277: 23965–23971 [DOI] [PubMed] [Google Scholar]
- Hill KL, Harfe BD, Dobbins CA, L'Hernault SW. (2000) dpy-18 encodes an α-subunit of prolyl-4-hydroxylase in Caenorhabditis elegans. Genetics 155: 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Xu WL, Gong SY, Li B, Wang XL, Xu D, Li XB. (2008) Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol Plant 134: 348–359 [DOI] [PubMed] [Google Scholar]
- Immerzeel P, Eppink MM, de Vries SC, Schols HA, Voragen AGJ. (2006) Carrot arabinogalactan proteins are interlinked with pectins. Physiol Plant 128: 18–28 [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. (2003a) The fasciclin-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiol 133: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Schultz CJ, Bacic A. (2003b) Non-enzymic cell wall (glyco)proteins. Rose J, , The Plant Cell Wall. Blackwell Publishing, Oxford, pp 111–154 [Google Scholar]
- Kawamoto T, Noshiro M, Shen M, Nakamasu K, Hashimoto K, Kawashima-Ohya Y, Gotoh O, Kato Y. (1998) Structural and phylogenetic analyses of RGD-CAP/big-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta 1395: 288–292 [DOI] [PubMed] [Google Scholar]
- Kelly J, Jarrell H, Millar L, Tessier L, Fiori LM, Lau PC, Allan B, Szymanski CM. (2006) Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J Bacteriol 188: 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Fujita M, Maeda Y. (2008) Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem 144: 287–294 [DOI] [PubMed] [Google Scholar]
- Knox JP. (1997) The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int J Cytol 171: 79–120 [DOI] [PubMed] [Google Scholar]
- Koski MK, Hieta R, Hirsilä M, Rönkä A, Myllyharju J, Wierenga RK. (2009) The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J Biol Chem 284: 25290–25301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GHH, Lilley KS, Dupree P, Grossniklaus U, Twell D. (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DT, Kieliszewski MJ, Showalter AM. (2006) Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol 169: 479–492 [DOI] [PubMed] [Google Scholar]
- Lee CB, Kim S, McClure B. (2009) A pollen protein, NaPCCP, that binds pistil arabinogalactan proteins also binds phosphatidylinositol 3-phosphate and associates with the pollen tube endomembrane system. Plant Physiol 149: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CB, Swatek KN, McClure B. (2008) Pollen proteins bind to the C-terminal domain of Nicotiana alata pistil arabinogalactan proteins. J Biol Chem 283: 26965–26973 [DOI] [PubMed] [Google Scholar]
- Lee KJD, Sakata Y, Mau SL, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. (2005) Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17: 3051–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé AC, Gomord V, Faye L. (1998) N-Glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38: 31–48 [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M. (2008) Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J 56: 351–363 [DOI] [PubMed] [Google Scholar]
- Li X, Wu H, Dillon S, Southerton S. (2009) Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don. BMC Genomics 10: 41–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu D, Tu L, Zhang X, Wang L, Zhu L, Tan J, Deng F. (2010) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep 29: 193–202 [DOI] [PubMed] [Google Scholar]
- Lim YS, McLaughlin T, Sung TC, Santiago A, Lee KF, O'Leary DDM. (2008) p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron 59: 746–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind JL, Bacic A, Clarke AE, Anderson MA. (1994) A style-specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. Plant J 6: 491–502 [DOI] [PubMed] [Google Scholar]
- Lind JL, Bonig I, Clarke AE, Anderson MA. (1996) A style-specific 120-kDa glycoprotein enters pollen tubes of Nicotiana alata in vivo. Sex Plant Reprod 9: 75–86 [Google Scholar]
- Liu D, Tu L, Li Y, Wang L, Zhu L, Zhang X. (2008) Genes encoding fasciclin-like arabinogalactan proteins are specifically expressed during cotton fiber development. Plant Mol Biol Rep 26: 98–113 [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG. (2010) Fascilin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Mahendran T, Williams PA, Phillips GO, Al-Assaf S, Baldwin TC. (2008) New insights into the structural characteristics of the arabinogalactan-protein (AGP) fraction of gum arabic. J Agric Food Chem 56: 9269–9276 [DOI] [PubMed] [Google Scholar]
- Mascara T, Fincher GB. (1982) Biosynthesis of arabinogalactan-protein in Lolium multiflorum (ryegrass) endosperm cells. 2. In vitro incorporation of galactosyl residues from UDP-galactose into polymeric products. Aust J Plant Physiol 9: 31–45 [Google Scholar]
- Malainine ME, inventor. September 7, 2007. An arabinogalactan protein having the property of absorbing fats and method for obtaining this arabinogalactan protein. World Intellectual Property Organization Publication No. WO/2007/100236 (A2) [Google Scholar]
- Mollet JC, Kim S, Jauh GY, Lord EM. (2002) Arabinogalactan proteins, pollen tube growth, and the reversible effects of Yariv phenylglycoside. Protoplasma 219: 89–98 [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878 [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Matthysse AG, Gelvin SB. (1999) Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet 261: 429–438 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Andeme-Onzighi C, Aboughe-Angone S, Bardor M, Ishii T, Lerouge P, Driouich A. (2006) The reb1-1 mutation of Arabidopsis: effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol 140: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Bannigan A, Chevalier L, Baskin TI, Driouich A. (2007) Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. Plant J 52: 240–251 [DOI] [PubMed] [Google Scholar]
- Nothnagel EA. (1997) Proteoglycans and related components in plant cells. Int Rev Cytol 174: 195–291 [DOI] [PubMed] [Google Scholar]
- Oka T, Saito F, Shimma Y, Yoko-o Y, Nomura Y, Matsuoka K, Jigami Y. (2010) Characterization of endoplasmic reticulum-localized UDP-D-galactose:hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol 152: 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveld A, Voragen AGJ, Schols HA. (2002) Characterization of hop pectins shows the presence of an arabinogalactan-protein. Carbohydr Polym 49: 407–413 [Google Scholar]
- Orlean P, Menon AK. (2007) GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res 48: 993–1011 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. (1999) Structure of the glycosyl-phosphatidylinositol anchor of an arabinogalactan-protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci USA 96: 14246–14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S, Kitamura S, inventors. November 10, 2005. Novel arabinogalactan, substance with antidiabetic activity and method of use thereof. World Intellectual Property Organization Publication No. WO/2005/105852 (A1) [Google Scholar]
- Park MH, Suzuki Y, Chono M, Knox JP, Yamaguchi I. (2003) CsAGP1, a gibberellin-responsive gene from cucumber hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol 131: 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LG, Coimbra S, Monteiro HOL, Sottomayor M. (2006) Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta 223: 374–380 [DOI] [PubMed] [Google Scholar]
- Pettolino F, Liao ML, Ying Z, Mau SL, Bacic A. (2006) Structure, function and cloning of arabinogalactan-proteins (AGPs): an overview. Food and Food Ingredients Journal of Japan 211: 12–25 [Google Scholar]
- Ponder GR, Richards GN. (1997) Arabinogalactan from Western larch. Part III. Alkaline degradation revisited, with novel conclusions on molecular structure. Carbohydr Polym 34: 251–261 [Google Scholar]
- Qi W, Fong C, Lamport DTA. (1991) Gum arabic glycoprotein is a twisted hairy rope: a new model based on O-galactosylhydroxyproline as the polysaccharide attachment site. Plant Physiol 96: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Wilson IW, Gan S, Washusen R, Moran GF, Southerton SG. (2008) Gene expression in Eucalyptus branch wood with marked variation in cellulose microfibril orientation and lacking G-layers. New Phytol 179: 94–103 [DOI] [PubMed] [Google Scholar]
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A. (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyl transferases. Plant Mol Biol 68: 43–59 [DOI] [PubMed] [Google Scholar]
- Sanchez C, Schmitt C, Kolodziejczyk E, Lapp A, Gaillard C, Renard D. (2008) The acacia gum arabinogalactan fraction is a thin oblate ellipsoid: a new model based on small-angle neutron scattering and ab initio calculation. Biophys J 94: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar HS, Showalter AM. (2007) A cellular networking model involving interactions among glycosyl-phosphatidylinositol (GPI)-anchored plasma membrane arabinogalactan proteins (AGPs), microtubules and F-actin in tobacco BY-2 cells. Plant Signal Behav 2: 8–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar HS, Yang J, Showalter AM. (2006) Molecular interactions of arabinogalactan proteins with cortical microtubules and F-actin in Bright Yellow-2 tobacco cultured cells. Plant Physiol 142: 1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Schaefer RM. (2010) Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res 339: 237–246 [DOI] [PubMed] [Google Scholar]
- Schibeci A, Pnjak A, Fincher GB. (1984) Biosynthesis of arabinogalactan-protein in Lolium multiflorum (Italian ryegrass) endosperm cells: subcellular distribution of galactosyltransferases. Biochem J 218: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A. (1998) GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci 3: 426–431 [Google Scholar]
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. (2004) Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana: endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J Biol Chem 279: 45503–45511 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Harrison MJ. (2008) Novel plant and fungal AGP-like proteins in the Medicago truncatula-Glomus intraradices arbuscular mycorrhizal symbiosis. Mycorrhiza 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Hauser K, Lind JL, Atkinson AH, Pu ZY, Anderson MA, Clarke AE. (1997) Molecular characterisation of a cDNA sequence encoding the backbone of a style-specific 120 kDa glycoprotein which has features of both extensins and arabinogalactan-proteins. Plant Mol Biol 35: 833–845 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. (2002) Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiol 129: 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Barber C, Wells B, Dolan L, Roberts K. (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol 12: 1840–1845 [DOI] [PubMed] [Google Scholar]
- Serpe MD, Nothnagel EA. (1999) Arabinogalactan-proteins in the multiple domains of the plant cell surface. Adv Bot Res 30: 207–289 [Google Scholar]
- Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Igasaki T, Yamada M, Yuasa K, Hasegawa J, Kato T, Tsukagoshi H, Nakamura K, Fukuda H, Matsuoka K. (2005) Experimental determination of proline hydroxylation and hydroxyproline arabinogalactosylation motifs in secretory proteins. Plant J 42: 877–889 [DOI] [PubMed] [Google Scholar]
- Showalter AM. (1993) Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. (2001) Arabinogalactan-proteins: structure, expression, and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ. (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA 96: 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A. (1996) A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J 9: 71–83 [DOI] [PubMed] [Google Scholar]
- Stenvik G-E, Butenko MA, Urbanowicz BR, Rose JKC, Aalena RB. (2006) Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glössl J, Léonard R, Stadlmann J, Altmann F, Steinkellner H, et al. (2007) A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Physiol 19: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Kieliszewski MJ, Showalter AM. (2004) Overexpression of tomato LeAGP-1 arabinogalactan-protein promotes lateral branching and hampers reproductive development. Plant J 40: 870–881 [DOI] [PubMed] [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA. (1999) Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem 274: 14724–14733 [DOI] [PubMed] [Google Scholar]
- Tan L, Qiu F, Lamport DTA, Kieliszewski MJ. (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J Biol Chem 279: 13156–13165 [DOI] [PubMed] [Google Scholar]
- Tiainen P, Myllyharju J, Koivunen P. (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem 280: 1142–1148 [DOI] [PubMed] [Google Scholar]
- Tsumauraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S. (1988) Arabinogalactan-proteins from primary and mature roots of radish (Raphanus sativus L.). Plant Physiol 86: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. (2002) Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J 32: 105–113 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. (2003) AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J 36: 256–270 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, van Kammen A, de Vries SC. (2002) A relationship between seed development, arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol Plant 114: 637–644 [DOI] [PubMed] [Google Scholar]
- Vuorela A, Myllyharju J, Nissi R, Pihlajaniemi T, Kivirikko KI. (1997) Assembly of human prolyl 4-hydroxylase and type III collagen in the yeast Pichia pastoris: formation of a stable enzyme tetramer requires coexpression with collagen and assembly of a stable collagen requires coexpression with prolyl 4-hydroxylase. EMBO J 16: 6702–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Wang H, Cheung AY. (1995) A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82: 395–403 [DOI] [PubMed] [Google Scholar]
- Wu HM, Wong E, Ogdahl J, Cheung AY. (2000) A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J 22: 165–176 [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. (2010) Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J Biol Chem 285: 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tan L, Lamport DT, Showalter AM, Kieliszewski MJ. (2008a) The O-Hyp glycosylation code in tobacco and Arabidopsis and a proposed role of Hyp-glycans in secretion. Phytochemistry 69: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. (2008b) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. (1994) Pectic polysaccharides from Chinese herbs: structure and biological activity. Carbohydr Polym 25: 269–276 [Google Scholar]
- Yang J, Sardar HS, McGovern KR, Zhang Y, Showalter AM. (2007) A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J 49: 629–640 [DOI] [PubMed] [Google Scholar]
- Yang J, Showalter AM. (2007) Expression and localization of AtAGP18, a lysine-rich arabinogalactan-protein in Arabidopsis. Planta 226: 169–179 [DOI] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E. (1967) Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem J 105: 1c–2c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J 41: 81–94 [DOI] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DTA, Kieliszewski MJ. (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]