Abstract
Heat shock response (HSR) is a universal mechanism in all organisms. It is under tight regulation by heat shock factors (HSFs) and heat shock proteins (HSPs) after heat shock (HS) to prevent stress damage. On the attenuation of HSR, HSP70 and HSF Binding Protein1 (HSBP1) interact with HSF1 and thus dissociate trimeric HSF1 into an inert monomeric form in humans. However, little is known about the effect of HSBP with thermal stress in plants. This report describes our investigation of the role of AtHSBP in Arabidopsis (Arabidopsis thaliana) by genetic and molecular approaches. AtHSBP was heat inducible and ubiquitously expressed in all tissues; AtHSBP was also crucial for seed development, as demonstrated by AtHSBP-knockout lines showing seed abortion. Thermotolerance results showed that AtHSBP participates in acquired thermotolerance but not basal thermotolerance and is a negative regulator of HSR. Subcellular localization revealed that the cytosol-localized AtHSBP translocated to the nucleus in response to HS. Protoplast two-hybrid assay results confirmed that AtHSBP interacts with itself and with the HSFs, AtHSFA1a, AtHSFA1b, and AtHSFA2. AtHSBP also negatively affected AtHSFA1b DNA-binding capacity in vitro. Quantitative polymerase chain reaction and western-blot analysis demonstrated that altered levels of AtHSBP lead to differential HSP expression, mainly during the recovery from HS. These studies provide a new insight into HSBP in plants and reveal that AtHSBP is a negative regulator of HSR and required for seed development.
When cells are exposed to elevated temperature, called heat stress or heat shock (HS), multiple mechanisms are activated to prevent stress-caused damage and to enhance survival by a phenomenon called the heat shock response (HSR; Lindquist, 1986; Lindquist and Craig, 1988). HSR is universally invoked in all organisms and is characterized by elevated synthesis of a specialized set of proteins called heat shock proteins (HSPs). Plants and other organisms have both basal thermotolerance (BT) and acquired thermotolerance (AT; Hong and Vierling, 2000, 2001). With BT, organisms have an innate potential to survive under thermal stress above the optimum for growth. AT is induced by prior exposure to moderately high but survivable temperature and provides resistance against a subsequent lethal HS (Parsell and Lindquist, 1993). Thermotolerance is coordinated by a signaling pathway that regulates heat tolerance to limit stress damage and to rebuild cellular homeostasis for survival and growth (Clarke et al., 2004; Larkindale et al., 2005).
HSPs include several conserved protein families, such as HSP100/ClpB, HSP90, HSP70/DnaK, HSP60/chaperonin, and small HSPs (sHSPs). HSPs primarily function as molecular chaperones to prevent aggregation and to promote appropriate refolding of denatured proteins caused by HS, which leads to increased thermotolerance (Parsell and Lindquist, 1993). AtHSP101 expression in response to HS is essential for thermotolerance in Arabidopsis (Arabidopsis thaliana; Hong and Vierling, 2000, 2001; Queitsch et al., 2000). HSP70 is one of the most conserved gene families in all organisms. It is developmentally regulated and can bind to denatured protein to promote protein refolding under HS (Parsell and Lindquist, 1993; Sung et al., 2001). In planta, sHSPs, with their ability to form large oligomers, are produced in response to HS. The conformational dynamics and aggregated state of sHSPs may be important in thermotolerance by preventing harmful stress damage (Miroshnichenko et al., 2005).
The heat-inducible expression of HSP genes is regulated by heat shock transcription factors (HSFs). The N-terminal helix-turn-helix DNA binding domain of HSFs is the most conserved functional domain, and the activation-induced trimerization is mediated by two arrays of hydrophobic heptad repeats (HR-A/B) characteristic of helical coiled-coil structures; suppression of HSF trimerization is likely mediated by another region of hydrophobic heptad repeats (HR-C) adjacent to the C-terminal domain (Rabindran et al., 1993; Harrison et al., 1994). HSFs bind to conserved cis-acting elements, heat shock elements (HSEs), defined as adjacent and inverse repeats of the motif 5′-nGAAn-3′, such as 5′-nGAAnnTTCnnGAAn-3′, located in the promoters of HSP genes (Schöffl et al., 1998). The number of HSFs varies among different eukaryotic organisms. In contrast to animals or yeast, plants possess more HSFs for adapting to the fluctuating environment. Experimental evidence shows that HSFA1a functions as a major HSF in tomato (Solanum lycopersicum; Mishra et al., 2002). In Arabidopsis, 21 HSFs form a complex HSF network, in which AtHSFA1a and AtHSFA1b play important roles in the induction of HSP genes in the early phase of HSR (Nover et al., 2001; Lohmann et al., 2004). In addition, a number of variant AtHSFA1a-binding sites are considered as the target genes of AtHSFA1a in vivo (Guo et al., 2008), and AtHSFA2 is a heat-induced HSF that maintains the HSP expression in extending AT in Arabidopsis (Charng et al., 2007).
Besides the feedback regulation of HSFs by HSPs, human HSF Binding Protein1 (HsHSBP1) is identified by yeast two-hybrid screening to specifically associate with the hydrophobic heptad repeats of HSF1 to regulate its activity. Transient expression shows that HsHSBP1 is a nucleus-localized protein and functions to negatively affect the DNA-binding capacity and transactivation activity of HSF1 during HSR (Satyal et al., 1998). Members of the HSBP family are small proteins (less than 10 kD) and are highly conserved among species (Tai et al., 2002). Comparison of the expressed sequence tags of HSBP homologs revealed only a single copy of HSBP in animals and dicots such as Arabidopsis but two HSBP isoforms in monocots such as maize (Zea mays) and rice (Oryza sativa; Satyal et al., 1998; Fu et al., 2002). EMPTY PERICARP2 (EMP2), encoding the first described HSBP-like protein in plants, is differentially regulated during early embryogenesis and postembryonic shoot development in maize (Fu et al., 2002; Fu and Scanlon, 2004). The emp2 mutant kernels are aborted at the coleoptile stage/stage 1, followed by necrosis and reabsorption of kernel contents. epm2 also leads an unattenuated HSR and shows greatly increased HSP expression. At the transcriptional level, overaccumulated emp2 mutant transcripts with a 5′ untranslated region truncation reduce endogenous EMP2 expression, and this indicates that the 5′ untranslated region is important for regulating its transcription (Fu et al., 2002). Furthermore, the expression and functions of EMP2 in maize shoot development are unaffected by HS or nonstress conditions, which suggests that the developmental defect of the emp2 mutation appears to be separated from its role in the attenuation of HSR (Fu and Scanlon, 2004). In addition to containing EMP2, maize has another HSBP paralog, ZmHSBP2, which is a heat-inducible gene during HSR. Both ZmHSBP paralogs interact nonredundantly with specific HSFs, and this reveals that EMP2 and ZmHSBP2 may have distinct functions during plant development and HSR. However, the functions of ZmHSBP2 need to be clarified (Fu and Scanlon, 2004; Fu et al., 2006).
To date, the functions of HSBP in thermotolerance remain unclear in plants. Therefore, we characterized AtHSBP, a single-copy gene in Arabidopsis, in response to HS and plant development. Thermotolerance tests showed that AtHSBP functions as a negative regulator of HSR, and seeds were aborted in AtHSBP-knockout plants. The cytosol-localized AtHSBP translocated to the nucleus during the recovery from HS, a pattern that differed from that of HsHSBP1, a predominantly nucleus-localized protein unaffected by HS (Satyal et al., 1998). Protoplast two-hybrid assay confirmed that AtHSBP interacts with itself and with HS-related HSFs. Electrophoretic mobility shift assay (EMSA) confirmed that AtHSBP negatively affects HSF DNA-binding capacity. These data demonstrate a new aspect of HSBP in plants by revealing that AtHSBP plays as a negative regulator of HSR and is required for plant development.
RESULTS
AtHSBP Is a Functional Homolog among Plants and Animals
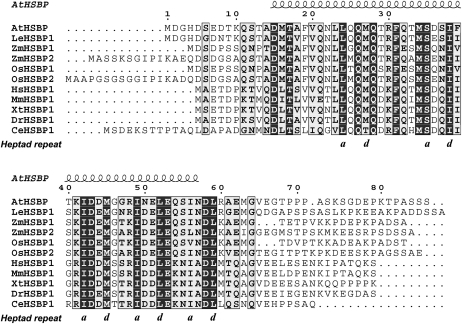
HsHSBP1 and EMP2 (ZmHSBP1) have been characterized as negative regulators during the attenuation of HSP transcription in human and maize, respectively (Satyal et al., 1998; Fu et al., 2002). The Arabidopsis Information Resource database (http://www.arabidopsis.org) shows AtHSBP (At4g15802) as a putative protein with five exons and four introns (Fig. 2A) that encodes 86 amino acid residues with predicted molecular mass and pI of 9.35 kD and 4.11, respectively (Fig. 1). Residues 15 to 59 of AtHSBP contain one continuous α-helix in the central region with hydrophobic heptad repeats, which corresponds to residues 15 to 49 of HsHSBP1 (Liu et al., 2009) and to residues 12 to 56 of EMP2 (Fig. 1). AtHSBP was predicted to have two potential coiled-coil regions: residues 20 to 34 (16% possibility) and 41 to 65 (60%–99% possibility). The α-helix region is highly conserved and shares at least 58% identity and greater than 78% similarity with the HSBP homologs in animals and plants (Supplemental Table S1). Thus, AtHSBP may have functional homology across plant and animal kingdoms and may act as a negative regulator of HSF transcriptional activity, as has been demonstrated for HsHSBP1 and EMP2.
Figure 2.
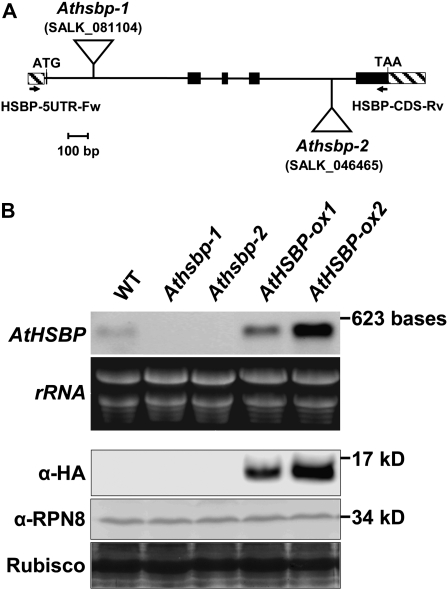
AtHSBP gene structure and characterization of AtHSBP-knockout and -overexpression lines. A, AtHSBP gene is composed of four introns and five exons (black boxes). The 5′ and 3′ untranslated regions are indicated by hatched boxes. The T-DNA insertions are in the first intron (Athsbp-1, SALK_081104) and fourth intron (Athsbp-2, SALK_046465), as indicated by triangles. The specific primers for genotyping are shown by black arrows. ATG and TAA are shown as initiation and stop codons, respectively. B, Expression of AtHSBP in AtHSBP-knockout (Athsbp-1 and -2) and AtHSBP-overexpression (AtHSBP-ox1 and -ox2) lines was analyzed by northern-blot analysis (top; full-length AtHSBP cDNA as probe) and western-blot analysis (bottom; α-HA or α-RPN8 antibody). RPN8 and Rubisco large subunit stained with Amido black are shown for equal loading. WT, Wild type.
Figure 1.
Sequence alignment of the HSBP homologs among different species. Sequences were aligned with ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and generated by ESPript 2.2 software (http://espript.ibcp.fr/ESPript/ESPript/). Residue numbering and secondary structure predictions correspond to AtHSBP. Identical residues are in black, and the other conserved residues are in gray and boldface. The predicted α-helix is depicted as a helix. The COILS program (http://www.ch.embnet.org/software/COILS_form.html; window of 14 residues) calculated the AtHSBP sequence to form two potential coiled-coil domains: residues 20 to 34 and 41 to 65. The hallmark of coiled-coil structure is a heptad repeat of seven amino acid residues, typically denoted by the letters a to g, with a predominance of hydrophobic residues at a and d positions, and residues at e and g positions are frequently charged. Positions a and d within a heptad repeat are marked.
Altered Levels of AtHSBP Regulate Thermotolerance
To investigate AtHSBP in response to HS, two defective AtHSBP homozygous T-DNA insertion lines, Athsbp-1 (SALK_081104) and Athsbp-2 (SALK_046465), were screened (Fig. 2A). Northern-blot analysis showed that both Athsbp-1 and -2 were null mutants (Fig. 2B). AtHSBP-overexpression lines (AtHSBP-ox1 and -ox2; in the T3 generation), which AtHSBP fused with a hemagglutinin (HA) epitope tag, were confirmed by northern- and western-blot analysis (Fig. 2B).
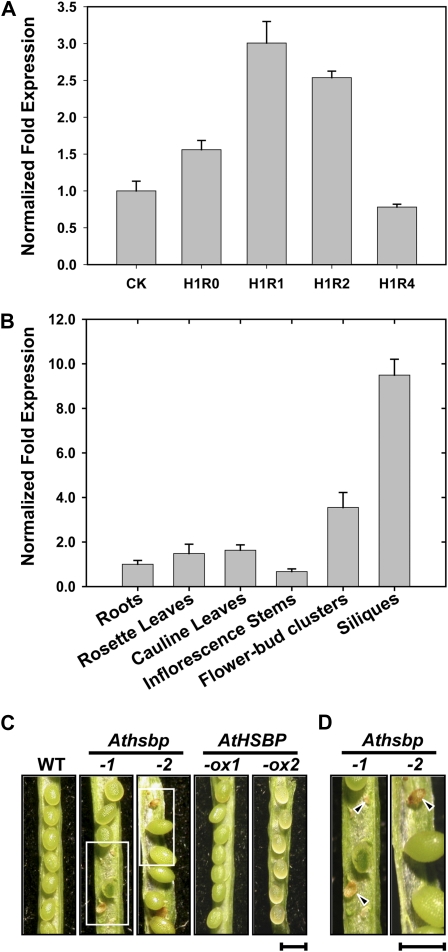
The phenotypes of AtHSBP-knockout (Athsbp), AtHSBP-overexpression (AtHSBP-ox), and AtHSP101-knockout (Athsp101; SALK_066374), a well-characterized heat-sensitive mutant (Hong and Vierling, 2000), lines were analyzed under normal growth conditions (Table I). The wild-type and Athsbp lines did not differ in phenotypes of rosette diameter, leaf index, and root length; however, the defective AtHSBP caused significantly earlier flowering, shorter siliques, and seed abortion, which resulted in less seed yield than the wild type. As noted, approximately 35% of seeds were aborted in Athsbp lines. The phenotypes of AtHSBP-ox lines were similar to those of the wild type.
Table I. Phenotypic characterization of AtHSBP mutant lines grown under normal conditions.
The wild-type, AtHSBP-knockout (Athsbp-1 and -2), AtHSBP-overexpression (AtHSBP-ox1 and -ox2), and AtHSP101-knockout (Athsp101) lines were grown under normal conditions, and phenotypes were analyzed. Rosette diameter was measured in 28-d-old plants. Leaf index was determined by the ratio of the length to width of the second pair of leaves. Root length was measured in 6-d-old seedlings. Flowering time was counted as the total number of leaves at anthesis when the first floral bud appeared. Silique length, seed yield, and seed abortion were measured from mature siliques. Data are means ± sd from at least 20 samples. *, Significant at P < 0.05 (Student's t test, as compared with the wild-type value).
| Line | Rosette Diameter | Leaf Index | Root Length | Flowering Time | Silique Length | Seed Yield | Seed Abortion |
| cm | length-width ratio | cm | no. of rosette leaves | mm | no. per silique | % per silique | |
| Wild type | 5.8 ± 0.2 | 2.1 ± 0.3 | 2.3 ± 0.4 | 7.4 ± 0.5 | 11.5 ± 0.3 | 38.3 ± 2.1 | 0 |
| Athsbp-1 | 5.8 ± 0.3 | 2.0 ± 0.2 | 2.1 ± 0.4 | 5.7 ± 0.5* | 7.8 ± 0.3* | 14.3 ± 0.6* | 37.5 ± 2.9 |
| Athsbp-2 | 5.5 ± 0.4 | 2.0 ± 0.3 | 2.0 ± 0.4 | 5.7 ± 0.5* | 7.5 ± 0.4* | 15.1 ± 2.8* | 33.9 ± 6.2 |
| AtHSBP-ox1 | 6.1 ± 0.4 | 1.8 ± 0.2 | 2.6 ± 0.4 | 7.2 ± 0.4 | 11.7 ± 0.4 | 35.5 ± 0.7 | 0 |
| AtHSBP-ox2 | 5.6 ± 0.3 | 1.9 ± 0.2 | 2.5 ± 0.3 | 7.4 ± 0.5 | 12.1 ± 0.5 | 36.5 ± 0.7 | 0 |
| Athsp101 | 6.0 ± 0.7 | 1.9 ± 0.2 | 2.1 ± 0.4 | 7.0 ± 0.4 | 10.8 ± 0.4 | 36.5 ± 2.8 | 0 |
We examined AtHSBP expression levels in response to thermal stress and in different tissues by real-time quantitative PCR (Fig. 3, A and B). Six-day-old wild-type seedlings were treated without HS (CK) or with HS at 37°C for 1 h (H1R0) and then recovered from HS for 1 h (H1R1) to 4 h (H1R4) to attenuate the HSR (Fig. 3A). With H1R0 and H1R1 treatments, the expression levels of AtHSBP were 1.5- and 3-fold higher, respectively, than that of the control (CK). After 4 h of recovery (H1R4), the levels returned to the basal level. Elongation factor1α (EF1α) transcript was assayed as a quantitative control (Supplemental Fig. S4A) and also showed similar results as when using AtACT2 (Fig. 3A). AtHSBP was ubiquitously expressed in roots, leaves, inflorescence stems, flower bud clusters, and siliques of 28-d-old wild-type plants under normal growth conditions. Especially in flowers and siliques, the expression was 3.5- and 9.5-fold higher, respectively, than in other tissues (Fig. 3B); meanwhile, seeds were aborted in Athsbp lines (Fig. 3, C and D). Therefore, heat-inducible AtHSBP is ubiquitously expressed in all tissues of Arabidopsis and is required in seed development.
Figure 3.
AtHSBP expression in response to HS and in different tissues, and seed abortion in AtHSBP-knockout lines under normal growth conditions. The expression patterns of AtHSBP were analyzed by real-time quantitative PCR. A, Six-day-old wild-type seedlings were treated without HS (CK) or with HS at 37°C for 1 h (H1R0) and then recovered from HS for 1 h (H1R1) to 4 h (H1R4). B, AtHSBP expression was analyzed in different tissues of 28-d-old wild-type plants under normal conditions, as indicated. C, Seed abortion occurred in AtHSBP-knockout lines (Athsbp-1 and -2). D, Magnification of frames in C. The arrowheads indicate the aborted seeds. WT, Wild type. Bars = 1 mm. [See online article for color version of this figure.]
The wild-type and mutant lines were tested for BT and AT in seed germination (Larkindale et al., 2005), hypocotyl elongation, and seedling survival rate (Charng et al., 2007; Table II). AtHSBP was not required for BT; however, during AT tests, the seedling survival rates of Athsbp lines were significantly increased (approximately 50% more), whereas those of AtHSBP-ox lines were significantly reduced (approximately 20% less), as compared with the wild type. Therefore, AtHSBP is a negative regulator in AT but not BT.
Table II. Basal and acquired thermotolerance tested in AtHSBP mutant lines.
Wild-type, AtHSBP-knockout (Athsbp-1 and -2), AtHSBP-overexpression (AtHSBP-ox1 and -ox2), and AtHSP101-knockout (Athsp101) lines were assayed for basal and acquired thermotolerance during seed germination, hypocotyl elongation, and seedling survival. Thermotolerance tests were calculated from 150 seedlings in each experiment. Data are means ± sd (n = 3). *, Significant at P < 0.05 (Student's t test, as compared with the wild-type value).
| Line | Basal Thermotolerance Test |
Acquired Thermotolerance Test |
|||
| Seed Germination, 45°C, 220 min | 2.5-d-Old Hypocotyl Elongation, 44°C, 20 min | 2.5-d-Old Hypocotyl Elongation, 37°C, 60 min, Recovery, 120 min, 44°C, 190 min | 3-d-Old Seedling Survival, 37°C, 60 min, Recovery, 120 min, 44°C, 190 min | 6-d-Old Seedling Survival, 37°C, 60 min, Recovery, 120 min, 44°C, 190 min | |
| % germination | % unheated growth | % wild-type survival | |||
| Wild type | 99.2 ± 0.8 | 49.7 ± 3.9 | 25.3 ± 0.4 | 100 | 100 |
| Athsbp-1 | 98.0 ± 1.8 | 45.7 ± 6.0 | 30.9 ± 1.0* | 148.8 ± 10.7* | 145.9 ± 9.6* |
| Athsbp-2 | 91.8 ± 4.7 | 49.1 ± 4.3 | 26.1 ± 0.7 | 156.4 ± 11.9* | 149.0 ± 10.3* |
| AtHSBP-ox1 | 97.0 ± 0.9 | 52.3 ± 3.1 | 25.2 ± 0.4 | 84.0 ± 10.2* | 83.5 ± 11.7* |
| AtHSBP-ox2 | 98.6 ± 1.3 | 46.2 ± 6.5 | 16.2 ± 0.1* | 76.2 ± 7.2* | 78.6 ± 11.1* |
| Athsp101 | 0 | 26.3 ± 4.3* | 5.34 ± 0.7* | 0* | 0* |
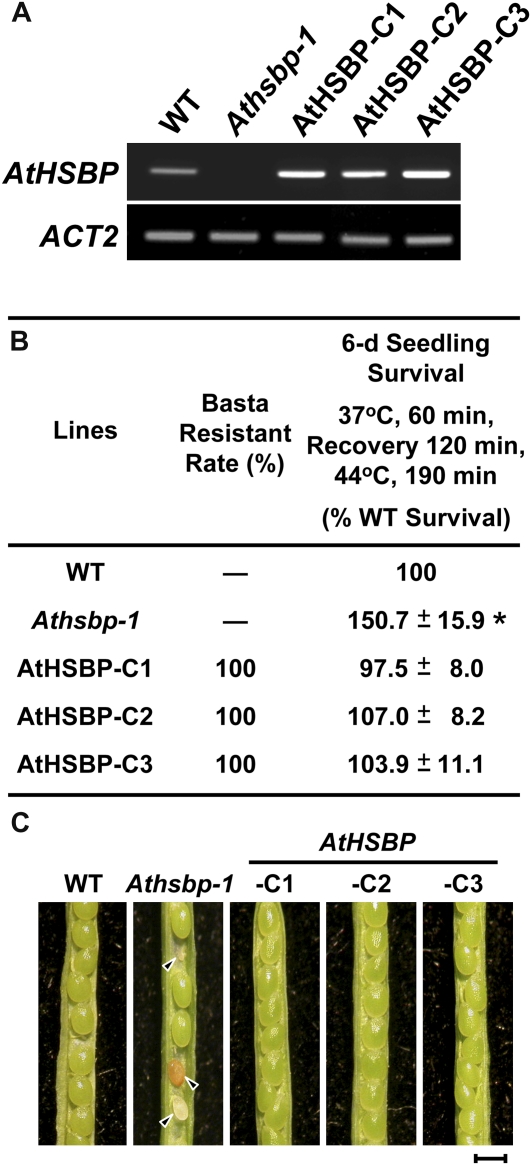
To examine whether the increased heat-resistant and aborted seed phenotypes of Athsbp lines resulted from the defect of AtHSBP, we transformed AtHSBP genomic DNA with its upstream 1-kb potential promoter region into an Athsbp-1 background for complementation. After selection by the herbicide Basta, we characterized three independent homozygous T-DNA transgenic lines (AtHSBP-C1, -C2, and -C3; in the T3 generation), and AtHSBP expression was determined by reverse transcription-PCR (Fig. 4A). The complementary lines restored the wild-type phenotypes of thermotolerance during AT test and seed set (Fig. 4, B and C).
Figure 4.
Increased heat-resistant and aborted seed phenotypes in AtHSBP-knockout lines were restored by complementation of AtHSBP transgene. A, Reverse transcription-PCR analysis of AtHSBP transcript levels in 6-d-old seedlings of the wild type (WT), Athsbp-1, and three independent complementary transgenic lines (AtHSBP-C1, -C2, and -C3; in the T3 generation) that harbored AtHSBP and bar genes in an Athsbp-1 background. AtACT2 (ACT2) is shown as a loading control. B, Basta resistance and seedling survival in acquired thermotolerance tests were calculated from 50 seedlings of each experiment, and 18.5 ± 1.5 wild-type seedlings survived after HS treatment. Data represent means ± sd (n = 3). *, Significant at P < 0.05 (Student's t test, as compared with the wild-type value). C, The complementary lines restored the wild-type seed set phenotype. The arrowheads indicate the aborted seeds. Bar = 1 mm. [See online article for color version of this figure.]
Cytosol-Localized AtHSBP Translocates to the Nucleus in Response to Thermal Stress
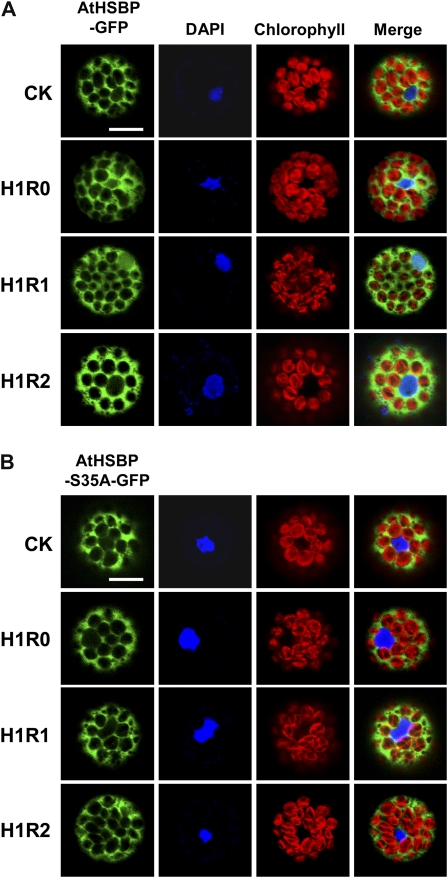
AtHSBP was fused to the N terminus of GFP (AtHSBP-GFP) driven by a 35S promoter and analyzed in wild-type Arabidopsis mesophyll protoplasts to assess subcellular localization during HSR (Fig. 5A). Protoplasts were treated without HS (CK) or with HS at 37°C for 1 h (H1R0) and then recovered from HS for 1 h (H1R1) or 2 h (H1R2) to attenuate the HSR. AtHSBP-GFP was predominantly expressed in the cytoplasm under normal conditions (CK). With H1R0 treatment, faint GFP signals were observed in the nucleus. During H1R1 treatment, AtHSBP-GFP translocated to the nucleus; however, with H1R2 treatment, the nucleus-localized GFP signals were undetectable. Supplemental Figure S1 shows two other independent results that confirmed those in Figure 5A. GUS-GFP fusion, as a control, showed the localization only in the cytoplasm, regardless of HS treatment (Supplemental Fig. S2). Therefore, these data supported that cytosol-localized AtHSBP translocates to the nucleus in response to thermal stress.
Figure 5.
Transient expression of AtHSBP in mesophyll protoplasts. Wild-type Arabidopsis protoplasts were transfected with the AtHSBP-GFP (A) or AtHSBP-S35A-GFP (B) construct and treated without HS (CK) or with HS at 37°C for 1 h (H1R0) and recovered from HS for 1 h (H1R1) or 2 h (H1R2). GFP signals were observed by confocal microscopy. Blue shows nucleus stained with 4′,6-diamino-phenylindole (DAPI), and red shows chlorophyll with autofluorescent light. Similar results were obtained from three independent replicates, and representative images are shown. Bars = 20 μm.
Analysis of the crystal structure of HsHSBP1 suggests that the conserved residue Ser-31 plays an important role in its function (Liu et al., 2009). Thus, AtHSBP Ser-35, corresponding to HsHSBP1 Ser-31 (Fig. 1), was mutated to Ala (S35A) and fused to GFP (AtHSBP-S35A-GFP), as shown in Figure 5A, and then its localization was analyzed. AtHSBP-S35A-GFP localized only in the cytoplasm, regardless of HS treatment (Fig. 5B). Thus, AtHSBP Ser-35 is important for nuclear localization during HSR.
AtHSBP Interacts with AtHSFs and Itself in Vivo
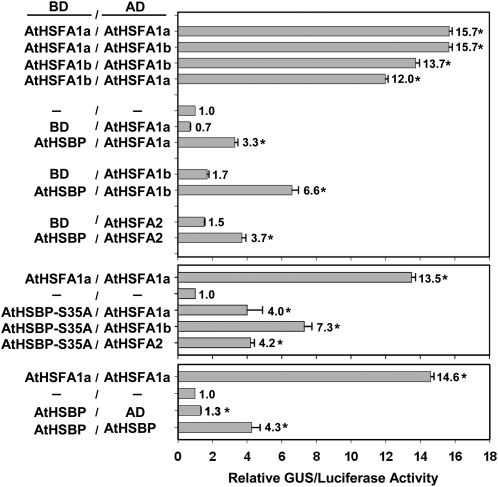
We used a protoplast two-hybrid assay (Ehlert et al., 2006) to investigate the interaction of AtHSBP with the well-studied and HS-related AtHSFs, AtHSFA1a, AtHSFA1b, and AtHSFA2 (Nover et al., 2001, Lohmann et al., 2004; Busch et al., 2005; Charng et al., 2007), and with itself in vivo. AtHSBP, AtHSBP-S35A, and AtHSFs were fused to a GAL4 DNA binding domain or activation domain (Fig. 6). Cotransfection with a reporter GUS (PGAL-UAS4:GUS) allowed for quantifying the interactions. The analysis of AtHSFA1a and AtHSFA1b, used as positive controls (Li et al., 2010), showed significant and strong interactions within themselves and with each other, as expected. AtHSBP and AtHSBP-S35A also showed significant interaction with AtHSFA1a, AtHSFA1b, or AtHSFA2, but was weaker than that in positive controls. HsHSBP1 has been suggested to form a homotrimer (Tai et al., 2002; Liu et al., 2009); thus, we tested the fusion proteins of AtHSBP binding domain and activation domain for interaction. AtHSBP was cross-interacted with itself, which implied the potential for its oligomerization.
Figure 6.
AtHSBP interacts with AtHSFs and itself by protoplast two-hybrid assay in mesophyll protoplasts. AtHSBP, AtHSBP-S35A, and AtHSFs were fused with the GAL4 DNA binding domain (BD) or activation domain (AD) and used for wild-type protoplast transfection, as indicated. Interactions between AtHSFA1a and AtHSFA1b were used as positive controls. Transfections without construct (−/−), with BD- and AD-AtHSFs (BD/AD-AtHSFs) constructs, and with BD-AtHSBP and AD (BD-AtHSBP/AD) constructs were used as references. The amount of relative GUS activity was normalized by luciferase luminescence. The fold expression was normalized relative to the amount of transfection without construct. Data are means ± sd (n = 3). *, Significant at P < 0.05 (Student's t test, as compared with the sample transfected without construct).
AtHSBP Negatively Affects AtHSFA1b HSE-Binding Capacity in Vitro
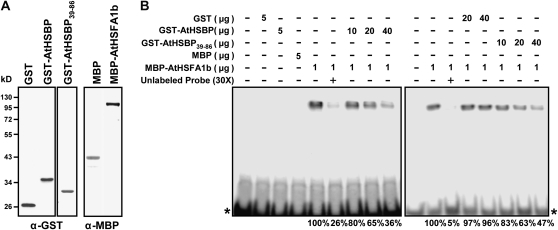
To examine whether AtHSBP represses HSF DNA-binding capacity, we performed EMSA. AtHSBP and AtHSFA1b were fused to glutathione S-transferase (GST) and maltose binding protein (MBP), respectively, and were affinity purified and characterized by western-blot analysis (Fig. 7A). Because AtHSBP translocated to the nucleus during the recovery from HS (Fig. 5A, H1R1), we preheated the protein at 37°C for 1 h before adding the [32P]dCTP-labeled HSE probe to mimic the physiological condition in vivo. GST, MBP, and GST-AtHSBP did not bind to the HSE probe, whereas MBP-AtHSFA1b bound to the HSE probe, which resulted in a band shift, as expected (Fig. 7B, left panel). Competition testing was conducted by adding 30× unlabeled HSE probes, which specifically titrated the DNA-binding activity of MBP-AtHSFA1b. Accordingly, the increased amount of GST-AtHSBP resulted in the decreased MBP-AtHSFA1b band-shift signal intensity, from 100% to 36%. Therefore, AtHSBP negatively affects AtHSFA1b DNA-binding capacity in vitro.
Figure 7.
AtHSBP negatively affects AtHSFA1b DNA binding in vitro. A, Affinity-purified GST, GST-AtHSBP, GST-AtHSBP39-86, MBP, and MBP-AtHSFA1b were characterized by western-blot analysis with α-GST or α-MBP antibody. B, EMSA experiment. The [α-32P]dCTP-labeled HSE probe was mixed with recombinant proteins or unlabeled probe, as indicated at the top. The asterisks indicate the free labeled probe. The MBP-AtHSFA1b DNA-binding activities in percentages are shown at the bottom.
AtHSBP was predicted to have two coiled-coil structures, with residues 47 to 64 having more than 95% possibility of forming a coiled-coil conformation (Fig. 1). Therefore, GST-AtHSBP39-86, containing residues 39 to 86 of AtHSBP, was affinity purified, characterized (Fig. 7A), and then examined for whether it affected MBP-AtHSFA1b DNA-binding capacity (Fig. 7B, right panel). The increased amount of GST, as controls, did not affect MBP-AtHSFA1b DNA-binding activity; however, with the increased amount of GST-AtHSBP39-86, the band-shift signal intensity decreased, from 100% to 47%. Thus, this potential coiled-coil domain can contribute to AtHSFA1b interaction in vitro.
AtHSBP Functions as a Negative Regulator of HSR
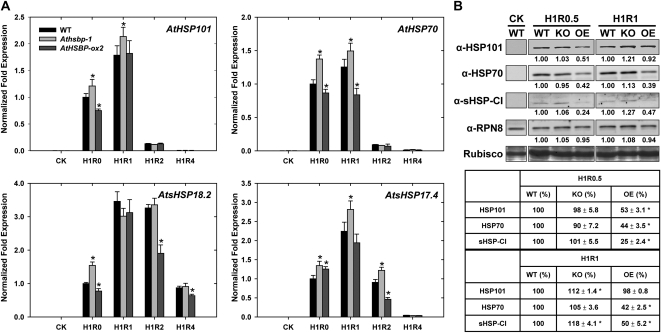
To verify that AtHSBP acts as a negative regulator of HSR, we analyzed HSP transcript and protein levels in the wild-type and mutant lines of Athsbp-1 and AtHSBP-ox2 by real-time quantitative PCR and western-blot analysis. The HSP genes of AtHSP101, AtHSP70, and class CI sHSP (sHSP-CI), AtsHSP18.2 and AtsHSP17.4, which have been well characterized during HSR and are regulated by AtHSFA1a or AtHSFA1b (Hong and Vierling, 2001; Lin et al., 2001; Zhang et al., 2003; Nishizawa et al., 2006), were used as candidates for the analyses. The wild type, Athsbp-1, and AtHSBP-ox2 showed no difference in accumulation of HSPs after 0.5 h of HS at 37°C (data not shown), so we analyzed HSP expression after 1 h of HS at 37°C (H1R0) and recovery from HS for 0.5 to 4 h (H1R0.5 to H1R4) during the recovery from HS.
HSP expression displayed diverse patterns from H1R0 to H1R2 (Fig. 8A). HSP expression was significantly up-regulated in Athsbp-1 but was significantly down-regulated in AtHSBP-ox2. After 4 h of recovery (H1R4), HSP expression returned to the basal level, as expected. EF1α transcript was assayed as a quantitative control (Supplemental Fig. S4B) and also showed similar results as when using AtACT2 (Fig. 8A). Western-blot analysis also confirmed that HSP protein level was significantly increased in Athsbp-1 but reduced in AtHSBP-ox2 (Fig. 8B, H1R0.5 and H1R1). These results agreed with the phenotype of AtHSBP-ox2, showing reduced thermotolerance as compared with the wild type in AT tests (Table II). These data support AtHSBP acting as a negative regulator during the attenuation of HSR.
Figure 8.
HSP gene transcript and protein levels in the wild-type, AtHSBP-knockout, and AtHSBP-overexpression lines during HSR. Six-day-old seedlings of the wild-type (WT), AtHSBP-knockout (Athsbp-1; KO), and AtHSBP-overexpression (AtHSBP-ox2; OE) lines were treated without HS (CK) or with HS at 37°C for 1 h (H1R0) and recovered from HS for 0.5 to 4 h (H1R0.5 to H1R4). A, Transcript levels of AtHSP101, AtHSP70, and AtsHSP-CI of AtsHSP18.2 and AtsHSP17.4 were determined by real-time quantitative PCR. The fold expression was normalized relative to the wild-type level at H1R0. B, Protein accumulation of HSP101, HSP70, sHSP-CI, and RPN8 was analyzed by western blot with the indicated antibodies, and the values of signal intensity relative to the wild-type sample are indicated. Similar results were obtained from three independent replicates, and one typical result is shown. RPN8 and Rubisco large subunit stained with Amido black are shown to ensure equal loading. The values of HSP, presented in percentages, were normalized to RPN8 and relative to the wild-type sample, as shown at the bottom of the panels. Transcript and protein levels were calculated from 100 seedlings of each experiment. Data are means ± sd (n = 3). *, Significant at P < 0.05 (Student's t test, as compared with the wild-type value).
DISCUSSION
In this report, we characterized AtHSBP in Arabidopsis in response to HS and showed that it is heat inducible, ubiquitously expressed, required for seed development, and acts as a negative regulator for the attenuation of HSR. Subcellular localization assay revealed that AtHSBP is a predominantly cytosol-localized protein and that its translocation to the nucleus depends on thermal stress. We also demonstrated that AtHSBP is sufficient to interact with AtHSFs to reduce HSF transactivation and the protein levels of HSP genes.
AtHSBP contains a single α-helix (residues 15–59) with two potential coiled-coil regions (residues 20–34 and 41–65) that show high similarity across all HSBP homologs (Fig. 1; Supplemental Table S1). Particularly, nearly all the heptad-repeat residues in the trimerization region, AtHSBP residues 19 to 53 corresponding to HsHSBP1 residues 15 to 49 (Liu et al., 2009), are almost conserved (Fig. 1). The GST-AtHSBP39-86, containing a potential coiled-coil region, showed decreased AtHSFA1b DNA-binding activity in vitro (Fig. 7B). This C-terminal coiled-coil domain could contribute to the interaction with HSFs. The heptad-repeat regions form coiled coils, as long as similarly sized hydrophobic residues are located at the a and d positions to ensure packing of the interhelical space, even if the overall sequence diverges. Likewise, AtHSBP could be cross-interacted with itself (Fig. 6), which suggested that the HSBP helical region may function to self-associate and may interact with proteins other than the trimerization domain of HSFs in the cell.
HSBPs exist as two isoforms in monocots but only one copy in dicots. Maize EMP2 required for shoot development has been demonstrated by Fu and Scanlon (2004); embryogenesis in emp2 mutant kernels is severely retarded at the time, 12 d after pollination, well before maize kernels become competent to invoke the HSR (Fu et al., 2002). These results suggested that EMP2 performs an important developmental function in embryogenesis; in addition, it serves as a positive regulator of HSR attenuation in seeds. We found that the expression of AtHSBP in flowers and siliques was higher than that in other tissues (Fig. 3B); meanwhile, AtHSBP-knockout lines also showed seed abortion (Fig. 3, C and D), which suggested that the defective AtHSBP may lead unattenuated HSR to trigger embryo abortion, similar to emp2 mutant kernels (Fu et al., 2002). However, which embryonic stage was affected to result in seed abortion of AtHSBP-knockout lines remains unclear. Besides, the expression of heat-inducible AtHSBP (Fig. 3A) was similar to that of ZmHSBP2 under thermal stress (Fu and Scanlon, 2004), and the roles of EMP2 and ZmHSBP2 in HSR were suggested to be nonredundant (Fu et al., 2006). These findings may support that one AtHSBP confers multiple functions required for embryo viability and regulation of the HSR.
HsHSBP1 has been demonstrated to be a nucleus-localized protein, regardless of HS (Satyal et al., 1998). EMP2 mainly localizes within the nucleus, and the signals are also detected in the cytoplasm of embryonic cells; nevertheless, the localization and function of ZmHSBP2 are still unclear (Fu and Scanlon, 2004). We found AtHSBP-GFP localized in the cytoplasm, a finding different from previous reports for maize and human (Satyal et al., 1998; Fu and Scanlon, 2004). Although AtHSBP was detected in the nucleus during a 1-h recovery from HS, the localization of the AtHSBP-S35A mutant and GUS-GFP control were unaffected by thermal stress (Fig. 5; Supplemental Fig. S2). These results demonstrate that the translocation of AtHSBP-GFP indeed depends on thermal stress. However, the nucleus-localized AtHSBP signals were undetectable during a 2-h recovery from HS (Fig. 5A). The cytoplasm showed highly expressed AtHSBP-GFP, so judging whether AtHSBP-GFP was relocated to the cytoplasm or decreased in level by a degradation process during a 2-h recovery from HS was difficult. Above all, the nuclear localization of AtHSBP occurred during the recovery from HS, which was associated with the attenuation of HSR.
As compared with AtHSBP-ox1, AtHSBP-ox2 showed approximately 2-fold higher AtHSBP protein accumulation (Fig. 2B), but the seedling survival rate in response to HS showed no statistically significant differences between them (Table II). The highly cytosol-expressed AtHSBP-GFP showed partial but not complete translocation to the nucleus during the HSR. The nucleus-localized signal of AtHSBP-GFP expressed in Athsbp-1 protoplasts also showed a similar intensity, as shown in Figure 5A during HSR (Supplemental Fig. S3, H1R1). Studies of HsHSBP1 structure demonstrate that HsHSBP1 has buried its polar side chain in the hydrophobic interior of the helix bundle; however, an exception, residue Ser-31, occurs abnormally at the a position of the heptad repeat, which implies that the residue Ser-31 may have important functions (Liu et al., 2009). We found the localization of AtHSBP-S35A mutant to be unaffected by HS (Fig. 5B), which suggested that AtHSBP Ser-35 is important for its function. These findings may imply that (1) a limited amount of the nucleus-localized AtHSBP is required for its function, and (2) the translocation of AtHSBP (without nuclear localization signal) may be mediated through HS-related factors that function during the attenuation of HSR. The S35A mutation may result in a loss of interacting ability, which is required for its nuclear localization, but additional studies are necessary to address this suggestion.
In animals, current research suggests that the inactive HSF1 monomer is in the cytoplasm, interacting with HSP70 chaperone in the control state. Following the activation of HSR, the nucleus-localized and activated HSF1 trimer acquires DNA-binding capacity and induces HSP expression. On the recovery of HS, HSBP1 and HSP70 bind directly to HSF1, which HSF1 dissociates into monomers and then returns to the cytoplasm (for review, see Wu, 1995; Morimoto, 1998; Satyal et al., 1998; Pirkkala et al., 2001). We confirmed that AtHSBP interacts with AtHSFs (Fig. 6) and decreases the HSF DNA-binding ability (Fig. 7B), and these findings were similar to HsHSBP1 functions in humans (Satyal et al., 1998). In plants, interactions of AtHSP70s and AtHSFA1a are demonstrated by yeast two-hybrid assay (Kim and Schöffl, 2002). HSP70 antisense assay reveals the requirement of an extended time for inactivation of HSF transcriptional activity, a longer shutoff time, which suggests that HSP70 family proteins may mediate HSF transcriptional activity (Lee and Schöffl, 1996). Although the defective AtHSBP led to unattenuated HSP expression during the recovery from HS (Fig. 8A), similar to the HSP70 antisense lines, whether AtHSBP coordinates with HSP70 in the attenuation of HSR in Arabidopsis remains unclear.
Modified protoplast two-hybrid assay investigates interacting proteins invoked in transcriptional regulation. HsHSBP1 is suggested to directly bind to the HSF1 trimerization domain for inactivation of HSF1 activity (Satyal et al., 1998). We found that AtHSBP significantly interacted with AtHSFA1a, AtHSFA1b, and AtHSFA2, although the interaction was weaker than that of positive controls (Fig. 6). We confirmed that AtHSBP decreased AtHSFA1b DNA-binding capacity in vitro (Fig. 7B); therefore, the interaction of AtHSBP and AtHSFs may be weak and transient in vivo. Furthermore, a high protein molar ratio of AtHSBP to AtHSFA1b (approximately 100-fold GST-AtHSBP higher than MBP-AtHSFA1b) did not totally reduce AtHSFA1b DNA-binding capacity in vitro, and these results also could not accurately reflect the AtHSBP activity in both the transcript and protein levels relative to AtHSFs in vivo, which suggested that AtHSBP may require other factors, such as HSP70, as reported in animals, to coordinate its activity.
In summary, we propose that the cytosol-localized AtHSBP translocates to the nucleus during recovery from HS (Fig. 5A, H1R1; localization data), where it negatively affects AtHSF DNA-binding capacity (Fig. 7B; EMSA data) and decreases the programmed HSP expression (Fig. 8; western-blot analysis) to attenuate HSR. In concert, AtHSBP expression was slightly up-regulated by HS (approximately 1.5-fold higher) and peaked during the 1-h recovery period (approximately 3-fold higher); the expression then returned to the basal level after a 4-h recovery from HS (Fig. 3A), which was in agreement with the AtHSBP behavior during the recovery from HS.
Numerous questions remain regarding the roles of the molecular chaperones HSP, HSF, and HSBP in regulating HSR. All of the HSF-interacting proteins reported to date act as negative regulators for HSF transcriptional activity (for review, see Morimoto, 1998; Pirkkala et al., 2001). The consequence of forming a molecular complex requires that HSF be kept in a state that can be readily activated, but how and when the proteins interact with HSF remain to be answered. Further study could evaluate any positive regulators of HSF or whether the fast activation mechanism of HSF is an intrinsic property that has been evolutionarily conserved to allow for the rapid activation of HSF with stress stimuli.
MATERIALS AND METHODS
Plant Material, Transformation, and Growth Conditions
Arabidopsis (Arabidopsis thaliana Columbia ecotype) was used as the wild type. AtHSBP (At4g15802) T-DNA insertion lines, SALK_081104 (Athsbp-1) and SALK_046465 (Athsbp-2), were obtained from the Arabidopsis Biological Resource Center (Ohio State University). For overexpression of AtHSBP, cDNA with NcoI sites was amplified and cloned into pPE1000 to confer the HA epitope (Hancock et al., 1997) and then subcloned into pCambia3300 harboring the Basta-resistant gene (bar; CAMBIA) through the SacI site. For complementation of the Athsbp-1 line, the wild-type copy of AtHSBP containing a 1-kb upstream promoter was amplified and cloned into pPE1000 by XhoI and NcoI and then subcloned into pCambia3300 through the SacI site. Agrobacterium tumefaciens-mediated genetic transformation of Arabidopsis was done by the floral dip method (Clough and Bent, 1998). Arabidopsis plants were grown in walk-in chambers at 24°C with 16 h of light at an intensity of 60 to 100 μmol m−2 s−1.
Thermotolerance Tests
Thermotolerance tests were performed according to Charng et al. (2006). Seeds were grown at 24°C with 16 h light for 0, 3, or 6 d before HS treatment. The plate was sealed with plastic electric tape and submerged in a water bath at the indicated temperature. Plates were directly heated at 45°C for 220 min or at 44°C for 20 min for testing BT in seed germination or hypocotyl elongation, respectively. For AT testing, plates were preheated at 37°C for 60 min and then recovered at 24°C for 120 min before 44°C HS for 190 min. Germinated seeds after 4 d and healthy-growing seedlings after 7 to 8 d from the end of 44°C HS treatment were counted. The hypocotyl elongation assay was conducted as described by Hong and Vierling (2000).
Transient Expression in Arabidopsis Mesophyll Protoplasts
Arabidopsis protoplast isolation was performed as described by Yoo et al. (2007). The S35A point mutation was performed by the Megapriming method (Landt et al., 1990). The cDNA clones of AtHSBP or AtHSBP-S35A were constructed into pRTL2-GFP driven by a 35S promoter (von Arnim et al., 1998). The GUS-GFP control was from pCambia1303 (CAMBIA). The constructs were transfected into protoplasts and incubated for 16 to 24 h. The GFP signals were observed by fluorescence confocal microscopy (TCS SP5 AOBS; Leica). The confocal planes were set to cover the nucleus, and the optical sectioning thickness was 1 μm. Each sequential section contained the nucleus, chloroplast, and cytoplasm.
Protoplast Two-Hybrid and GUS Activity Assays
The protoplast two-hybrid assay was performed as described by Ehlert et al. (2006). For the effector constructions, cDNA of AtHSFA1a, AtHSFA1b, AtHSFA2, AtHSBP, or AtHSBP-S35A was constructed into p35S-GBD-GW or p35S-GAD-GW to confer the GAL4 DNA binding domain or activation domain, respectively. The transactivation assay was cotransfected with a mixture of 25 μg of effector, 10 μg of PGAL4-UAS4:GUS reporter, and 5 μg of MTC301 normalization plasmid. The samples were incubated for 40 h, and then both the GUS and luciferase activities were measured. Luciferase luminescence was measured by the luciferase assay buffer according to the technical manual (Promega). GUS activity assay was performed according to Yoo et al. (2007). The GUS activities of all samples were normalized against the luciferase internal control.
Recombinant Protein Purification and EMSA
AtHSBP and AtHSBP39-86 were cloned into pGEX-6P-1 (Amersham), and AtHSFA1b was cloned into pMAL-p2 (New England Biolabs). The resulting plasmids were transformed into Escherichia coli BL21 (DE3) cells. The GST- and MBP-recombinant proteins were purified with glutathione resin (Sigma) and amylose resin (New England Biolabs), respectively, according to the technical manual. The oligonucleotides of HSE-Fw and HSE-Rv were annealed and then labeled with [α-32P]dCTP as an HSE probe. For EMSA, the recombinant proteins were incubated at 37°C for 1 h before adding 30 ng of [32P]dCTP-labeled HSE probe and further incubated for 30 min at room temperature. The reaction mixtures were analyzed by 6.5% (w/v) native-PAGE in 0.5× Tris-borate/EDTA buffer (pH 8.3). The dried gel was exposed to a storage phosphor screen and scanned with Typhoon 9400 (Amersham).
Real-Time Quantitative PCR
Total RNA was prepared with Trizol reagent (Invitrogen) and the TURBO DNA-free Kit (Applied Biosystems). cDNA synthesis was performed using high-capacity cDNA Reverse Transcription Kits (Applied Biosystems). Real-time PCR primers were designed by Primer3 software (http://frodo.wi.mit.edu). Real-time quantitative PCR results were analyzed by a MyiQTM thermocycler (Bio-Rad) with the iQ SYBR Green Supermix (Bio-Rad). The data were analyzed by iQ5 Optical System Software (Bio-Rad). The internal control for tissue-specific expression comparison (Fig. 3B) was AtPP2A (At1g13320; Czechowski et al., 2005), and AtACT2 (At3g18780) was used for heat treatments (Figs. 3A and 8A; Volkov et al., 2003; Charng et al., 2007).
Western-Blot Analysis
Proteins were extracted with the buffer containing 50 mm Tris-HCl (pH 8.0), 2% (w/v) SDS, 2% (v/v) β-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride. The proteins were quantified with the use of Bio-Rad protein assay reagent. Horseradish peroxidase was detected with the use of the WEST-ZOL Plus Western Blot Detection System (iNtRON Biotechnology). RPN8 (Yang et al., 2004) and Rubisco large subunit stained with Amido black were used as internal controls.
Primers and Oligomers
All the primers and oligomers used in this work are listed in Supplemental Table S2.
Sequence data from this article can be found in GenBank as follows: AtHSFA1a (Arabidopsis), NM_117884; AtHSFA1b (Arabidopsis), NM_180501; AtHSFA2 (Arabidopsis), NM_001124916; LeHSBP1 (tomato), AW624356; ZmHSBP1 (maize), AAM15929; ZmHSBP2 (maize), AAR18070; OsHSBP1 (rice), AU075659; OsHSBP2 (rice), BE040146; HsHSBP1 (Homo sapiens), NP_001528; MmHSBP1 (Mus musculus), NP_077181; XtHSBP1 (Xenopus tropicalis), NP_001011422; DrHSBP1 (Danio rerio), AAH59566; CeHSBP1 (Caenorhabditis elegans), NP_502406.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Two independent transient expression experiments of AtHSBP-GFP in mesophyll protoplasts.
Supplemental Figure S2. Transient expression of a control construct, GUS-GFP, in mesophyll protoplasts.
Supplemental Figure S3. Transient expression of AtHSBP-GFP in Athsbp-1 mutant mesophyll protoplasts.
Supplemental Figure S4. AtHSBP and HSP gene expression were analyzed by real-time quantitative PCR and normalized by the internal control, EF1α.
Supplemental Table S1. Comparison (percent identity, percent similarity) of the amino acid sequences of the α-helix region for AtHSBP and other organisms.
Supplemental Table S2. Primers for genotyping, cloning, mutation, real-time quantitative PCR, and EMSA.
Supplementary Material
Acknowledgments
We thank Drs. Hsu-Liang Hsieh and Su-May Yu for providing pRTL2-GFP and MTC301 luciferase plasmids, respectively. We are also grateful to Drs. Chu-Yung Lin, Hong-Yong Fu, and Yee-Yung Charng for supplying antibodies against HSP70, RPN8, and HSP101, respectively. Dr. Yee-Yung Charng is thanked for critically reading the manuscript and commenting, and the TC5 Bio-Image Center at National Taiwan University is acknowledged for confocal laser scanning microscopy.
References
- Busch W, Wunderlich M, Schöffl F. (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS. (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Mur LA, Wood JE, Scott IM. (2004) Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J 38: 432–447 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W. (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46: 890–900 [DOI] [PubMed] [Google Scholar]
- Fu S, Meeley R, Scanlon MJ. (2002) Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Rogowsky P, Nover L, Scanlon MJ. (2006) The maize heat shock factor-binding protein paralogs EMP2 and HSBP2 interact non-redundantly with specific heat shock factors. Planta 224: 42–52 [DOI] [PubMed] [Google Scholar]
- Fu S, Scanlon MJ. (2004) Clonal mosaic analysis of EMPTY PERICARP2 reveals nonredundant functions of the duplicated HEAT SHOCK FACTOR BINDING PROTEINs during maize shoot development. Genetics 167: 1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Chen S, Liu K, Liu Y, Ni L, Zhang K, Zhang L. (2008) Isolation of heat shock factor HsfA1a-binding sites in vivo revealed variations of heat shock elements in Arabidopsis thaliana. Plant Cell Physiol 49: 1306–1315 [DOI] [PubMed] [Google Scholar]
- Hancock KR, Phillips LD, White DW, Ealing PM. (1997) pPE1000: a versatile vector for the expression of epitope-tagged foreign proteins in transgenic plants. Biotechniques 22: 861–862, 865 [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. (1994) Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263: 224–227 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Kim BH, Schöffl F. (2002) Interaction between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J Exp Bot 53: 371–375 [DOI] [PubMed] [Google Scholar]
- Landt O, Grunert HP, Hahn U. (1990) A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96: 125–128 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Schöffl F. (1996) An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Mol Gen Genet 252: 11–19 [DOI] [PubMed] [Google Scholar]
- Li M, Doll J, Weckermann K, Oecking C, Berendzen KW, Schöffl F. (2010) Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur J Cell Biol 89: 126–132 [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. (1986) The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. (1988) The heat-shock proteins. Annu Rev Genet 22: 631–677 [DOI] [PubMed] [Google Scholar]
- Liu X, Xu L, Liu Y, Tong X, Zhu G, Zhang XC, Li X, Rao Z. (2009) Crystal structure of the hexamer of human heat shock factor binding protein 1. Proteins 75: 1–11 [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Miroshnichenko S, Tripp J, Nieden U, Neumann D, Conrad U, Manteuffel R. (2005) Immunomodulation of function of small heat shock proteins prevents their assembly into heat stress granules and results in cell death at sublethal temperatures. Plant J 41: 269–281 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. (1993) Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259: 230–234 [DOI] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. (1998) Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev 12: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A. (1998) Regulation of the heat-shock response. Plant Physiol 117: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. (2001) Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 126: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LJ, McFall SM, Huang K, Demeler B, Fox SG, Brubaker K, Radhakrishnan I, Morimoto RI. (2002) Structure-function analysis of the heat shock factor-binding protein reveals a protein composed solely of a highly conserved and dynamic coiled-coil trimerization domain. J Biol Chem 277: 735–745 [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Schöffl F. (2003) Heat-stress-dependency and developmental modulation of gene expression: the potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J Exp Bot 54: 2343–2349 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Wu C. (1995) Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD. (2004) Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279: 6401–6413 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lohmann C, Prändl R, Schöffl F. (2003) Heat stress-dependent DNA binding of Arabidopsis heat shock transcription factor HSF1 to heat shock gene promoters in Arabidopsis suspension culture cells in vivo. Biol Chem 384: 959–963 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.