Abstract
Glycerate 3-kinase (GLYK) is the terminal enzyme of the photorespiratory cycle in plants and many cyanobacteria. For several C4 plants, notably grasses of the NADP-malic enzyme (ME) subtype, redox regulation of GLYK has been reported, but the responsible molecular mechanism is not known. We have analyzed the enzyme from the NADP-ME C4 plant maize (Zea mays) and found that maize GLYK, in contrast to the enzyme from C3 plants and a dicotyledonous NADP-ME C4 plant, harbors a short carboxy-terminal extension. In its oxidized (night) form, a disulfide bridge is formed between the two cysteine residues present in this extra domain, and GLYK activity becomes inhibited. Cleavage of this bond by thioredoxin f produces the fully active thiol form, releasing autoinhibition. Fusion of the maize GLYK redox-regulatory domain to GLYK from C3 plants confers redox regulation to these otherwise unregulated enzymes. It appears that redox regulation of GLYK could be an exclusive feature of monocotyledonous C4 plants of the NADP-ME type, in which linear electron transport occurs only in the mesophyll chloroplasts. Hence, we suggest that GLYK, in addition to its function in photorespiration, provides glycerate 3-phosphate for the accelerated production of triose phosphate and its export from the mesophyll. This could facilitate the activation of redox-regulated Calvin cycle enzymes and the buildup of Calvin cycle intermediates in the bundle sheath of these particular C4 plants during the dark/light transition.
Glycerate kinases are important enzymes of primary metabolism in all organisms. Glycerate 2-kinases are involved in the degradation of Glc and other metabolic pathways in animals and bacteria (Guo et al., 2006; Kehrer et al., 2007) but have not been reported for plants. Instead, a glycerate 3-kinase (GLYK) occurs in plant chloroplasts and many cyanobacteria, where it produces glycerate 3-phosphate (3PGA) from glycerate in the last step of the photorespiratory C2 cycle (Boldt et al., 2005; Bartsch et al., 2008). This pathway is an indispensable auxiliary component of the photosynthetic CO2 assimilation network in cyanobacteria and plants (Eisenhut et al., 2008), and its operation is most evident in higher plants with the C3 pathway of photosynthesis. Although it was long known that photorespiration also occurs in C4 plants (Osmond and Harris, 1971), the indispensability of this pathway for the survival of plants of this particular photosynthetic type in normal air was proven only recently (Dever et al., 1995; Zelitch et al., 2009).
Based on differences in the bundle sheath-located decarboxylation step of their CO2 concentration cycle, C4 plants are separated into the three subgroups of NADP-malic enzyme (ME), NAD-ME, and phosphoenolpyruvate carboxykinase types (Kanai and Edwards, 1999). Biochemical studies have shown that most enzymes of the photorespiratory cycle occur with highest activities in the bundle sheath of C4 plants (Woo and Osmond, 1977; Ohnishi and Kanai, 1983). GLYK is an exception from this rule, since it occurs exclusively in the mesophyll of C4 plants, irrespective of their C4 photosynthetic subtype (Osmond and Harris, 1971; Usuda and Edwards, 1980a). Hence, photorespiratory glycerate is produced in the bundle sheath but phosphorylated to 3PGA in the mesophyll of all C4 plants (Usuda and Edwards, 1980b). Another study reported thiol-dependent regulation of GLYK in several C4 plants, and it was proposed that GLYK, in addition to its function in the final step of photorespiration, could also serve as part of the intercellular transport system for 3PGA (Kleczkowski and Randall, 1986).
Thiol-dependent activation is a well-known feature of several Calvin cycle enzymes, and thioredoxins (Trxs) are important components of this process (for review, see Buchanan and Balmer, 2005; Meyer et al., 2009). On the other hand, knowledge about the regulation of photorespiratory enzymes is scarce. We found that GLYK (ZmGLYK) from the C4 plant maize (Zea mays), in contrast to GLYKs from most other plants, carries a small C-terminal extension including two Cys residues that spontaneously combine to form a disulfide under nonreducing conditions, resulting in the inhibition of the enzyme. Cleavage of the disulfide bond by Trx f reestablished full activity. Artificial fusion of this redox-regulatory domain to GLYK from C3 plants conferred autoinhibition and Trx activation to these otherwise unregulated enzymes.
RESULTS
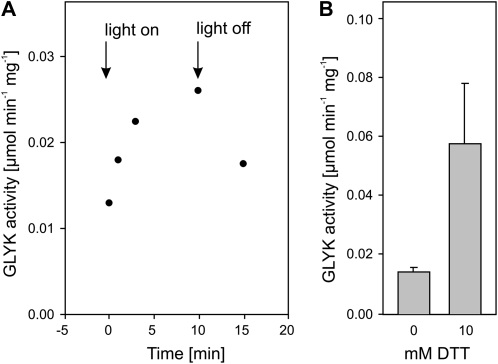
In accordance with previous studies (Kleczkowski and Randall, 1985), initial experiments confirmed that leaf ZmGLYK activity is subject to day/night regulation in planta (Fig. 1A). Enzyme activity in leaf extracts was low without the addition of 1,4-dithio-dl-threitol (DTT) but could be significantly enhanced by its presence during extraction and in the activity assay (Fig. 1B). Absence of DTT from the assay mixture led to a rapid drop in activity, which further confirms that ZmGLYK activity is thiol regulated. We also observed that leaf glycerate levels after 12 h of darkness (1.13 ± 0.21 μmol g−1 fresh weight) were more than 10-fold higher than those of 3PGA (0.09 ± 0.01 μmol g−1 fresh weight).
Figure 1.
Light and thiol activation of GLYK in desalted maize leaf soluble crude extracts. A, Light activation after 4 h of dark adaptation of leaves. After 10 min of illumination, leaves were darkened again. B, Thiol activation by addition of 10 mm DTT to extracts from dark-adapted leaves. Means ± se from three independent experiments are shown.
At the beginning of our study, GLYK sequences from C4 plants were not known. Hence, we used the Arabidopsis (Arabidopsis thaliana) GLYK sequence (Boldt et al., 2005) for similarity searches in maize EST libraries and identified several overlapping ESTs, which were combined to a continuous nucleotide sequence encoding the full-length putative ZmGLYK. This sequence was recently deposited in GenBank, and the gene product was tentatively annotated as hypothetical protein LOC100274052 (Alexandrov et al., 2009). The predicted amino acid sequence is highly similar to GLYKs from other plants, for example rice (Oryza sativa; 87% sequence identity with Os01g0682500) and Arabidopsis (69% sequence identity with At1g80380) over its whole length (alignment in Supplemental Fig. S1). In order to find out whether the hypothetical maize protein LOC100274052 is a functional GLYK, the nucleotide sequence encoding the predicted mature protein was ligated into the Escherichia coli protein expression vector pBAD/His-B. The affinity-purified recombinant protein showed GLYK activity, which allowed the functional annotation of LOC100274052 as ZmGLYK.
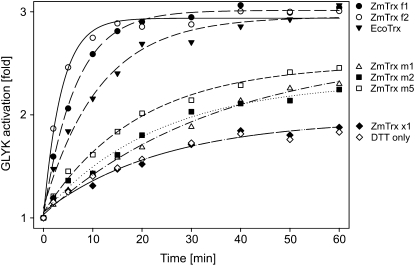
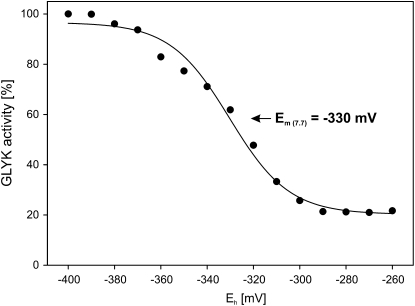
Addition of 10 mm DTT had a distinct but relatively slow activating effect on the recombinant ZmGLYK. Activation showed biphasic kinetics and could be significantly accelerated by the simultaneous addition of reduced E. coli Trx (EcoTrx), which resulted in about 3-fold activation within 40 min (Fig. 2). On the other hand, the addition of 10 mm 4,5-dihydroxy-1,2-dithiane (DTTox) did not result in any further reduction of the basal GLYK activity of the recombinant enzyme from maize. The redox midpoint potential Em of the activation of ZmGLYK was −330 ± 2 mV at pH 7.7 (Fig. 3).
Figure 2.
Trx-dependent activation kinetics of ZmGLYK. Recombinant ZmGLYK (0.14 μm) was incubated at 23°C in 100 mm Tricine-KOH, pH 7.7, with 10 μm of different maize and E. coli Trxs plus 10 mm DTT or with DTT alone as a control. Aliquots were assayed for time-dependent activation.
Figure 3.
Redox titration of ZmGLYK activity. Recombinant ZmGLYK was incubated with varying ratios of DTTox/DTT and 0.9 μm EcoTrx at pH 7.7 and assayed for activity. Data points represent means from three independent experiments. The redox midpoint potential Em was calculated by fitting of the data set to the Nernst equation (solid line).
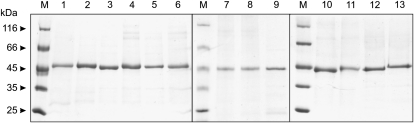
Since ZmGLYK was activated by EcoTrx, which has broad substrate specificity, we aimed to identify the maize Trx (ZmTrx) that is responsible for its regulation in vivo. As f-type and m-type Trxs are known to redox regulate enzymes in the chloroplast (Schürmann and Buchanan, 2008), we searched databases for nucleotide sequences encoding these proteins in the maize genome, using the sorghum (Sorghum bicolor) Trxs annotated by Chibani et al. (2009) as a starting point. This allowed identifying coding sequences for two ZmTrx f and three ZmTrx m, which were named according to their sorghum equivalents (Supplemental Fig. S2). We also identified a ZmTrx x-encoding sequence. This particular Trx is believed to be involved in peroxide detoxification but not in enzyme regulation (Collin et al., 2003) and was included in our study as a negative control. These six maize Trxs, without their plastidial targeting sequences as predicted by ChloroP and sequence comparisons, were overexpressed in E. coli and affinity purified (Fig. 4) to test their potential to activate ZmGLYK. We found that ZmGLYK was activated fastest by ZmTrx f1 and f2, leading to full activation of the enzyme after about 20 min (Fig. 2). Activation by the three m-type ZmTrxs was much less efficient and only slightly faster than activation by DTT alone. Moreover, these Trxs did not fully activate ZmGLYK, even with longer incubation times than those shown in Figure 2. ZmTrx x had no activating effect in addition to that observed with DTT alone.
Figure 4.
SDS-PAGE gel with recombinant maize Trxs and commercial E. coli Trx. About 7 μg of each recombinant protein was applied per lane. Lane 1, ZmTrx f1; lane 2, ZmTrx f2; lane 3, ZmTrx m1; lane 4, ZmTrx m2; lane 5, ZmTrx m5; lane 6, ZmTrx x; lane 7, EcoTrx; lane M, size markers.
The apparent Km values of ZmGLYK for d-glycerate and ATP were only slightly affected by thiol activation (Table I) and were similar to published data for recombinant GLYKs (AtGLYK and OsGLYK) from the dicotyledonous C3 plant Arabidopsis and the monocotyledonous C3 plant rice (Bartsch et al., 2008). In addition, the specific activity of activated ZmGLYK was also close to the activities of the C3 plant GLYKs. These latter enzymes were neither activated by DTT/EcoTrx nor inhibited by oxidized DTT (data not shown). In contrast, the specific activity of ZmGLYK decreased to about one-third in nonreducing conditions. This alteration corresponds to the results obtained with leaf extracts (Fig. 1) and the observed time-dependent changes in activity (Fig. 2).
Table I. Kinetic parameters of oxidized and reduced recombinant ZmGLYK (maize) in comparison with recombinant AtGLYK (Arabidopsis) and OsGLYK (rice).
Reduction was achieved by incubation with 10 mm DTT and 0.9 μm EcoTrx for 60 min, and oxidation was achieved by incubation without reducing agent. Kinetic parameters ± se were calculated by nonlinear regression analysis. Parameters for AtGLYK and OsGLYK are from Bartsch et al. (2008).
| GLYK | Km (d-Glycerate) | Km (ATP) | Vmax |
| mm | μmol min−1 mg−1 | ||
| ZmGLYK (oxidized) | 0.358 ± 0.049 | 0.839 ± 0.070 | 115 ± 3.8 |
| ZmGLYK (reduced) | 0.399 ± 0.033 | 0.707 ± 0.055 | 379 ± 14 |
| AtGLYK (oxidized) | 0.266 ± 0.003 | 0.834 ± 0.138 | 364 ± 54 |
| OsGLYK (oxidized) | 0.450 ± 0.054 | 0.812 ± 0.116 | 295 ± 65 |
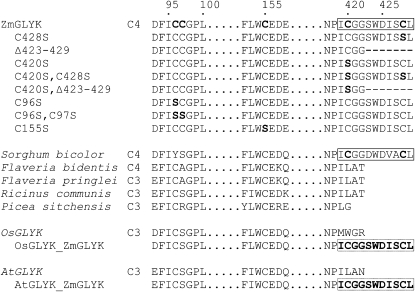
The most distinct structural difference between ZmGLYK and the enzymes from C3 plants, in addition to the typical variability in the chloroplast import sequences, was a C-terminal extension of seven amino acids in ZmGLYK (Fig. 5). A highly similar extension exists in the predicted primary structure of SbGLYK (currently annotated as hypothetical protein with similarity to phosphoribulokinase/uridine kinase) from sorghum, a species that also belongs to the NADP-ME subgroup of C4 plants (Paterson et al., 2009). It was not present, however, in any of the other available GLYK amino acid sequences, neither that of the dicotyledonous NADP-ME C4 plant Flaveria bidentis (Drincovich et al., 1998) nor those from C3 plants.
Figure 5.
GLYK sequence stretches showing amino acids potentially involved in redox regulation of ZmGLYK and variations introduced by site-specific mutagenesis. For comparison, corresponding sequences are shown for GLYK from the C4 plants sorghum (XP_002439104) and F. bidentis and the C3 plants Flaveria pringlei, Ricinus communis (XP_002517532), Picea sitchensis (ABR16413), rice (NP_001043886), and Arabidopsis (NP_849912). Flaveria sequences are not published (U. Gowick, personal communication). Numbers indicate the positions in the full-length protein (including the chloroplast-targeting peptide) of ZmGLYK. Mutated amino acids are printed in boldface, and the regulatory C-terminal extension is boxed.
The C-terminal extension of ZmGLYK harbors a Cys residue at its penultimate position. Another Cys residue resides nine amino acids upstream in ZmGLYK and SbGLYK but does not occur in any of the examined C3 plant GLYKs. Similar to other redox-regulated enzymes, such as GAPDH (Sparla et al., 2002) and NADP-MDH (Krimm et al., 1999), these two Cys residues could reversibly form a disulfide bond and the C terminus of ZmGLYK could function as a redox regulation domain. In a nonreducing cellular environment, formation of the disulfide bond would result in inhibition and reducing conditions would reactivate the enzyme.
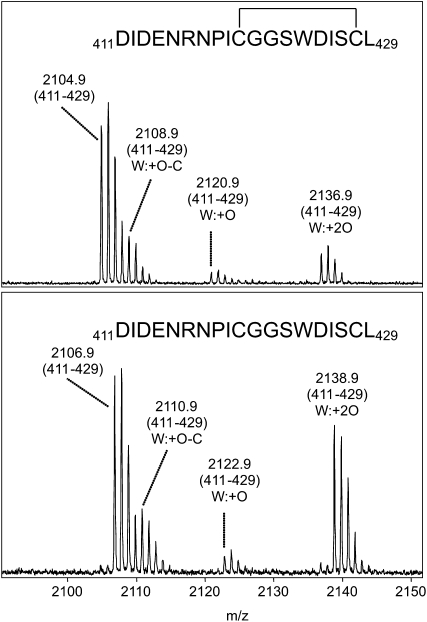
This possibility was investigated by different treatments of ZmGLYK samples excised from a nonreducing polyacrylamide gel and subsequent peptide analysis by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS; Fig. 6). After digestion with AspN, the observed mass difference of 2 D between the respective peptides covering the last 19 amino acids (positions 411–429; untreated/oxidized, mass-to-charge ratio [m/z] 2,104.9; reduced, m/z 2,106.9) clearly established that Cys-420 and Cys-428 form a disulfide bond under nonreducing conditions. This bond becomes cleaved upon reduction with thiols. Alkylation by iodoacetamide of the Cys residues inside the C-terminal part of ZmGLYK was only observed after reduction of the sample (e.g. ion signal at m/z 1,632.7 corresponding to amino acids 411–424 with carbamidomethylated Cys-420; data not shown). In addition, peptide mapping of tryptic as well as AspN digests indicated that the adjacent Cys residues Cys-96 and Cys-97 may also form a disulfide bond (data not shown). Following alkylation with iodoacetamide to identify free thiol groups, an ion signal at m/z 2,457.2 corresponding to amino acids 138 to 156 with carbamidomethylated Cys-155 was detected, showing that Cys-155 exists in the thiol form even under nonreducing conditions.
Figure 6.
Sections of MALDI mass spectra of an AspN digest of recombinant ZmGLYK before (top) and after (bottom) on-target reduction with 10 mm DTT. The labeled ion signals correspond to the C-terminal peptide encompassing amino acid residues 411 to 429. Due to the occurrence of a Trp residue (W424), the typical oxidation pattern of Trp-containing peptides (Suckau et al., 2003) appeared. The mass increase of 2 D after reduction corresponds to cleavage of the disulfide bond of the oxidized peptide.
To more accurately locate the size of the autoinhibitory domain and identify possible interactions with more N-terminal Cys residues, we generated a series of site-specific mutants (Figs. 5 and 7) and analyzed the enzymatic activities of the enzyme variants before and after activation (Table II). The unmodified ZmGLYK was again about 3-fold activated under reducing conditions. Very similar to the findings shown in Table I, maximum activity of ZmGLYK was similar to those of AtGLYK and OsGLYK, and the activity of the two C3 plant enzymes was only marginally enhanced under reducing conditions. Mutant ZmGLYKC428S showed reduced autoinhibition, while autoinhibition was nearly entirely absent in ZmGLYKC420S and ZmGLYKC420S,C428S, in which both C-terminal Cys residues are exchanged for Ser. Autoinhibition was also not observed with mutant ZmGLYKC420S,Δ423-429. On the other hand, it was not fully absent although distinctly reduced in ZmGLYKΔ423-429, in which Cys-420 was still present. Notably, Vmax was not significantly affected by any of these C-terminal mutations under reducing conditions. Exchange of the three far more N-terminal Cys residues in ZmGLYKC96S, ZmGLYKC96S,C97S, and ZmGLYKC155S consistently reduced maximum activities to about 50% under reducing conditions and even less under oxidizing conditions. These particular enzyme variants can still be severalfold activated by thiols, indicating that the respective Cys residues are not necessary for interaction with the C-terminal domain. We conclude that the last 10 amino acids present in the C terminus of ZmGLYK act as an autoinhibitory domain, in which Cys-420 and Cys-428 are of central functional importance.
Figure 7.
SDS-PAGE gel with recombinant GLYK variants from maize, rice, and Arabidopsis (for details of introduced mutations, see Fig. 5). About 3 μg of each recombinant protein was applied per lane. Lane M, Size markers; lane 1, ZmGLYK; lane 2, ZmGLYKC428S; lane 3, ZmGLYKΔ423-429; lane 4, ZmGLYKC420S; lane 5, ZmGLYKC420S,C428S; lane 6, ZmGLYKC420S,Δ423-429; lane 7, ZmGLYKC96S; lane 8, ZmGLYKC96S,C97S; lane 9, ZmGLYKC155S; lane 10, OsGLYK; lane 11, OsGLYKZmGLYK-Cterm; lane 12, AtGLYK; lane 13, AtGLYKZmGLYK-Cterm.
Table II. Enzymatic activities and Trx activation of GLYK variants (see Fig. 5).
Reduction of the enzymes was achieved by incubation with 10 mm DTT and 0.9 μm EcoTrx for 60 min, and oxidation was achieved by incubation without reducing agent. Enzyme activities were measured with 2 mm each of the two substrates, d-glycerate and ATP. Values represent means ± se from three independent experiments.
| Enzyme Variant | GLYK Activity (Oxidized) | GLYK Activity (Reduced) | Activation |
| μmol min−1 mg−1 | fold | ||
| ZmGLYK | 82.7 ± 4.0 | 258.2 ± 7.7 | 3.12 |
| ZmGLYKC428S | 119.1 ± 5.5 | 258.7 ± 4.1 | 2.17 |
| ZmGLYKC420S | 240.2 ± 12.0 | 256.9 ± 17.5 | 1.07 |
| ZmGLYKC420S,C428S | 210.0 ± 1.4 | 245.6 ± 11.9 | 1.17 |
| ZmGLYKΔ423-429 | 165.3 ± 6.6 | 240.4 ± 8.3 | 1.45 |
| ZmGLYKC420S,Δ423-429 | 252.0 ± 4.7 | 282.0 ± 3.5 | 1.12 |
| ZmGLYKC96S | 31.9 ± 0.8 | 140.5 ± 3.5 | 4.40 |
| ZmGLYKC96S,C97S | 19.7 ± 0.4 | 127.5 ± 6.4 | 6.46 |
| ZmGLYKC155S | 34.1 ± 5.1 | 119.1 ± 3.4 | 3.50 |
| OsGLYK | 236.6 ± 11.8 | 276.5 ± 17.4 | 1.17 |
| OsGLYKZmGLYK-Cterm | 22.5 ± 1.6 | 302.6 ± 19.2 | 13.4 |
| AtGLYK | 212.0 ± 12.7 | 247.6 ± 5.7 | 1.16 |
| AtGLYKZmGLYK-Cterm | 76.3 ± 1.2 | 236.1 ± 9.5 | 3.09 |
While the interacting “internal” domain is not yet known, we finally wanted to find out whether the autoinhibitory domain of ZmGLYK can interact with GLYKs from C3 plants, which are not redox regulated. To this end, we replaced the original four C-terminal amino acids of AtGLYK and OsGLYK with the last 11 amino acids from the C terminus of ZmGLYK. Activity measurements with these engineered C3 GLYKs confirmed that they were distinctly inhibited under oxidizing conditions and that autoinhibition was fully relieved under reducing conditions (Table II). This result further substantiates that the C-terminal extension is responsible for redox regulation of ZmGLYK.
DISCUSSION
At the beginning of our studies, GLYK sequences from C4 plants as well as the mechanism of the earlier reported thiol activation of the maize enzyme were not known. Similarity searches pointed to potentially GLYK-encoding maize ESTs, and overexpression of these nucleotide sequences confirmed the functionality of the recombinant protein as a thiol-regulated GLYK. The protein has been tentatively annotated as hypothetical protein LOC100274052 (Alexandrov et al., 2009) and, on the basis of our functional data, can now be annotated as ZmGLYK.
Similar to GLYK extracted from maize leaves (Fig. 1), the activity of recombinant ZmGLYK was high under reducing (day) conditions and decreased to less than one-third under nonreducing (night) conditions (Table I). We cannot exclude the possibility that the enzyme could be even more inhibited in vivo; however, our data show that autoxidation of ZmGLYK does not require interacting proteins and hence represents an inherent property of the enzyme. ZmGLYK activation under reducing conditions was strongly accelerated by EcoTrx and even more so by ZmTrx f1 and f2 (Fig. 2). In comparison, incubation with ZmTrx m1, m2, or m5 did not fully activate the enzyme and activation was distinctly slower. As expected, ZmTrx x had no activating effect at all. This shows that light-dependent activation of ZmGLYK is mediated by Trx f, which is also the most prevalent activator of redox-regulated Calvin cycle enzymes (Buchanan and Balmer, 2005; Meyer et al., 2009). The midpoint redox potential Em of ZmGLYK of −330 ± 2 mV at pH 7.7 (Fig. 3), assuming a slope of −59 mV per pH unit, corresponds to −289 mV at pH 7.0. This is very similar to reported Em,7.0 values of redox-regulated Calvin cycle enzymes, which range from −290 to −305 mV (Buchanan and Balmer, 2005), and strongly indicates that ZmGLYK is coactivated with these enzymes.
While the overall primary structure of ZmGLYK is very similar to that of previously characterized unregulated GLYKs from Arabidopsis and rice (Boldt et al., 2005; Bartsch et al., 2008), it differs from these C3 plant enzymes by a C-terminal extension of seven amino acids, including Cys-428 at the penultimate position (Fig. 5). A second Cys residue is located closely upstream at position 420 of ZmGLYK and also at the corresponding position in SbGLYK but not in known GLYKs without C-terminal extension. Our data show that this domain, comprising 10 amino acids, is essential for redox regulation of ZmGLYK. This conclusion is based on three lines of evidence.
First, the C-terminal domain alters (inhibits) the enzyme's activity (Tables I and II) after formation of a disulfide bond between Cys-420 and Cys-428 by autoxidation under nonreducing conditions, as shown by MALDI-TOF-MS analyses (Fig. 6). We did not precisely assess the rate of autoxidation, but since inactivation occurs rapidly after extraction of the enzyme from leaves, it seems to be a relatively fast process.
Second, artificially introduced mutations in the C-terminal domain abolished autoinhibition (Table II). Exchange of Cys-420 for Ser resulted in a constitutively active enzyme. We would have expected the same effect after exchange of Cys-428 for Ser; however, autoinhibition was only reduced but not completely abolished in this particular mutant. Similar observations were made with GAPDH (Sparla et al., 2002) and Fru-1,6-bisP phosphatase (Jacquot et al., 1997), where the exchange of only one of the two Cys residues involved in regulatory disulfide formation did not result in a complete loss of redox dependency. Simultaneous exchange of both Cys-420 and Cys-428 also completely abolished autoinhibition. Exchange of the more N-terminal Cys-96, Cys-97, and Cys-155 for Ser considerably reduced maximum activity, but these enzyme variants can still be activated by thiols.
Third, C-terminal fusion of the ZmGLYK autoinhibitory domain to AtGLYK and OsGLYK made these naturally thiol-insensitive C3 plant enzymes redox sensitive (Table II). The modified OsGLYK was even more inhibited under oxidizing conditions than ZmGLYK, while inhibition of the AtGLYK fusion protein was similar to that of ZmGLYK. After reduction with EcoTrx, the chimerical mutant enzymes showed activities that were similar to the unmodified wild-type forms and to activated ZmGLYK.
With these data, the function of the C terminus of ZmGLYK as an autoinhibitory redox regulation domain is clearly established; however, the mechanistic details remain to be investigated. Since the substrate affinities are not significantly altered by redox regulation (Table I), the inhibition appears to be caused mainly by reduction of the turnover number (kcat). C-terminal extensions are known components of redox regulation in other chloroplast enzymes such as glyceraldehyde-3-phosphate dehydrogenase subunit B, Rubisco activase, or NADP-malate dehydrogenase (for review, see Schürmann and Buchanan, 2008). For example, upon formation of a disulfide bond between two C-terminal Cys residues of the regulatory subunit of Rubisco activase, the C terminus impairs binding of ATP to the enzyme's P-loop domain (Portis et al., 2008). A similar mechanism operates in NADP-malate dehydrogenase, in which the disulfide form of the C-terminal extension comes close to the active site, where it acts as an internal inhibitor (Johansson et al., 1999; Krimm et al., 1999). Notably, transfer of the regulatory peptide from sorghum NADP-malate dehydrogenase to Thermus flavus NAD-malate dehydrogenase conveyed redox regulation to the constitutively active bacterial enzyme (Issakidis-Bourguet et al., 2006). In contrast to these oligomeric enzymes, size-exclusion chromatography revealed a monomeric structure of recombinant ZmGLYK both in the oxidized and the reduced states (data not shown), confirming an earlier observation with the enzyme purified from maize leaves (Kleczkowski and Randall, 1985). Crystal structures for GLYK from plants are not available. A preliminary model, based on the structure of GLYK from the yeast Saccharomyces cerevisiae (de La Sierra-Gallay et al., 2004; Kelley and Sternberg, 2009), indicates that the redox-regulatory C-terminal extension of ZmGLYK is located close to the enzyme's ATP-binding P-loop domain (Supplemental Fig. S3).
While we can currently only speculate upon the detailed physiological function of redox regulation of GLYK, it is clearly related to specific requirements of C4 photosynthetic metabolism in the respective species. We regard two aspects as most relevant. First, in contrast to the other photorespiratory enzymes, GLYK is confined to the mesophyll rather than to the bundle sheath in leaves of all C4 plants, irrespective of their C4 photosynthetic type (Osmond and Harris, 1971; Usuda and Edwards, 1980a). Hence, the photorespiratory C2 cycle spans over two cell types in C4 plants: glycerate is produced from glycolate 2-phosphate in the bundle sheath but becomes phosphorylated to 3PGA in the mesophyll (Usuda and Edwards, 1980b). Second, available primary structures (Fig. 5) and an earlier survey of GLYK activation in a number of C3 and C4 species (Kleczkowski and Randall, 1986) strongly suggest that redox regulation of GLYK could be an exclusive feature of monocotyledonous C4 plants of the NADP-ME subgroup. In grasses of this C4 photosynthetic type, in contrast to all other C4 plants, PSII is confined to the mesophyll chloroplasts and does not occur in the bundle sheath (Woo et al., 1970). As a consequence, bundle sheath chloroplasts depend on the import of reducing equivalents from the mesophyll. This occurs via the triose phosphate/3PGA shuttle and requires adequately large gradients in the concentration of the respective metabolites. Inhibition of GLYK during the night and its activation at dawn would ensure that, at the onset of illumination, sufficient glycerate is available to “quick-start” 3PGA formation for reduction and subsequent export of reducing equivalents in the form of triose phosphate to the bundle sheath. Indeed, maize leaves rapidly build up 3PGA and triose phosphates during photosynthetic induction after darkness even in the absence of CO2 (Furbank and Leegood, 1984; Leegood, 2000). Recent proteomic data suggest that the enzymes involved in triose phosphate reduction are primarily located in the mesophyll chloroplasts of maize, indicating that the mesophyll-localized triose phosphate shuttle should be viewed as part of the bundle sheath-localized Calvin cycle in this species rather than as a parallel pathway (Majeran et al., 2005). In light of this hypothesis, the GLYK-accelerated production and export of triose phosphate during the dark/light transition could facilitate the activation of redox-regulated Calvin cycle enzymes and the buildup of Calvin cycle intermediates in C4 plants with PSII-deficient bundle sheath cells.
MATERIALS AND METHODS
Determination of Enzyme Activities and Kinetic Parameters
GLYK activity was determined using two different assays described elsewhere (Bartsch et al., 2008). Assay 1 followed ADP formation and was used with the affinity-purified recombinant enzyme variants. Assay 2 followed the rate of 3PGA formation and was used with leaf extracts. DTT and DTTox (Sigma-Aldrich) were included or not included as specified for the individual experiments. Blank values without d-glycerate were subtracted. Kinetic parameters were determined by nonlinear regression analysis using GraphPad Prism 5 software.
GLYK Activity in Maize Leaves
Maize (Zea mays) plants were grown for 3 weeks on a 5:1 mixture of soil (type VM; Einheitserdewerk, Uetersen) and vermiculite in controlled environment chambers (12-h-light/12-h-dark cycle, 22°C/18°C, 150–200 μE m−2 s−1) and regularly watered with 0.2% (v/v) Wuxal liquid fertilizer (Aglukon). For in vivo light activation, maize leaves were kept in the dark for 4 h and samples were taken before and after illumination as indicated in the legend to Figure 1. The leaf samples (about 0.5 g) were quickly ground in a chilled mortar using 1 mL of ice-cold 100 mm Tricine-KOH, pH 7.8, 100 mm KCl, and 5 mm DTT. Addition of DTT was necessary to prevent immediate autoxidation of ZmGLYK but did not significantly enhance activity over a short period of time at 4°C. Leaf extracts were centrifuged (14,000g, 1 min, 4°C), the supernatants were rapidly desalted by passage through a PD-10 column (GE Healthcare), and GLYK activity was immediately measured in assay 2 with 5 mm DTT. For in vitro thiol activation, the extract was incubated for 30 min with or without 10 mm DTT at room temperature prior to assay 2.
Glycerate and 3PGA Contents of Maize Leafs
Leaf samples (0.2 g) were taken from the fourth and fifth leaves of 5-week-old maize plants after 12 h of darkening. Metabolites were extracted with perchloric acid (Häusler et al., 2000). Glycerate was determined using assay 1 above (omitting glycerate). The reaction was initiated by the addition of 0.5 units of Escherichia coli glycerate kinase 1, which has a very low Km for glycerate (Bartsch et al., 2008). 3PGA was determined using assay 2 above (omitting glycerate) and 0.5 units of 3PGA kinase to start the reaction.
Constructs for Mutagenesis and Overexpression
Coding sequences for ZmGLYK (NP_001141904.1, LOC100274052) and six plastidial Trxs (ZmTrx f1, NP_001150158, LOC100283787; ZmTrx f2, ACF79373, LOC100273021; ZmTrx m1, NP_001150752, LOC100284385; ZmTrx m2, NP_001150752, LOC100284385; ZmTrx m5, NP_001105330, PCO147105; ZmTrx x, ACG33853, LOC100280852) were PCR amplified from leaf cDNA produced by standard protocols from RNA using proofreading Pfu polymerase (MBI Fermentas) in combination with the Taq-PCR Master Mix Kit (Qiagen) and gene-specific primers as listed in Supplemental Table S1. Forward primers were designed to exclude sequences encoding the protein chloroplast-targeting peptides as predicted by ChloroP. PCR products were first cloned into pGEM-T (Promega), excised via the introduced flanking restriction sites, and then ligated into the expression vectors pBAD/His-A (Invitrogen; AtGLYK and OsGLYK), pBAD/His-B (ZmGLYK), and pET-28a (Novagen; ZmTrx f1, f2, m1, m2, m5, and x). Site-specific mutants of ZmGLYK with modifications at the C terminus (for Cys-to-Ser conversions or deletions, see Fig. 5) were generated by PCR using reverse mutagenesis primers as listed in Supplemental Table S1. Mutants of central Cys residues (Cys-96, Cys-97, and Cys-155) were generated by PCR amplification of the pBAD vector containing the ZmGLYK-encoding sequence with Phusion High-Fidelity Polymerase (Finnzymes) and the respective mutagenesis primers. For the PCR-based generation of the fusion constructs of AtGLYK and OsGLYK with the C terminus of ZmGLYK, we used previously cloned cDNA (Bartsch et al., 2008) in combination with a reverse primer encoding the C-terminal amino acids of ZmGLYK. The correctness of all constructs was verified by sequencing.
Overexpression and Purification of Recombinant Proteins
These constructs and two earlier produced overexpression constructs for AtGLYK and OsGLYK (Bartsch et al., 2008) were transformed into E. coli strain TOP10 (Invitrogen). Cells were grown in 100 mL of Luria-Bertani medium containing 50 μg mL−1 ampicillin (pBAD/His) or 50 μg mL−1 kanamycin (pET-28a). After an optical density of 0.6 (600 nm) was reached, gene expression was induced by adding 0.02% (w/v) l-Ara (pBAD/His) or 1 mm isopropylthio-β-galactoside (pET-28a). After further incubation for 16 h at 30°C, cells were harvested by centrifugation (10,000g, 10 min, 4°C). Cells were resuspended in 20 mm sodium phosphate, pH 7.8, 500 mm sodium chloride, and broken by sonication (six 10-s bursts, 90 W, ice cooling). Supernatants obtained after centrifugation (20,000g, 30 min, 4°C) were used for His-tag affinity purification of the recombinant enzymes according to the manufacturer's protocol. GLYK protein concentrations were determined with Roti-Nanoquant (Roth) using bovine serum albumin as a standard (Bradford, 1976). Trx concentrations were estimated by densitometry of SDS-PAGE gels using EcoTrx (Sigma-Aldrich) as a standard.
Redox Titration
The midpoint oxidation-reduction potential of ZmGLYK activity was examined using defined ratios of DTT and DTTox to establish a range of ambient potentials (Eh; Hutchison and Ort, 1995). Eh was calculated using the Nernst equation, Eh = Em + (RT/nF) ln([DTTox]/[DTTred]), with n = 2 for a two-electron reaction, RT/F = 25.693 mV, and an Em, DTT value of −327 mV (Lees and Whitesides, 1993). In brief, samples were incubated in 300 μL of 100 mm Tricine-KOH, pH 7.7, with 10 mm total DTTred/DTTox for 90 min at room temperature and assayed for GLYK activity with assay 1 above. The midpoint redox potential Em was determined by fitting GLYK activity data to the Nernst equation using Sigmaplot software (Systat).
Trx Effects and Redox Sensitivity of Recombinant GLYKs
Details of the time-dependent activation of ZmGLYK by different Trxs are given in the legend to Figure 2. Redox sensitivity of recombinant GLYKs was determined by comparing activities of the enzyme variants in their reduced and oxidized states. For reduction, about 0.9 μm purified GLYK protein was incubated with 10 mm DTT and 0.9 μm EcoTrx (Sigma-Aldrich) in 20 mm sodium phosphate, pH 7.8, for 60 min at room temperature. For complete autoxidation of C-terminal Cys residues, the enzymes were incubated without DTT and Trx for 60 min. GLYK activity was measured with enzyme assay 1 (plus 10 mm DTT for the activity of the reduced enzyme). Activation experiments were repeated at least two times for each GLYK variant, and means ± se were calculated.
MALDI-TOF-MS
Protein bands were excised from Coomassie Brilliant Blue-stained SDS-polyacrylamide gels, washed twice with 30% (v/v) acetonitrile in 25 mm ammonium bicarbonate and with 50% (v/v) acetonitrile in 10 mm ammonium bicarbonate, and dehydrated in acetonitrile. Prior to digestion, triplicate samples were (1) reduced and alkylated with 20 mm tributylphosphine and 40 mm iodoacetamide in 25 mm ammonium bicarbonate, using the ProteoPrep Reduction and Alkylation Kit (Sigma-Aldrich), (2) only alkylated with 40 mm iodoacetamide, and (3) not further treated. Treated samples were washed twice with 25 mm ammonium bicarbonate and once with 50% (v/v) acetonitrile in 10 mm ammonium bicarbonate, dehydrated in acetonitrile, and dried at 37°C. The dried gel pieces were reswollen with 5 μL of protease solution, either 10 ng μL−1 sequencing-grade trypsin (Promega) or 3.6 ng μL−1 endoproteinase AspN (Roche), in 3 mm Tris-HCl, pH 8.5, and incubated for 5 to 8 h at 37°C. Thereafter, 5 μL of 0.3% (w/v) trifluoroacetic acid in 50% (v/v) acetonitrile was added, and the samples were agitated at room temperature for 30 to 60 min. The resulting peptide-containing solution was applied onto a MALDI target plate with α-cyano-4-hydroxycinnamic acid as matrix and analyzed by MALDI-TOF-MS as described (Mikkat et al., 2004). On-target reduction was performed by incubating 0.6 μL of peptide sample with 0.6 μL of DTT solution (20 mm in 54 mm NH4OH) for 5 to 8 min before 0.6 μL of matrix solution was added.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of GLYK protein sequences.
Supplemental Figure S2. Alignment of maize and E. coli Trx sequences.
Supplemental Figure S3. Ribbon model of ZmGLYK.
Supplemental Table S1. Oligonucleotides used for PCR amplification and mutation.
Supplementary Material
Acknowledgments
We thank Udo Gowick and Peter Westhoff (University of Duesseldorf) for providing unpublished GLYK sequences from Flaveria and Stefan Timm (University of Rostock) for the rice cDNA.
References
- Alexandrov N, Brover V, Freidin S, Troukhan M, Tatarinova T, Zhang H, Swaller T, Lu YP, Bouck J, Flavell R, et al. (2009) Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch O, Hagemann M, Bauwe H. (2008) Only plant-type (GLYK) glycerate kinases produce D-glycerate 3-phosphate. FEBS Lett 582: 3025–3028 [DOI] [PubMed] [Google Scholar]
- Boldt R, Edner C, Kolukisaoglu Ü, Hagemann M, Weckwerth W, Wienkoop S, Morgenthal K, Bauwe H. (2005) D-Glycerate 3-kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 17: 2413–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Chibani K, Wingsle G, Jacquot JP, Gelhaye E, Rouhier N. (2009) Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol Plant 2: 308–322 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- de La Sierra-Gallay IL, Collinet B, Graille M, Quevillon-Cheruel S, Liger D, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, et al. (2004) Crystal structure of the YGR205w protein from Saccharomyces cerevisiae: close structural resemblance to E. coli pantothenate kinase. Proteins 54: 776–783 [DOI] [PubMed] [Google Scholar]
- Dever LV, Blackwell RD, Fullwood NJ, Lacuesta M, Leegood RC, Onek LA, Pearson M, Lea PJ. (1995) The isolation and characterization of mutants of the C4 photosynthetic pathway. J Exp Bot 46: 1363–1376 [Google Scholar]
- Drincovich MF, Casati P, Andreo CS, Chessin SJ, Franceschi VR, Edwards GE, Ku MSB. (1998) Evolution of C4 photosynthesis in Flaveria species: isoforms of NADP-malic enzyme. Plant Physiol 117: 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. (2008) The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiotically to plants. Proc Natl Acad Sci USA 105: 17199–17204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Leegood RC. (1984) Carbon metabolism and gas exchange in leaves of Zea mays L. Planta 162: 457–462 [DOI] [PubMed] [Google Scholar]
- Guo JH, Hexige S, Chen L, Zhou GJ, Wang X, Jiang JM, Kong YH, Ji GQ, Wu CQ, Zhao SY, et al. (2006) Isolation and characterization of the human D-glyceric acidemia related glycerate kinase gene GLYCTK1 and its alternatively splicing variant GLYCTK2. DNA Seq 17: 1–7 [DOI] [PubMed] [Google Scholar]
- Häusler RE, Fischer KL, Flügge UI. (2000) Determination of low-abundant metabolites in plant extracts by NAD(P)H fluorescence with a microtiter plate reader. Anal Biochem 281: 1–8 [DOI] [PubMed] [Google Scholar]
- Hutchison RS, Ort DR. (1995) Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxin-activated chloroplast enzymes. Methods Enzymol 252: 220–228 [DOI] [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Lavergne D, Trivelli X, Decottignies P, Miginiac-Maslow M. (2006) Transferring redox regulation properties from sorghum NADP-malate dehydrogenase to Thermus NAD-malate dehydrogenase. Photosynth Res 89: 213–223 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Lopez-jaramillo J, Miginiac-Maslow M, Lemaire S, Cherfils J, Chueca A, LopezGorge J. (1997) Cysteine-153 is required for redox regulation of pea chloroplast fructose-1,6-bisphosphatase. FEBS Lett 401: 143–147 [DOI] [PubMed] [Google Scholar]
- Johansson K, Ramaswamy S, Saarinen M, Lemaire-Chamley M, Issakidis-Bourguet E, Miginiac-Maslow M, Eklund H. (1999) Structural basis for light activation of a chloroplast enzyme: the structure of Sorghum NADP-malate dehydrogenase in its oxidized form. Biochemistry 38: 4319–4326 [DOI] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. (1999) The biochemistry of C4 photosynthesis. Sage RF, Monson RK, , C4 Plant Biology. Academic Press, San Diego, pp 49–87 [Google Scholar]
- Kehrer D, Ahmed H, Brinkmann H, Siebers B. (2007) Glycerate kinase of the hyperthermophilic archaeon Thermoproteus tenax: new insights into the phylogenetic distribution and physiological role of members of the three different glycerate kinase classes. BMC Genomics 8: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD. (1985) Light and thiol activation of maize leaf glycerate kinase: the stimulating effect of reduced thioredoxins and ATP. Plant Physiol 79: 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD. (1986) Thiol-dependent regulation of glycerate metabolism in leaf extracts. Plant Physiol 81: 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, Goyer A, Issakidis-Bourguet E, Miginiac-Maslow M, Lancelin JM. (1999) Direct NMR observation of the thioredoxin-mediated reduction of the chloroplast NADP-malate dehydrogenase provides a structural basis for the relief of autoinhibition. J Biol Chem 274: 34539–34542 [DOI] [PubMed] [Google Scholar]
- Leegood RC. (2000) Transport during C4 photosynthesis. Leegood RC, Sharkey TD, von Caemmerer S, , Advances in Photosynthesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 459–469 [Google Scholar]
- Lees WJ, Whitesides GM. (1993) Equilibrium-constants for thiol-disulfide interchange reactions: a coherent, corrected set. J Org Chem 58: 642–647 [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. (2009) Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Mikkat S, Koy C, Ulbrich M, Ringel B, Glocker MO. (2004) Mass spectrometric protein structure characterization reveals cause of migration differences of haptoglobin α chains in two-dimensional gel electrophoresis. Proteomics 4: 3921–3932 [DOI] [PubMed] [Google Scholar]
- Ohnishi J, Kanai R. (1983) Differentiation of photorespiratory activity between mesophyll and bundle sheath cells of C4 plants. I. Glycine oxidation by mitochondria. Plant Cell Physiol 24: 1411–1420 [Google Scholar]
- Osmond CB, Harris B. (1971) Photorespiration during C4 photosynthesis. Biochim Biophys Acta 234: 270–282 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Portis AR, Li C, Wang D, Salvucci ME. (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Sparla F, Pupillo P, Trost P. (2002) The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J Biol Chem 277: 44946–44952 [DOI] [PubMed] [Google Scholar]
- Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. (2003) A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem 376: 952–965 [DOI] [PubMed] [Google Scholar]
- Usuda H, Edwards GE. (1980a) Localization of glycerate kinase and some enzymes for sucrose synthesis in C3 and C4 plants. Plant Physiol 65: 1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda H, Edwards GE. (1980b) Photosynthetic formation of glycerate in isolated bundle sheath cells and its metabolism in mesophyll cells of the C4 plant Panicum capillare L. Aust J Plant Physiol 7: 655–662 [Google Scholar]
- Woo KC, Downton WJS, Osmond CB, Anderson JM, Boardman NK, Thorne SW. (1970) Deficient photosystem II in agranal bundle sheath chloroplasts of C4 plants. Proc Natl Acad Sci USA 67: 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Osmond CB. (1977) Participation of leaf mitochondria in the photorespiratory carbon oxidation cycle: glycine decarboxylation activity in leaf mitochondria from different species and its intra-mitochondrial location. Plant Cell Physiol 3: 315–323 [Google Scholar]
- Zelitch I, Schultes NP, Peterson RB, Brown P, Brutnell TP. (2009) High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol 149: 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.