Abstract
The human Paf1 complex (Paf1C) subunit Parafibromin assists in mediating output from the Wingless/Int signaling pathway, and dysfunction of the encoding gene HRPT2 conditions specific cancer-related disease phenotypes. Here, we characterize the organismal and molecular roles of PLANT HOMOLOGOUS TO PARAFIBROMIN (PHP), the Arabidopsis (Arabidopsis thaliana) homolog of Parafibromin. PHP resides in an approximately 670-kD protein complex in nuclear extracts, and physically interacts with other known Paf1C-related proteins in vivo. In striking contrast to the developmental pleiotropy conferred by mutation in other plant Paf1C component genes in Arabidopsis, loss of PHP specifically conditioned accelerated phase transition from vegetative growth to flowering and resulted in misregulation of a very limited subset of genes that included the flowering repressor FLOWERING LOCUS C. Those genes targeted by PHP were distinguished from the bulk of Arabidopsis genes and other plant Paf1C targets by strong enrichment for trimethylation of lysine-27 on histone H3 (H3K27me3) within chromatin. These findings suggest that PHP is a component of a plant Paf1C protein in Arabidopsis, but has a more specialized role in modulating expression of a subset of Paf1C targets.
The Paf1 complex (Paf1C) has been best characterized in budding yeast (Saccharomyces cerevisiae) as a transcriptional cofactor participating in initiation and elongation, with various specific roles including mediating ubiquitination of histone H2B by Rad6/Bre1, promoting interaction of the SET1 and SET2 histone methyltransferases with chromatin at active genes, and assisting in pre-mRNA processing through 3′ end formation (Krogan et al., 2003a, 2003b; Ng et al., 2003; Penheiter et al., 2005; Sheldon et al., 2005). Although Paf1C proteins interact with a variety of active genes in yeast, Paf1C is required for correct expression of only a subset of genes (Chang et al., 1999; Penheiter et al., 2005). Phenotypic analysis of deletion mutants for the five core components of Paf1C—Paf1, Ctr9, Leo1, Rtf1, and Cdc73—suggests that Paf1 and Ctr9 have important but identical function, while the remaining three factors may contribute variable and limited activities to Paf1C (Betz et al., 2002).
In humans, homologs of Paf1, Ctr9, Leo1, and Cdc73 participate in a Paf1C-like complex (hPAF) that similarly interacts with PolII at transcriptionally active genes (Rozenblatt-Rosen et al., 2005; Yart et al., 2005; Zhu et al., 2005). hPAF additionally contains hSki8, a protein distantly related to yeast Ski8, required for 3′-5′ mRNA degradation by the exosome (Anderson and Parker, 1998) and meiotic recombination (Arora et al., 2004). Loss of human Ctr9 was found to attenuate interleukin-6 (IL-6)-responsive gene expression mediated through the transcription factor STAT3 (Youn et al., 2007). The human Cdc73 homolog, called Parafibromin, is required for correct 3′ transcript processing for at least some genes, likely through direct interaction with the CPSF-CstF RNA processing factor (Rozenblatt-Rosen et al., 2009). Disruption of HRPT2 gene encoding Parafibromin is associated with hyperparathyroidism-jaw tumor syndrome (Carpten et al., 2002), an autosomal dominant disorder typified by adenoma of the parathyroid gland, and leading to hyperparathyroidism, ossifying fibroma of the jaw, and a variety of renal and endocrine tumors (Jackson et al., 1990; Haven et al., 2000). Parafibromin is a probable tumor suppressor as tumor occurrence is frequently associated with somatic loss of heterozygosity at the HRPT2 locus, or inactivating HRPT2 mutation in familial hyperparathyroidism-jaw tumor syndrome (Carpten et al., 2002). Consistent with this role as a tumor suppressor, knockdown of HRPT2 in human cells derepressed the c-myc proto-oncogene and led to cell proliferation (Lin et al., 2008). In addition, expression of cyclin D1 was suppressed in Parafibromin-transfected cells, and overexpressed in parathyroid carcinomas lacking Parafibromin immunoreactivity (Woodard et al., 2005). Whether this resulted from direct regulation of the cyclin D1 gene by Parafibromin/hPAF or is an indirect effect of disruption of the cell cycle is unknown. Parafibromin is also important for transcriptional output of the Wingless/Int (Wnt) pathway through its direct physical interaction with nuclear β-catenin, a role that is conserved with its Drosophila homolog Hyrax (Mosimann et al., 2006).
In the reference plant Arabidopsis (Arabidopsis thaliana), the VIP4, VIP5, VIP6 (also called ELF8), and ELF7 genes encode obvious homologs of Leo1, Rtf1, Ctr9, and Paf1, respectively, whereas VIP3 encodes a protein closely related to hSki8 (Zhang and van Nocker, 2002; Zhang et al., 2003; He et al., 2004; Oh et al., 2004). Strong mutations in each of these genes condition similar pleiotropic developmental phenotypes characterized by weak growth, defects in leaf and floral development, and acceleration of the natural phase transition from vegetative growth to flowering (Zhang and van Nocker, 2002; Zhang et al., 2003; He et al., 2004; Oh et al., 2004). Transcriptional profiling of plants dysfunctional for VIP3, VIP5, or VIP6 revealed substantial overlap of misexpressed genes, and at least VIP3, VIP4, and VIP6 physically interact in vivo, suggesting that these proteins define a plant Paf1C (Oh et al., 2004, 2008). Unlike yeast Paf1C components, Arabidopsis Paf1C proteins are not required to maintain global levels of H3K4 or H3K36 methylation, but directly or indirectly promote these modifications at transcriptionally active genes (He et al., 2004; Oh et al., 2004, 2008). In addition, at least VIP3 is required to maintain H3 density within highly expressed genes (Oh et al., 2008). Plant Paf1C promotes expression of the MADS-box gene FLOWERING LOCUS C (FLC), which acts as an integrator of several flowering regulatory pathways. FLC is subject to epigenetic silencing through a mechanism related to the Polycomb-group (PcG) PRC2 complex, and this is associated with accumulation of H3K27me3 within FLC chromatin (Bäurle and Dean, 2006). Loss of VIP3 also results in hypermethylation of H3K27 within FLC chromatin, suggesting that VIP3, and perhaps Paf1C generally, antagonizes PcG-mediated silencing (Oh et al., 2008).
In this study, we characterized the Arabidopsis homolog of the remaining conserved subunit of Paf1C, Parafibromin/Cdc73, and present evidence that this Arabidopsis protein participates in a plant Paf1C but has a more specialized role in modulating expression of a subset of Paf1C targets.
RESULTS
PLANT HOMOLOGOUS TO PARAFIBROMIN Represents the Plant Homolog of Parafibromin
Analysis of the predicted proteome and whole-genome conceptual translation revealed that Arabidopsis contains a single predicted gene encoding a Parafibromin-related protein. A catalog of ESTs showed that this gene, At3g22590, produces one major polyadenylated transcript encoding for a 415-amino acid protein sharing approximately 24% identity and approximately 39% similarity with Parafibromin (Fig. 1; Supplemental Fig. S1). Based on this homology, we designated this gene PLANT HOMOLOGOUS TO PARAFIBROMIN (PHP). The PHP protein shares sequence homology with Parafibromin and Cdc73 within three distinct domains, with the highest level of conservation in an approximately 200-amino acid, carboxyl-terminal segment that has been characterized in Parafibromin as a site for interaction with PolII and hPAF components (Yart et al., 2005) and that is truncated by several disease-associated HRPT2 alleles (Carpten et al., 2002). PHP is lacking amino-terminal regions that are strongly conserved among metazoan Parafibromin homologs, consistent with the definition of this region as devoted to interaction with β-catenin (Mosimann et al., 2006), and the absence of a canonical Wnt signaling pathway in plants. Analysis of public microarray data showed that PHP is expressed ubiquitously, but preferentially in domains enriched for mitotically active tissues, including the shoot, root, and floral apices (data not shown).
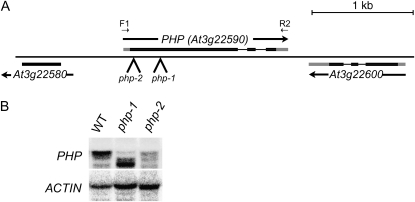
Figure 1.
Structure and expression of Arabidopsis PHP. A, Region of chromosome III containing the PHP locus. Exons are shown as black (translated region) or gray (untranslated region) boxes. The positions of the insertions corresponding to the php-1 and php-2 alleles are indicated. Annealing positions of oligonucleotide primers are shown. B, RNA gel-blot analysis of PHP RNAs in wild-type (WT), php-1, and php-2 seedlings. The entire transcribed region of PHP (F1/R2) was used as probe.
PHP Interacts Physically with Other Paf1C Components in Vivo and Participates in an Approximately 670-kD Protein Complex
To evaluate potential physical interaction between PHP protein and other Arabidopsis Paf1C subunits, we carried out coimmunoprecipitation experiments (Fig. 2A). A genomic copy of PHP was engineered to be expressed with a carboxyl-terminal FLAG peptide and was introduced into the php-1 background. Protein extracts from lysates of purified nuclei were subjected to immunoprecipitation employing anti-FLAG antibodies, and immunoprecipitates were analyzed with antibodies generated against VIP3, VIP4, and VIP6. All antibodies readily detected proteins of the anticipated size in immunoprecipitates from PHP-FLAG plants, but not from wild-type nontransgenic plants (Fig. 2A). This suggests that PHP physically associates with each of these proteins in plant nuclei.
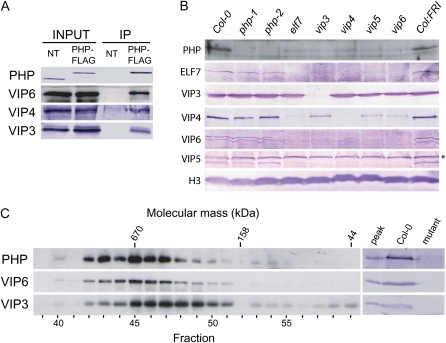
Figure 2.
Characterization of PHP and PHP-associated proteins. A, Nuclear extracts prepared from nontransgenic wild-type (NT) and php-1:PHP-FLAG plants were subjected to immunoprecipitation using anti-FLAG antibody. Immunoprecipitates (IP) were analyzed by SDS-PAGE and immunoblotting with antisera generated against the indicated proteins. B, Total cellular proteins were prepared from wild type (Col-0 or Col:FRI as indicated), php mutants, or mutants for the plant Paf1C components ELF7, VIP3, VIP4, VIP5, and VIP6 as indicated, and were analyzed by SDS-PAGE and immunoblotting using antisera raised against the indicated proteins. An unrelated, immunoreactive protein species detected by anti-VIP5 antisera is indicated by an asterisk (*). C, Nuclear extracts were subjected to gel filtration chromatography using a Superose 6 column with an effective fractionation range of 5 to 5,000 kD, and fractions were analyzed by SDS-PAGE and immunoblotting using antisera raised against the indicated proteins. Elution positions of molecular mass standards (kD) and chromatographic fraction numbers are indicated at the top and bottom, respectively. In the sections at right, the peak PHP fraction (no. 45) was analyzed together with total cellular extracts from wild type (Col-0) and php, vip3, or vip6 mutants to demonstrate specificity of the antibodies. [See online article for color version of this figure.]
We previously discovered that VIP6 is required for stability of Paf1C (Oh et al., 2004). To gain additional insight into the relationship between PHP and other Paf1C subunits, we analyzed the effect of loss of ELF7, VIP3, VIP4, VIP5, and VIP6 on PHP protein expression as measured in total cell lysates (Fig. 2B). We found that PHP protein accumulated to substantially lower levels in each of these mutant backgrounds, suggesting that Paf1C integrity is important for PHP stability. In contrast, loss of PHP in either php-1 or php-2 had no discernable effect on intracellular levels of ELF7, VIP3, VIP4, VIP5, or VIP6 proteins (Fig. 2B).
To further characterize PHP protein interactions, we used gel filtration chromatography to determine the in vivo mass of PHP and associated proteins (Fig. 2C). Nuclear extracts from PHP-FLAG plants were fractionated, and the elution profile of PHP-FLAG, as well as VIP6 and VIP3, was monitored by immmunoblotting. PHP coeluted precisely with VIP6 over a broad range, with peak elution near a 670-kD mass marker (Fig. 2C). We did not detect PHP in late-eluting fractions containing low-molecular-mass proteins, indicating that PHP is not abundant as a monomer in nuclei. VIP3 also coeluted over this range, but was also strongly enriched in fractions corresponding to 400- to 670-kD proteins, and was also observed in very-low-mass fractions (Fig. 2C). Considered together with the results of our coimmunoprecipitation experiments, this result suggests that VIP3 additionally exists in a smaller protein complex and also possibly as a monomer.
Dysfunction of PHP Leads to Alteration of Flowering
To analyze the cellular and organismal function of PHP, we documented phenotypic and molecular defects associated with PHP mutation. We identified two Arabidopsis lines, designated php-1 and php-2, carrying insertion mutations in the 5′ region of the first exon of PHP (Fig. 1A). We were unable to detect full-length PHP mRNAs in either line by reverse transcription-PCR. However, RNA gel blotting using most of the PHP transcribed region as a probe showed that a truncated PHP RNA species accumulated in php-1 plants (Fig. 1B). Immunoblot analysis using antibodies directed against the PHP protein detected a single protein in wild-type plants but did not detect a similar sized or smaller protein in either php-1 or php-2 plants (Fig. 2B; data not shown).
Under long-day, photoperiodically inductive growth conditions, early development of php plants was indistinguishable from that of wild-type plants. However, php mutants exhibited an abbreviated vegetative phase and accelerated onset of flowering, producing significantly fewer vegetative nodes (leaves) than wild-type plants. This phenotypic defect was fully rescued by the transgenic expression of the FLAG epitope-modified, genomic copy of PHP (Fig. 3A). Extended-light photoperiods, which promote the phase transition to flowering in wild-type Arabidopsis, was similarly effective in promoting flowering of php-2 plants, indicating that PHP does not play an important role in the well-characterized photoperiodic induction mechanisms of flowering. Constitutive, antisense expression of PHP RNA in transgenic Arabidopsis accelerated flowering, but did not noticeably affect other aspects of development in the 100 independent lines observed (data not shown). Under a variety of conditions, plants homozygous for the php-2 allele flowered earlier than php-1 homozygous plants (Fig. 3B). Considering also the substantial accumulation of truncated PHP RNA in php-1 plants, we conclude that the php-1 allele is hypomorphic.
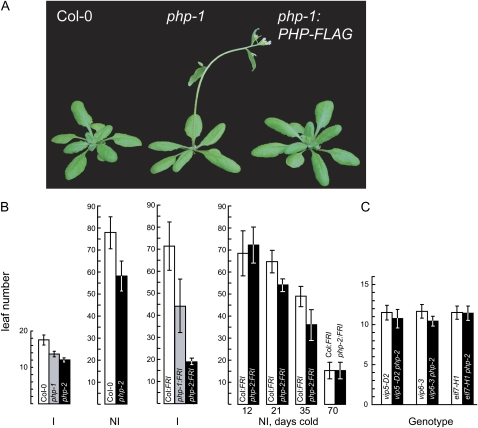
Figure 3.
Flowering phenotype of php plants. A, Wild-type ecotype Col-0 (left), php-1 (center), and php-1 expressing a PHP-FLAG transgene (right) after 4 weeks of growth. B, Summary of total leaves formed on the main shoot for wild-type ecotype Col-0, Col-0 carrying an introgressed, dominant FRI allele (Col:FRI), and php mutants. Plants were grown under flowering-inductive (16 h light/8 h dark; I) or noninductive (8 h light, 16 h dark; NI) photoperiods. To evaluate the vernalization response, germinating seeds were maintained in the cold for 12, 21, 35, or 70 d. Values represent the mean and sd for at least 12 plants. C, Analysis of flowering in double mutants. Plants were grown under inductive photoperiods. [See online article for color version of this figure.]
The plant Paf1C complex subunit genes VIP3, VIP4, VIP5, VIP6, and ELF7 also repress the phase transition from vegetative growth to flowering, at least in part by promoting expression of the MADS-box flowering inhibitor gene FLC (Zhang and van Nocker, 2002; Zhang et al., 2003; He et al., 2004). Strong FLC expression is additionally dependent on activity of the FRIGIDA (FRI) gene, which is dysfunctional in the common laboratory strain Columbia-0 (Col-0). To assess the phenotypic consequences of loss of PHP activity in a genetic background that expresses FLC to high levels, we introduced the php alleles into a Col-0 strain carrying introgressed, functional FRI alleles from the wild ecotype San Feliu-2. Whereas this Col:FRI strain flowered only after producing approximately 70 leaves in photoperiodically inductive conditions, php mutants in this genetic background transitioned to flowering after producing approximately 45 (php-1) or approximately 20 (php-2) leaves. Extended cold exposure at the germinating seed stage had a promotive effect on flowering in php plants that was similar to that seen for wild type, suggesting that PHP does not act directly in the documented cold-promotion flowering pathway (Fig. 3B). The effect of the php-2 mutation to largely suppress the late flowering conferred by FRI suggested that, as documented for the characterized VIP genes, PHP is required for efficient expression of FLC. Indeed, we found that FLC mRNA expression was substantially reduced in a php-2 background (see Fig. 4 below). Unlike the characterized VIP genes, loss of PHP in the Col:FRI background did not result in substantial loss of expression of MAF1, MAF2, or MAF3, three additional FLC-related genes that may similarly act to repress flowering (Ratcliffe et al., 2001, 2003; data not shown). To investigate epistatic relationships between PHP and other Paf1C component genes, we analyzed phenotypes of mutants carrying php-2 in combination with strong mutations in VIP5, VIP6, or ELF7. Loss of PHP did not strongly enhance either the early flowering phenotype or developmental pleiotropy of these mutants, suggesting that the activity of PHP does not extend beyond that of these other genes (Fig. 3C; data not shown).
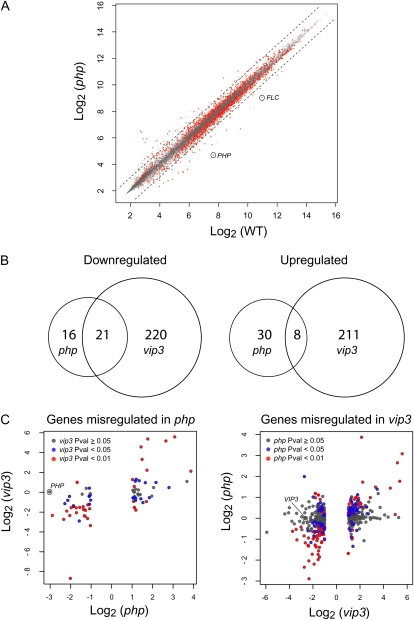
Figure 4.
Gene expression profiling of php mutants. A, Scatter plot of wild type (WT; Col:FRI) versus php-2:FRI datasets composed with log-transformed signal intensities. Genes with P value <0.01 are shown in red, and 2-fold increase or decrease is indicated with a dotted line. Signal positions for FLC and PHP are circled. B, Venn diagram indicating the number and overlap of up-regulated or down-regulated genes in php-2:FRI and vip3:FRI mutants. C, Genes that were determined to be misexpressed in php-2:FRI mutants relative to wild-type plants, based on a criteria of >2-fold change and P < 0.01, were analyzed for misregulation in vip3:FRI mutants, as previously determined (left section; Oh et al., 2008). Genes misexpressed in the vip3:FRI mutant were analyzed for misregulation in the php-2:FRI mutant (right section). In each section, the P-value statistic for expression change is indicated as shown in the key. The x and y axes indicate expression change in each mutant relative to wild type. Mutants and wild type were in the Col:FRI background. Microarray data were derived from two independent biological replicates.
PHP Participates in Regulation of a Small Number of Genes Strongly Enriched in H3K27me3
To identify cryptic genetic functions of PHP, we compared transcriptional profiles of wild-type and php-2 seedlings grown under long-day photoperiods (Fig. 4). Consistent with the lack of obvious phenotypic defects in php backgrounds, we identified only a very small subset of genes that were misregulated in the php-2 mutant, relative to wild-type plants: 37 down-regulated genes and 38 up-regulated genes (Fig. 4; Supplemental Table S1). These subsets substantially overlapped those identified as misregulated in a vip3 mutant analyzed at an equivalent developmental stage (Fig. 4B; P < 1.4E-32 and P < 2.4E-09, respectively; Fisher's Exact test). Nearly all of the genes down-regulated in php-2 were also down-regulated in vip3 (Fig. 4C). However, genes up-regulated in the php mutant were more heterogeneous with respect to dependence on VIP3. Based on current annotation, none of the misregulated genes identified were obviously involved in cell division (Supplemental Table S1), suggesting that, unlike HRPT2/Parafibromin, PHP activity is not linked with cell cycle regulation. Analysis of publicly available microarray data for PHP-regulated genes did not identify obvious commonalities among expression patterns. However, both up-regulated and down-regulated genes were characterized by generally low levels of expression throughout most of the plant with a high degree of spatial regulation (Supplemental Fig. S2).
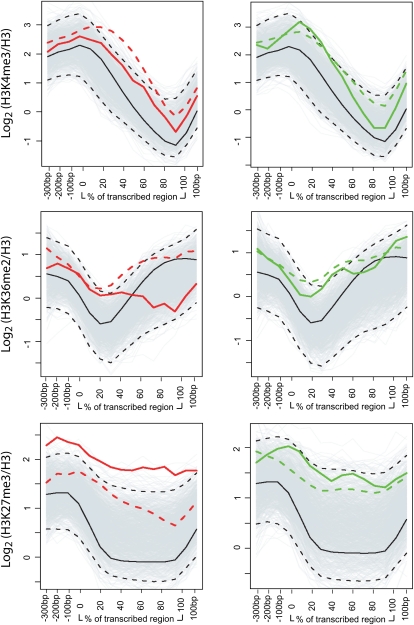
We previously showed that genes subject to regulation by VIP3 were distinguished by high levels of H3K27me3 across the extent of the transcribed region, in combination with strong enrichment for H3K4me3 and H3K36me2 (Oh et al., 2008). Analysis of the chromatin signatures of genes misregulated in php-2 relative to wild type revealed that PHP targets were enriched for H3K4me3 or H3K36me2 relative to the genomic average, but that this enrichment was weaker than that observed for vip3 (Fig. 5). In contrast, PHP targets were enriched for H3K27me3 to levels exceeding those of VIP3 (Fig. 5). This was especially striking for those genes that were up-regulated in php relative to wild type. Thus, the subset of PHP-regulated genes appears to be distinguished from the bulk of Paf1C targets by very high enrichment for H3K27me3.
Figure 5.
Histone methylation profiles within PHP-targeted genes. Mean genic positional signals for H3K4me3, H3K36me2, and H3K27me3 within the subsets of genes up-regulated (solid red) or down-regulated (solid green) in the php-2 mutant were calculated from the data set reported previously (Oh et al., 2008) and are shown relative to the genomic average (black). Signals are depicted across the promoter, transcribed, and 3′ regions as indicated on the x axes. The 95th percentile confidence intervals (dashed black lines) were determined for the mean positional signals within 1,000 randomly resampled gene sets (background gray lines). Mean positional signals for those genes up-regulated or down-regulated in the vip3 mutant are indicated as dashed red or dashed green lines, respectively, as previously determined (Oh et al., 2008).
PHP Is Required for Histone Modifications on FLC Chromatin
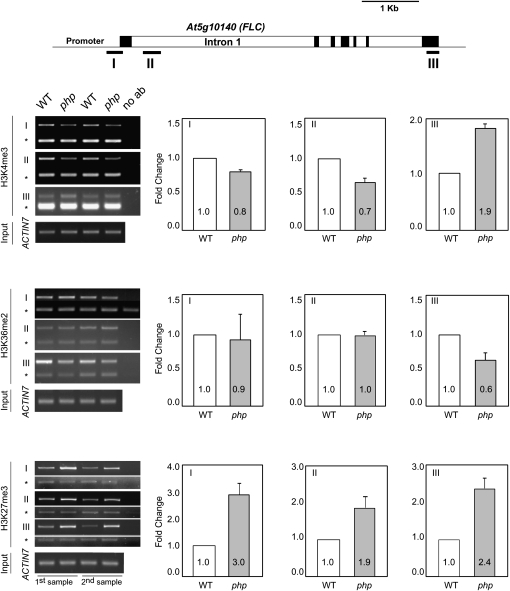
FLC expression is promoted by a mechanism associated with H3K4 and/or H3K36 methylation of FLC chromatin, and repressed through PcG activity associated with H3K27me3 (Dennis and Peacock, 2007). Recently, we showed that loss of the plant Paf1C gene VIP3 was associated with decreased enrichment for H3K4me3 within 5′ regions of the FLC gene concomitant with increased enrichment for H3K4me3 within FLC 3′ regions. In addition, H3K36me2 was decreased and H3K27me3 increased throughout the FLC gene in vip3 mutants (Oh et al., 2008). To determine if PHP might similarly participate in chromatin modifications at the FLC locus, we carried out chromatin immunoprecipitation (ChIP) with antibodies recognizing H3K4me3, H3K36me2, and H3K27me3 and amplified selected FLC genomic regions (Fig. 6). Compared with wild type, H3K4me3 in the strong php-2 background was decreased within the proximal promoter and 5′ segment of the first long intron, and substantially increased at a 3′ region. H3K36me2 was essentially unchanged within promoter and intronic regions, but significantly decreased at the 3′ end, whereas H3K27me3 was increased at all three regions tested. These changes in all three chromatin modifications due to loss of PHP are thus comparable to those observed with loss of VIP3, and further connect function of PHP with plant Paf1C.
Figure 6.
ChIP analysis of histone H3 methylations within FLC chromatin. ChIP was carried out using antibodies recognizing H3K4me3, H3K36me2, or H3K27me3 within a promoter segment (I), intronic region near the 5′ end (II), or 3′ region (III) of the FLC gene from wild-type (WT) Col-0 or php-2 plants. Band intensities from gel images were quantified and normalized based on those for ACTIN7 (lower band in each gel image marked with asterisk). ChIP analysis was performed twice using biologically independent samples and yielded essentially identical results. A depiction of the FLC gene, indicating the position of amplified segments and with exons depicted as black boxes, is shown above. No ab, No antibody control. Error bars show the sd from the mean.
DISCUSSION
The cellular and organismal functions of the conserved Paf1C transcriptional cofactor in higher eukaryotes have not been well studied. We sought to gain insight into potentially conserved functions of Parafibromin and Paf1C through analyses of the organismal and molecular activities of the homologous PHP protein from Arabidopsis. Here we show that, in vivo, PHP protein associates with the Paf1C-related proteins VIP3, VIP4, VIP6, and other unknown proteins in an approximately 670-kD complex. Our finding that VIP6, PHP, VIP4, and ELF7 proteins are destabilized in a vip5 mutant background, and that VIP5 is destabilized in vip3/4/6 and elf7 mutant backgrounds, also suggests that VIP5 is physically linked with these proteins. Our additional observation that ELF7 protein is lost in vip3/4/5/6 backgrounds, and that VIP4/5/6 are lost in an elf7 background, implicates ELF7 as a Paf1C subunit. In contrast, loss of PHP has no apparent affect of the other Paf1C-related proteins, perhaps suggesting that PHP is peripherally associated with Paf1C. The combined predicted mass of VIP3/4/5/6, ELF7, and PHP (413 kD) is substantially less than the approximately 670 kD estimated from gel filtration chromatography, suggesting additional stable interactions with unknown proteins. VIP3 is also found in a somewhat smaller protein that does not appear to include PHP or VIP6 (Fig. 2) and thus could represent a stable Paf1C-associated subcomplex. This is consistent with our observation that, unlike other Paf1C proteins, at least a fraction of VIP3 protein is stable in php and other paf1C mutant backgrounds (Fig. 2). Excellent candidates for additional VIP3/Paf1C-associated proteins, given the homology between VIP3 and hSki8, are the Arabidopsis homologs of Ski2 and Ski3, proteins that are functionally associated with Ski8 (Orban and Izaurralde, 2005).
The interdependence of VIP3/4/5/6 and ELF7 proteins observed here can explain the substantially similar developmental pleiotropy and patterns of gene misregulation seen among the respective mutants (Zhang and van Nocker, 2002; Zhang et al., 2003; He et al., 2004; Oh et al., 2004), and precludes meaningful analysis of individual functions of these subunits. In contrast, loss of PHP in the stronger php-2 background, which does not affect cellular levels of other Paf1C proteins, leads to only limited effect on the phenotype and transcriptome. This mild effect probably is not due to genetic redundancy, as PHP is the only gene encoding a Parafibromin-like protein in Arabidopsis. An obvious possibility is that the php-2 allele (as well as php-1) is hypomorphic, and that weak PHP activity is sufficient to carry out most functions. However, we favor the alternative possibility, that the stronger php-2 allele is essentially null, both because we were unable to detect PHP-immunoreactive protein in extracts from php-2 plants, and because antisense expression of PHP RNA did not lead to developmental defects more severe than the disruption of flowering time observed for php-2 plants. This interpretation is consistent with a scenario where PHP serves as an accessory subunit to assist Paf1C in the regulation of a specific subset of genes, potentially with distinct cofactor requirements, or with unusual chromatin features. Our finding that those genes whose expression was promoted by PHP were essentially a subset of those genes promoted by VIP3 (Fig. 4) supports this. This is evocative of the proposed role of Parafibromin in recruiting hPAF to genes targeted for activation through the Wnt signaling pathway (Mosimann et al., 2006), or the proposed role for human Ctr9 in recruitment of STAT3 to promote IL-6-responsive transcription (Youn et al., 2007). In an analogous manner, PHP might mediate Paf1C function at the FLC gene as an effector of one of the several charted pathways of FLC regulation (Bäurle and Dean, 2006). This pathway would be distinct from the well-characterized pathway associated with FLC silencing during extended growth in cold, which appears to be carried out independently of PHP (Fig. 3B). Loss of PHP completely suppressed the repressive effect of FRI on flowering (Fig. 3B), suggesting that PHP and FRI might interact functionally, although the observation that PHP protein is expressed to similar levels in the Col-0 and Col:FRI backgrounds (Fig. 2B) indicates such a mechanism would not involve modulation of PHP levels by FRI.
That PHP might assist Paf1C at a specific subset of genes is also supported by our observation that genes misregulated in the strong php-2 mutant tend to be distinguished from the bulk of genes misregulated in a strong mutant for the Paf1C-related gene VIP3. Specifically, PHP-regulated genes, especially those that are subject to repression by PHP, are more strongly enriched for H3K27me3 (Fig. 5), a posttranslational histone modification conserved among most higher eukaryotes and associated with transcriptional repression through the PcG protein PRC2 (Schwartz and Pirrotta, 2007). The observation that both PHP-repressed and PHP-promoted genes show high levels of H3K27me3 is unanticipated, but might be trivially explained by previous observations that genes with high levels of H3K27me3 tend to show very tissue-specific expression patterns (Zhang et al., 2007; Oh et al., 2008). Thus, if genes that are directly targeted by PHP are developmentally regulated, then indirect (downstream) targets should also tend to be developmentally regulated and be enriched with H3K27me3. A more precise interpretation of function will require identification of those genes that are direct transcriptional targets of PHP.
While PHP might discriminate some Paf1C targets, loss of PHP was associated with chromatin defects at the FLC gene that were similar to those observed in plants disrupted for the Paf1C-associated protein VIP3. First, H3K27me3 accumulated further across FLC chromatin, suggesting PHP in concert with Paf1C might antagonize PcG/PRC2 activity. Second, H3K4me3 accumulated ectopically at the 3′ end of FLC (Fig. 6), suggesting that PHP/Paf1C limits this modification. Potential mechanisms, as previously suggested for Paf1C in general (Oh et al., 2008), include mediating interaction of chromatin with histone H3K4 demethylases, or participating in transcription-coupled, nucleosomal histone exchange.
In summary, we present evidence that PHP is a subunit of a plant Paf1C, and that the structure of the Paf1C cofactor is well conserved between human and Arabidopsis. As observed for Parafibromin, PHP appears to have a specialized role in Paf1C activities. Our data suggests that PHP has a special role in regulating expression of genes highly enriched in H3K27me3, including the flowering switch FLC. The mechanism of this specialization, and whether this mechanism is shared with its human homolog Parafibromin, is an unresolved yet intriguing question.
MATERIALS AND METHODS
Identification of PHP
The PHP gene was identified using the HRPT2 open reading frame translation (Parafibromin, accession Q6P1J9) as a query in BLASTP analysis with derived protein sequences from Arabidopsis (Arabidopsis thaliana), and in TBLASTN analysis with the sequenced Arabidopsis genome (The Arabidopsis Information Resources version 8). These analyses identified only PHP as related to Parafibromin (Expect [E] statistical values for matches of 1e-10 and 1e-22, respectively, with all other matches showing E values >2). BLASTP analysis of derived human protein sequences (RefSeq version 27) using PHP as a query identified only Parafibromin (E = 3e-27, with all other matches showing E values >1). Analysis of microarray data for expression of PHP and other genes utilized the AtGenExpress Development data set (Schmid et al., 2005).
Plant Material and Growth Conditions
Arabidopsis lines containing the php-1 and php-2 insertion alleles were obtained from the Arabidopsis Biological Resource Center (SALK_008357 and SALK_150644, respectively). Homozygous mutants were identified within segregating populations using PCR with primers (PHP-F1, 5′-TTCTAGAAACCATGGATCCATTATCAG-3′; PHP-R2, 5′-GAATGCGACCGAGATCGCACAAAC-3′) flanking the insertion site. Plants were grown at 23°C in photoperiodically inductive (16 h light/8 h dark) or noninductive (8 h light/16 h dark) conditions with fluorescent lighting. Cold treatments were as previously described (Zhang and van Nocker, 2002). The Col:FRI introgression line has been described previously (Lee et al., 1994). All experiments utilized plants of the Col-0 genetic background unless otherwise indicated.
Nuclear Extract Preparation: Gel Filtration Chromatography and Coimmunoprecipitation Analyses
Arabidopsis nuclear proteins were prepared using a modification of the methods described previously (Green et al., 1987; Bowler et al., 2004). Briefly, 50 g of inflorescences including stems and young siliques were collected, rinsed with prechilled water, and homogenized in 200 mL of ice-cold buffer A (1 m hexylene glycol, 10 mm PIPES/KOH [pH 7.0], 10 mm MgCl2, and 0.2% Triton X-100, containing 5 mm 2-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, and 0.1 μg/mL pepstatin A). The homogenized material was filtered through four layers of Miracloth (Calbiochem), and centrifuged at 2,000g for 10 min at 4°C. The nuclear pellets were resuspended in 20 mL of buffer B (buffer A substituting 0.5 m hexylene glycol), and centrifuged at 3,000g for 10 min at 4°C. The nuclear pellets were washed with buffer B two more times. After the final wash, the pellets were resuspended in 2 mL of buffer C (10 mm Tris-HCl [pH 8.0], 10 mm NaCl, 2 mm EDTA, and 0.2% Triton X-100, containing 1 μg/mL leupeptin, 1 μg/mL pepstatin A, 1 mm phenylmethylsulfonyl fluoride, 10 mm NaF, 5 mm sodium pyrophosphate, and 1% phosphatase inhibitor cocktail 1 [Sigma]). Then, 80 μL of 5 m NaCl was added and the suspension was incubated on ice for 20 min with gentle agitation. Finally, after centrifugation at 30,000g for 1 h at 4°C, the supernatant containing nuclear proteins was recovered.
For chromatographic analysis, freshly prepared nuclear extracts (200 μL) were applied to a Superose 6 column (Amersham-Pharmacia) in buffer D (30 mm Tris-HCl [pH 7.5], 30 mm NaCl, 0.1 mm EDTA, 0.1 mm 2-mercaptoethanol, 10% glycerol, 0.2% Triton X-100) at a flow rate of 0.3 mL/min. Fractions (300 μL) were collected and TCA was added to a final concentration of 10% (v/v). Precipitated material was collected by centrifugation, resuspended in SDS sample buffer, and analyzed by SDS-PAGE. Protein mass was determined by calibrating the gel filtration column with molecular mass standards (Bio-Rad, cat. no. 151-1901).
For coimmunoprecipitation analyses, 500 μL of the nuclear extracts prepared as described above were diluted by adding an equal volume of buffer C. The solution was then mixed with Anti-FLAG M2 monoclonal antibody (Sigma; catalog no. F-3165) and incubated at 4°C overnight. Protein A-agarose beads (30 μL) were then added, and the mixture was incubated a further 3 h at 4°C. Protein A-agarose beads were collected by centrifugation and washed with 1 mL ice-cold washing buffer (buffer C containing 0.05% Triton X-100) five times. After the final wash, the proteins bound on the beads were released by boiling with 70 μL of sample buffer.
Immunoblotting was done as described previously (Harlow and Lane, 1988), using polyvinylidene difluoride (Bio-Rad) or nitrocellulose membranes and alkaline-phosphatase- or horseradish peroxidase-conjugated antibodies. Rabbit IgG polyclonal antibodies recognizing VIP3 and VIP6 were described previously (Oh et al., 2004). Antisera detecting PHP, VIP5, and ELF7 were generated using as antigens the PHP protein (amino acids 2–415), VIP5 carboxyl-terminal portion (amino acids 356–643), and ELF7 amino-terminal portion (amino acids 14–281) expressed in Escherichia coli as hexahistidine fusions and purified using Ni2+-affinity chromatography.
Transcriptional Profiling and Analysis of Histone H3 Methylation Profiles within PHP-Dependent Genes
RNA gel-blot analysis of PHP expression utilized the entire transcribed region of PHP as a probe; the probe was amplified by reverse transcription-PCR from seedling-derived RNAs using primers PHP-F1 and PHP-R2. Expression of approximately 22,600 genes was analyzed in aerial tissues from 14-d-old soil-grown wild-type (Col:FRI) and php mutant (php-2:FRI) plants using the Affymetrix ATH1 GeneChip, and two independent biological replicates. Data from CEL files were adjusted for background and normalized using the Bioconductor GCRMA package (http://www.bioconductor.org). Statistically significant differences (P < 0.01 and 2-fold difference) in gene expression between wild type and php were detected using the Bioconductor LIMMA package (Smyth, 2004). Microarray data have been deposited with the Gene Expression Omnibus database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE13913. Of 38 up-regulated genes and 37 down-regulated genes in php-2 relative to wild type, 29 (up-regulated) and 25 (down-regulated) were included in the gene set evaluated for histone H3 methylations as described previously (Oh et al., 2008), and were utilized for the analysis of H3 methylation profiles. Genic profiles were derived by analyzing probe signals for 100-bp windows within the proximal promoter (−350 to −50 relative to the transcriptional start site), transcriptional start site region (−49 to 0 bp to 5% of transcribed region), transcribed region (intervals of 10% of transcribed region from 5%–95%), 3′ end region (from 95%–100% of transcribed region to +50 bp relative to the 3′ end), and 3′ flanking region (51–150 bp relative to the 3′ end). To assess statistical significance of the chromatin signatures for PHP-regulated genes, we computed the 95th percentile confidence intervals for mean positional signals within 1,000 randomly resampled gene sets, each containing 29 (for up-regulated) or 25 (for down-regulated) genes. ChIP for the analysis of H3 methylation within FLC chromatin was carried out using techniques and antibodies as described previously (Oh et al., 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of PHP, Parafibromin, Hyrax, and Cdc73.
Supplemental Figure S2. Developmental regulation of PHP-targeted genes.
Supplemental Table S1. List of genes misregulated in the php-2 mutant.
Supplementary Material
Acknowledgments
We thank members of the Gene Expression in Disease and Development working group (Michigan State University) for helpful discussions.
References
- Anderson JS, Parker RP. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanisms for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora C, Kee K, Maleki S, Keeney S. (2004) Antiviral protein Ski8 is a direct partner of Spo11 in meotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559 [DOI] [PubMed] [Google Scholar]
- Bäurle I, Dean C. (2006) The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. (2002) Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, lipid and nuclei acid metabolism. Mol Genet Genomics 268: 272–285 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, et al. (2002) HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet 32: 676–680 [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol 19: 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ. (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10: 520–527 [DOI] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua NH. (1987) Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Haven CJ, Wong FK, van Dam EW, van der Juijt R, van Asperen C, Jansen J, Rosenberg C, de Wit M, Roijers J, Hoppener J, et al. (2000) A genotypic and histopathological study of a large Dutch kindred with hyperparathyroidism-jaw tumor syndrome. J Clin Endocrinol Metab 85: 1449–1454 [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CE, Norum RA, Boyd SB, Talpos GB, Wilson SD, Taggart RT, Mallette LE. (1990) Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: a clinically and genetically distinct syndrome. Surgery 108: 1006–1012 [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. (2003a) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. (2003b) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. (1994) The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6: 903–909 [Google Scholar]
- Lin L, Zhang JH, Panicker LM, Simonds WF. (2008) The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci USA 105: 17420–17425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. (2006) Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. (2003) The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278: 33625–33628 [DOI] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S. (2008) Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet 4: e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Zhang H, Ludwig P, van Nocker S. (2004) A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. (2005) Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, the exosome. RNA 11: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol 126: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. (2005) The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol 25: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Maley JL, Meyerson M. (2009) The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA 106: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. (2005) A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. (2005) Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene 24: 1272–1276 [DOI] [PubMed] [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. (2005) The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol 25: 5052–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn MY, Yoo HS, Kim MJ, Hwang SY, Choi Y, Desiderio SV, Yoo JY. (2007) hCTR9, a component of Paf1 complex, participates in the transcription of interleukin 6-responsive genes through regulation of STAT3-DNA interactions. J Biol Chem 282: 34727–34734 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ransom C, Ludwig P, van Nocker S. (2003) Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch FLOWERING LOCUS C. Genetics 164: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, van Nocker S. (2002) The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 31: 663–673 [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. (2005) The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 19: 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.