Abstract
There exist four members of family GT43 glycosyltransferases in the Arabidopsis (Arabidopsis thaliana) genome, and mutations of two of them, IRX9 and IRX14, have previously been shown to cause a defect in glucuronoxylan (GX) biosynthesis. However, it is currently unknown whether IRX9 and IRX14 perform the same biochemical function and whether the other two GT43 members are also involved in GX biosynthesis. In this report, we performed comprehensive genetic analysis of the functional roles of the four Arabidopsis GT43 members in GX biosynthesis. The I9H (IRX9 homolog) and I14H (IRX14 homolog) genes were shown to be specifically expressed in cells undergoing secondary wall thickening, and their encoded proteins were targeted to the Golgi, where GX is synthesized. Overexpression of I9H but not IRX14 or I14H rescued the GX defects conferred by the irx9 mutation, whereas overexpression of I14H but not IRX9 or I9H complemented the GX defects caused by the irx14 mutation. Double mutant analyses revealed that I9H functioned redundantly with IRX9 and that I14H was redundant with IRX14 in their functions. In addition, double mutations of IRX9 and IRX14 were shown to cause a loss of secondary wall thickening in fibers and a much more severe reduction in GX amount than their single mutants. Together, these results provide genetic evidence demonstrating that all four Arabidopsis GT43 members are involved in GX biosynthesis and suggest that they form two functionally nonredundant groups essential for the normal elongation of GX backbone.
Secondary walls constitute the bulk of cellulosic biomass produced by vascular plants. Cellulosic biomass in the form of fibers and wood is an important raw material for a myriad of industrial uses, such as timber, pulping, papermaking, and textiles. Due to the dwindling of nonrenewable fossil fuels and the detrimental effects of burning fossil fuels on the global environment, there has been an urgent call to develop alternative renewable energy sources, and the lignocellulosic biomass from plants is considered to be an attractive renewable source for biofuel production (Somerville, 2006). However, lignocellulosic biomass is recalcitrant to the enzymatic conversion of cellulose into sugars, because cellulose is embedded in a complex mixture of polysaccharides and lignin polymers that block the accessibility of degrading enzymes. It has been shown that reduction of lignin and xylan by chemical or enzymatic treatment or by the transgenic approach reduces the recalcitrance of the lignocellulosic biomass to saccharification (Chen and Dixon, 2007; Himmel et al., 2007; Lee et al., 2009a). Therefore, a complete understanding of how individual components of lignocellulosic biomass are biosynthesized will potentially allow us to design novel strategies for genetic modification of cell wall composition and, hence, reduction in biomass recalcitrance to biofuel production.
Xylan is the main hemicellulose that cross-links with cellulose in the secondary walls of dicot plants (Carpita and McCann, 2000). It is made of a linear backbone of β-(1,4)-linked xylosyl residues, about 10% of which are attached with side chains of single residues of glucuronic acid (GlcA) and/or 4-O-methylglucuronic acid (MeGlcA) via α-(1,2)-linkages. The backbone xylosyl residues may also be substituted with the arabinosyl group and acetylated. Based on the nature of the side chains, xylan is generally grouped as (methyl)glucuronoxylan (GX), which is the main hemicellulose in dicots, and arabinoxylan and glucuronoarabinoxylan, which are the most abundant hemicelluloses in grass cell walls (Ebringerová and Heinze, 2000). In addition to the xylosyl backbone, the reducing end of xylan from birch (Betula verrucosa), spruce (Picea abies), Arabidopsis (Arabidopsis thaliana), and poplar (Populus alba × Populus tremula) contains a unique tetrasaccharide sequence β-d-Xylp-(1→3)-α-l-Rhap-(1→2)-α-d-GalpA-(1→4)-d-Xylp (Shimizu et al., 1976; Johansson and Samuelson, 1977; Andersson et al., 1983; Peña et al., 2007; Lee et al., 2009a).
The biosynthesis of xylan requires multiple glycosyltransferases and other modifying enzymes. Early biochemical studies revealed the activities of xylosyltransferases, glucuronosyltransferases, arabinosyltransferases, methyltransferases, and acetyltransferases that are likely involved in the biosynthesis of xylan (Baydoun et al., 1983, 1989; Kuroyama and Tsumuraya, 2001; Gregory et al., 2002; Porchia et al., 2002; Urahara et al., 2004; Zeng et al., 2008). However, none of the genes corresponding to these xylan biosynthetic enzymes have been identified. Recent molecular and genetic studies in Arabidopsis and poplar have led to the identification of a number of glycosyltransferases that are essential for GX biosynthesis. Among them, several members of the families GT47 and GT8 from Arabidopsis (FRA8, F8H, IRX8, and PARVUS) and poplar (GT47C, GT8D, and GT8E/8F) are implicated in the biosynthesis of the GX reducing end sequence (Aspeborg et al., 2005; Brown et al., 2005, 2007; Zhong et al., 2005; Zhou et al., 2006, 2007; Lee et al., 2007b, 2009b, 2009c; Peña et al., 2007; Persson et al., 2007). These glycosyltransferase genes are specifically expressed in vessels and fibers, and their encoded proteins are targeted to Golgi, where GX is synthesized, except for PARVUS and GT8E/8F, which are predominantly located in the endoplasmic reticulum (Lee et al., 2007b, 2009c). Mutations of the Arabidopsis FRA8, IRX8, and PARVUS genes all led to a near loss of the reducing end tetrasaccharide sequence and a reduction in GX amount (Brown et al., 2007; Lee et al., 2007b; Peña et al., 2007), indicating their essential roles in the biosynthesis of the GX reducing end sequence, although their exact enzymatic activities are still unknown.
The genetic studies have also identified roles of two members of family GT43 glycosyltransferases, IRX9 and IRX14, from Arabidopsis and GT43B from poplar in the biosynthesis of the GX xylosyl backbone (Brown et al., 2007; Peña et al., 2007; Zhou et al., 2007). The expression of IRX9 has been shown to be associated with cells undergoing secondary wall biosynthesis, and its encoded protein is targeted to the Golgi. Mutation of the IRX9 gene causes a drastic reduction in xylan xylosyltransferase activity (Brown et al., 2007; Lee et al., 2007a) and concomitantly a substantial decrease in the GX chain length and GX amount (Peña et al., 2007). Mutation of IRX14 was shown to result in a reduction in the GX level and the xylosyltransferase activity (Brown et al., 2007). In addition, two functionally redundant glycosyltransferases, IRX10 and IRX10-like, which belong to family GT47, were also demonstrated to be required for the normal GX level and xylan xylosyltransferase activity, suggesting their involvement in the biosynthesis of the GX xylosyl backbone (Brown et al., 2009; Wu et al., 2009).
In this report, we performed comprehensive molecular and genetic studies of the roles of all members of the Arabidopsis family GT43 glycosyltransferases in GX biosynthesis. We show that, like IRX9, the other three GT43 members, I9H (IRX9 homolog), IRX14, and I14H (IRX14 homolog), are expressed in secondary wall-containing cells and that their encoded proteins are targeted to the Golgi. We have found that the GX defects in the irx9 mutant can be rescued by overexpression of I9H but not IRX14 and I14H. Similarly, overexpression of I14H but not IRX9 and I9H is able to complement the GX defects caused by the irx14 mutation. Furthermore, genetic analysis of an array of double mutants revealed redundant and nonredundant roles of GT43 members in GX biosynthesis. Our findings demonstrate that the Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the normal elongation of GX backbone.
RESULTS
Family GT43 Glycosyltransferase Genes Are Expressed in Cells Undergoing Secondary Wall Thickening
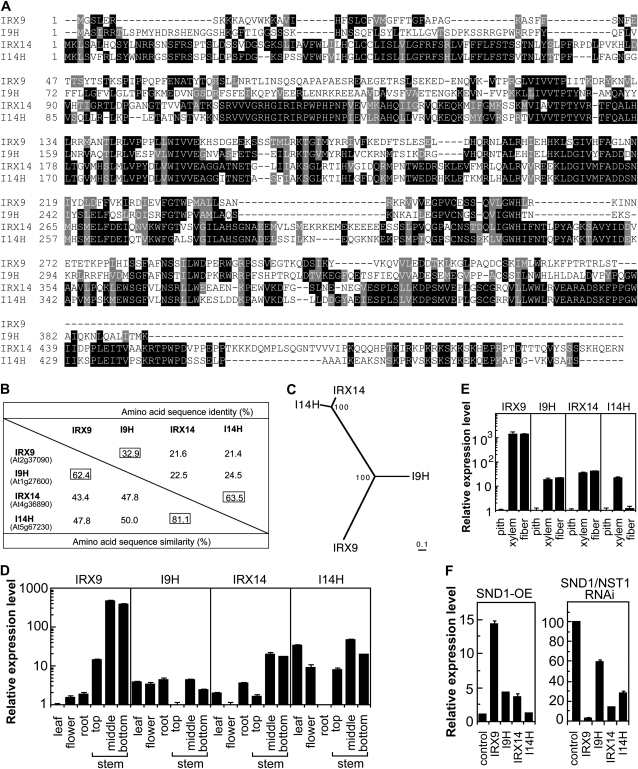
The previous findings that two members of the Arabidopsis family GT43 glycosyltransferases are involved in GX biosynthesis raised the question of whether other GT43 members perform a similar function. There exist four GT43 members, IRX9 (At2g37090), I9H (IRX9 homolog; At1g27600), IRX14 (At4g36890), and I14H (IRX14 homolog; At5g67230), in the Arabidopsis genome. Among them, IRX14 and I14H share the highest sequence similarity (81.1%), and I9H shares a relatively higher sequence similarity with IRX9 than with IRX14 and I14H (Fig. 1, A and B). Phylogenetic analysis further indicates that IRX14 and I14H are more closely related to each other than to IRX9 and I9H, and vice versa (Fig. 1C). IRX9 was previously shown to be specifically expressed in vessels and fibers (Peña et al., 2007), but the expression patterns of other GT43 members are not known. Quantitative PCR analysis showed that I9H and I14H were ubiquitously expressed in most of the organs examined, whereas IRX9 and IRX14 were preferentially expressed in the inflorescence stems (Fig. 1D). Further expression analysis in the isolated cells from stems revealed that IRX9, I9H, IRX14, and I14H were preferentially expressed in xylem and/or interfascicular fibers, the cell types undergoing secondary wall thickening (Fig. 1E). We next tested whether the expression of IRX9, I9H, IRX14, and I14H was regulated by SND1, a master transcriptional switch activating the biosynthetic pathways of xylan, cellulose, and lignin (Zhong et al., 2006, 2007). It was found that their expression was reduced in the SND1/NST1 RNA interference line and that SND1 overexpression induced the expression of IRX9, I9H, and IRX14 (Fig. 1F).
Figure 1.
Sequence comparison and expression analysis of family GT43 members. A, Amino acid sequence alignment of the four Arabidopsis GT43 proteins. The numbers shown at the left of each sequence are the positions of amino acid residues in the corresponding proteins. Identical and similar residues are shaded with black and gray, respectively. B, Percentage of identity and similarity of GT43 proteins. C, Phylogenetic relationship of Arabidopsis GT43 members. The GT43 sequences were aligned using the ClustalW program (Thompson et al., 1994), and the phylogenetic tree was displayed using the TREEVIEW program (Page, 1996). The 0.1 scale denotes 10% change, and the bootstrap values are presented in percentages at the nodes. D, Quantitative PCR analysis of the expression of IRX9, I9H, IRX14, and I14H in different Arabidopsis organs. The lowest expression level among the organs analyzed was set to 1. E, Quantitative PCR analysis of the expression of IRX9, I9H, IRX14, and I14H in pith cells, xylem cells, and interfascicular fibers that were laser microdissected from wild-type Arabidopsis stems. The expression level in pith cells was taken as 1. F, Quantitative PCR analysis of the expression of IRX9, I9H, IRX14, and I14H in the SND1 overexpressor (SND1-OE; left) and the SND1/NST1 RNA interference line (right). The expression of each gene in the wild type was used as a control. Error bars in D to F denote se of three biological replicates.
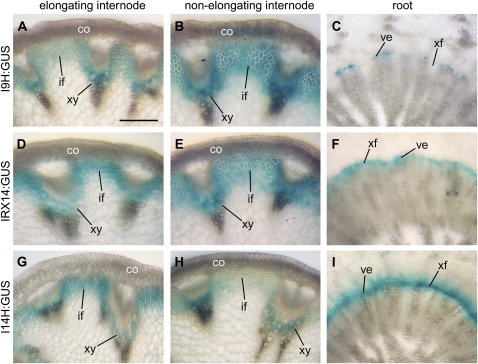
We further examined the expression patterns of I9H, IRX14, and I14H in transgenic Arabidopsis plants using the GUS reporter gene. Consistent with the quantitative PCR analysis, these genes were found to be specifically expressed in interfascicular fibers and xylem cells in the inflorescence stems (Fig. 2, A, B, D, E, G, and H). In the roots, IRX14 and I14H were expressed in both developing vessels and xylary fibers in the secondary xylem (Fig. 2, F and I), whereas I9H expression was only evident in developing vessels but not in xylary fibers (Fig. 2C). These expression studies demonstrate a close association of I9H and I14H with cells undergoing secondary wall thickening, suggesting their possible involvement in secondary wall biosynthesis.
Figure 2.
Expression patterns of I9H, IRX14, and I14H in elongating and nonelongating internodes of inflorescence stems and in roots. Cross sections of stems and roots from transgenic Arabidopsis plants expressing the I9H, IRX14, and I14H genes fused with the GUS reporter gene were stained for GUS activity (shown as blue). co, Cortex; if, interfascicular fiber; ve, vessels; xf, xylary fiber; xy, xylem. Bar in A = 130 mm for A to I.
Family GT43 Glycosyltransferases Are Targeted to the Golgi
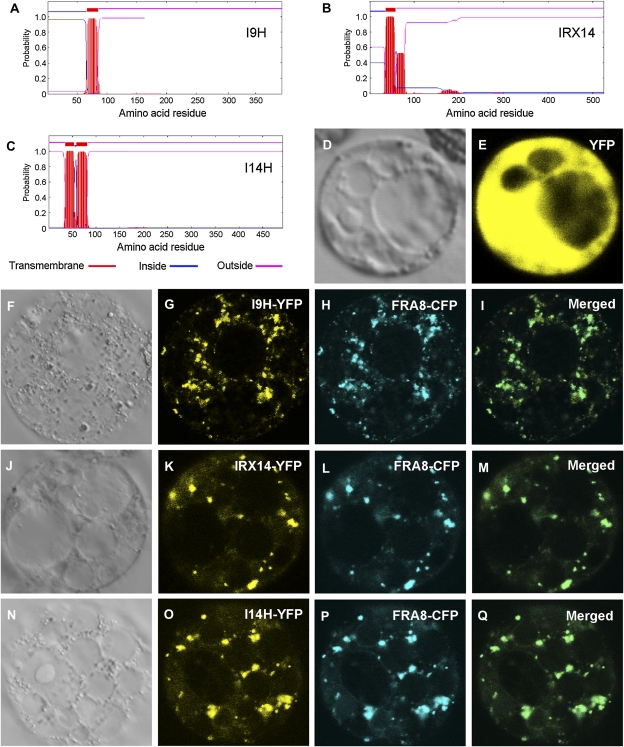
IRX9 was previously demonstrated to reside in the Golgi (Peña et al., 2007), but the subcellular locations of other GT43 members have not yet been examined. Sequence analysis of I9H, IRX14, and I14H using the TMHMM2.0 program for the prediction of transmembrane helices in proteins (http://www.cbs.dtu.dk/service/TMHMM-2.0/) predicts that they are membrane proteins with a single (I9H and IRX14) or double (I14H) transmembrane helices (Fig. 3, A–C). To study their actual subcellular locations, yellow fluorescent protein (YFP)-tagged GT43 members were coexpressed with cyan fluorescent protein (CFP)-tagged FRA8 in carrot (Daucus carota) cells. Examination of the fluorescent signals revealed that I9H, IRX14, and I14H exhibited a punctate distribution, a pattern matched with FRA8 (Fig. 3, F–Q), which is known to be localized in the Golgi (Zhong et al., 2005). The control cells expressing YFP alone showed signals throughout the cytoplasm (Fig. 3, D and E). These results demonstrate that I9H, IRX14, and I14H are Golgi-localized proteins.
Figure 3.
Subcellular localization of I9H, IRX14, and I14H. Fluorescent protein-tagged fusion proteins were expressed in carrot protoplasts, and the signals were visualized with a laser confocal microscope. A to C, I9H, IRX14, and I14H are membrane proteins as predicted by the TMHMM2.0 program. Inside indicates the cytoplasmic side of the membrane, and outside indicates the noncytoplasmic side of the membrane. D and E, A carrot protoplast (D; differential interference contrast image) expressing YFP alone showing the fluorescent signals throughout the cytoplasm (E). F to I, A carrot protoplast (F) coexpressing I9H-YFP (G) and the Golgi-localized FRA8-CFP (H). J to M, A carrot protoplast (J) coexpressing IRX14-YFP (K) and FRA8-CFP (L). N to Q, A carrot protoplast (N) coexpressing I14H-YFP (O) and FRA8-CFP (P). Note the colocalization of I9H-YFP (I), IRX14-YFP (M), and I14H-YFP (Q) with the Golgi marker FRA8-CFP.
Overexpression of I9H But Not IRX14 and I14H Complements the irx9 Mutant Defects
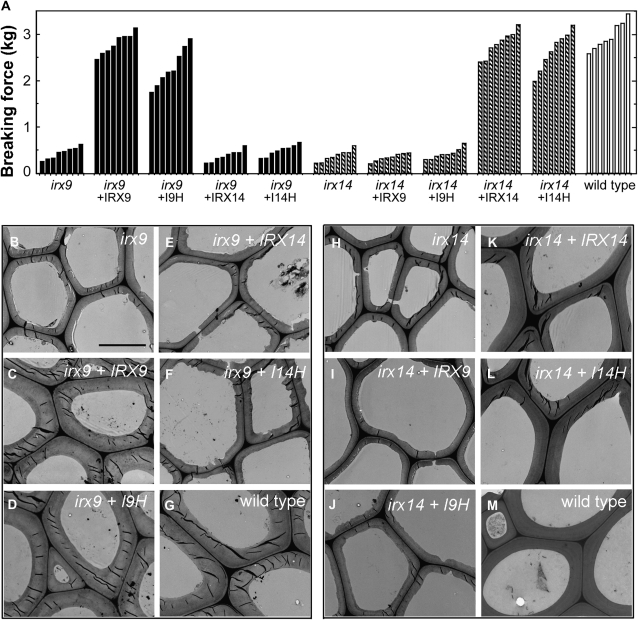
Previous genetic studies showed that mutation of either IRX9 or IRX14 caused a partial reduction in the xylosyltransferase activity and GX content (Brown et al., 2007; Lee et al., 2007a; Peña et al., 2007). Because all the GT43 members are expressed in cells undergoing secondary wall thickening, we reasoned that they might be functionally redundant and that the partial defects in GX biosynthesis in irx9 or irx14 could be due to the partial compensation by other GT43 members. To test this possibility, we investigated whether the defects caused by either irx9 or irx14 could be rescued by overexpression of GT43 members driven by the cauliflower mosaic virus (CaMV) 35S promoter. One of the prominent phenotypes of irx9 was the weakened stem strength due to the reduced secondary wall thickness (Fig. 4, A and B). Overexpression of I9H and IRX9 in the irx9 mutant effectively restored the stem breaking strength and the secondary wall thickness of fibers to the wild-type levels (Fig. 4, A, C, D, and G), indicating that I9H is a functional homolog of IRX9. In addition, overexpression of I9H and IRX9 rescued the collapsed vessel phenotype caused by the irx9 mutation (Supplemental Fig. S2, A–C). It was noted that not all I9H-complemented irx9 plants exhibited the same breaking strength as that of the wild type, which is likely due to the variation of the degree of complementation among different individuals. In contrast, overexpression of IRX14 and I14H did not complement the defects in stem strength, secondary wall thickness, and vessel morphology conferred by irx9 (Fig. 4, A, E, and F; Supplemental Fig. S2, D and E).
Figure 4.
Complementation of the irx9 and irx14 mutants by overexpression of GT43 members. The full-length cDNAs of the four Arabidopsis GT43 members driven by the CaMV 35S promoter were introduced into irx9 or irx14, and the bottom parts of inflorescence stems of 10-week-old transgenic plants were examined for the stem breaking strength (A) and the secondary wall thickness in interfascicular fibers (B–M). Bar in B = 5.4 mm for B to M. A, Measurement of the breaking strength of stems of the wild type, irx9, irx14, and the mutants overexpressing GT43 members. Each bar represents the breaking force of the inflorescence stem of individual plants. It was evident that the stem breaking strength of irx9 was restored by overexpression of IRX9 or I9H and that of irx14 was restored by overexpression of IRX14 or I14H. B to M, Transmission electron microscopy showing the restoration of secondary wall thickness of the interfascicular fibers in the stems of the irx9 mutant by overexpression of IRX9 and I9H and that of the irx14 mutant by overexpression of IRX14 and I14H.
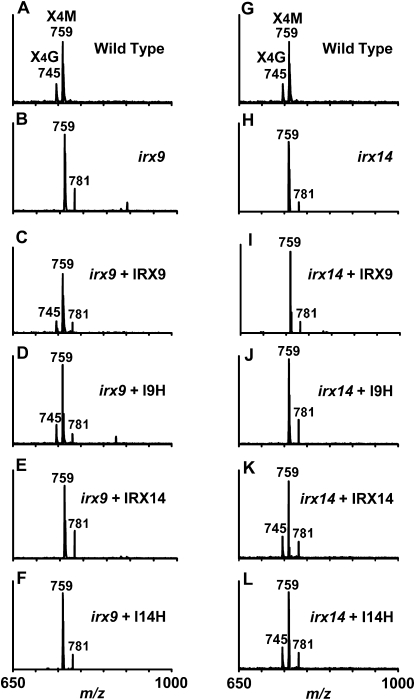
The reduced secondary wall thickness in irx9 was previously found to be caused by GX defects, including a decrease in GX level, a loss of nonmethylated GlcA, and a reduced GX chain length (Peña et al., 2007). Analysis of the GX content revealed that I9H overexpression in the irx9 mutant restored the cell wall Xyl level to that of the wild type (Table I), indicating that excess I9H could complement the loss of IRX9 to restore the GX level. It should be noted that similar to fra8, irx9 and irx14 cause not only a reduction in Xyl but also a reduction in Glc, which might be attributed to an indirect impediment of cellulose synthesis caused by a reduction in GX (Zhong et al., 2005). The observed increase in the levels of some other cell wall sugars might be simply due to the relative decrease in the levels of cellulose and GX or an increased synthesis of other wall polysaccharides in response to the reduced level of cellulose, a phenotype commonly observed in cellulose-deficient mutants (Fagard et al., 2000; His et al., 2001). Overexpression of GT43 members in irx9 also resulted in a change in the levels of other sugars in addition to Xyl, which might reflect the complexity of cell wall metabolism. The structure of GX in the complemented plants was examined by analyzing the acidic xylooligosaccharides using matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Acidic xylooligosaccharides from the wild type contain two prominent ion peaks [M+Na]+ at mass-to-charge ratios (m/z) of 745 and 759, which correspond to xylotetrasaccharides substituted with one GlcA and one MeGlcA, respectively (Fig. 5A). The loss of GlcA side chains caused by the irx9 mutation (Fig. 5B) was rescued by overexpression of I9H and IRX9 (Fig. 5, C and D) but not by IRX14 and I14H (Fig. 5, E and F).
Table I. Monosaccharide composition of cell walls from inflorescence stems of the wild type, irx9, irx14, and the irx9 and irx14 plants overexpressing GT43 members.
The wall residues for cell wall composition analysis were prepared from stems. The data are means (mg g−1 dry cell wall) ± se of duplicate assays.
| Sample | Xyl | Glc | Man | Gal | Ara | Rha |
| Wild type | 108.7 ± 4.6 | 412.1 ± 4.7 | 18.9 ± 1.3 | 17.5 ± 1.3 | 12.6 ± 2.2 | 11.1 ± 1.2 |
| irx9 | 60.8 ± 0.3 | 337.9 ± 0.4 | 39.0 ± 0.1 | 38.4 ± 1.3 | 28.6 ± 0.2 | 16.4 ± 0.6 |
| irx9 + IRX9 | 91.6 ± 3.1 | 370.4 ± 3.0 | 19.1 ± 3.2 | 16.7 ± 0.8 | 16.7 ± 0.9 | 14.9 ± 0.9 |
| irx9 + I9H | 107.4 ± 2.8 | 446 ± 3.8 | 24.1 ± 0.9 | 18.2 ± 0.8 | 18.2 ± 0.6 | 12.7 ± 0.6 |
| irx9 + IRX14 | 60.2 ± 5.9 | 366.9 ± 35.0 | 44.2 ± 5.0 | 25.8 ± 3.4 | 28.8 ± 3.2 | 19.3 ± 2.9 |
| irx9 + I14H | 52.7 ± 11.6 | 393.7 ± 42.2 | 43.1 ± 5.6 | 24.9 ± 3.6 | 23.9 ± 2.2 | 16.4 ± 1.2 |
| irx14 | 49.4 ± 5.0 | 334 ± 8.4 | 23.6 ± 0.7 | 32.9 ± 2.9 | 12.0 ± 1.5 | 12.2 ± 0.9 |
| irx14 + IRX9 | 45.9 ± 3.9 | 338.5 ± 11.7 | 23.6 ± 2.2 | 28.6 ± 0.8 | 13.7 ± 1.3 | 17.1 ± 0.8 |
| irx14 + I9H | 56.0 ± 2.0 | 372.1 ± 9.6 | 31.5 ± 2.6 | 29.1 ± 0.2 | 14.6 ± 0.1 | 18.2 ± 0.9 |
| irx14 + IRX14 | 98.5 ± 4.3 | 385.4 ± 3.8 | 27.7 ± 1.3 | 24.9 ± 1.4 | 13.9 ± 0.2 | 12.7 ± 0.8 |
| irx14 + I14H | 96.9 ± 1.2 | 397.5 ± 6.0 | 18.0 ± 1.3 | 22.7 ± 3.3 | 11.5 ± 0.1 | 15.2 ± 0.2 |
Figure 5.
MALDI-TOF mass spectra of acidic xylooligosaccharides generated by β-endoxylanase digestion of GX from the stems of the wild type, irx9, irx14, and the mutants overexpressing GT43 members. The ions at m/z 745 and 759 correspond to xylotetrasaccharides bearing a GlcA residue (X4G) or a methylated GlcA residue (X4M). The ion at m/z 781 corresponds to the doubly sodiated X4M. Note that the missing ion at m/z 745 corresponding to X4G in irx9 was restored by overexpression of IRX9 or I9H and that in irx14 was restored by overexpression of IRX14 or I14H.
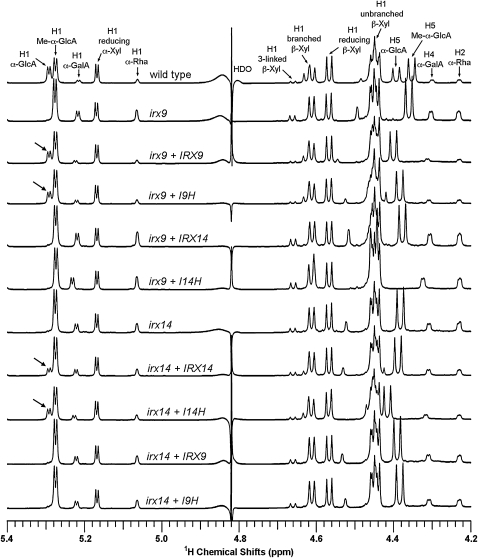
The structure of GX from the complemented plants was further analyzed using NMR spectroscopy. The resonances that are assigned to the GX reducing end tetrasaccharide sequence, β-d-Xyl-(1→3)-α-l-Rha-(1→2)-α-d-GalA-(1→4)-d-Xyl, include H1 of α-d-GalA, H1 of α-l-Rha, H1 of 3-linked β-d-Xyl, H4 of α-d-GalA, and H2 of α-l-Rha (Peña et al., 2007). It was previously found that the resonances for the GX reducing end tetrasaccharide sequence in irx9 were highly elevated compared with those of the wild type, which is caused by a significant reduction in the GX chain length and thereby a relative increase in the ratio of the GX reducing end sequence over the backbone xylosyl residues (Peña et al., 2007). It was evident that overexpression of I9H but not IRX14 and I14H in irx9 restored the resonance intensity for the GX reducing end tetrasaccharide sequence to the wild-type level (Fig. 6; Table III), indicating that excess I9H is sufficient to complement the GX chain length defect conferred by irx9. Consistent with the MALDI-TOF MS results showing that I9H overexpression restores the α-d-GlcA side chains in GX (Fig. 5), the resonances assigned to H1 of α-d-GlcA residues, which were absent in irx9, were restored to the wild-type level in the I9H-complemented irx9 plants (Fig. 6).
Figure 6.
1H NMR spectra of acidic xylooligosaccharides generated by β-endoxylanase digestion of GX from the stems of the wild type, irx9, irx14, and the mutants overexpressing GT43 members. Resonances are labeled with the position of the assigned proton and the identity of the residue containing that proton. Note the restoration of the resonance of H1 of α-GlcA in irx9 complemented with IRX9 and I9H and in irx14 complemented with IRX14 and I14H (arrows). The resonances of H1 of α-d-GalA, H1 of α-l-Rha, H1 of 3-linked β-d-Xyl, H4 of α-d-GalA, and H2 of α-l-Rha are from the GX reducing end tetrasaccharide sequence. Note the elevated relative resonance intensity of the GX reducing end tetrasaccharide sequence in irx9 and irx14 and the reduction in its resonance intensity in irx9 complemented with IRX9 and I9H and in irx14 complemented with IRX14 and I14H. The doublet resonance peak at 4.39 ppm in the wild type is assigned to H5 α-GlcA and that at 4.35 ppm is assigned to H5 Me-α-GlcA. The resonance shift of H5 proton in other samples is likely attributed to its high sensitivity to a slight variation of pH and concentration of the samples because of its close proximity to the carboxyl group. A similar cause may lead to a slight resonance shift of H1 and H4 α-GalA. HDO, Hydrogen deuterium oxide.
Table III. Relative abundance of the reducing end tetrasaccharide sequence and side chains, and the DP of GX from the wild type and GT43 mutants overexpressing various GT43 members.
| Sample | Frequency of Occurrence of GlcA and MeGlcA Side Chainsa | Relative Abundance of the Reducing End Tetrasaccharide Sequenceb | Average DPc |
| Wild type | 21.6% | 100% | 93 |
| irx9 | 21.3% | 31.9% | 29 |
| irx9 + IRX9 | 20.3% | 88.1% | 84 |
| irx9 + I9H | 19.0% | 79.2% | 72 |
| irx9 + IRX14 | 20.3% | 23.1% | 23 |
| irx9 + I14H | 20.8% | 34.0% | 30 |
| irx14 | 22.0% | 37.6% | 36 |
| irx14 + IRX9 | 23.4% | 40.9% | 37 |
| irx14 + I9H | 23.3% | 40.9% | 38 |
| irx14 + IRX14 | 21.5% | 70.3% | 68 |
| irx14 + I14H | 20.4% | 82.3% | 77 |
The frequency of occurrence of the GlcA and MeGlcA side chains was calculated from the ratios of the NMR resonance of the branched β-Xyl to that of total β-Xyl.
The relative abundance of the reducing end tetrasaccharide sequence was determined from the ratios of the NMR resonance of the reducing end sequence to that of the branched β-Xyl. The abundance of the reducing end tetrasaccharide sequence in the wild type was taken as 100%.
The average DP was calculated according to Peña et al. (2007).
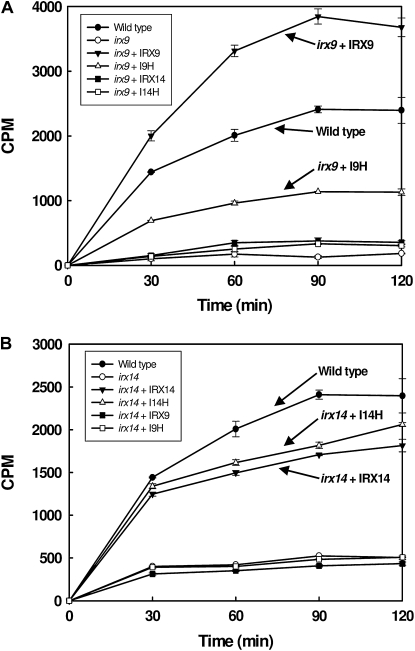
The restoration of GX level and structure in I9H-complemented irx9 plants was accompanied by a partial rescue of the GX xylosyltransferase activity (Fig. 7A). Overexpression of IRX14 and I14H did not restore the xylosyltransferase activity in the irx9 mutant. It was interesting that overexpression of IRX9 in irx9 led to an increased xylosyltransferase activity compared with the wild type. Together, these results suggest that I9H can perform the same biochemical function as IRX9 in the biosynthesis of GX backbone and that they are functionally distinct from IRX14 and I14H.
Figure 7.
Time course of the xylosyltransferase activity in the wild type, irx9, irx14, and the mutants overexpressing GT43 members. A, Overexpression of I9H or IRX9 but not IRX14 or I14H in irx9 rescued the deficiency in the xylosyltransferase activity caused by the irx9 mutation. Note the increased xylosyltransferase activity in irx9 overexpressing IRX9. B, Overexpression of I14H or IRX14 but not IRX9 or I9H in irx14 rescued the deficiency in the xylosyltransferase activity caused by the irx14 mutation. CPM, Counts per minute as a measurement of radioactivity.
Overexpression of I14H But Not IRX9 and I9H Complements the irx14 Mutant Defects
Our finding that overexpression of IRX14 and I14H does not complement the irx9 mutant defects prompted us to test whether IRX14 and I14H form another pair of functional homologs involved in GX backbone biosynthesis. It was found that overexpression of I14H and IRX14 but not IRX9 and I9H in the irx14 mutant rescued the defects in the stem strength, secondary wall thickness, vessel morphology, the GX level, the GlcA side chains, and the GX xylosyltransferase activity (Figs. 4, A and H–M, 5, G–L, and 7; Table I; Supplemental Fig. S2, G–L). The irx14 mutation was previously shown to cause a reduction in the GX level (Brown et al., 2007), but it is unknown whether the GX chain length is affected in the mutant. NMR analysis showed that similar to those in irx9, the resonances for the GX tetrasaccharide reducing end sequence in irx14 were highly increased compared with those in the wild type (Fig. 6), indicating that the GX chain length in irx14 is significantly reduced (Table III). Overexpression of I14H in irx14 apparently restored the resonance signals for the GX tetrasaccharide reducing end sequence to the wild-type level, indicating its ability to complement the GX chain length defect conferred by irx14 (Fig. 6; Table III). These results demonstrate that I14H and IRX14 form another group of functional homologs involved in GX biosynthesis.
Double Mutant Analysis Reveals Redundant Roles of GT43 Members in GX Biosynthesis
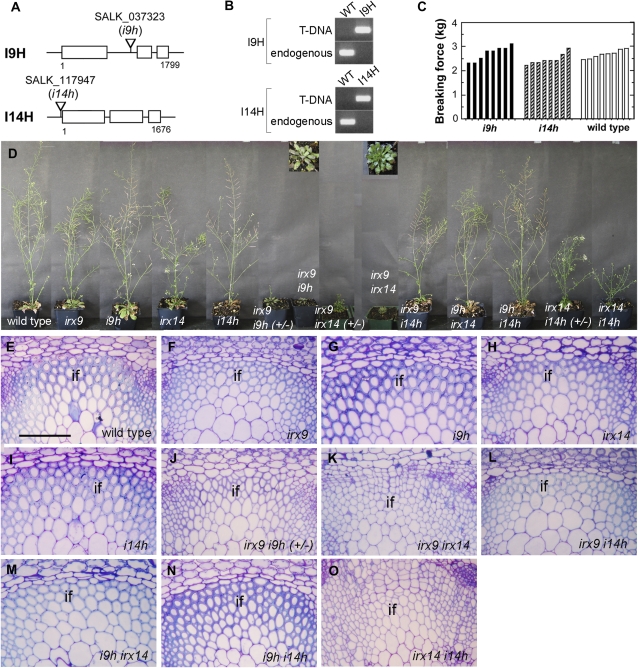
The findings that I9H is expressed in the same cell types as IRX9 and excess I9H is able to complement the irx9 mutant defects indicate that they function redundantly in GX biosynthesis. Likewise, I14H might function redundantly with IRX14 in GX biosynthesis. To test these hypotheses, we generated an array of double mutants with all possible combinations of the GT43 members (Fig. 8; Supplemental Fig. S1) and examined their effects on plant growth and GX biosynthesis. Although the irx9 or irx14 mutant exhibited reductions in stem strength, secondary wall thickness, and GX level (Brown et al., 2007; Peña et al., 2007), T-DNA insertion mutation of I9H or I14H alone did not have any discernible effects (Figs. 8 and 9; Table II; Supplemental Fig. S3). However, homozygous double mutations of IRX9 and I9H led to a strong retardation in plant growth (Fig. 8D). Even after prolonged growth (4 months), no inflorescence stems grew out from the rosettes, which prevented us from further analysis of the double homozygous mutants. The double mutant that is homozygous for irx9 and heterozygous for i9h (+/−) did grow out short inflorescence stems after prolonged growth. Examination of the irx9 i9h (+/−) inflorescence stems revealed that the interfascicular fibers had thinner walls (Figs. 8J and 9F) than did irx9 (Figs. 8F and 9B) and that the xylem vessels were severely collapsed (Supplemental Fig. S3F). In addition, cell wall composition analysis showed that the Xyl content in irx9 i9h (+/−) was more severely reduced than that in irx9 (Table II). These results demonstrate that IRX9 and I9H play redundant roles in GX biosynthesis and secondary wall thickening.
Figure 8.
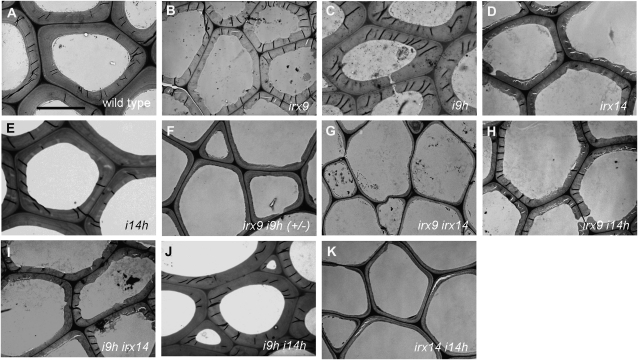
Effects of single and double mutations of GT43 members on plant growth and fiber wall thickening. A, Diagrams showing the sites of T-DNA insertions in the I9H and I14H genes. B, PCR identification of homozygous i9h and i14h mutants. T-DNA indicates the amplified DNA fragment with a T-DNA left border primer and an I9H or I14H primer flanking the T-DNA insertion site. Endogenous indicates the amplified I9H or I14H DNA fragment with primers spanning the T-DNA insertion site. WT, Wild type. C, Breaking strength measurement showing that the stem strength of i9h and i14h is similar to that of the wild type. Each bar represents the breaking force of the inflorescence stem of individual plants. D, Morphology of 10-week-old single and double GT43 mutants. Insets show the enlarged images of the rosette plants of irx9 i9h and irx9 irx14. E to O, Cross sections of stems showing the interfascicular fibers in single and double GT43 mutants. Note the extremely thin fiber walls in irx9 irx14 and irx14 i14h. The bottom parts of inflorescence stems of 10-week-old plants except for irx9 i9h (+/−) and irx9 irx14, which were from 4- and 6-month-old plants, respectively, were sectioned and stained with toluidine blue for anatomy. if, Interfascicular fiber. Bar in E = 93 mm for E to O.
Figure 9.
Transmission electron micrographs of the cell walls of interfascicular fibers in the stems of the wild type and GT43 mutants. The bottom parts of the inflorescence stems of 10-week-old plants except for irx9 i9h (+/−) and irx9 irx14, which were from 4- and 6-month-old plants, respectively, were examined for fiber wall thickness. Bar in A = 7.2 mm for A to K.
Table II. Monosaccharide composition of cell walls from inflorescence stems of the wild type and GT43 mutants.
The wall residues for cell wall composition analysis were prepared from stems. The data are means (mg g−1 dry cell wall) ± se of duplicate assays.
| Sample | Xyl | Glc | Man | Gal | Ara | Rha |
| Wild type | 110.4 ± 1.4 | 402.0 ± 0.2 | 18.0 ± 0.4 | 14.6 ± 0.2 | 11.0 ± 0.2 | 11.7 ± 0.4 |
| irx9 | 57.2 ± 0.3 | 336.9 ± 0.4 | 35.1 ± 0.1 | 39.9 ± 1.3 | 23.8 ± 0.2 | 13.9 ± 0.6 |
| i9H | 106.6 ± 3.8 | 418.1 ± 4.5 | 17.5 ± 0.7 | 13.9 ± 0.6 | 9.0 ± 0.1 | 10.4 ± 0.1 |
| irx14 | 59.5 ± 2.1 | 346.5 ± 7.5 | 24.6 ± 2.5 | 16.2 ± 1.3 | 11.2 ± 0.1 | 10.9 ± 0.5 |
| i14h | 102.3 ± 4.8 | 391.8 ± 7.6 | 17.9 ± 2.3 | 15.1 ± 1.1 | 10.3 ± 0.5 | 10.6 ± 0.5 |
| irx9 i9h (+/−) | 40.2 ± 1.1 | 329.7 ± 8.4 | 40.4 ± 2.1 | 43.4 ± 1.4 | 35.8 ± 1.1 | 19.4 ± 2.0 |
| irx9 irx14 | 26.5 ± 0.5 | 292.4 ± 5.3 | 20.2 ± 4.8 | 49.3 ± 3.4 | 26.0 ± 1.9 | 12.1 ± 0.2 |
| irx9 i14h | 49.2 ± 1.7 | 337.9 ± 13.4 | 30.6 ± 1.5 | 23.1 ± 1.8 | 15.2 ± 1.3 | 13.7 ± 0.9 |
| i9h irx14 | 59.3 ± 3.9 | 358.0 ± 15.7 | 25.7 ± 2.9 | 15.6 ± 1.7 | 11.2 ± 0.5 | 11.9 ± 0.3 |
| i9h i14h | 94.2 ± 21.9 | 366.7 ± 24.3 | 12.6 ± 0.4 | 12.3 ± 1.0 | 8.9 ± 0.4 | 9.5 ± 0.5 |
| irx14 i14h | 29.1 ± 1.8 | 251.7 ± 11.1 | 24.3 ± 1.8 | 26.9 ± 2.2 | 19.3 ± 1.1 | 13.9 ± 0.6 |
Similar to irx9 i9h, double homozygous mutations of IRX14 and I14H cause a much stronger effect on plant growth compared with irx14 (Fig. 8D). The irx14 i14h grew out short inflorescence stems, allowing us to examine the secondary wall thickening and the GX level. The irx14 i14h stems had little secondary wall thickening in the interfascicular fibers (Figs. 8O and 9K), a severely collapsed vessel morphology (Supplemental Fig. S3K), and a much lower Xyl content (Table II) than those of irx14 (Figs. 8H and 9D). The double mutant that is homozygous for irx14 and heterozygous for i14h (+/−) also had a more severe retardation in plant growth compared with irx14 (Fig. 8D).
We next examined the plant growth and secondary wall thickening in the double mutants irx9 i14h, i9h irx14, and i9h i14h. The plant growth (Fig. 8D), secondary wall thickness (Figs. 8 and 9), and vessel morphology (Supplemental Fig. S3) of the irx9 i14h and the i9h irx14 double mutants were similar to those of irx9 and irx14, respectively. Cell wall composition analysis revealed that the Xyl content in irx9 i14h had a slight reduction compared with irx9, whereas the Xyl content in i9h irx14 was similar to that in irx14 (Table II). Although no significant alterations in plant growth, secondary wall thickness, and vessel morphology were seen in the i9h i14h double mutant (Figs. 8, D and N, and 9J; Supplemental Fig. S3J) compared with the wild type (Figs. 8E and 9A; Supplemental Fig. S3A), the Xyl content in i9h i14h was slightly reduced (Table II).
We further investigated the effects of double mutations of IRX9 and IRX14 on plant growth and GX biosynthesis. Both irx9 irx14 and irx9 irx14 (+/−) exhibited strong retardation in plant growth (Fig. 8D). The irx9 irx14 plants grew out small inflorescence stems after 6 months of growth, and examination of these stems revealed the absence of discernible secondary walls in the interfascicular fibers (Figs. 8K and 9G) and collapsed vessels in the xylem (Supplemental Fig. S3G). These phenotypes were much more severe than those in the irx9 (Fig. 8F; Supplemental Fig. S3B) or irx14 (Fig. 8H; Supplemental Fig. S3D) single mutant. Further cell wall composition analysis of irx9 irx14 stems revealed a drastic reduction in the Xyl content (Table II), indicating a more severe effect on GX biosynthesis than the irx9 or irx14 mutation alone.
The results from these comprehensive genetic analyses demonstrate that all four GT43 members are involved in GX biosynthesis, although IRX9 and IRX14 play major roles. Together with the mutant complementation studies, our findings suggest that the family GT43 glycosyltransferases form two functionally nonredundant groups, IRX9 together with I9H and IRX14 together with I14H, both of which are essential for GX biosynthesis.
DISCUSSION
IRX9 and IRX14 Function Nonredundantly in the Elongation of GX Backbone
Our findings that IRX9 cannot functionally complement the loss of IRX14 and vice versa suggest that IRX9 and IRX14 are two functionally nonredundant GT43 members essential for GX biosynthesis. Considering the fact that both IRX9 and IRX14 are required for the elongation of GX backbone (Peña et al., 2007; Fig. 6), it is reasonable to propose that IRX9 and IRX14 function cooperatively in the elongation of GX backbone. Such a cooperative function might have important implications in understanding the biochemical mechanism of GX backbone biosynthesis. The GX backbone is composed of β-(1,4)-linked xylosyl residues, and its biosynthesis needs to overcome a steric problem because each xylosyl residue is flipped nearly 180° with respect to its neighboring residues. Similar to a model proposed for cellulose biosynthesis, in which two glycosyltransferase activities operate cooperatively from opposite sites to add cellobiosyl units to the β-(1,4)-glucan chain (Carpita and Vergara, 1998), the biosynthesis of GX backbone might also require two enzyme activities that operate in such a mechanism. Although their exact biochemical functions await further investigation, IRX9 and IRX14 are strong candidates for such two glycosyltransferase activities involved in the elongation of the β-(1,4)-xylan chain. This hypothesis is consistent with the facts that mutation of either irx9 or irx14 results in a defect in the GX chain length and a reduction in the xylosyltransferase activity and that IRX9 and IRX14 are nonredundant in their functions.
The chemical analysis of GX showed that the irx9 and irx14 mutants exhibited a similar severity of defects in the amount and the degree of polymerization (DP) of GX (Tables II and III), which is congruent with their proposed cooperative functions in the elongation of GX backbone. However, the irx9 mutant appears to have a more severe retardation in plant growth than the irx14 mutant. It has been demonstrated that a deformation in vessels in cell wall mutants could lead to a strong retardation in plant growth (Lee et al., 2009b; McCarthy et al., 2009), which is likely due to defects in solute transport. It is possible that the GX defect in the vessels in irx9 is more severe than that in irx14, thereby causing a stronger effect on plant growth. However, chemical analysis did not reveal a significant difference in the amount and the DP of GX between irx9 and irx14. This could be due to the fact that the total cell walls used for analysis are from stems in which fibers are the most abundant secondary wall-containing cells; therefore, any possible difference of GX defects in the vessels between irx9 and irx14 could not be detected.
Redundancy of Family GT43 Genes in GX Biosynthesis
We have demonstrated that I9H and I14H are redundant in function with IRX9 and IRX14, respectively, based on the following lines of evidence. First, all four GT43 members are expressed in the same cell types that undergo secondary wall thickening (Fig. 2), and their proteins are targeted into the Golgi (Fig. 3), where GX is synthesized. Second, overexpression of I9H and I14H can effectively rescue the GX defects, including the GX chain length, caused by the mutations of IRX9 and IRX14, respectively (Figs. 4–6; Tables I and III). Third, double mutation of IRX9 and I9H results in a more severe defect in the GX level and a strong retardation of plant growth, and likewise, double mutation of IRX14 and I14H leads to similar phenotypes (Fig. 8; Table II). Together, these results suggest that IRX9 and I9H are a pair of functional homologs and IRX14 and I14H are another pair of functional homologs. This finding explains the early observations that the irx9 or irx14 mutant only exhibits partial reductions in GX content and secondary wall thickness (Brown et al., 2005, 2007; Peña et al., 2007). However, the i9h or i14h mutation alone does not cause any observable GX defects, indicating that IRX9 and IRX14 are functionally dominant in GX biosynthesis in vivo. The functional dominance of IRX9 and IRX14 could be caused by their higher expression levels or their higher specific enzymatic activities. It should be pointed out that although all the GT43 members are expressed in cells undergoing secondary wall biosynthesis, there are some differences in their expression levels in different organs and secondary wall-forming cell types (Figs. 1 and 2). For example, I9H is specifically expressed in vessels in the secondary xylem of roots (Fig. 2C), and I14H has a relatively higher expression level in the xylem than in the interfascicular fibers in nonelongating stems (Fig. 2H). The differences in their expression patterns might account for the differential involvement of GT43 members in GX biosynthesis in different secondary wall-forming cell types.
The gene redundancy appears to be common in GX biosynthesis in Arabidopsis. For example, FRA8 and F8H are a pair of redundant genes required for GX reducing end sequence biosynthesis (Lee et al., 2009b), and IRX10 and IRX10-L are another pair involved in GX backbone biosynthesis (Brown et al., 2009; Wu et al., 2009). In both of these cases, single mutation of FRA8 or IRX10 results in a defect in GX biosynthesis but single mutation of its homolog does not. Double mutations of these genes, either FRA8 and F8H or IRX10 and IRX10-L, cause a strong retardation in plant growth, a phenotype that is also observed in the double mutants irx9 i9h and irx14 i14h. Since GX is a critical component required for cross-linking cellulose microfibrils in secondary walls, gene redundancy in GX biosynthesis likely presents an evolutionary advantage to safeguard the proper making of secondary walls, which are the key structure of vessels essential for water transport in the plant body.
It was noticed that the DP of GX in irx9 or irx14 was not completely rescued by overexpression of itself or its corresponding functional homolog (Table III). This could be attributed to the fact that the cell walls used for GX analysis were isolated from a pool of individual transgenic plants that likely had different degrees of complementation in the xylan backbone length. Therefore, the average DP of GX in the rescued mutants appears slightly shorter than that in the wild type. However, cell wall composition analysis showed a nearly complete restoration of the total Xyl content (Table I), secondary wall thickness (Fig. 4), and vessel morphology (Supplemental Fig. S2) in the rescued mutants compared with the wild type. This finding indicates that a substantial restoration of the DP of GX is sufficient to rescue the defects in GX level, leading to the restoration of secondary wall thickness and vessel morphology.
Genes Involved in GX Backbone Biosynthesis
It was previously proposed that some of the cellulose synthase-like (CSL) genes might encode xylan synthase (Richmond and Somerville, 2001), which is congruent with the findings that several hemicellulose backbones, such as xyloglucan β-1,4-glucan (Cocuron et al., 2007), β-1,4-mannan (Dhugga et al., 2004; Liepman et al., 2005), and β-(1,3;1,4)-mix-linked glucan (Burton et al., 2006), are synthesized by members of the CSL gene families. However, no recombinant CSL proteins were shown to exhibit any xylan synthase activity (Liepman et al., 2005). Transcription profiling analysis of wood formation in poplar revealed no correlations between secondary wall synthesis and the expression of CSL genes with the exception of the CSLA (a mannan synthase) gene (Aspeborg et al., 2005; Suzuki et al., 2006). Although mutation of CSLD5 has been reported to cause a decrease in xylosyltransferase activity and xylan immunolabeling, the CSLD5 gene is not expressed in cells undergoing secondary wall thickening; thus, the observed phenotypes were considered to be indirect (Bernal et al., 2007). Therefore, genes responsible for the biosynthesis of GX backbone remain elusive. Because GT43 members are specifically expressed in cells undergoing secondary wall thickening and GX biosynthesis (Peña et al., 2007; Fig. 2) and IRX9 and IRX14 are required for the normal GX xylosyltransferase activity and the normal elongation of GX chains (Brown et al., 2007; Lee et al., 2007a; Peña et al., 2007; Fig. 6), GT43 members are apparently strong candidates for GX xylosyltransferases or part of the complexes required for the xylosyltransferase activities. However, an attempt to assay the xylosyltransferase activity of recombinant IRX9 proteins was not successful (Peña et al., 2007). Our finding that IRX9 and IRX14 are nonredundant in function suggests that expression of both IRX9 and IRX14 would be required for the GX xylosyltransferase activity. Furthermore, the recent finding that IRX10 and IRX10-L are also required for the normal GX xylosyltransferase activity (Brown et al., 2009; Wu et al., 2009) indicates that the biosynthesis of GX backbone is much more complicated than expected. As suggested previously, a complex composed of multiple glycosyltransferases and other proteins might be involved in GX backbone synthesis (Peña et al., 2007). Further biochemical characterization of GT43 members together with other players will be necessary to unravel the biochemical process of GX biosynthesis.
MATERIALS AND METHODS
Plant Growth Conditions
Plants were grown in a greenhouse under 14-h-light/10-h-dark cycles. Common garden potting soil was used for growing plants with biweekly application of plant fertilizers.
Gene Expression Analysis
Total RNA from Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants was isolated with a Qiagen RNA isolation kit. The organs used were from 7-week-old plants grown in a greenhouse. RNA from different cell types (interfascicular fibers, xylem, and pith cells) was isolated and amplified as described previously (Zhong et al., 2006). First-strand cDNA was synthesized from total RNA treated with DNase I and then used as a template for real-time quantitative PCR analysis with the QuantiTect SYBR Green PCR kit (Clontech). The PCR primers for IRX9 are 5′-TGGCCAATGGCATTGTTATCGGCT-3′ and 5′-GGTGCTTAAACGTGTTCTTGTG-3′, those for I9H are 5′-TCCAGGAGACATCATTTATAGAGC-3′ and 5′-CTATTTCATAGTTATGAGAGCCTG-3′, those for IRX14 are 5′-AAGATCAAATGCCATTATCCCAAG-3′ and 5′-TCAGTTTCTTTCTTGATGCTTAGACGA-3′, and those for I14H are 5′-CCTGGCTGGATCATAAAGTCACC-3′ and 5′-TCAGCTAGTTGCTGACACTTTGAC-3′. The relative expression level was calculated by normalizing the PCR threshold cycle number of each gene with that of the EF1a reference gene. The data were averages of three biological replicates. Reverse transcription (RT)-PCR was applied to examine the transcript level in the GT43 mutants as described previously (Zhong et al., 2005). The expression of the EF1a gene was used as an internal control for determining the RT-PCR amplification efficiency among different samples.
GUS Reporter Gene Analysis
To ensure the inclusion of all sequences required for the expression of the endogenous genes, the I9H, IRX14, or I14H gene containing a 3-kb 5′ upstream sequence, the entire coding region, and a 2-kb 3′ downstream sequence was PCR-amplified using gene-specific primers (Supplemental Table S1) and used for the GUS reporter gene analysis. The GUS reporter gene was inserted in-frame right before the stop codon of these genes and then cloned into the binary vector pBI101 (Clontech) to create the GUS reporter constructs. The constructs were transformed into wild-type Arabidopsis plants by Agrobacterium tumefaciens-mediated transformation (Bechtold and Bouchez, 1994) to generate the GUS reporter transgenic plants. Inflorescence stems and roots from 7-week-old first-generation transgenic plants were examined for the GUS activity as described previously (Zhong et al., 2005). At least 30 independent transgenic plants were tested, and a consistent GUS staining pattern was observed for each GUS reporter construct.
Subcellular Localization
The subcellular localization of I9H, IRX14, and I14H was performed by cotransfecting YFP-tagged fusion proteins and the CFP-tagged Golgi marker (FRA8) into carrot (Daucus carota) protoplasts (Liu et al. 1994). The full-length cDNAs of I9H, IRX14, and I14H were fused in-frame with the YFP cDNA and ligated between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 (Clontech). Fluorescence signals in transfected protoplasts were visualized using a TCs SP2 spectral confocal microscope (Leica Microsystems). At least 20 fluorescence-positive protoplasts were examined, and the same subcellular localization pattern was observed for each construct. Images were saved and processed with Adobe Photoshop version 7.0 (Adobe Systems).
Complementation of the irx9 and irx14 Mutants
The full-length cDNAs of GT43 members driven by the CaMV 35S promoter were cloned into the pGPTV binary vector. The constructs were introduced into the Arabidopsis irx9 or irx14 mutant by Agrobacterium-mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on hygromycin, and the first-generation transgenic plants were used for breaking strength and anatomical analyses. Basal parts of the main inflorescence of 10-week-old plants were measured for breaking force using a digital force/length tester (Zhong et al., 1997). The breaking force was calculated as the force needed to break apart a stem segment. For each construct, at least 96 transgenic plants were generated and examined.
Histology
Tissues were fixed in 2% formaldehyde and embedded in low-viscosity (Spurr's) resin (Electron Microscopy Sciences) as described (Burk et al., 2006). For light microscopy, 1-μm-thick sections were cut with a microtome and stained with toluidine blue. For transmission electron microscopy, 85-nm-thick sections were cut, poststained with uranyl acetate and lead citrate, and observed using a Zeiss EM 902A transmission electron microscope (Carl Zeiss). For each construct, stems from at least eight transgenic plants (first generation) with representative phenotypes were sectioned, and representative data are shown.
Cell Wall Sugar Composition Analysis
Inflorescence stems from at least 20 independent lines of Arabidopsis plants were used for cell wall isolation according to Zhong et al. (2005). Cell wall sugars (as alditol acetates) were determined following the procedure described by Hoebler et al. (1989). The alditol acetates of the hydrolyzed cell wall sugars were analyzed on an Agilent 6890N gas-liquid chromatograph equipped with a 30-m × 0.25-mm (i.d.) silica capillary column (DB 225; Alltech Associates).
MALDI-TOF MS
Xylooligosaccharides were prepared by digestion of 4 n KOH solubilized wall preparations with β-xylanase M6 (Megazyme) as described (Zhong et al., 2005). The acidic xylooligosaccharides were analyzed using a MALDI-TOF MS device operated in the positive-ion mode with an accelerating voltage of 30 kV, an extractor voltage of 9 kV, and a source pressure of approximately 8 × 10−7 torr. The aqueous sample was mixed (1:1, v/v) with the MALDI matrix (0.2 m 2,5-dihydroxbenzoic acid and 0.06 m 1-hydroxyisoquinoline in 50% acetonitrile) and dried on the stainless steel target plate. Spectra are averages of 100 laser shots.
1H NMR Spectroscopy
NMR spectra of the acidic xylooligosaccharides from β-xylanase digestion were acquired at 20°C on a Varian Inova 600-MHz spectrometer (599.7 MHz, 1H) using a 5-mm cryogenic triple resonance probe (Varian). All NMR samples were prepared using 100% D2O in 3-mm standard NMR tubes. 1H chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt. For all experiments, 64 transients were collected using a spectral width of 6,000 Hz and an acquisition time of 5 s. The residual water resonance was suppressed by a 1-s presaturation pulse at a field strength of 40 Hz. One-dimensional spectra were processed using MestReC (MestReC Research) with 0.2-Hz apodization followed by zero filling to 128 k points. The 1H NMR assignments were done by comparison with the NMR spectra data for Arabidopsis acidic xylooligosaccharides (Zhong et al., 2005; Peña et al., 2007).
Assay of Xylosyltransferase Activity
Microsomes were isolated from the inflorescence stems of at least four individual 8-week-old plants for each genotype following the procedure of Kuroyama and Tsumuraya (2001) and stored at −80°C until used. For assay of the xylosyltransferase activity, microsomes (100 mg per 30-mL reaction) were incubated with the reaction mixture containing 50 mm HEPES-KOH, pH 6.8, 5 mm MnCl2, 1 mm dithiothreitol, 0.5% Triton X-100, 0.1 mm cold UDP-Xyl (CarboSource Service), 0.2 mg mL−1 Xyl6 (Megazyme), and UDP-[14C]Xyl (0.1 mCi; American Radiolabeled Chemical). The reaction products were separated from UDP-[14C]Xyl by paper chromatography according to Ishikawa et al. (2000) and counted for the amount of radioactivity with a Perkin-Elmer scintillation counter.
The Arabidopsis Genome Initiative locus identifiers for the Arabidopsis genes investigated in this study are IRX9 (At2g37090), I9H (At1g27600), IRX14 (At4g36890), and I14H (At5g67230).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR detection of GT43 transcripts in various GT43 mutants.
Supplemental Figure S2. Complementation of the collapsed vessel phenotype in the irx9 and irx14 mutants by overexpression of GT43 members.
Supplemental Figure S3. Vessel morphology in single and double mutants of GT43 genes.
Supplemental Table S1. Primers used in this work.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the T-DNA knockout lines of IRX9, I9H, IRX14, and I14H and the editor and reviewers for their constructive comments and suggestions.
References
- Andersson SI, Samuelson O, Ishihara M, Shimizu K. (1983) Structure of the reducing end-groups in spruce xylan. Carbohydr Res 111: 283–288 [Google Scholar]
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al. (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baydoun EAH, Usta JAR, Waldron KW, Brett CT. (1989) A methyltransferase involved in the biosynthesis of 4-O-methylglucuronoxylan in etiolated pea epicotyls. J Plant Physiol 135: 81–85 [Google Scholar]
- Baydoun EAH, Waldron KW, Brett CT. (1983) The interaction of xylosyltransferase and glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem J 257: 853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Bouchez D. (1994) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Potrykus I, Spangenberg G, , Gene Transfer to Plants. Springer-Verlag, Berlin, pp 19–23 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller HV, et al. (2007) Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J 52: 791–802 [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacreb R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Burk DH, Zhong R, Morrison WH III, Ye ZH. (2006) Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J Integr Plant Biol 48: 85–98 [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-b-D-glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M. (2000) The cell wall. Buchanan BB, Gruissem W, Jones RL, , Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 52–108 [Google Scholar]
- Carpita N, Vergara C. (1998) A recipe for cellulose. Science 279: 672–673 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al. (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Ebringerová A, Heinze T. (2000) Xylan and xylan derivatives: biopolymers with valuable properties. 1. Naturally occurring xylan structures, isolation procedures and properties. Macromol Rapid Commun 21: 542–556 [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory ACE, Smith C, Kerry ME, Wheatley ER, Bolwell GP. (2002) Comparative subcellular immunolocation of polypeptides associated with xylan and callose synthases in French bean (Phaseolus vulgaris) during secondary wall formation. Phytochemistry 59: 249–259 [DOI] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Höfte H. (2001) Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-β-glucanase. Planta 212: 348–358 [DOI] [PubMed] [Google Scholar]
- Hoebler C, Barry JL, David A, Delort-Laval J. (1989) Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J Agric Food Chem 37: 360–367 [Google Scholar]
- Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y. (2000) Characterization of pectin methyltransferase from soybean hypocotyls. Planta 210: 782–791 [DOI] [PubMed] [Google Scholar]
- Johansson MH, Samuelson O. (1977) Reducing end groups in birch xylan and their alkaline degradation. Wood Sci Technol 11: 251–263 [Google Scholar]
- Kuroyama H, Tsumuraya Y. (2001) A xylosyltransferase that synthesizes β-(1→4)-xylans in wheat (Triticum aestivum L.) seedlings. Planta 213: 231–240 [DOI] [PubMed] [Google Scholar]
- Lee C, O'Neill MA, Tsumuraya Y, Darvill AG, Ye ZH. (2007a) The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 48: 1624–1634 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. (2009a) Down-regulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol 50: 1075–1089 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. (2009b) The F8H glycosyltransferase is a functional paralog of FRA8 involved in glucuronoxylan biosynthesis in Arabidopsis. Plant Cell Physiol 50: 812–827 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye ZH. (2009c) The poplar GT8E and GT8F glycosyltransferases are functional orthologs of Arabidopsis PARVUS involved in glucuronoxylan biosynthesis. Plant Cell Physiol 50: 1982–1987 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH. (2007b) The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48: 1659–1672 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ. (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Page RDM. (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchia AC, Sorensen SO, Scheller HV. (2002) Arabinoxylan biosynthesis in wheat: characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol 130: 432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47: 131–143 [PubMed] [Google Scholar]
- Shimizu K, Ishihara M, Ishihara T. (1976) Hemicellulases of brown rotting fungus, Tyromyces palustris. II. The oligosaccharides from the hydrolysate of a hardwood xylan by the intracellular xylanase. Mokuzai Gakkaishi 22: 618–625 [Google Scholar]
- Somerville C. (2006) The billion-ton biofuel vision. Science 312: 1277. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL. (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urahara T, Tsuchiya K, Kotake T, Tohno-oka T, Komae K, Kawada N, Tsumuraya Y. (2004) A β-(1→4)-xylosyltransferase involved in the synthesis of arabinoxylans in developing barley endosperms. Physiol Plant 122: 169–180 [Google Scholar]
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- Zeng W, Chatterjee M, Faik A. (2008) UDP-xylose-stimulated glucuronyltransferase activity in wheat microsomal membranes: characterization and role in glucurono(arabino)xylan biosynthesis. Plant Physiol 147: 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Pena MJ, Zhou G-K, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH, Darvill AG, York WS, Ye ZH. (2005) Arabidopsis Fragile Fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH. (1997) Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GK, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH. (2007) Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiol 48: 689–699 [DOI] [PubMed] [Google Scholar]
- Zhou GK, Zhong R, Richardson EA, Morrison WH, Nairn CJ, Wood-Jones A, Ye ZH. (2006) The poplar glycosyltransferase GT47C is functionally conserved with Arabidopsis Fragile Fiber8. Plant Cell Physiol 47: 1229–1240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.